Abstract
A molecular database for all clinically important Zygomycetes was constructed from nucleotide sequences from the nuclear small-subunit (18S) ribosomal DNA and domains D1 and D2 of the nuclear large-subunit (28S) ribosomal DNA. Parsimony analysis of the aligned 18S and 28S DNA sequences was used to investigate phylogenetic relationships among 42 isolates representing species of Zygomycetes reported to cause infections in humans and other animals, together with commonly cultured contaminants, with emphasis on members of the Mucorales. The molecular phylogeny provided strong support for the monophyly of the Mucorales, exclusive of Echinosporangium transversale and Mortierella spp., which are currently misclassified within the Mucorales. Micromucor ramannianus, traditionally classified within Mortierella, and Syncephalastrum racemosum represent the basal divergences within the Mucorales. Based on the 18S gene tree topology, Absidia corymbifera and Rhizomucor variabilis appear to be misplaced taxonomically. A. corymbifera is strongly supported as a sister group of the Rhizomucor miehei-Rhizomucor pusillus clade, while R. variabilis is nested within Mucor. The aligned 28S sequences were used to design 13 taxon-specific PCR primer pairs for those taxa most commonly implicated in infections. All of the primers specifically amplified DNA of the size predicted based on the DNA sequence data from the target taxa; however, they did not cross-react with phylogenetically related species. These primers have the potential to be used in a PCR assay for the rapid and accurate identification of the etiological agents of mucormycoses and entomophthoromycoses.
The number of opportunistic species reported to be involved in fungal infections in humans is increasing rapidly (37). Of these, members of the Zygomycetes represent excellent examples of fungi that are generally regarded as nonpathogenic. They are widespread in nature and subsist on decaying vegetation. Zygomycetes, however, are becoming more commonly involved in disease complexes as secondary infections of immunocompromised human immunodeficiency virus patients (41). Transplantation patients, who are artificially immunosuppressed by medication, are also exposed to the risk of zygomycoses (34). Other common risk factors for acquiring these infections include hematologic malignancy, renal failure, and diabetes mellitus (12). Zygomycoses are classified as either mucormycoses or entomophthoromycoses depending on whether the etiological agent is a member of the Mucorales or the Entomophthorales (9). Mucormycoses are most frequently caused by species within the genera Rhizopus, Rhizomucor, Absidia, Cunninghamella, and Mucor (6). Although these fungi show minimal intrinsic pathogenicity for normal, healthy individuals, they initiate acute, aggressive, fulminant, and rapidly progressive disease in debilitated and immunocompromised patients (1, 11, 21, 38, 43). Entomophthoromycoses or subcutaneous zygomycoses, in contrast, are chronic, slowly progressing subcutaneous infections most frequently observed in individuals living in tropical climates (36). This disease is typically characterized by an insidious onset of massive induration of subcutaneous soft tissue involving the limbs, trunk, or buttocks. Deeply invasive infections of the gastrointestinal, rhinofacial, pulmonary, pericardial, or retroperitoneal tract have been reported (29, 35, 46) but are rare.
Zygomycete fungi pose difficult diagnostic and therapeutic challenges because (i) the spectrum of opportunistic zygomycoses is expanding (9, 46), (ii) their clinical manifestations can be fatal without rapid diagnosis and treatment (21), and (iii) strains that fail to sporulate under normal laboratory conditions may be encountered, thereby making morphological identification difficult (e.g., Saksenaea vasiformis [25]). While zygospore production has been used as a diagnostic tool for the identification of rare, unusual, or atypical heterothallic zygomycetes (47), practical considerations limit this and other time-consuming morphological methods to major medical mycology reference laboratories. Although immunological approaches have been developed for the diagnosis of Rhizopus arrhizus (49) and Basidiobolus sp. infections (19), cross-reactivity of antibodies is often observed (7). Given these problems, DNA-based molecular typing techniques show enormous potential for rapidly and accurately identifying the etiological agents of zygomycoses (34). To address this problem, we constructed a data set based on 18S and 28S ribosomal DNA (rDNA) sequences of 42 isolates of Zygomycetes, including every species reported to cause infections in humans and other animals. By using the aligned 28S rDNA sequences, 13 taxon-specific PCR primer pairs that specifically amplify DNA for the most commonly reported taxa were designed. Given their specificity, these primer pairs represent valuable diagnostic tools for the rapid and accurate identification of species causing mucoromycoses and entomophthoromycoses.
MATERIALS AND METHODS
Fungal strains and cultivation.
Of the 42 strains of Zygomycetes studied (Table 1), 20 were isolated from clinical sources. All strains are stored by lyophilization or in liquid nitrogen vapor (−175°C) in the Agricultural Research Service (ARS) Culture Collection (NRRL), Peoria, Ill., and the Fungal Reference Center in Jena, Germany (FRC Jena). Mycelium for DNA isolation was grown in YM broth (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 2% dextrose; Difco, Detroit, Mich.) at room temperature for 2 to 5 days. Strains grown on YM agar (2% Difco agar) were examined morphologically according to the method of O'Donnell (30) to confirm their identity.
TABLE 1.
