Abstract
Circular RNAs (circRNAs) are covalently closed RNA molecules that play important regulatory roles in various tumors. Prostate cancer (PCa) is one of the most common malignant tumors in the world, with high morbidity and mortality. In recent years, more and more circRNAs have been found to be abnormally expressed and involved in the occurrence and development of PCa, including cell proliferation, apoptosis, invasion, migration, metastasis, chemotherapy resistance, and radiotherapy resistance. Most of the circRNAs regulate biological behaviors of cancer through a competitive endogenous RNA (ceRNA) regulatory mechanism, and some can exert their functions by binding to proteins. circRNAs are also associated with many clinicopathological features of PCa, including tumor grade, lymph node metastasis, and distant metastasis. In addition, circRNAs are potential diagnostic and prognostic biomarkers for PCa. Considering their critical regulatory roles in the progression of PCa, circRNAs would be the potential therapeutic targets. In this paper, the current research status of circRNAs in PCa is briefly reviewed.
Keywords: circular RNA, prostate cancer, function, ceRNA, clinical significance
Graphical abstract

This paper reviews the role and clinical significance of circRNA in the occurrence and development of prostate cancer, so as to provide a comprehensive understanding of the relationship between circRNA and prostate cancer.
Introduction
Prostate cancer (PCa) is the most common type of cancer and the second leading cause of cancer mortality in men.1 In recent years, the incidence of PCa has significantly increased around the world, which seriously menaces men's health.1, 2, 3 Clinically, PCa can be defined as local or advanced stage, and treatment methods include surgery, radiotherapy, androgen deprivation therapy (ADT), and chemotherapy. Although these therapies for PCa have been successful to a certain extent, the effects of these treatments are still limited. Once patients progress to metastatic castration-resistant PCa (mCRPC), overall survival (OS) will be significantly reduced.4 Therefore, it is of great significance to find new molecular targets applied on the diagnosis and treatment of PCa.
Circular RNAs (circRNAs) are a novel class of non-coding RNA molecules with a single closed covalent loop. Unlike the linear RNA molecule, it lacks a 5′ cap structure and a 3′ polyadenylated tail structure.5 In 1976, circRNA was first identified in RNA viruses and was thought to have no biological function.6,7 Until recent years, with the development of RNA deep sequencing technology and bioinformatics, more and more circRNAs have been identified in normal and malignant human tissues or cells.8, 9, 10, 11, 12 Salzman et al. have confirmed that circRNAs are the major transcripts of a variety of human cell types.13 Moreover, accumulating evidence suggests that circRNAs have been found to regulate gene expression at transcriptional, posttranscriptional, and translational levels, which are involved in multiple pathological processes of diseases, such as neurological dysfunction,14 diabetes,15 cardiovascular diseases,16 and tumors.17
A series of studies suggest that circRNAs play pivotal roles in the occurrence and progression of PCa. In this article, we review the relevant literature and summarize the expression, biological functions, and clinical significance of circRNAs associated with PCa.
Biogenesis of circRNAs
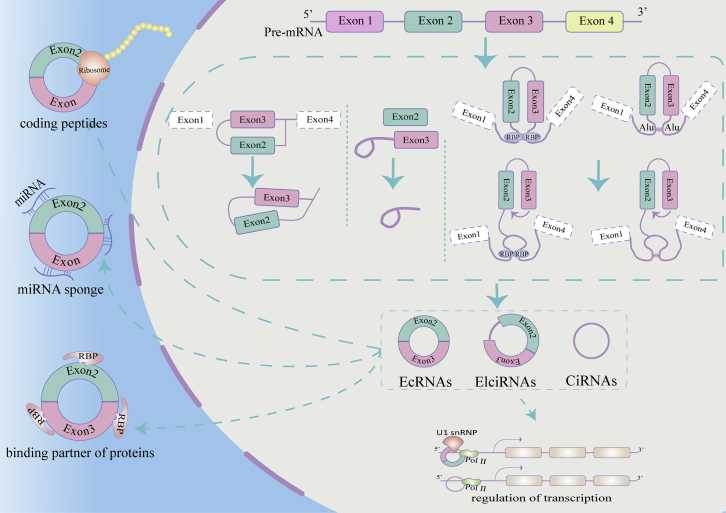
According to different origins, circRNAs can be classified into three main types: exonic circRNAs (ecRNAs),18 exon-intron circRNAs (EIciRNAs),19 and circular intronic RNAs (ciRNAs).20 circRNAs are produced from precursor messenger RNAs (mRNAs) via a unique back-splicing process. To explain the back-splicing processes, Jeck et al. proposed two widely accepted circRNA cycle models: lariat-driven circularization and intron-pairing-driven circularization.21 The lariat-driven circularization mode means that the splicing donor covalently binds to the splicing acceptor, which generate an exon-containing lariat and eventually form ecRNAs.22 In the intron-pairing-driven circularization model, the two exons are brought close by complementary base pairs between introns, and spliceosome cuts away the exons and introns to form ecRNAs or EIciRNAs.19 ciRNAs are produced by intron chains, which escape from the debranching and degradation in the process of back splicing.20 RNA-binding proteins (RBPs) play an important role in the biogenesis of circRNAs by promoting or inhibiting intron pairing. Quaking (QKI) and myoblindness (MBL) proteins can promote circRNA circulation by binding to specific sequence sites in flanking introns and linking two flanking introns together.9,23,24 In contrast, adenosine deaminase acting on RNA 1 (ADAR1) can bind to double-stranded RNA, destabilize RNA pairing, and thereby inhibit circRNAs biogenesis.25,26 In short, the biogenesis of circRNAs is regulated by many factors, such as enzymes, intronic sequences, and transcription factors.23,26, 27, 28
Functions of circRNAs
Plenty of studies have explored the potential functions of circRNAs. Five main functions of circRNAs have been revealed: sponging microRNAs (miRNAs),8 acting as protein sponges or decoys,10,12 adjusting alternative splicing,9 encoding a peptide or protein,11 and regulating transcription of parental genes.29 Serving as miRNAs sponges is the most common function of circRNAs.8 With multiple miRNA response elements, circRNA can competitively bind miRNA acting as intracellular competitive endogenous RNA (ceRNA) to regulate the expression of related genes. For instance, ciRS-7 targets miR-7 and upregulates HOXB13, the target of oncogenic miR-7, thereby promoting growth and metastasis of cancer.30 Interestingly, a miRNA stabilization mechanism has also been suggested for circRNAs.31,32 In addition, acting as protein sponges or decoys is also an important function for circRNAs. In 2014, Ashwal-Fluss et al. found that circMbl could sponge out the excess MBL protein by binding to it.9 Some circRNAs have also been identified to regulate alternative splicing. A circRNA from the SEPALLATA3 gene promotes the recruitment of splicing factors to the transcripts and regulates splicing of its cognate mRNA.33 Furthermore, increasing studies indicate that circRNAs participate in the different biological processes by translating into proteins (Figure 1). Although circRNAs lack the 7-methylguanosine (m7G) cap at the 5′ end and poly(A) tail at the 3′ end, they could complete the translation through an internal ribosome entry site (IRES) element-dependent or m6A modification-dependent manner.11,34,35 For example, circ-ZNF609 has an open reading frame (ORF) and is translated into a polypeptide chain in a splicing-dependent and cap-independent manner.11 Some circRNAs are found to regulate gene transcription. For example, circEIF3J and circPAIP2 can promote expression of their parental genes in a cis-acting manner. Mechanistically, an EIciRNA-U1 snRNP complex interacts with polymerase II (pol II) at the promoters of parental genes to enhance transcription.29
Figure 1.
Biogenesis and functions of circRNAs
The non-canonical back-splicing process generates three types of circRNAs: ecircRNAs, EIciRNAs, and ciRNAs. circRNAs regulate the transcription by interacting with RNA pol II. circRNAs can be translated into polypeptides. circRNAs serve as miRNA sponges. circRNAs mediate RBP actions.
Detection methods of circRNAs
The determination of circRNAs presents challenges. A two-step strategy is currently recommended for the validation of circRNAs, which has been proved successful in the application of miRNAs.36 This approach should include the following steps: (1) discovery and identification of circRNAs using RNA sequencing (RNA-seq) or microarray technology; (2) validation of circRNAs using reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Sanger sequencing.