Strains analyzed in this study
| Species and straina | Equivalent strain designationb | Clinical source | 18S/28S GenBank accession no.c |
|---|---|---|---|
| Absidia coerulea NRRL 1315NT | CBS 104.08 | No | AF113405/AF113443 |
| Absidia coerulea NRRL A-9483 | CBS 100.32 | Yes; aborted bovine fetus | AF113406/AF113444 |
| Absidia corymbifera NRRL 2982 | CBS 100.31 | Yes; aborted bovine fetus | AF113407/AF113445 |
| Absidia corymbifera NRRL 28639 | CBS 101.55 | Yes; human cornea | AF113408/AF113446 |
| Absidia glauca NRRL 1329 | CBS 102.08 | AF113409/AF113447 | |
| Absidia repens NRRL 1336 | ATCC 14849 | AF113410/AF113448 | |
| Apophysomyces elegans NRRL 22325T | CBS 476.78 | No | AF113411/AF113449 |
| Apophysomyces elegans NRRL 28632 | CBS 658.93 | Yes; human (osteomyelitis) | AF113412/AF113450 |
| Basidiobolus haptosporus NRRL 28635 | CBS 358.65 | Yes; human (creeping granuloma) | AF113413/AF113451 |
| Basidiobolus ranarum NRRL 20525 | UK 10281 | AF113414/AF113452 | |
| Chlamydoabsidia padenii NRRL 2977T | CBS 172.67 | No | AF113415/AF113453 |
| Cokeromyces recurvatus NRRL 2243T | CBS 158.50 | No | AF113416/AF113454 |
| Conidiobolus coronatus NRRL 1912 | ATCC 32801 | AF113417/AF113455 | |
| Conidiobolus coronatus NRRL 28638 | CBS 209.66 | AF113418/AF113456 | |
| Conidiobolus incongruus NRRL 28636 | CBS 108.84 | Yes; human epidermal subcutaneous tissue | AF113419/AF113457 |
| Conidiobolus lamprauges NRRL 28637T | CBS 153.56 | No | AF113420/AF113458 |
| Cunninghamella bertholletiae NRRL 6436 | CBS 190.84 | Yes; human heart (lymphosarcoma) | AF113421/AF113459 |
| Cunninghamella elegans NRRL 28624 | CBS 151.80 | Yes; human lung (leukemia) | AF113422/AF113460 |
| Cunninghamella polymorpha NRRL 6441NT | CBS 693.68 | No | AF113423/AF113461 |
| Echinosporangium transversale NRRL 3116T | CBS 357.67 | No | AF113424/AF113462 |
| Micromucor ramannianus NRRL 5844 | IMI 150942 | No | X89435/AF113463 |
| Mortierella polycephala NRRL 22890 | X89436/AF113464 | ||
| Mortierella wolfii NRRL 28640 | CBS 611.70 | Yes; bovine lung (mycotic pneumonia) | AF113425/AF113465 |
| Mucor amphibiorum NRRL 28633T | CBS 763.74 | Yes; amphibian | AF113426/AF113466 |
| Mucor circinelloides f. lusitanicus NRRL 3631 | CBS 277.49 | AF113427/AF113467 | |
| Mucor hiemalis f. hiemalis NRRL 3624NT | CBS 201.65 | AF113428/AF113468 | |
| Mucor indicus NRRL 28634T | CBS 226.29 | AF113429/AF113469 | |
| Mucor mucedo NRRL 3635 | CBS 144.24 | X89434/AF113470 | |
| Mucor racemosus NRRL 3640T | CBS 260.68 | AF113430/AF113471 | |
| Mucor ramosissimus NRRL 3042NT | CBS 135.65 | Yes; human nasal lesion | AF113431/AF113472 |
| Rhizomucor miehei NRRL 28774 | CBS 370.71 | Yes; human sputum | AF113432/AF113473 |
| Rhizomucor pusillus NRRL 2543 | ATCC 22064 | Yes; animal lung | AF113433/AF113474 |
| Rhizomucor pusillus NRRL 28626 | CBS 245.58 | Yes; aborted bovine fetus | AF113434/AF113475 |
| Rhizomucor variabilis NRRL 28773T | CBS 103.93 | Yes; human wrist and hand | AF113435/AF113476 |
| Rhizopus azygosporus NRRL 28627 | CBS 359.92 | Yes; liver of a premature baby (necrotizing enterocolitis) | AF113436/AF113477 |
| Rhizopus microsporus var. chinensis NRRL 28629T | CBS 294.31 | Yes; bovine fetus | AF113437/AF113478 |
| Rhizopus microsporus var. microsporus NRRL 28775 | CBS 308.87 | Yes; skin of human hand | AF113438/AF113479 |
| Rhizopus microsporus var. rhizopodiformis NRRL 28630 | CBS 220.92 | Yes; human lung | AF113439/AF113480 |
| Rhizopus oryzae NRRL 28631 | CBS 146.90 | Yes; human pallatum molle | AF113440/AF113481 |
| Rhizopus stolonifer NRRL 1477 | ATCC 12938 | AF113441/AF113482 | |
| Saksenaea vasiformis NRRL 2443 | ATCC 44101 | No | AF113442/AF113483 |
| Syncephalastrum racemosum NRRL 2496 | CBS 440.59 | No | X89437/AF113484 |
All of the species listed have been reported as causing human and other animal infections (6, 15, 23, 40, 50) except for A. glauca, A. repens, C. polymorpha, E. transversale, and M. ramannianus. NRRL numbers designate strains from the ARS Culture Collection, National Center for Agricultural Utilization Research (formerly the Northern Regional Research Laboratory), Peoria, Ill. NT, ex-neotype strain; T, ex-type strain.
Culture collection abbreviations: ATCC, American Type Culture Collection, Manassas, Va.; CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands; IMI, CABI Bioscience (formerly the International Mycological Institute), Egham, United Kingdom; UK, University of Kansas, Lawrence.
DNA isolation.
Total genomic DNA was isolated from lyophilized mycelium according to the CTAB (hexacetyltrimethylammonium bromide; Sigma Chemical Co., St. Louis, Mo.) miniprep protocol described by O'Donnell et al. (33). Approximately 50 mg of pulverized mycelium was resuspended in 700 μl of CTAB extraction buffer (100 mM Tris-Cl [pH 8.4], 1.4 M NaCl, 25 mM EDTA, 2% CTAB) and vortexed for 10 s. Following extraction, an equal volume of chloroform was added to each tube, vortexed for 5 s, and then spun for 10 min at 12,300 × g in a Savant (Holbrook, N.Y.) microcentrifuge. A 500-μl portion of the upper phase was removed to a new 1.5-ml tube, and DNA was precipitated by the addition of an equal volume of −20°C isopropanol. After the DNA was pelleted at 12,300 × g in a Savant microcentrifuge for 1 min, the supernatant was discarded and the pellet was gently washed with 70% ethanol and resuspended in 200 μl of TE buffer (10 mM Tris-Cl [pH 8.0]–1 mM EDTA [pH 8.0]). For PCR amplifications, 8 μl of the genomic DNA stock was diluted in 1 ml of deionized water and stored at −20°C when not in use. For PCR experiments 25 μl of the diluted genomic DNA was added to an equal volume of a 2× PCR master mix (see below).
PCR.
PCR amplification mixtures typically contained approximately 10 to 20 ng of genomic DNA, 0.225 mM each deoxynucleotide (Boehringer, Mannheim, Germany), 25 pmol of each primer, 50 mM KCl, 10 mM Tris-Cl (pH 8.4), 2.5 mM MgCl2, 0.1 mg of gelatin/ml, and 1.25 U of AmpliTaq polymerase (Perkin-Elmer, Foster City, Calif.) in a reaction volume of 50 μl. PCR products were amplified in a Perkin-Elmer 9600 thermal cycler by using the fastest ramp times. The temperature profile included an initial denaturing step of 2 min at 94°C; 40 cycles of 30 s at 94°C for DNA denaturation, 30 s at 52°C for primer annealing, and 90 s at 72°C for primer extension; a final extension of 10 min at 72°C; and a 4°C soak. All amplicons were separated electrophoretically in 1.5% agarose gels (FMC, Rockland, Maine). Amplification of the 28S rDNA with taxon-specific PCR primer pairs was performed by the PCR method listed above, except that the annealing temperature was increased to 60°C. PCR fragments amplified with the 13 taxon-specific primer pairs were size-fractionated in 2% NuSieve GTG–1% agarose gels (length, 25 cm; width, 20 cm) (FMC).
Primers.
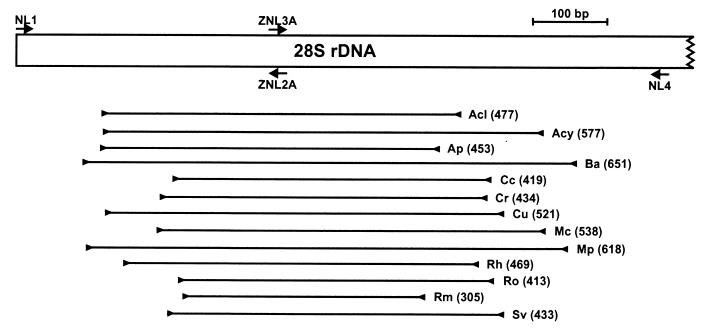
To generate templates for sequencing, primer pairs PNS1–NS41 and NS51–NS8Z or NS5–NS8Z were used to amplify the 18S rDNA as two overlapping fragments, and primer pair NL1–NL4 was used to amplify the 5′ end of 28S rDNA spanning domains D1 and D2. The following primers were used to sequence the 18S rDNA: NS2, NS3, NS5, NS7, and NS8 (48); PNS1, NS6Z, and NS8Z (32); and NS41 and NS51 (5, 33). Sequencing of the 5′ end of the 28S rDNA was conducted by using primers NL1 and NL4 (31) and primers ZNL2A (5′-CTTTTCATCTTTCCCTCACGG-3′) and ZNL3A (5′-GTACCGTGAGGGAAAGATGAAAAG-3′). The positions of the NL primers are given in Fig. 3.
FIG. 3.
Map of the 5′ end of the nuclear large-subunit 28S rDNA showing positions of primers (labeled arrows) used for PCR amplification and DNA sequencing. The positions of the fragments amplified by the 13 taxon-specific PCR primer pairs are indicated by lines below the map. The length of each PCR product in base pairs and the target taxon are given at the right of each amplicon. Acl, Absidia coerulea; Acy, A. corymbifera; Ap, Apophysomyces elegans; Ba, Basidiobolus haptosporus and B. ranarum; Cc, Conidiobolus coronatus; Cr, Cokeromyces recurvatus; Cu, Cunninghamella bertholletiae, Cunninghamella elegans, and Cunninghamella polymorpha; Mc, Mucor circinelloides and Mucor ramosissimus; Mp, Mortierella polycephala; Rh, Rhizopus azygosporus and Rhizopus microsporus; Ro, Rhizopus oryzae; Rm, R. miehei and R. pusillus; Sv, S. vasiformis.
Cycle sequencing.
Amplicons were purified with a GeneClean kit (Bio 101, Buena Vista, Calif.). Cycle sequencing was conducted in a Perkin-Elmer 9600 thermal cycler with “FS” or “Bigdye” fluorescent-labeled DyeDeoxy protocols (Perkin-Elmer) by using the following temperature profile: 15 s at 96°C and 4 min at 55°C for 25 cycles, followed by a 4°C soak. All sequencing reaction mixtures were run on an Applied Biosystems model 377 automated DNA sequencer after purification via gel chromatography through Sephadex G-50 (SuperFine; Pharmacia, Piscataway, N.J.) spin columns.
Analysis of DNA sequences.