The total RNA samples treated with RNase R after the removal of ribosomal RNA and polyadenosylated mRNA are the preferred samples for the preparation of circRNA-seq library.21,37, 38, 39 However, in many circRNA databases, RNase R was not processed.38,40, 41, 42, 43 Next-generation sequencing is a powerful way to map the whole genome of circRNAs. However, next-generation sequencing may have transcription mistakes produced from the reverse transcriptase or ligation, while the bioinformatics threshold or algorithm also affects the specificity and sensitivity.44,45 For circRNAs with longer sequences (>300–500), the commonly used sequencing by synthesis technique (Illumina) is less reliable due to random transcriptional errors.46 The novel third-generation sequencing, based on single-molecule sensing technology, has significantly longer read lengths and shorter sequencing times. Although there is still a certain error rate, they have the potential to promote the detection of circRNAs in the future.46 In fact, the error rate of the newest single-molecule sequencing is less than five errors per billion base pairs.47 RNA-seq can be used to find potential new circRNAs, but microarray can only detect characteristic circRNAs with the corresponding probes. On the other hand, microarray technology has a higher detection efficiency of circRNA than RNA-seq.48
The validation of circRNA candidates is the process of the identification between circRNA and linear RNA. The first step in the validation of circRNA candidates is to demonstrate the circularity of the transcript, which requires the design of specific divergent primers to detect circRNAs with RT-qPCR.49 A further verification step is to observe the stability of circRNA via digesting the RNA samples with RNase R.50 Notably, some linear RNAs are incompletely digested by RNase R.51 Sanger sequencing can also be used to verify the specific back splicing of circRNAs. For Sanger sequencing, it is noted that trans-spliced linear RNA might display the same sequence as back splicing of circRNA.52 Northern blotting is a gel electrophoresis method used for circRNA validation, which can separate circRNAs from their normal counterparts.8,53,54 The specificity of the results in the Northern blotting is optimized by quantifying the entire circRNA length. In addition to the previously mentioned methods, complete functional proof of circRNAs can be provided through cell experiments.49,55,56
Research of circRNAs in PCa
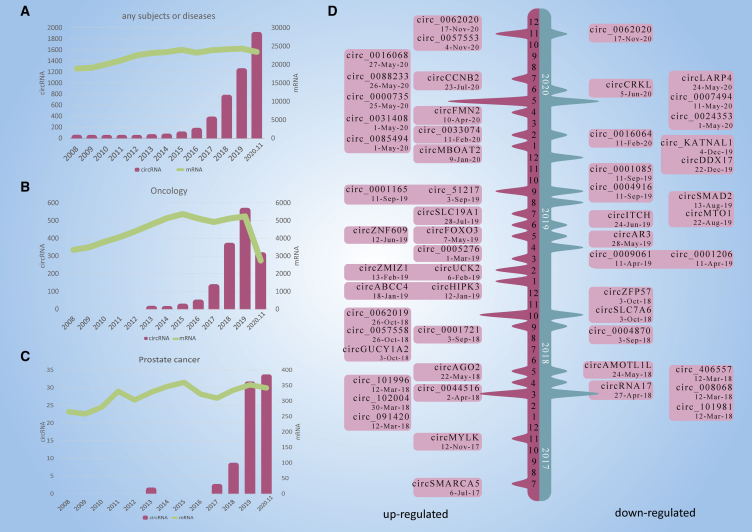
We systematically searched the Web of Science, PubMed, and Embase to identify all relevant literature published in English. The following search terms were used to identify any relevant studies: "circular RNA," "circRNA," "prostate cancer," "prostate neoplasm," "prostate tumor," and "prostate adenocarcinoma." We also used the combined Boolean operators "AND" or “OR” in the title/abstract. The amount of research on circRNAs has increased rapidly, while that on protein-coding genes has remained stable (Figure 2A). Similar trends are observed in the context of general oncology (Figure 2B) and PCa (Figure 2C). Most of these studies were published in the past 4 years. Besides, many circRNAs related to PCa were further verified by RT-qPCR (Figure 2D). These findings indicate that circRNAs and their roles in the development of PCa are attracting increasing attention.
Figure 2.
Research of circRNAs related to PCa
(A–C) The amount of research, as quantified by the annual number of peer-reviewed publications, has been relatively stable for mRNAs (celadon line), but not for circRNAs (purple bars), in the following categories: (A) overall, for any subject or disease; (B) oncology; and (C) PCa. (D) Increasing numbers of novel circRNAs were identified from 2017 to December 2020.
circRNA expression profiles in PCa
The advancement of high-throughput sequencing and bioinformatics has contributed to the discovery of circRNAs in PCa. Quantities of circRNAs that are dysregulated in PCa cell lines and tissues have been identified (Table 1). For example, 177 differentially expressed circRNAs were identified via a genome-wide circRNA-based microarray analysis of six pairs of PCa and adjacent normal tissues, among which 134 circRNAs were downregulated and 43 circRNAs were upregulated.57 Through circRNA chromatin immunoprecipitation, Ge et al. found 88,750 circRNAs in five pairs of PCa and adjacent normal tissues, among which 749 were differentially expressed.58 Xia et al. identified 1,021 circRNAs differentially expressed in four pairs of PCa and adjacent normal prostate tissues, and the combination of prostate specific antigen (PSA) levels and the two differentially expressed circRNAs significantly increased the sensitivity and specificity (84.5% and 90.9%, respectively) compared with PSA alone.59 Moreover, high-throughput sequencing was used to identify 827 upregulated and 1,279 downregulated circRNAs associated with PCa mesenchymal transformation in the PCa cell line.60 In another study, Zhang et al. analyzed the difference of circRNAs in three PCa cells (RWPE-1, 22RV1, and PC-3) by high-throughput circRNAs sequencing. A total of 9,545 circRNAs were detected and hundreds of differentially expressed circRNAs were identified.61 Some databases are often used to identify differentially expressed circRNAs, like the Gene Expression Synthesis Database (GEO). Using the GSE140927 dataset, Wu et al. identified 60 circRNAs differentially expressed between PCa tissues and normal tissues, and used bioinformatics to predict three circRNA-miRNA-mRNA interaction axes.62
Table 1.
Overview of circRNAs identified by RNA-seq and microarray in PCa
| Sample | Special treatment | Detection method | GEO database | Data source | Total circRNA | Number of circRNA differently expressed (fold change ≥2) | circRNAs validated by qRT-PCR | Reference | Year |
|---|---|---|---|---|---|---|---|---|---|
| One pair PCa and PCN tissues | – | circRNA microarray | GSE155792 | circRNA microarray | 170,340 | – | five | Deng et al.63 | 2020 |
| Three pair PCa and PCN tissues | RNase R | circRNA microarray | – | circRNA microarray | – | 15 (three upregulated, 12 downregulated) | one | Li et al.64 | 2020 |
| Four pair PCa and PCN tissues | RNase R | ceRNA microarray | – | ceRNA microarray | 88,371 | 1,021 (117 upregulated, 904 downregulated) | three | Xia et al.59 | 2018 |
| Five pairs HGT and LGT | – | circRNA microarray | – | circRNA microarray | – | 2,238(1,238 upregulated, 1,000 downregulated) | five | Yang et al.65 | 2019 |
| Five pair PCa and PCN tissues | – | circRNA microarray | – | circRNA microarray | 12,532 | 95 (71 upregulated, 24 downregulated) | three | Song et al.66 | 2019 |
| Five pair PCa and PCN tissues | RNase R | circRNA microarray | – | circRNA microarray | 7,385 | 988 (436 upregulated, 552 downregulated) | five | Shang et al.67 | 2020 |
| Five pair PCa and PCN tissues | – | circRNA microarray | – | circRNA microarray | 88,750 | 749 (261 upregulated, 487 downregulated) | – | Ge et al.58 | 2020 |
| Six pair PCa and PCN tissues | – | circRNA microarray | – | circRNA microarray | 9,599 | 177 (43 upregulated, 134 downregulated) | six | Rochow et al.57 | 2020 |
| Blood samples from the six patients and six healthy persons | RNase R | circRNA microarray | – | circRNA microarray | – | 109 (69 upregulated, 42 downregulated) | eight | Li et al.68 | 2020 |
| H-k LNCaP cells and normal LNCaP cells | RNase R | Sanger sequencing | GSE72844 | RNA-seq | 4,389 | 139 upregulated and 93 downregulated | six | Fei et al.28 | 2017 |
| LNCaP cell line | RNase R | circRNA microarray | GSE118959 | circRNA microarray | 13,617 | 930 (278 upregulated, 588 downregulated) | 10 | Greene et al.69 | 2019 |
| LNCaP-miR-145 cells and LNCaP-NC | – | circRNA microarray | – | circRNA microarray | 10,743 | 267 upregulated, 149 upregulated | five | He et al.70 | 2018 |
| PC-3M IE8 cells | RNase R | circRNA microarray | – | bioinformatics analysis | 4,788 | 827 upregulated and 1,279 downregulated | four | Yan et al.60 | 2020 |
| RWPE-1, 22RV1 and PC3 cell lines | RNase R | RNA-seq | – | RNA-seq and bioinformatics analysis | 9,545 | – | 20 | Zhang et al.61 | 2018 |
HGT, high-grade (Gleason > 8) PCa tissues; LGT low-grade (Gleason < 6) PCa tissues; PCN, PCa tissues paired adjacent normal prostate tissues; H-k, HNRNPL-knockdown; NC, normal control.
HGT and LGT taken from the two different areas within the same PCa sample.
In general, there are a large number of aberrantly expressed circRNAs between PCa tissues and normal tissues. In most studies, the number of circRNAs downregulated was higher than those upregulated.
Functions and mechanisms of circRNAs in PCa
circRNAs and hallmarks of cancer
In 2011, some scholars proposed 10 cancer characteristics that lead to the gradual transformation of normal cells into cancer cells, with important and positive implications for cancer research.71 We briefly summarized the circRNAs involved in the critical phases of PCa tumorigenesis and progression (Table 2). These circRNAs get involved in diverse processes of PCa, including cell proliferation, apoptosis, invasion, migration, metastasis, chemical resistance, and radiation resistance.
Table 2.