Following initial alignment with CLUSTAL W (version 1.60) (18), sequence alignments were manipulated visually with TSE, a DOS text software program (SemWare; Marietta, Ga.). Unweighted phylogenetic analyses were performed on the individual and combined data sets by using the heuristic search option in PAUP*4.0b1 (44), with 1,000 stepwise random addition sequences. The partition-homogeneity test (PHT) implemented with PAUP was used to evaluate the concordance of the 18S and 28S rDNA data sets, by using 1,000 replicates with MAXTREES set to 5,000. Uninformative characters were excluded from the PHT. A second incongruence test, the Wilcoxon signed-ranks Templeton test, was implemented with PAUP, by using the most parsimonious tree (MPT) and a 70% majority rule bootstrap tree as constraints in a separate analysis. Clade stability was estimated from 1,000 bootstrap replications (10) with PAUP and by decay indices (4) calculated with TreeRot (42).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 18S and 28S rDNA sequences of the 42 isolates analyzed in this study are given in Table 1 (see below).
RESULTS
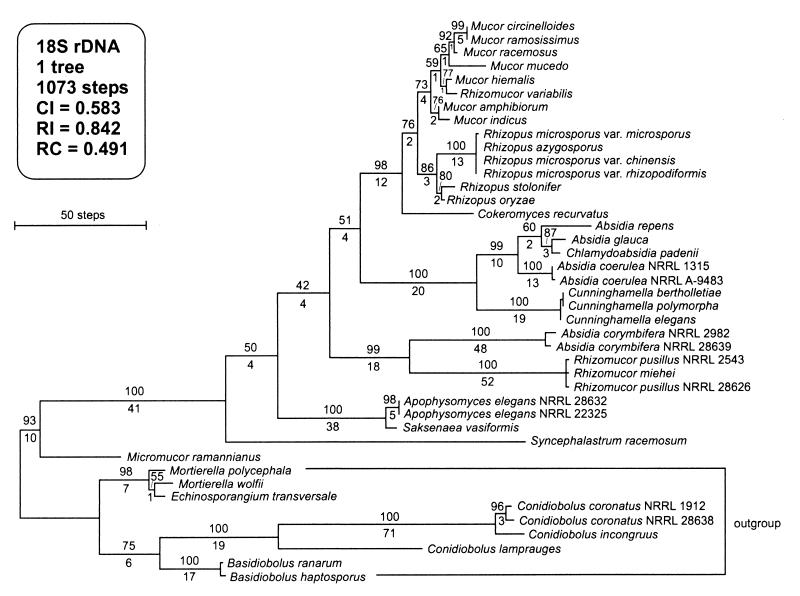
In order to design PCR primer pairs specific for taxa representing the most important opportunistic Zygomycetes, we obtained nuclear small-subunit (18S) rDNA and nuclear large-subunit (28S) rDNA sequences for 42 isolates representing all species reported to cause infections in humans and other animals plus the most common contaminants (Table 1). All of these sequences were generated in the present study except for four 18S rDNA sequences obtained from GenBank. Except for 31 and 36 bp at the 5′ and 3′ ends, respectively, the 18S rDNA sequences were complete. The aligned 18S rDNA data set consisted of 1,881 characters, of which 1,377 were unambiguously aligned and included in the phylogenetic analysis. Unweighted maximum-parsimony analysis of the 18S rDNA data, using the heuristic search option with 1,000 random stepwise addition sequences implemented with PAUP 4.0b1 (44), yielded a single MPT 1,073 steps long (Fig. 1). Based on phylogenetic results obtained by Jensen et al. (20) and Gehrig et al. (14), sequences of the Entomophthorales (i.e., Basidiobolus and Conidiobolus spp.) and Echinosporangium transversale-Mortierella spp. were used as outgroups to root the tree. Clinically important species are nested in all lineages. Phylogenetic analysis provided strong support for the monophyly of the Mucorales. Micromucor ramannianus (bootstrap = 93%; decay index = 10; formerly classified as Mortierella ramanniana within the Micromucor subgenus of Mortierella, 13) and Syncephalastrum racemosum (bootstrap = 100%; decay index = 41) represent the two basal taxa within this order.
FIG. 1.
Single most parsimonious phylogram inferred from the 18S rDNA sequence data, showing phylogenetic relationships of Zygomycetes. Sequences of Mortierella, Echinosporangium, Conidiobolus, and Basidiobolus spp. were chosen as outgroups to root the tree based on previous phylogenetic analyses of 18S rDNA sequence data (14, 20). Numbers above nodes represent bootstrap frequencies; numbers below nodes are decay indices calculated with TreeRot (42). Note that neither Absidia spp. nor Rhizomucor spp. form exclusive groups within the 18S gene tree.
Visual inspection of the aligned 18S rDNA sequences indicated that they were too highly conserved to be used in the design of taxon-specific PCR primer pairs. Therefore we sequenced domains D1 and D2 at the 5′ end of the 28S rDNA. Of the 772 aligned nucleotide characters, 425 were coded as ambiguous and excluded from the phylogenetic analysis. Parsimony analysis of the 347 included characters, using the same search options indicated above, yielded seven equally MPTs 640 steps long. Figure 2 is a phylogram of the first tree. The other six MPTs are topologically concordant with the phylogram shown in Fig. 2 except for six nodes within the Mucor-Rhizopus-Cokeromyces lineage that received decay scores of 0. Only these six nodes collapsed in a strict consensus of the seven MPTs. The 18S rDNA gene tree (Fig. 1), with 20 nodes receiving bootstrap scores of ≥90%, is more robust than the 28S rDNA tree (Fig. 2), in which only 14 nodes received this measure of clade support. To assess whether the 18S and 28S rDNA data could be analyzed as a combined data set, these data were subjected to the PHT and the Wilcoxon signed-ranks Templeton test implemented in PAUP (44). Results of the two incongruence tests, the PHT (P < 0.006, excluding ambiguous and uninformative characters) and the Templeton test (P < 0.0001 and P = 0.0411 by constraining the 18S rDNA data onto the 28S rDNA MPT and the 70% majority rule bootstrap consensus trees, respectively), statistically rejected combining these gene data sets.
FIG. 2.