Dysregulated circRNAs in PCa
| Name | CircBase ID | Expression change | Target | Gene | Function | Types of PCa tissues and PCa cell lines | Reference | Year |
|---|---|---|---|---|---|---|---|---|
| circSMARCA5 | hsa_circ_0001445 | up | – | – | promoted cell proliferation and inhibited cell apoptosis | 21 pairs of PCa tissues and adjacent normal tissues; LNCaP, LNCaP-AI, 22RV1, DU145, PC-3, and WPMY-1 cell lines | Kong et al.72 | 2017 |
| circRNA-MYLK | hsa_circ_0141940 | up | miR-29a | – | promoted proliferation, invasion, and migration | 17 pairs of PCa tissues and adjacent normal tissues; DU145, LNCaP, PC-3, PC-3MIE8, and WPMY-1 cell lines | Dai et al.73 | 2018 |
| circ-102004 | – | up | – | – | promoted cell proliferation, migration and invasion; decreased cell apoptosis | 16 PCa samples and six normal prostate tissues; PC3 and 22RV1 cell lines | Si-Tu et al.74 | 2018 |
| circCSNK1G3 | hsa_circ_0001522 | up | miR-181b/day | – | promoted cell proliferation | LNCaP, 22Rv1, PC-3, and V16A cell lines | Chen et al.32 | 2019 |
| circABCC4 | hsa_circ_0030586 | up | miR-1182 | FOXP4 | promoted cell proliferation, migration, and invasion | 47 pairs of PCa tissues and adjacent normal tissues; PC3 and DU145 cells | Huang et al.75 | 2019 |
| circ0005276 | hsa_circ_0005276 | up | – | – | promoted cell proliferation and migration | 90 pairs of PCa tissues and adjacent normal tissues; DU145, LNCaP, PC-3, VCaP, and RWPE-1 cell lines | Feng et al.76 | 2019 |
| circAGO2 | hsa_circ_0135889 | up | – | – | promoted proliferation, invasion, and metastasis | PC-3 cell line | Cheng et al.77 | 2019 |
| circHIPK3 | hsa_circ_0000284 | up | miR-193a-3p | MCL1 | promoted cell proliferation, migration, and invasion | 26 pairs of PCa tissues and adjacent normal tissues; LNCaP, PC3, DU145, 22Rv1, and RWPE-1 cell lines | Chen et al.78 | 2019 |
| circHIPK3 | hsa_circ_0,000284 | up | miR-338-3p | ADAM17 | promoted cell proliferation and invasion | 60 pairs of PCa tissues and adjacent normal tissues; 22RV1, PC-3, DU145, LNCaP, and RWPE-1 cell lines | Cai et al.79 | 2019 |
| circZNF609 | hsa_circ_0000615 | up | miR-186-5p | – | promoted cell proliferation and metastasis | 25 pairs of PCa tissues and adjacent normal tissues; PC-3 and LNCaP cell lines | Jin et al.80 | 2019 |
| circRNA-51217 | – | up | miR-646 | TGFβ1 | promoted cell invasion | C4-2, PC3, DU145, LNCaP, and HEK293T cell lines | Xu et al.81 | 2020 |
| circ_0044516 | hsa_circ_0044516 | up | miR-29a-5p | – | promoted cell proliferation and metastasis | blood samples from the six patients and six healthy persons; PC3, 2B4, LNcap, and 5637 cell lines | Li et al.68 | 2020 |
| hsa_circ_0000735 | hsa_circ_0000735 | up | miR-7 | – | promoted cell viability, migration, invasion; inhibited cell apoptosis and cell resistance to docetaxel | 50 pairs of PCa tissues and adjacent normal tissues; PC3, DU145, PC-3/DTX, DU145/DTX, and RWPE-1 cell lines | Gao et al.82 | 2020 |
| circ-ZNF609 | hsa_circ_0000615 | up | miR-501-3p | HK2 | promoted cell proliferation, migration and glycolysis; inhibited cell apoptosis | 30 pairs of PCa tissues and adjacent normal tissues; 22RV1, VCaP, DU145, LNCap, and RWPE-1 cell lines | Du et al.83 | 2020 |
| circ_CCNB2 | hsa_circ_0035483 | up | miR-30b-5p | KIF18A | promoted cell proliferation, migration and autophagy; inhibited apoptosis | 25 pairs of PCa tissues and adjacent normal tissues; DU145, LNCaP, DU145/IR, and LNCaP/IR cell lines | Cai et al.84 | 2020 |
| circMBOAT2 | hsa_circ_0007334 | up | miR-1271-5p | mTOR | promoted cell proliferation, migration, and invasion | 50 pairs of PCa tissues and adjacent normal tissues; LNCaP, VCaP, PC3, DU145, C4-2B, and RWPE-1 cell lines | Shi et al.85 | 2020 |
| circFOXO3 | hsa_circ_0006404 | up | miR-29a-3p | SLC25A15 | promoted cell proliferation and inhibited cell apoptosis | 53 pairs of PCa tissues and adjacent normal tissues; blood samples from the 26 patients and 19 healthy persons; LNCaP, 22Rv1, DU145, PC-3, and WPMY-1 cell lines | Kong et al.86 | 2020 |
| circZMIZ1 | hsa_circ_0005844 | up | – | – | promoted cell proliferation | blood samples from the 14 patients and 14 healthy persons; DU145, C4-2, LNCaP, 22RV1, and RWPE-1 cell lines | Jiang et al.87 | 2020 |
| circ_0057553 | hsa_circ_0057553 | up | miR-515-5p | YES1 | promoted cell viability, migration, invasion, and glycolysis; inhibited cell apoptosis | 37 pairs of PCa tissues and adjacent normal tissues; 22RV1, PC3, DU145, LNCap, and RWPE-1 cell lines | Zhang et al.88 | 2020 |
| circ_0088233 | hsa_circ_0088233 | up | miR-185-3p | E2F1 and WNT2B | promoted cell proliferation, migration, invasion, and inhibited apoptosis | 46 pairs of PCa tissues and adjacent normal tissues; LNCaP, PC3, DU145, 22Rv1, and RWPE-1 cell lines | Deng et al.63 | 2020 |
| circ-0016068 | hsa_circ_0016068 | up | miR-330-3p | BMI-1 | promoted cell proliferation, migration, and invasion | 42 pairs of PCa tissues and adjacent normal tissues; VCaP, PC3, DU145, 22RV1, and RWPE-1 cell lines | Li et al.64 | 2020 |
| circ_SLC19A1 | hsa_circ_0062019 | up | miR-497 | SEPT2 | promotes cell growth and invasion | PC3, 22Rv1, DU145, LNCaP, and WPMY-1 cell lines | Zheng et al.89 | 2020 |
| circFMN2 | hsa_circ_0005100 | up | miR-1238 | LHX2 | promoted cell proliferation, migration, and invasion; inhibited apoptosis and EMT | 90 pairs of PCa tissues and adjacent normal tissues; blood samples from the 30 patients and 30 healthy persons; VCaP, PC3, DU145, LNCaP, and RWPE-1 cell lines | Shang et al.67 | 2020 |
| circHIPK3 | hsa_circ_0000284 | up | miR-338-3p | Cdc25B | promoted cell proliferation and inhibited apoptosis | 45 PCa samples and 25 normal prostate tissues; LNCaP, DU145, PC3, 22RV1, and RWPE-1 cell lines | Liu et al.90 | 2020 |
| circSMARCA5 | hsa_circ_0071043 | up | miR-432 | PDCD10 | accelerated the proliferation, metastasis, and glycolysis of cells | 30 pairs of PCa tissues and adjacent normal tissues; DU145, 22RV1, and RWPE-1 cell lines | Dong et al.91 | 2020 |
| circSLC19A1 | hsa_circ_0062019 | up | miR-326 | MAPK1 | promoted the proliferation, migration, and invasion of PCa cells | 48 pairs of PCa tissues and adjacent normal tissues; DU145, PC3, LNCaP, 22RV1, and RWPE-1 cell lines | Huang92 | 2020 |
| circ_0062020 | hsa_circ_0062020 | up | miR-615-5p | – | promoted cell proliferation, migration, and invasion; inhibited apoptosis and radiosensitivity | 30 radiosensitive PCa samples, 30 radioresistant PCa samples and 30 normal prostate tissues; DU145, LNCaP, and WPMY-1cell lines | Li et al.93 | 2020 |
| circRNA17 | hsa_circ_0001427 | down | miR-181c-5p | ARv7 | inhibited cell enzalutamide resistance and invasion | 26 PCa samples and 13 normal prostate tissues; C4–2, CWR22Rv1, and 293T cell lines | Wu et al.94 | 2019 |
| circSMAD2 | hsa_circ_0047603 | down | miR-9 | STAT and MEK/ERK pathways | inhibited cell proliferation and migration; blocked EMT process | 20 pairs of PCa tissues and adjacent normal tissues; LNCaP and PC3 cell lines | Han et al.95 | 2019 |
| circ_ITCH | hsa_circ_0001141 | down | miR-197 | – | inhibited cell proliferation and promoted cell apoptosis | DU 145, 22RV1, VCaP, PC-3, and RWPE-1 cell lines | Yuan et al.96 | 2019 |
| hsa_circ_0001206 | hsa_circ_0001206 | down | miR-1285-5p | Smad4 | inhibited cell proliferation, migration, and invasion | 50 pairs of PCa tissues and adjacent normal tissues; DU145, PC-3, LNCaP, and RWPE-1 cell lines | Song et al.66 | 2019 |
| circ_ITCH | hsa_circ_0001141 | down | miR-17-5p | HOXB13 | suppressed cell growth, invasion and metastasis; induced cell apoptosis | 52 pairs of PCa tissues and adjacent normal tissues; C4-2, LNCaP, DU145, 22Rv1, PC-3, VcaP, and RWPE-1 cell lines | Wang et al.97 | 2019 |
| circUCK2 | hsa_circ_001,357 | down | miR-767-5p | TET1 | inhibited cell proliferation and invasion | C4-2 cell line | Xiang et al.98 | 2019 |
| hsa_circ_0004870 | hsa_circ_0004870 | down | miR-145 | RBM39 | – | LNCaP cell line | Greene et al.69 | 2019 |
| circAMOTL1L | hsa_circ_000,350 | down | miR-193a-5p | PCDHA8 | inhibited cell migration and invasion | 62 PCa samples, and 35 normal prostate tissues; Du145, LNCaP, PC3, and RWPE-1 cell lines | Yang et al.65 | 2019 |
| circDDX17 | hsa_circ_0063308 | down | miR-346 | LHPP | inhibited cell proliferation, invasion, and EMT | 20 pairs of PCa tissues and adjacent normal tissues; 22Rv1 and PC-3 cell lines | Lin et al.99 | 2020 |
| circCRKL | hsa_circ_0001206 | down | miR-141 | KLF5 | repressed cell cycle, migration, and invasion, and boosted apoptosis | 45 pairs of PCa tissues and adjacent normal tissues; LNCaP, DU145, C4-2, 22RV1, and RWPE-1 cell lines | Nan et al.100 | 2020 |
| hsa_circ_0007494 | hsa_circ_0007494 | down | miR-616 | PTEN | inhibited cell proliferation, migration, and invasion | 49 pairs of PCa tissues and adjacent normal tissues; C4-2, PC3, 22Rv1, DU145, LNCaP, and RWPE-1 cell lines | Zhang et al.101 | 2020 |