One of seven equally most-parsimonious phylograms inferred by maximum parsimony analysis of 347 nucleotides of the 28S rDNA from 42 strains of Zygomycetes and common contaminants, by using sequences of Mortierella, Echinosporangium, Conidiobolus, and Basidiobolus spp. to root the tree. Bootstrap intervals (above internodes) from 1,000 replications and decay indices (below internodes) are indicated.
Results of the phylogenetic analyses indicate that Absidia corymbifera and Rhizomucor variabilis appear to be misplaced taxonomically. Based on the 18S gene tree topology, A. corymbifera is strongly supported as a sister group of a Rhizomucor miehei-Rhizomucor pusillus clade (bootstrap = 99%; decay index = 18) while R. variabilis is deeply nested within Mucor. Because Absidia and Rhizomucor appear to be polyphyletic in the 18S gene tree, and several genera appear to be either paraphyletic (i.e., Mucor, Rhizopus, and Absidia) or polyphyletic (i.e., Rhizomucor) within the 28S rDNA gene tree, various monophyly constraints were subjected to the Kishino-Hasegawa likelihood test implemented in PAUP 4.0b1 (44). Trees found by forcing Rhizomucor spp. or Absidia spp. to form monophyletic groups were significantly longer (Rhizomucor and Absidia constraint trees were 43 and 56 steps longer, respectively) than the MPT and statistically worse (P, defined as the probability of obtaining a more extreme t value with the two-tailed test under the null hypothesis of no difference between the two trees, is <0.0001). Constraints forcing the monophyly of Mucor spp. or Rhizopus spp., using the 28S rDNA data, were equal in length and not statistically worse than the MPT. However, the Absidia and Rhizomucor monophyly constraints were 14 and 19 steps longer, respectively, and significantly worse than the MPT at P < 0.05 by the two-tailed test.
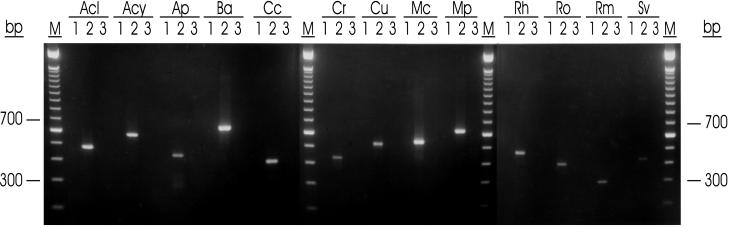
In contrast to the 18S rDNA data, visual inspection of the aligned 28S rDNA sequences readily identified unique regions that we used to design 13 taxon-specific PCR primer pairs for the most important opportunistic Zygomycetes (Fig. 3; Table 2). Based on the 28S rDNA gene tree topology (Fig. 2), when species most closely related to the clinical taxa were tested as negative controls to test for primer cross-reactions, all 13 primer pairs specifically amplified PCR products of the expected sizes from the target taxa (Fig. 4; Table 2), by use of an annealing temperature of 60°C.
TABLE 2.
Thirteen taxon-specific primer pairs that amplify a fragment of the 28S rDNA from Zygomycetes
| PCR primer pair (5′ to 3′) | Zygomycetes species identified | Size of PCR product (bp) |
|---|---|---|
| Acl1 (ATCATGCGTTTGCCCTTTAGC) | Absidia coerulea | 477 |
| Acl2 (CTAAGCGAGAAAAAAGAGAAAC) | ||
| Acy1 (CGGATTGTAAACTAAAGAGCG) | Absidia corymbifera | 577 |
| Acy2 (CCAAAGTAGATTACAGTTCTAG) | ||
| Ap1 (GAATTGTAAACTTTAGAGTCGTTG) | Apophysomyces elegans | 453 |
| Ap2 (TGAACCACAGTATTTCGCGAA) | ||
| Ba1 (AAAATCTGTAAGGTTCAACCTTG) | Basidiobolus haptosporus | 651 |
| Ba2 (TGCAGGAGAAGTACATCCGC) | Basidiobolus ranarum | |
| Cc1 (TCTCTTAACTTGCTTCTATGCC) | Conidiobolus coronatus | 419 |
| Cc2 (CTTTAATTAAGCTAATCAACATG) | ||
| Cr1 (GTGAGAATCCCGTGAATTCAC) | Cokeromyces recurvatus | 434 |
| Cr2 (CAAAGCACTCAACTATTTCGC) | ||
| Cu1 GGATTGTAAACTAAAGTTTTC | Cunninghamella bertholletiae | 521 |
| Cu2 AAATTCTCTAATTATTCCCTC | Cunninghamella elegans | |
| Cunninghamella polymorpha | ||
| Mc1 ATTTTCCTGGCACACCAGATT | Mucor circinelloides | 538 |
| Mc2 GCGAATAAAAAATATACTAGATGT | Mucor ramosissimus | |
| Mp1 TGGCCGGTTTACTGGTCCGAA | Mortierella polycephala | 618 |
| Mp2 CGAGTATAAAAAGGACACGGC | ||
| Rh1 TTTTCCAGGCAAGCCGGACCG | Rhizopus azygosporus | 469 |
| Rh2 TATTCCCAGCCAACTCGCCAAAT | Rhizopus microsporus | |
| Ro1 AGCATTTGCCTTTTGTGATACGC | Rhizopus oryzae | 413 |
| Ro2 ACCGTAGTACCTCAGAAAACC | ||
| Rm1 TCTATTGCGATGCATGCTCC | Rhizomucor miehei | 305 |
| Rm2 GGTCTCTTTAGACTCCAAAGC | Rhizomucor pusillus | |
| Sv1 CTTTGGCTTGAGCATTGGAC | Saksenaea vasiformis | 433 |
| Sv2 AGACTAAATCAATGACTTCTGG |
FIG. 4.