| circ_KATNAL1 | hsa_circ_0008068 | down | miR-145-3p | WISP1 | inhibited cell proliferation and invasion; promoted cell apoptosis | 22Rv1, DU145, LNCaP, PC3, and WPMY-1 cell lines | Zheng et al.102 | 2020 |
| cir-ITCH | hsa_circ_0001141 | down | miR-17 | Wnt/β-catenin and PI3K/AKT/mTOR pathways | inhibited cell proliferation, migration, and invasion | 10 pairs of PCa tissues and adjacent normal tissues; LNCaP, PC-3, and RWPE-1 cell lines | Li et al.103 | 2020 |
| circ-MTO1 | hsa_circ_0076979 | down | miR-17-5p | – | inhibited cell proliferation and invasion | 298 pairs of PCa tissues and adjacent normal tissues; DU145, PC-3, VCaP, and RWPE-1 cell lines | Hu et al.104 | 2020 |
| circ-LARP4 | hsa_circ_0026224 | down | – | FOXO3A | inhibited cell migration and invasion | 55 pairs of PCa tissues and adjacent normal tissues; 22RV1, DU145, LNCap, and P69 cell lines | Weng et al.105 | 2020 |
| circFoxo3 | hsa_circ_0006404 | down | – | FOXO3 | inhibited cell survival, migration, invasion, and chemoresistance to docetaxel | 22 low-grade PCa samples, 24 high-grade PCa samples, and 18 normal prostate tissues; VCaP, LNCaP, Du145, PC3, and PWPE1 cell lines | Shen et al.106 | 2020 |
Cell proliferation
Tumor cells can maintain an active proliferative state by activating the cell proliferation signaling pathway.71 The PI3K/Akt signaling pathway plays an important role in the regulatory of cell proliferation, and its over-activation facilitates the proliferation, metastasis, and recurrence of PCa.107,108 Alterations of the PI3K/Akt pathway occur in 42% of localized prostate tumors and 100% of metastatic prostate tumors.109,110 The mammalian target of rapamycin (mTOR) kinase is a downstream signaling molecule of the PI3K/Akt pathway.111,112 circMBOAT2 upregulates mTOR expression by sponging miR-1271-5p, further activates the PI3K/Akt signaling pathway, and thereby promotes PCa cell proliferation and metastasis.85 A study found that circ_ITCH can inhibit the activation of the PI3K/AKT/mTOR pathway, thus inhibiting the proliferation and progression of PCa cells.103 It is noted that circ_ITCH exerts similar function in various cancers.113, 114, 115, 116 In addition, phosphatase and tensin homologues (PTEN) is a well-known negative regulator of the PI3K/Akt pathway.117 A study found the loss of PTEN in 44% of primary prostate carcinomas.118 Zhang et al. found that PTEN is low expressed in PCa tissues, while hsa_circ_0007494 could suppress the proliferation of PCa cells by upregulating the expression of PTEN.101 Moreover, Huang et al. proposed that circABCC4 promotes proliferation and invasion of PCa cells through circABCC4/miR-1182/FOXP4 axis.75 Chen et al. also found that circHIPK3/miR-193A-3P/MCL1 axis contributes to the proliferation and migration of PCa cells.78 It is worth noting that circRNAs can promote the proliferation of cancer cells through multiple signaling pathways. For example, circ_SLC19A1 can promote proliferation and metastasis of PCa cells through two signaling pathways, circ_SLC19A/miR-497/SEPT2 and circ_SLC19A/miR-326/MAPK1.89,92 This phenomenon suggests that the regulatory role of circRNA in tumor progression is more likely to be reticular than linear. Besides, circ-102004 was verified to inhibit the proliferation and migration of PCa cells.74 It has also been reported that circZMIZ1 can promote the proliferation of PCa cells.87
In addition, dysregulation of cell cycle regulators contributes to the unrestricted growth and proliferation of tumor cells.71 CDC25B activates the cyclin-dependent kinase (CDK) complex and plays a crucial role in cell cycle control.119 For example, circHIPK3 promotes G2/M transition and induces PCa cell proliferation by sponging miR-338-3p and increasing CDC25B expression, thus playing a carcinogenic role in PCa.90 Another study also confirmed that circHIPK3 can promote PCa cell proliferation through the cirHIPK3/miRNA-338-3P/ADAM17 axis, thus promoting the occurrence of PCa cell malignancy.79 KLF5 is a tumor suppressor in the progression of PCa.120 circCRKL can inhibit the cell cycle progression in PCa by regulating miR-141/KLF5 and plays a tumor suppressor role in PCa.100 Besides, Kong et al. also found that circSMARCA5 promotes cell cycle, inhibits cell apoptosis, and acts as an oncogene in PCa.72 Further studies showed that circSMARCA5 could promote glycolysis and proliferation of PCa cells through circSMARCA5/miR-432/PDCD10 axis, thus accelerating the process of PCa.91 Moreover, the overexpression of circFMN2 can reduce the percentage of G0/G1 phase of PCa cells, increase the percentage of PCa cells in G2 phase, and inhibit cell apoptosis.67
Apoptosis
Apoptosis and autophagy are the main mechanisms of controlled cell death.121 Caspase protein is a key regulator of apoptosis pathways. circ_KATNAL1 can regulate the activity of caspase through the miR-145-3P/WISP1 pathway, thus regulating apoptosis.102 Forkhead box transcription factor class O3 (FOXO3) is another key factor involved in the process of apoptosis.122 Shen et al. have shown that circFoxo3 enhances Foxo3 expression through sponging miRNAs in PCa, promoting FoxO3-mediated apoptosis.106 Similarly, Weng et al. found that circ-LARP4 inhibits cell migration of PCa by upregulating FOXO3A expression.105 In addition, circ_Foxo3 can also promote apoptosis in other cancers, such as breast cancer and bladder cancer.123,124 However, circFoxo3 was found to act in a carcinogenic role in PCa through the circFoxo3/miR-29a-3P/SLC25A15 axis in another study.86 Therefore, the role of circFOXO3 in PCa is still debatable. Moreover, Zhang et al. found that circ_0057553 increases cell proliferation and inhibits cell apoptosis through miR-515-5P/YES1 signaling pathway.88 Wang et al. confirmed that the circ_ITCH/miR-17-5p/HOXB13 signaling pathway contributes to the apoptosis of PCa cells.97 Furthermore, another study found that the expression of circ_ITCH is decreased in PCa tissues, and upregulation of circ_ITCH could promote apoptosis by targeting miR-197 × 96
Invasion, migration, and metastasis
Tumor cells invade and migrate into lymphatic vessels and blood vessels, eventually resulting in metastasis to distant organs.125 Some circRNAs can promote the invasion, migration, and metastasis of PCa. Argonaute 2 (AGO2) is a component of the mammalian AGO protein family, which is widely expressed and involved in posttranscriptional gene silencing.126 AGO2 has been verified as having expression levels that are aberrant in various types of cancers, including gastric cancer,127 colorectal cancer,128 and neuroblastoma,129 meanwhile AGO2 regulates proliferation, migration, and invasion.130 Chen et al. reported that circAGO2 interacts physically with human antigen R (HuR) protein, inhibits AGO2/miRNA-mediated gene silencing, and promotes tumor genesis and aggression.77 TGFβ1 plays a crucial role in the migration and invasion of many types of tumors.131 TGFβ1 has been reported to induce PCa cell invasion in vitro by Smad and non-Smad signaling.132, 133, 134 A study indicated that circRNA-51217 can sponge miRNA-646 to increase TGFβ1 expression and thereby induces TGFβ1/P-SMAD signaling to increase PCa cell invasion.81 Feng et al. revealed that the hsa-circ-0005276/FUS axis promotes the proliferation, migration, and epithelial-mesenchymal transition (EMT) of PCa cells by upregulating XIAP.76 Similarly, Jin et al. proposed that circZNF609 promotes invasion and metastasis of PCa cells by downregulating miR-186-5p.80 Li et al. found that circ_0044516 promotes tumor invasion and metastasis through interaction with miR-29a-5P.68
There is increasing evidence that circRNAs can also act as inhibitors of PCa invasion or metastasis. EMT is closely related to tumor metastasis.135 E-cadherin and N-cadherin are important proteins in the EMT process and are considered as markers of EMT.136 Han et al. proved that overexpression of circSMAD2 can upregulate E-cadherin and downregulate N-cadherin by sponging miR-9, thus inhibiting the EMT process of PCa.95 In addition, Lin et al. proposed that circDDX17 could inhibit proliferation, invasion, and EMT of PCa through the circDDX17/miR-346/LHPP signaling pathway.99 Similarly, circAMOTL1L can upregulate the expression of E-cadherin by targeting the miR-193A-5P/PCDHA8 axis to prevent the metastasis of PCa.65 In fact, the inhibition of EMT in PCa cells by p53 is also achieved through the circAMOTL1L/miR-193a-5P/PCDHA8 signaling pathway.65
Chemotherapy resistance
Most PCa cells are sensitive to androgen castration therapy (ADT), which makes ADT a first-line treatment for patients with advanced PCa.137 However, some studies have shown that ADT only slightly improves the survival rate of patients, and one-third of patients will relapse and eventually develop castration-resistant PCa (CRPC).138 Chemotherapy drug resistance is a troublesome problem in the therapy of CRPC. Tumor response to various chemotherapies is a complex process involving multiple circRNAs.