Gel (2% NuSieve GTG–1% agarose) showing taxon-specific amplification of a fragment of the 28S rDNA from Zygomycetes by using 13 taxon-specific primer pairs (Table 2). PCR products were amplified as described in Materials and Methods by using a uniform annealing temperature of 60°C. Lanes 1, no DNA (negative control); lanes 2, the 13 clinically important taxa (positive controls); lanes 3, taxa phylogenetically related to the 13 investigated taxa (negative controls demonstrating primer pair specificity); lanes M, 100-bp molecular weight marker (Gibco BRL, Detroit, Mich.); Acl 2, Absidia coerulea NRRL 1315; Acl 3, Absidia repens NRRL 1336; Acy 2, A. corymbifera NRRL 28639; Acy 3, R. pusillus NRRL 28626; Ap 2, Apophysomyces elegans NRRL 28632; Ap 3, S. vasiformis NRRL 2443; Ba 2, Basidiobolus haptosporus NRRL 28635; Ba 3, Conidiobolus lamprauges NRRL 28637; Cc 2, Conidiobolus coronatus NRRL 28638; Cc 3, Conidiobolus incongruus NRRL 28636; Cr 2, Cokeromyces recurvatus NRRL 2243; Cr 3, Rhizopus oryzae NRRL 28631; Cu 2, Cunninghamella bertholletiae NRRL 6436; Cu 3, A. coerulea NRRL 1315; Mc 2, Mucor ramosissimus NRRL 3042; Mc 3, Mucor racemosus NRRL 3640; Mp 2, Mortierella polycephala NRRL 22890; Mp 3, Mortierella wolfii NRRL 28640; Rh 2, Rhizopus microsporus NRRL 28775; Rh 3, Rhizopus stolonifer NRRL 1477; Ro 2, Rhizopus oryzae NRRL 28631; Ro 3, R. stolonifer NRRL 1477; Rm 2, R. pusillus NRRL 28626; Rm 3, A. corymbifera NRRL 28639; Sv 2, S. vasiformis NRRL 2443; Sv 3, A. elegans NRRL 28632.
DISCUSSION
Using isolates representing species of Zygomycetes reported in the literature as causing human or animal disease (9, 46), we constructed a DNA sequence database that we used to investigate phylogenetic relationships within the Mucorales and to develop species-specific PCR primer pairs for the rapid and accurate detection and identification of these medically important fungi. The single MPT topology inferred from the 18S rDNA data (Fig. 1) is interpreted as the best current hypothesis of phylogenetic relationships within the Mucorales because it is generally concordant with traditional morphologically based generic-level classification schemes and, relative to the 28S gene tree, more nodes within the 18S gene tree received higher measures of support from bootstrapping and decay analysis. Both gene trees show that clinically important species are nested in all lineages. Consistent with the phylogenetic results of Jensen et al. (20) and Gehrig et al. (14), sequences of the Entomophthorales and E. transversale-Mortierella spp. proved to be excellent outgroups for purposes of rooting the ribosomal gene trees. One surprising result of the molecular phylogeny was the basal split between M. ramannianus and S. racemosum and the other ingroup taxa, suggesting that these lineages may have descended from the earliest divergences within the Mucorales. Results of the molecular phylogeny help resolve the problematic systematic position of M. ramannianus, which Gams (13) provisionally placed in the Micromucor subgenus of Mortierella with the note that this subgenus is not closely related to other species of Mortierella.
Although the PHT and Templeton test results indicated that the nuclear ribosomal data sets should not be combined, no significant conflict in the 18S and 28S gene tree topologies that involved nodes strongly supported by bootstrapping in both phylograms was observed. With either data set, hypotheses of the monophyly of Absidia and Rhizomucor were strongly rejected by the Kishino-Hasegawa likelihood test implemented in PAUP (44). This result could have been predicted for A. corymbifera because it is atypical of the genus in that it produces nonappendaged zygospore suspensors, sporangiophores that arise singly from stolons rather than in whorls, and it is thermophilic. Emphasizing the systematic importance of nonappendaged suspensors, Beauverie (2) erected the genus Mycocladus to accommodate this taxon. Hesseltine and Ellis (17), however, recognized Mycocladus as a subgenus within Absidia, but this taxonomy is not supported by the likelihood tests, which indicate that Mycocladus does not form a monophyletic group with Absidia. Results of the molecular phylogeny also support the transfer of R. variabilis to Mucor. As noted by Zheng and Chen (50), R. variabilis is phenotypically unlike any other species of Rhizomucor in that it is not thermophilic and it produces rhizoids from hyphae, stolons, and sporangia. Based on the available data, R. variabilis appears to be most closely related phylogenetically to Mucor hiemalis and Mucor mucedo, which also have been reported to cause mycoses.
While numerous DNA-based systems using oligonucleotides as hybridization probes (3, 8, 39) or as PCR primers (16, 22, 24, 27, 28, 45) are available for the identification of medically important fungi, the present study represents the first successful amplification of Zygomycetes using taxon-specific PCR primer pairs. The experimental strategy used to develop a sensitive and comprehensive PCR-based system for the identification of these fungi has taken advantage of the following: (i) the primer pairs were designed and tested within a phylogenetic context based on discrete DNA sequence data from a broad sample of Zygomycetes, including all taxa reported as pathogens of humans and other animals, (ii) it is technically simple in that it only requires the ability to amplify DNA fragments via PCR, and (iii) it uses the highly repetitive 28S rDNA gene as a target which should increase its sensitivity when this system is tested on infectious agents from clinical samples. Experiments are under way to modify this system for fragment analysis using GeneScan and Genotyper software on a 377 automated DNA sequencer (Applied Biosystems, Perkin-Elmer) so that amplicons can be sized rapidly and accurately. Although it is unnecessary to identify the infection-causing Zygomycete for the purpose of treatment, precise identification of the species is highly desirable in order to more fully characterize the etiology. With minor modifications, the molecular tools described should make it possible to provide more data about the establishment and manifestation of infections caused by Zygomycetes and to monitor their persistence during antifungal therapy (45).