Docetaxel (DTX)-based chemotherapy is the standard first-line treatment for CRPC patients and can prolong the survival time of patients.139 However, repeated DTX treatment will produce DTX resistance and reduce the therapeutic effect.140 Understanding the underlying mechanisms of DTX resistance is critical to improve the prognosis of patients with CRPC. miR-7 plays a crucial role in the chemical resistance of various tumors, such as small-cell lung cancer,141 glioblastoma,142 and hepatocellular carcinoma.143 Gao et al. found that hsa_circ_0000735 is upregulated in DTX-resistant PCa tissues and cells, and inhibition of the expression of hsa_circ_0000735 could improve the sensitivity of PCa to DTX by sponging miR-7.82 In another study, circFoxo3 was demonstrated to be low expressed in PCa tissues, and circFoxo3 could improve the sensitivity of PCa to DTX.106
Although CRPC is resistant to ADT, it still relies on androgen through the androgen receptor (AR) pathway.144 Enzalutamide, as a new oral drug targeting androgen receptor signaling pathway, can competitively inhibit the binding of AR.145 It is a first-line drug for patients with CRPC who have not received chemotherapy.146 Its affinity to androgen receptors is five to eight times higher than that of previous antiandrogen drugs such as bicalutamide.147 However, about 20%–40% of patients will show inherent resistance to enzalutamide, and patients with initial objective response will eventually develop secondary drug resistance.148 Androgen receptor splicing variants (AR-Vs) in tumor cells of CRPC patients are associated with resistance to both abiraterone and enzalutamide.149,150 In a study, the researchers found that upregulating the expression of circRNA17 can inhibit the growth and invasion of enzalutamide-resistant cells, and confirmed that the circRNA17/miR-181c-5p/ARv7 signaling pathway plays a crucial role in the process of enzalutamide resistance.94 In addition, the expression level of circRNA17 is positively correlated with that of miR-181c-5p, indicating that circRNA17 functions as a reservoir or a stabilizing factor for miR-181c-5p, rather than sponging the miRNAs.94 Greene et al. reported that the circRNA expression profile is associated with enzalutamide-resistant PCa. Compared with the control, there are 278 upregulated circRNAs and 588 downregulated circRNAs in enzalutamide-resistant cell lines.69
Radiotherapy resistance
Radiation therapy has made exciting breakthroughs in the metastasis of PCa and postoperative repairing.151, 152, 153 However, a large proportion of PCa patients receiving radiotherapy relapse, which may be due to the development of radiation resistance.154 Understanding the anti-radiation mechanism of PCa is helpful to confront recurrent PCa and block its metastasis. Several studies have shown that circRNAs are connected with the regulation of radiosensitivity of many cancers.155, 156, 157 Cai et al. discovered that circ_CCNB2 is over-expressed in radiation-resistant PCa tissues and cells, and inhibition of circ_CCNB2 expression could increase the radiosensitivity of PCa through the miR-30b-5P/KIF18A axis.84 Therefore, inhibition of circ_CCNB2 can improve the radiotherapy efficacy of patients with recurrent PCa. In another study, circ-ZNF609 was abnormally upregulated in PCa tissues and cells. circ-ZNF609 accelerates glycolysis through miR-501-3P/HK2 axis, promoting the progression of PCa cells and radiation therapy resistance.83 It has also been reported that circ_0062020 can inhibit radiotherapy sensitivity through circ_0062020/miR-615-5P/TRIP13 signaling pathway.93 In general, the association between circRNA imbalance and the development of radioresistance of PCa remains to be further studied.
Mechanisms of circRNAs in PCa
circRNAs mainly play regulatory roles in the pathological process of PCa by acting as miRNA sponges. Some studies have also found that circRNAs in PCa can bind to proteins to play regulatory roles. It should be noted that circRNAs can be translated into polypeptides, but the existence of this kind of mechanism has not been found in studies related to PCa.
The ceRNA hypothesis indicates that circRNAs can compete with mRNA for binding to miRNAs, thus positively regulating target genes of miRNAs.8 For example, circHIPK3 acts as a molecular sponge for miR-193a-3P in PCa. Since miR-193A-3P usually inhibits MCL1 expression by targeting its 3′ UTR, circHIPK3 ultimately upregulates the expression of MCL1 in PCa.78 However, there are a few reports that circRNAs binding with miRNAs can enhance the inhibition of miRNA on downstream target genes. For example, Wu et al. revealed that circRNA17 inhibits the expression of ARv7 by sponging to miR-181c-5.94 Similarly, it has been reported that circ_KATNAL1 binding with miR-145-3p inhibits the expression of downstream target gene WISP1.102 The specific mechanism of this phenomenon remains to be further studied.
circRNAs play crucial roles in regulating gene expression at the transcriptional level by interacting with RBPs. Feng et al. found that the RNA-binding protein FUS interacts with circ0005276 to promote XIAP expression, thus promoting the occurrence and development of PCa.76 In another study, circAGO2 inhibits AGO2/miRNA-mediated gene silencing by binding to HuR, thereby promoting tumorigenesis and invasiveness in the PCa.77
Clinical significance of circRNAs in PCa
Localized PCa shows a relatively long-term survival, but metastatic PCa remains largely incurable even after intensive multimodal therapy.158 Therefore, early diagnosis, prognosis judgment, and the development of new therapies are of great significance for PCa.
Since its introduction in 1987 as a serum tumor marker, PSA has been the standard biomarker used to screen, diagnose, and monitor PCa.159 However, the application of PSA was thought to offer a small benefit for reducing PCa morbidity, but with flaws of overdiagnosis and overtreatment.160, 161, 162 Moreover, the level of PSA could be affected by multiple factors, such as trauma, prostatitis, and age.163,164 Prostate biopsy is the gold standard method to confirm the presence of cancer, but this technique is invasive and has some complications, such as infection and bleeding.165 circRNAs show great potential in the diagnosis and prognosis as biomarkers of PCa. First of all, as unique endogenous non-coding RNAs, circRNAs are highly conserved and widely expressed in a variety of tissues.166 Second, circRNA has a covalent closed-loop structure and high stability against RNA exonuclease degradation.21 Finally, in addition to physical tissues, PCa-related circRNAs can also be detected in the human bodily fluids, such as blood and urine.167,168 More and more circRNAs have been identified as diagnosis and prognosis biomarkers of PCa (Figure 3).
Figure 3.
The clinical significance of circRNAs in PCa
circRNAs could be detected from various clinical samples using RNA-seq or microarray. Besides, circRNAs can be used as diagnosis or prognosis biomarkers, and circRNAs are also promising therapeutic targets.
circRNAs and clinicopathologic characteristics in PCa
Many circRNAs have been validated to be related or unrelated to some clinicopathological parameters of PCa patients (Table 3). Xia et al. found that circ_SLC19A1 is related to the patient's age, Gleason score, plasma PSA level, and tumor invasion, while circ_0057558 is only related to Gleason score.59 A study found that circAR3 is significantly upregulated in the serum of PCa patients. Except for Gleason score, circAR3 is also related to lymph node metastasis (LNM), but not serum PSA levels or tumor invasion.169 Moreover, the expression of circAMOTL1L is significantly lower in PCa tissues than in normal prostate tissue, and its expression is related to Gleason score and serum PSA level.65 circFOXO3 can be detected in PCa tissues and serum, and it is irrelevant to the patient's age or PSA level, but is related to the Gleason score.86 Wang et al. found that circ_ITCH is downregulated in PCa tissues and closely related to Gleason score, serum PSA level, T stage, and OS rate, but not related to age, LNM, and number of tumor.97 Besides, Huang et al. studied the circ_ITCH and had some different findings. They found that circ_ITCH is downregulated and closely related to T stage, LNM, surgical margin, disease-free survival (DFS), and OS, but has nothing to do with Gleason score and serum PSA level. In fact, circ_ITCH is the only circRNA related to surgical margins in PCa-related studies and is the circRNA related to the largest number of clinicopathological parameters in the currently available research.170 circ_HIPK3 is one of the most studied circRNAs in PCa. Cai et al. found that it is upregulated in PCa tissues and has relationships with T stage, LNM, and distant metastasis (DM).79 A study carried out by Chen et al. showed circ_HIPK3 is upregulated in PCa tissues and related to T stage.78 Moreover, Liu et al. revealed that circ_HIPK3 is upregulated in PCa tissues and has a relationship with Gleason score.90
Table 3.
Relationship between circRNAs level and clinicopathologic characteristics in PCa
| circRNA name | CircBase ID | Host gene | Cutoff value | Internal reference | Dysregulation | Number of patients |
|---|---|---|---|---|---|---|
| circ_SLC19A1 | hsa_circ_0062019 | SLC19A1 | – | 18S rRNA | upregulated | 173 |
| circ_0057558 | hsa_circ_0057558 | SLC19A1 | – | 18S rRNA | upregulated | 173 |
| circAR3 | – | AR | – | GAPDH | upregulated | 91 |
| circAMOTL1L | hsa_circ_000,350 | PCHAD8 | – | GAPDH | Downregulated | 97 |
| circFOXO3 | hsa_circ_0006404 | FOXO3 | MEL | β-actin | upregulated | 53 |
| circABCC4 | hsa_circ_0030586 | ABCC4 | – | GAPDH & U6 | upregulated | 47 |
| circ_ITCH | – | ITCH | MEL | GAPDH & U6 | downregulated | 52 |
| circ-MTO1 | hsa_circ_0076979 | MTO1 | MEL | GAPDH | downregulated | 298 |
| hsa_circ_0001206 | hsa_circ_0001206 | CRKL | – | β-actin | downregulated | 50 |
| hsa_circ_0009061 | hsa_circ_0009061 | KDM1A | – | β-actin | downregulated | 50 |
| circRNA17 | hsa_circ_0001427 | PDLIM5 | – | GAPDH | downregulated | 39 |
| circHIPK3 | hsa_circ_0000284 | HIPK3 | – | GAPDH or U6 | upregulated | 26 |
| circ_ITCH | – | ITCH | MEL | GAPDH | downregulated | 324 |
| circ_0088233 | hsa_circ_0088233 | PAPPA | – | GAPDH | upregulated | 46 |
| circMBOAT2 | hsa_circ_0007334 | MBOAT2 | – | GAPDH | upregulated | 112 |
| circ-0016068 | hsa_circ_0016068 | BTG2 | MEL | GAPDH | upregulated | 42 |
| circFMN2 | hsa_circ_0005100 | FMN2 | MEL | 18S rRNA | upregulated | 88 |
| circ_HIPK3 | – | HIPK3 | – | – | upregulated | 45 |
| hsa_circ_0007494 | hsa_circ_0007494 | ROCK2 | – | GAPDH | downregulated | 49 |
| circ-LARP4 | – | LARP4 | – | GAPDH | downregulated | 55 |
| hsa_circ_0000735 | hsa_circ_0000735 | P2RX1 | MEL | GAPDH | upregulated | 50 |
| Age | Tumor size | Gleason score | PSA | T stage | LNM | Number of tumors | DM | Invasion | Surgical margin | DFS | OS | Reference | Year |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes | – | yes | yes | – | – | – | – | yes | – | – | – | Xia et al.59 | 2018 |
| No | – | yes | no | – | – | – | – | no | – | – | – | Xia et al.59 | 2018 |
| – | – | yes | no | – | yes | – | – | no | – | – | – | Luo et al.169 | 2019 |
| No | – | yes | yes | – | – | – | – | – | – | – | – | Yang et al.65 | 2019 |
| No | – | yes | no | – | – | – | – | – | – | – | – | Kong et al.86 | 2020 |
| No | – | – | – | yes | yes | – | yes | – | – | – | yes | Huang et al.75 | 2019 |
| No | – | yes | yes | yes | no | no | – | – | – | – | yes | Wang et al.97 | 2019 |
| No | – | no | no | yes | yes | – | – | – | no | yes | yes | Hu et al.104 | 2020 |
| No | – | yes | no | yes | no | – | – | – | – | – | – | Song et al.66 | 2019 |
| No | – | no | no | yes | no | – | – | – | – | – | – | Song et al.66 | 2019 |
| – | – | yes | – | – | – | – | – | – | – | – | – | Wu et al.94 | 2019 |
| – | – | – | – | yes | – | – | – | – | – | – | – | Chen et al.78 | 2019 |
| No | – | no | no | yes | yes | – | – | – | yes | yes | yes | Huang et al.170 | 2019 |
| No | – | – | – | yes | – | – | – | – | – | – | – | Deng et al.63 | 2020 |
| No | – | yes | no | yes | no | – | – | – | – | yes | – | Shi et al.85 | 2020 |
| No | – | yes | – | yes | yes | – | – | – | – | – | yes | Li et al.64 | 2020 |
| No | – | – | – | yes | yes | – | yes | – | – | – | – | Shang et al.67 | 2020 |
| No | – | yes | – | – | no | – | no | – | – | – | – | Liu et al.90 | 2020 |
| No | yes | no | – | yes | no | – | yes | – | – | – | – | Zhang et al.101 | 2020 |
| – | – | – | – | – | – | – | – | – | – | – | yes | Weng et al.105 | 2020 |
| – | – | – | – | – | – | – | – | – | – | – | yes | Gao et al.82 | 2020 |
MEL, median expression level.