Although we have developed a comprehensive DNA sequence database that includes all Zygomycetes reported to be medically important, we anticipate that additional species may be identified as agents of infection. For this reason, we are presently expanding the database of Zygomycetes to include 18S and 28S rDNA sequence data for representatives of all mucoralean genera. In addition, because the rDNA-based database is not robust enough to resolve closely related species of some genera such as Mucor, Rhizopus, and Cunninghamella (Fig. 1 and 2), we have begun to investigate species limits within the Mucorales using several intron-containing nuclear genes (e.g., actin and translation elongation factor EF-1α. As noted by Maiden et al. (26), the overwhelming advantage of discrete DNA sequence data is that they are electronically portable between laboratories worldwide and can be extended to as many loci as required to identify strains objectively, independently of morphology and mating tests. To promote this end, the aligned 18S and 28S sequences analyzed in the present study are available from TreeBASE (44a) as matrix accession numbers M564 and M563 (study accession number S396), respectively.
ACKNOWLEDGMENTS
We thank the Centraalbureau voor Schimmelcultures (Baarn, The Netherlands) for providing many of the strains used in this study, Robert W. Lichtwardt (University of Kansas, Lawrence) for supplying the culture of Basidiobolus ranarum, Larry Tjarks for the oligonucleotides, and Steve Prather and Sam Sylvester for the illustrations. This work was performed during a visit at NCAUR-USDA in Peoria, Ill.
K.V. thanks Johannes Wöstenmeyer (University of Jena, Jena, Germany) for financial support.
REFERENCES
- 1.Amin S B, Ryan R M, Metlay L A, Watson W J. Absidia corymbifera infections in neonates. Clin Infect Dis. 1998;26:990–992. doi: 10.1086/513940. [DOI] [PubMed] [Google Scholar]
- 2.Beauverie J. Études sur le polymorphisme des champignons. Ann Univ Lyon. 1900;3:162–180. [Google Scholar]
- 3.Bowman B H. A model PCR/probe system for the identification of fungal pathogens. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 423–430. [Google Scholar]
- 4.Bremer K. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution. 1988;42:795–803. doi: 10.1111/j.1558-5646.1988.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 5.Bruns, T. Personal communication.
- 6.De Hoog G S, Guarro J. Zygomycota. In: de Hoog G S, Guarro J, editors. Atlas of clinical fungi. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures; 1995. pp. 41–58. [Google Scholar]
- 7.De Ruiter G A, Van Bruggen-van der Lugt A W, Nout M J, Middelhoven W J, Soentoro P S, Notermans S H, Rombouts F M. Formation of antigenic extracellular polysaccharides by selected strains of Mucor spp., Rhizopus spp., Rhizomucor spp., Absidia corymbifera and Syncephalastrum racemosum. Antonie Leeuwenhoek. 1992;62:189–199. doi: 10.1007/BF00582579. [DOI] [PubMed] [Google Scholar]
- 8.Einsele H, Hebart H, Roller G, Loffler J, Rothenhofer I, Muller C A, Bowden R A, van Burik J, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elgart M L. Zygomycosis. Dermatol Clin. 1996;14:141–146. doi: 10.1016/s0733-8635(05)70334-x. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Fermanis G G, Matar K S, Steele R. Endobronchial zygomycosis. Aust N Z J Surg. 1991;61:391–393. doi: 10.1111/j.1445-2197.1991.tb00242.x. [DOI] [PubMed] [Google Scholar]
- 12.Fridkin S K, Jarvis W R. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gams W. A key to the species of Mortierella. Persoonia. 1977;9:381–391. [Google Scholar]
- 14.Gehrig H, Schüßler A, Kluge M. Geosiphon pyriforme, a fungus forming endocytobiosis with Nostoc (cyanobacteria), is an ancestral member of the Glomales: evidence by SSU rRNA analysis. J Mol Evol. 1996;43:71–81. doi: 10.1007/BF02352301. [DOI] [PubMed] [Google Scholar]
- 15.Guarro J, de Hoog G S, Figueras M J, Gené J. Rare opportunistic fungi, part 2. In: de Hoog G S, Guarro J, editors. Atlas of clinical fungi. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures; 1995. pp. 241–272. [Google Scholar]
- 16.Haynes K A, Westerneng T J, Fell J W, Moens W. Rapid detection and identification of pathogenic fungi by polymerase chain reaction amplification of large subunit ribosomal DNA. J Med Vet Mycol. 1995;33:319–325. doi: 10.1080/02681219580000641. [DOI] [PubMed] [Google Scholar]
- 17.Hesseltine C W, Ellis J J. The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia. 1964;56:568–601. [Google Scholar]
- 18.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL W: improved software for multiple sequence alignment. Cabios. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 19.Imwidthaya P, Srimuang S. Immunodiffusion test for diagnosing basidiobolomycosis. Mycopathologia. 1992;118:127–131. doi: 10.1007/BF00437144. [DOI] [PubMed] [Google Scholar]
- 20.Jensen A B, Gargas A, Eilenberg J, Rosendahl S. Relationships of the insect-pathogenic order Entomophthorales (Zygomycota, Fungi) based on phylogenetic analyses of nuclear small subunit ribosomal DNA sequences (SSU rDNA) Fungal Genet Biol. 1998;24:325–334. doi: 10.1006/fgbi.1998.1063. [DOI] [PubMed] [Google Scholar]
- 21.Kambham N, Heller D S, Weitzman I. Incidental finding of zygomycetes-like hyphae in the placenta: a case report. J Reprod Med. 1998;43:230–232. [PubMed] [Google Scholar]