Summarizing the current research data, common PCa parameters associated with circRNAs include Gleason score, serum PSA level, tumor T stage, LNM, DM, OS, and DFS. Other parameters include age, tumor size, surgical margin, and invasion. circRNAs related to the tumor number of PCa have not been found.
circRNAs as diagnostic biomarkers for PCa
Several studies have explored the promising value of circRNA being a diagnostic biomarker for PCa. There are 15, eight, and five circRNAs that are related to T stage, LNM, and DM, respectively (Table 3). For diagnosis, receiver operating characteristic (ROC) curves were applied to evaluate the clinical diagnostic value of circRNAs. Xia et al. studied the circ_0062019 and circ_0057558 and the area under the ROC curve (AUC) of these two circRNA. This study revealed that the AUC of circ_0062019 is 0.828 with 95% confidence interval (CI) from 0.764 to 0.893, and the sensitivity and specificity are 80.0% and 74.6%, respectively. Correspondingly, the AUC of circ_0057558 is 0.729 with 95% CI from 0.651 to 0.807, and the sensitivity and specificity are 74.1% and 63.4%, respectively. Moreover, when the two differential circRNAs are combined, the AUC is better (0.861) than PSA alone (0.854).59 Huang et al. verified that the AUC of circ_ITCH is 0.812, and the sensitivity and specificity are 88.3% and 61.7%, respectively.170 Therefore, circRNAs are potential diagnostic biomarkers for PCa, and the combination of multiple circRNAs often contributes to better diagnostic value.
circRNAs as prognostic biomarkers for PCa
circRNA levels can be used to predict patient survival parameters (Table 4). Univariate or multivariate Cox regression analysis is often used to explore the relationship between circRNA and PCa prognosis. Here, we summarize the results found by the researchers up to now. For prognosis, there are six and three circRNAs that are related to OS and DFS, respectively. Among them, two downregulated circRNAs (circ_MTO1 and circ_ITCH) as well as one upregulated circRNA (circMBOAT2) are related to poor DFS.85,104,170 Besides, three downregulated circRNAs (circ_ITCH, circ-MTO1, circ-LARP4) as well as three upregulated circRNAs (circABCC4, circ-0016068, circ_0000735) are related to poor OS.75,82,97,104,105,170 The above circRNAs can be detected in PCa tissues. Wang et al. first constructed an eight-circRNA risk score model, which could reliably predict the biochemical recurrence of PCa patients. The AUC for the model (AUC = 0.799) was better than the AUC of clinical factors (AUC of PSA = 0.557, AUC of Gleason score = 0.626, and AUC of pathological stage = 0.569).171
Table 4.
circRNAs as prognostic biomarkers for PCa
| circRNA | Cutoff value | Number of circRNA expression |
p value | HR | 95% CI |
Follow-up (months) | Prognosis | Reference | Year | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Lower | Higher | ||||||||
| circMBOAT2 | MEL | 25 | 25 | 0.024 | 0.33 (KMA) | 0.13 | 0.86 | 96 | DFS | Shi et al.85 | 2020 |
| MEL | 31 | 31 | 0.037 | 0.39 (KMA) | 0.1 | 0.98 | 96 | DFS | |||
| circ_ITCH | MEL | 162 | 162 | <0.001 | 0.476 (MCRA) | 0.329 | 0.690 | – | DFS | Huang et al.170 | 2019 |
| circMTO1 | MEL | 149 | 149 | 0.005 | 0.541 (UCRA) | 0.351 | 0.832 | 48 | DFS | Hu et al.104 | 2019 |
| circ_0000735 | MEL | 25 | 25 | 0.02 | – | – | – | – | OS | Gao et al.82 | 2020 |
| circ_0016068 | MEL | 21 | 21 | 0.036 | – | – | – | – | OS | Li et al.64 | 2020 |
| circLARP4 | – | 55 | 55 | 0.009 | – | – | – | – | OS | Weng et al.105 | 2020 |
| circABCC4 | – | 23 | 24 | <0.05 | – | – | – | – | OS | Huang et al.75 | 2019 |
| circ_ITCH | MEL | 162 | 162 | 0.001 | 0.415 (MCRA) | 0.245 | 0.703 | – | OS | Huang et al.170 | 2019 |
| circ_ITCH | MEL | 26 | 26 | <0.01 | – | – | – | – | OS | Wang et al.97 | 2019 |
| circMTO1 | MEL | 149 | 149 | 0.016 | 0.444 (UCRA) | 0.229 | 0.860 | 48 | OS | Hu et al.104 | 2019 |
HR, hazard ratio; KMA, Kaplan-Meyer analysis; MCRA, multivariate Cox regression analysis; UCRA, univariate Cox regression analysis.
circRNAs as therapeutic targets for PCa
Considering the important roles of circRNAs in tumorigenesis and development of PCa, circRNAs definitely could be the potential therapeutic targets. Novel therapies will be based on the individual changes of circRNAs in PCa, bringing the tumor therapy into the era of precision therapy. So far, two strategies have been proposed to treat tumors by targeting circRNAs. The first is to restore the function of circRNAs with tumor suppressor activities to modulate native disease-linked circRNAs, while the second is to block the actions of non-coding RNAs with oncogenic function.172 In general, the study of circRNAs as potential therapeutic targets has attracted extensive attention in oncology.
Challenges and future perspective
Currently, there is a better understanding about the role of circRNAs in PCa, but they are still a long way from clinical application. First of all, the correct analysis and detection is the primary condition for the use of circRNAs as biomarkers. However, the detection techniques for circRNAs still have limitations, especially in terms of sensitivity and specificity. Future studies must document the standard operating procedures to analyze all variables that may affect circRNA detection, such as sample processing, RNA isolation protocol, and data normalization strategies.173,174 To avoid these potential errors, basic and applied researchers need to collaborate with clinicians to translate circRNA research findings to clinical applications. Laboratory scientists are the link between the basic science and clinical applications. During the phases of clinical validation based on retrospective or prospective studies, the primary task of the laboratory scientist is to monitor the quality of the measurements and make a practice-oriented evaluation of the data.
In addition, safe and effective delivery of the oligonucleotides into the cancer tissue remains a challenge. Although modifications could increase the stability and affinity to targets, most of the oligonucleotide therapies need additional optimal delivery systems to achieve the desired biological effects.175 To improve delivery efficiency, viral and non-viral vectors have been developed. The virus-based delivery system has high transduction efficiency and can effectively deliver genetic material to target cells. However, viral vectors have some disadvantages, such as immunogenic/inflammatory responses, low loading capacity, and quality control. Compared with viral vectors, non-viral delivery systems are relatively safe, and can effectively avoid the problems faced by viral vectors through rational design and appropriate modifications. Therefore, non-viral delivery systems have emerged as promising intracellular biomolecule carriers. RNA nanoparticles are representative of the non-viral vectors. RNA nanoparticles are modular, so functional modules composed of RNA can self-assemble into the multifunctional architectures. There are two main approaches for RNA nanoparticle construction. The first approach is the sequence-dependent self-folding of RNA nanostructures based on computational algorithms and secondary or tertiary structure prediction.176,177 The second approach is to assemble RNA nanoparticles using the naturally occurring RNA motifs as core building blocks, such as three-way junction, four-way junction, and kissing loops.178, 179, 180 In short, the field of RNA vectors has developed rapidly over the last decade, which will facilitate the application of circRNAs in targeted therapy.
Conclusion
PCa is a complex disease, and its specific pathogenesis is still unclear. Elucidating circRNA biology has been crucial to understanding tumorigenesis over the past decade. As outlined in this review, the functions of circRNAs are involved in various physiological and pathological processes, such as cell proliferation, migration, invasion, apoptosis, chemotherapy resistance, and radiotherapy resistance. It has been reported that circRNAs are significantly correlated with many clinicopathological features of PCa and survival parameters of PCa patients, which make them potential diagnostic and prognostic biomarkers of PCa. The characteristics of circRNAs, such as conservation, stability, specificity, and detectability, make them potential diagnostic and prognostic biomarkers for PCa. In addition, circRNAs play important regulatory roles in the carcinogenesis and progression of PCa and are potential targets for the treatment of PCa.