- 22.Kappe R, Fauser C N, Maiwald M, Sonntag H G. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J Med Microbiol. 1998;47:811–820. doi: 10.1099/00222615-47-9-811. [DOI] [PubMed] [Google Scholar]
- 23.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea and Febiger; 1992. [Google Scholar]
- 24.LoBuglio K F, Taylor J W. Phylogeny and PCR identification of the human pathogenic fungus Penicillium marneffei. J Clin Microbiol. 1995;33:85–89. doi: 10.1128/jcm.33.1.85-89.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lye G R, Wood G, Nimmo G. Subcutaneous zygomycosis due to Saksenaea vasiformis: rapid isolate identification using a modified sporulation technique. Pathology. 1996;28:364–365. doi: 10.1080/00313029600169364. [DOI] [PubMed] [Google Scholar]
- 26.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makimura K, Murayama S Y, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell T G, Freedman E A, Meyer W, White T J, Taylor J W. PCR identification of Cryptococcus neoformans. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 431–436. [Google Scholar]
- 29.Nazir Z, Hasan R, Pervaiz S, Alam A, Moazam F. Invasive retroperitoneal infection due to Basidiobolus ranarum with response to potassium iodide–case report and review of the literature. Ann Trop Paediatr. 1997;17:161–164. doi: 10.1080/02724936.1997.11747880. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell K. Zygomycetes in culture. Palfrey contributions in botany. no. 2. Athens: Department of Botany, University of Georgia; 1979. [Google Scholar]
- 31.O'Donnell K. Fusarium and its near relatives. In: Reynolds D R, Taylor J W, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, Conn: CAB International; 1993. pp. 225–233. [Google Scholar]
- 32.O'Donnell K, Cigelnik E, Benny G L. Phylogenetic relationships among the Harpellales and Kickxellales. Mycologia. 1998;90:624–639. [Google Scholar]
- 33.O'Donnell K, Cigelnik E, Weber N S, Trappe J M. Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia. 1997;89:48–65. [Google Scholar]
- 34.Oliver M R, Van Voorhis W C, Boeckh M, Mattson D, Bowden R A. Hepatic mucormycosis in a bone marrow transplant recipient who ingested naturopathic medicine. Clin Infect Dis. 1996;22:521–524. doi: 10.1093/clinids/22.3.521. [DOI] [PubMed] [Google Scholar]
- 35.Pasha T M, Leighton J A, Smilack J D, Heppell J, Colby T V, Kaufman L. Basidiobolomycosis: an unusual fungal infection mimicking inflammatory bowel disease. Gastroenterology. 1997;112:250–254. doi: 10.1016/s0016-5085(97)70242-7. [DOI] [PubMed] [Google Scholar]
- 36.Restrepo A. Treatment of tropical mycoses. J Am Acad Dermatol. 1994;31:S91–S102. doi: 10.1016/s0190-9622(08)81277-7. [DOI] [PubMed] [Google Scholar]
- 37.Richardson M D. Opportunistic and pathogenic fungi. J Antimicrob Chemother. 1991;28(Suppl. A):1–11. doi: 10.1093/jac/28.suppl_a.1. [DOI] [PubMed] [Google Scholar]
- 38.Rinaldi M G. Zygomycosis. Infect Dis Clin N Am. 1989;3:19–41. [PubMed] [Google Scholar]
- 39.Sandhu G S, Kline B C, Stockman L, Roberts G D. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 1995;33:2913–2919. doi: 10.1128/jcm.33.11.2913-2919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schipper M A, Maslen M M, Hogg G G, Chow C W, Samson R A. Human infection by Rhizopus azygosporus and the occurrence of azygospores in zygomycetes. J Med Vet Mycol. 1996;34:199–203. [PubMed] [Google Scholar]
- 41.Scully C, de Almeida O P, Sposto M R. The deep mycoses in HIV infection. Oral Dis. 1997;3(Suppl. 1):S200–S207. doi: 10.1111/j.1601-0825.1997.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 42.Sorenson M D. TreeRot. Ann Arbor: University of Michigan; 1996. [Google Scholar]
- 43.St-Germain G, Robert A, Ishak M, Tremblay C, Claveau S. Infection due to Rhizomucor pusillus: report of four cases in patients with leukemia and review. Clin Infect Dis. 1993;16:640–645. doi: 10.1093/clind/16.5.640. [DOI] [PubMed] [Google Scholar]
- 44.Swofford D L. PAUP: phylogenetic analysis using parsimony, version 4.0b1. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 44a.TreeBase: a database of phylogenetic information. 16 July 1999, posting date. TreeBASE [Online.] http://www.herbaria.harvard.edu/treebase/. [19 August 1999, last date accessed.]
- 45.Van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh T J, Renshaw G, Andrews J, Kwon-Chung J, Cunnion R C, Pass H I, Taubenberger J, Wilson W, Pizzo P A. Invasive zygomycosis due to Conidiobolus incongruus. Clin Infect Dis. 1994;19:423–430. doi: 10.1093/clinids/19.3.423. [DOI] [PubMed] [Google Scholar]
- 47.Weitzman I, Whittier S, McKitrick J C, Della-Latta P. Zygospores: the last word in identification of rare or atypical zygomycetes isolated from clinical specimens. J Clin Microbiol. 1995;33:781–783. doi: 10.1128/jcm.33.3.781-783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 49.Wysong D R, Waldorf A R. Electrophoretic and immunoblot analyses of Rhizopus arrhizus antigens. J Clin Microbiol. 1987;25:358–363. doi: 10.1128/jcm.25.2.358-363.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng R-Y, Chen G-Q. A non-thermophilic Rhizomucor causing human primary cutaneous mucormycosis. Mycosystema. 1991;4:45–57. [Google Scholar]