In summary, the use of circRNAs in the diagnosis and treatment of PCa is promising and attractive. However, further studies are needed before circRNAs can be incorporated into clinical practice.
Acknowledgments
We thank all other researchers in our laboratory. This project was supported by the National Natural Science Foundation of China (no. 81900645, no. 82170779), the Yellow Crane Talents Outstanding Youth Foundation of Wuhan in 2019, and the Outstanding Youth Foundation of Tongji Hospital affiliated to Tongji Medical College, Huazhong University of Science and Technology in 2020 (no. 2020YQ15).
Author contributions
K.T., Y.T., and X.L. designed the study. X.L., T.Y., Y.H., and Y.T. collected data. Z.Y., Z.C., D.X., H.L., E.P., and X.Y. analyzed the data. Y.T. and X.L. made the figures and tables. K.T., Y.T., and X.L. drafted the article. All authors revised the paper and approved the final version of the manuscript, and agreed to be accountable for all aspects of the work.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Zhiqiang Chen, Email: zhqchen_8366@163.com.
Kun Tang, Email: tangsk1990@163.com.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Center M.M., Jemal A., Lortet-Tieulent J., Ward E., Ferlay J., Brawley O., Bray F. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Nuhn P., De Bono J.S., Fizazi K., Freedland S.J., Grilli M., Kantoff P.W., Sonpavde G., Sternberg C.N., Yegnasubramanian S., Antonarakis E.S. Update on systemic prostate cancer therapies: management of metastatic castration-resistant prostate cancer in the era of precision oncology. Eur. Urol. 2019;75:88–99. doi: 10.1016/j.eururo.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 8.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 9.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A., et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhter R. Circular RNA and Alzheimer's disease. Adv. Exp. Med. Biol. 2018;1087:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 15.Stoll L., Sobel J., Rodriguez-Trejo A., Guay C., Lee K., Venø M.T., Kjems J., Laybutt D.R., Regazzi R. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol. Metab. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T., et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 17.Geng Y., Jiang J., Wu C. Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen I., Chen C.Y., Chuang T.J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Chenard C.A., Richard S. New implications for the QUAKING RNA binding protein in human disease. J. Neurosci. Res. 2008;86:233–242. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P., Cotter M.B., Bowden M., Lis R.T., Zhao S.G., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolisetty M.T., Graveley B.R. Circuitous route to transcription regulation. Mol. Cell. 2013;51:705–706. doi: 10.1016/j.molcel.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R.C., Ke S., Meng F.K., Lu J., Zou X.J., He Z.G., Wang W.F., Fang M.H. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P., et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 32.Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., Zeng Y., Hua J.T., Petricca J., Guo H., et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843 e22. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C., et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y., Jia X., Xu J. The new function of circRNA: translation. Clin. Transl. Oncol. 2020;22:2162–2169. doi: 10.1007/s12094-020-02371-1. [DOI] [PubMed] [Google Scholar]
- 36.Fendler A., Stephan C., Yousef G.M., Kristiansen G., Jung K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat. Rev. Urol. 2016;13:734–752. doi: 10.1038/nrurol.2016.193. [DOI] [PubMed] [Google Scholar]
- 37.Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X.O., Dong R., Zhang Y., Zhang J.L., Luo Z., Zhang J., Chen L.L., Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panda A.C., Grammatikakis I., Munk R., Gorospe M., Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip. Rev. RNA. 2017;8 doi: 10.1002/wrna.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maass P.G., Glazar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C., et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. (Berl). 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y.C., Li J.R., Sun C.H., Andrews E., Chao R.F., Lin F.M., Weng S.L., Hsu S.D., Huang C.C., Cheng C., et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia S., Feng J., Lei L., Hu J., Xia L., Wang J., Xiang Y., Liu L., Zhong S., Han L., et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 44.Houseley J., Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen T.B., Veno M.T., Damgaard C.K., Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franz A., Rabien A., Stephan C., Ralla B., Fuchs S., Jung K., Fendler A. Circular RNAs: a new class of biomarkers as a rising interest in laboratory medicine. Clin. Chem. Lab. Med. 2018;56:1992–2003. doi: 10.1515/cclm-2018-0231. [DOI] [PubMed] [Google Scholar]
- 47.Abascal F., Harvey L.M.R., Mitchell E., Lawson A.R.J., Lensing S.V., Ellis P., Russell A.J.C., Alcantara R.E., Baez-Ortega A., Wang Y., et al. Somatic mutation landscapes at single-molecule resolution. Nature. 2021;593:405–410. doi: 10.1038/s41586-021-03477-4. [DOI] [PubMed] [Google Scholar]
- 48.Li S., Teng S., Xu J., Su G., Zhang Y., Zhao J., Zhang S., Wang H., Qin W., Lu Z.J., et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20:1420–1433. doi: 10.1093/bib/bby006. [DOI] [PubMed] [Google Scholar]
- 49.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., CircInteractome Gorospe M. A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki H., Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 53.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 54.Veno M.T., Hansen T.B., Veno S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boeckel J.N., Jae N., Heumuller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 57.Rochow H., Jung M., Weickmann S., Ralla B., Stephan C., Elezkurtaj S., Kilic E., Zhao Z., Jung K., Fendler A., et al. Circular RNAs and their linear transcripts as diagnostic and prognostic tissue biomarkers in prostate cancer after prostatectomy in combination with clinicopathological factors. Int. J. Mol. Sci. 2020;21:7812. doi: 10.3390/ijms21217812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ge S., Sun C., Hu Q., Guo Y., Xia G., Mi Y., Zhu L. Differential expression profiles of circRNAs in human prostate cancer based on chip and bioinformatic analysis. Int. J. Clin. Exp. Pathol. 2020;13:1045–1052. [PMC free article] [PubMed] [Google Scholar]
- 59.Xia Q., Ding T., Zhang G., Li Z., Zeng L., Zhu Y., Guo J., Hou J., Zhu T., Zheng J., et al. Circular RNA expression profiling identifies prostate cancer- specific circRNAs in prostate cancer. Cell Physiol. Biochem. 2018;50:1903–1915. doi: 10.1159/000494870. [DOI] [PubMed] [Google Scholar]
- 60.Yan Z., Xiao Y., Chen Y., Luo G. Screening and identification of epithelial-to-mesenchymal transition-related circRNA and miRNA in prostate cancer. Pathol. Res. Pract. 2020;216:152784. doi: 10.1016/j.prp.2019.152784. [DOI] [PubMed] [Google Scholar]
- 61.Zhang C., Xiong J., Yang Q., Wang Y., Shi H., Tian Q., Huang H., Kong D., Lv J., Liu D., et al. Profiling and bioinformatics analyses of differential circular RNA expression in prostate cancer cells. Future Sci. OA. 2018;4:FSOA340. doi: 10.4155/fsoa-2018-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y.P., Lin X.D., Chen S.H., Ke Z.B., Lin F., Chen D.N., Xue X.Y., Wei Y., Zheng Q.S., Wen Y.A., et al. Identification of prostate cancer-related circular RNA through bioinformatics analysis. Front. Genet. 2020;11:892. doi: 10.3389/fgene.2020.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng Z.H., Yu G.S., Deng K.L., Feng Z.H., Huang Q., Pan B., Deng J.Z. Hsa_circ_0088233 alleviates proliferation, migration, and invasion of prostate cancer by targeting hsa-miR-185-3p. Front. Cell Dev. Biol. 2020;8:528155. doi: 10.3389/fcell.2020.528155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q., Wang W., Zhang M., Sun W., Shi W., Li F. Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front. Cell Dev. Biol. 2020;8:827. doi: 10.3389/fcell.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Yang Z., Qu C.B., Zhang Y., Zhang W.F., Wang D.D., Gao C.C., Ma L., Chen J.S., Liu K.L., Zheng B., et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516–2532. doi: 10.1038/s41388-018-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song Z., Zhuo Z., Ma Z., Hou C., Chen G., Xu G. Hsa_Circ_0001206 is downregulated and inhibits cell proliferation, migration and invasion in prostate cancer. Artif. Cells Nanomed. Biotechnol. 2019;47:2449–2464. doi: 10.1080/21691401.2019.1626866. [DOI] [PubMed] [Google Scholar]
- 67.Shan G., Shao B., Liu Q., Zeng Y., Fu C., Chen A., Chen Q. circFMN2 sponges miR-1238 to promote the expression of LIM-homeobox gene 2 in prostate cancer cells. Mol. Ther. Nucleic Acids. 2020;21:133–146. doi: 10.1016/j.omtn.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Li T., Sun X., Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J. Cell. Biochem. 2020;121:2118–2126. doi: 10.1002/jcb.28239. [DOI] [PubMed] [Google Scholar]
- 69.Greene J., Baird A.M., Casey O., Brady L., Blackshields G., Lim M., O'Brien O., Gray S.G., McDermott R., Finn S.P. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci. Rep. 2019;9:10739. doi: 10.1038/s41598-019-47189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He J.H., Han Z.P., Zhou J.B., Chen W.M., Lv Y.B., He M.L., Li Y.G. MiR-145 affected the circular RNA expression in prostate cancer LNCaP cells. J. Cell. Biochem. 2018;119:9168–9177. doi: 10.1002/jcb.27181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 72.Kong Z., Wan X., Zhang Y., Zhang P., Zhang Y., Zhang X., Qi X., Wu H., Huang J., Li Y. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem. Biophys. Res. Commun. 2017;493:1217–1223. doi: 10.1016/j.bbrc.2017.07.162. [DOI] [PubMed] [Google Scholar]
- 73.Dai Y., Li D., Chen X., Tan X., Gu J., Chen M., Zhang X. Circular RNA myosin light chain kinase (MYLK) promotes prostate cancer progression through modulating Mir-29a expression. Med. Sci. Monit. 2018;24:3462–3471. doi: 10.12659/MSM.908009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Si-Tu J., Cai Y., Feng T., Yang D., Yuan S., Yang X., He S., Li Z., Wang Y., Tang Y., et al. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int. J. Biol. Macromol. 2019;122:1235–1243. doi: 10.1016/j.ijbiomac.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 75.Huang C., Deng H., Wang Y., Jiang H., Xu R., Zhu X., Huang Z., Zhao X. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J. Cell. Mol. Med. 2019;23:6112–6119. doi: 10.1111/jcmm.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng Y., Yang Y., Zhao X., Fan Y., Zhou L., Rong J Yu Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10:792. doi: 10.1038/s41419-019-2028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M., et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen D., Lu X., Yang F., Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag. Res. 2019;11:1415–1423. doi: 10.2147/CMAR.S190669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai C., Zhi Y., Wang K., Zhang P., Ji Z., Xie C., Sun F. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi: 10.2147/OTT.S196931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin C., Zhao W., Zhang Z., Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif. Cells Nanomed. Biotechnol. 2019;47:3350–3358. doi: 10.1080/21691401.2019.1648281. [DOI] [PubMed] [Google Scholar]
- 81.Xu H., Sun Y., You B., Huang C.P., Ye D., Chang C. Androgen receptor reverses the oncometabolite R-2-hydroxyglutarate-induced prostate cancer cell invasion via suppressing the circRNA-51217/miRNA-646/TGFbeta1/p-Smad2/3 signaling. Cancer Lett. 2020;472:151–164. doi: 10.1016/j.canlet.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 82.Gao Y., Liu J., Huan J., Che F. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020;20:334. doi: 10.1186/s12935-020-01421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du S., Zhang P., Ren W., Yang F., Du C. Circ-ZNF609 accelerates the radioresistance of prostate cancer cells by promoting the glycolytic metabolism through miR-501-3p/HK2 Axis. Cancer Manag. Res. 2020;12:7487–7499. doi: 10.2147/CMAR.S257441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai F., Li J., Zhang J., Huang S. Knockdown of circ_CCNB2 sensitizes prostate cancer to radiation through Repressing autophagy by the miR-30b-5p/KIF18A Axis. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2019.3538. [DOI] [PubMed] [Google Scholar]
- 85.Shi J., Liu C., Chen C., Guo K., Tang Z., Luo Y., Chen L., Su Y., Xu K. Circular RNA circMBOAT2 promotes prostate cancer progression via a miR-1271-5p/mTOR axis. Aging (Albany NY) 2020;12:13255–13280. doi: 10.18632/aging.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kong Z., Wan X., Lu Y., Zhang Y., Huang Y., Xu Y., Liu Y., Zhao P., Xiang X., Li L., et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell. Mol. Med. 2020;24:799–813. doi: 10.1111/jcmm.14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang H., Lv D.J., Song X.L., Wang C., Yu Y.Z., Zhao S.C. Upregulated circZMIZ1 promotes the proliferation of prostate cancer cells and is a valuable marker in plasma. Neoplasma. 2020;67:68–77. doi: 10.4149/neo_2019_190213N116. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y., Shi Z., Li Z., Wang X., Zheng P., Li H. Circ_0057553/miR-515-5p regulates prostate cancer cell proliferation, apoptosis, migration, invasion and aerobic glycolysis by targeting YES1. Onco Targets Ther. 2020;13:11289–11299. doi: 10.2147/OTT.S272294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng Y., Li J.X., Chen C.J., Lin Z.Y., Liu J.X., Lin F.J. Extracellular vesicle-derived circ_SLC19A1 promotes prostate cancer cell growth and invasion through the miR-497/septin 2 pathway. Cell Biol. Int. 2020;44:1037–1045. doi: 10.1002/cbin.11303. [DOI] [PubMed] [Google Scholar]
- 90.Liu F., Fan Y., Ou L., Li T., Fan J., Duan L., Yang J., Luo C., Wu X. CircHIPK3 facilitates the G2/M transition in prostate cancer cells by sponging miR-338-3p. Onco Targets Ther. 2020;13:4545–4558. doi: 10.2147/OTT.S242482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dong C., Fan B., Ren Z., Liu B., Wang Y. CircSMARCA5 facilitates the progression of prostate cancer through miR-432/PDCD10 Axis. Cancer Biother. Radiopharm. 2021;36:70–83. doi: 10.1089/cbr.2019.3490. [DOI] [PubMed] [Google Scholar]
- 92.Huang B., Zhou D., Huang X., Xu X., Xu Z. Silencing circSLC19A1 inhibits prostate cancer cell proliferation, migration and invasion through regulating miR-326/MAPK1 Axis. Cancer Manag. Res. 2020;12:11883–11895. doi: 10.2147/CMAR.S267927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H., Zhi Y., Ma C., Shen Q., Sun F., Cai C. Circ_0062020 knockdown strengthens the radiosensitivity of prostate cancer cells. Cancer Manag. Res. 2020;12:11701–11712. doi: 10.2147/CMAR.S273826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu G., Sun Y., Xiang Z., Wang K., Liu B., Xiao G., Niu Y., Wu D., Chang C. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019;10:37. doi: 10.1038/s41419-018-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han N., Ding L., Wei X., Fan L., Yu L. circSMAD2 governs migration and epithelial-mesenchymal transition by inhibiting microRNA-9. J. Cell. Biochem. 2019 doi: 10.1002/jcb.29638. [DOI] [PubMed] [Google Scholar]
- 96.Yuan Y., Chen X., Huang E. Upregulation of circular RNA itchy E3 ubiquitin protein ligase inhibits cell proliferation and promotes cell apoptosis through targeting MiR-197 in prostate cancer. Technol. Cancer Res. Treat. 2019;18 doi: 10.1177/1533033819886867. 1533033819886867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X., Wang R., Wu Z., Bai P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019;19:328. doi: 10.1186/s12935-019-0994-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Xiang Z., Xu C., Wu G., Liu B., Wu D. CircRNA-UCK2 increased TET1 inhibits proliferation and invasion of prostate cancer cells via sponge MiRNA-767-5p. Open Med. (Wars) 2019;14:833–842. doi: 10.1515/med-2019-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin Q., Cai J., Wang Q.Q. The significance of circular RNA DDX17 in prostate cancer. Biomed. Res. Int. 2020;2020:1878431. doi: 10.1155/2020/1878431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nan C., Wang Y., Yang S., Chen Y. circCRKL suppresses the progression of prostate cancer cells by regulating the miR-141/KLF5 axis. Pathol. Res. Pract. 2020;216:153182. doi: 10.1016/j.prp.2020.153182. [DOI] [PubMed] [Google Scholar]
- 101.Zhang S., Zhang X., Chen G., Zheng X., Zhu X., Shan L. Hsa_circ_0007494 suppresses prostate cancer progression via miR-616/PTEN axis. Exp. Cell Res. 2020;395:112233. doi: 10.1016/j.yexcr.2020.112233. [DOI] [PubMed] [Google Scholar]
- 102.Zheng Y., Chen C.J., Lin Z.Y., Li J.X., Liu J., Lin F.J., Zhou X. Circ_KATNAL1 regulates prostate cancer cell growth and invasiveness through the miR-145-3p/WISP1 pathway. Biochem. Cell Biol. 2020;98:396–404. doi: 10.1139/bcb-2019-0211. [DOI] [PubMed] [Google Scholar]
- 103.Li S., Yu C., Zhang Y., Liu J., Jia Y., Sun F., Zhang P., Li J., Guo L., Xiao H., et al. Circular RNA cir-ITCH is a potential therapeutic target for the treatment of castration-resistant prostate cancer. Biomed. Res. Int. 2020;2020:7586521. doi: 10.1155/2020/7586521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu Y., Guo B. Circ-MTO1 correlates with favorable prognosis and inhibits cell proliferation, invasion as well as miR-17-5p expression in prostate cancer. J. Clin. Lab. Anal. 2020;34:e23086. doi: 10.1002/jcla.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weng X.D., Yan T., Liu C.L. Circular RNA_LARP4 inhibits cell migration and invasion of prostate cancer by targeting FOXO3A. Eur. Rev. Med. Pharmacol. Sci. 2020;24:5303–5309. doi: 10.26355/eurrev_202005_21312. [DOI] [PubMed] [Google Scholar]
- 106.Shen Z., Zhou L., Zhang C., Xu J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi: 10.1016/j.canlet.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 107.Blattner M., Liu D., Robinson B.D., Huang D., Poliakov A., Gao D., Nataraj S., Deonarine L.D., Augello M.A., Sailer V., et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell. 2017;31:436–451. doi: 10.1016/j.ccell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen C., Cai Q., He W., Lam T.B., Lin J., Zhao Y., Chen X., Gu P., Huang H., Xue M., et al. AP4 modulated by the PI3K/AKT pathway promotes prostate cancer proliferation and metastasis of prostate cancer via upregulating L-plastin. Cell Death Dis. 2017;8:e3060. doi: 10.1038/cddis.2017.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Millis S.Z., Jardim D.L., Albacker L., Ross J.S., Miller V.A., Ali S.M., Kurzrock R. Phosphatidylinositol 3-kinase pathway genomic alterations in 60,991 diverse solid tumors informs targeted therapy opportunities. Cancer. 2019;125:1185–1199. doi: 10.1002/cncr.31921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scott K.L., Kabbarah O., Liang M.C., Ivanova E., Anagnostou V., Wu J., Dhakal S., Wu M., Chen S., Feinberg T., et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wan L., Zhang L., Fan K., Cheng Z.X., Sun Q.C., Wang J.J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed. Res. Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang C., Yuan W., Yang X., Li P., Wang J., Han J., Tao J., Li P., Yang H., Lv Q., et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer. 2018;17:19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morgensztern D., McLeod H.L. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer Drugs. 2005;16:797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 118.Yoshimoto M., Cunha I.W., Coudry R.A., Fonseca F.P., Torres C.H., Soares F.A., Squire J.A. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br. J. Cancer. 2007;97:678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sur S., Agrawal D.K. Phosphatases and kinases regulating CDC25 activity in the cell cycle: clinical implications of CDC25 overexpression and potential treatment strategies. Mol. Cell. Biochem. 2016;416:33–46. doi: 10.1007/s11010-016-2693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y., Liu K., Zhang Y., Qi J., Lu B., Shi C., Yin Y., Cai W., Li W. ABL-N may induce apoptosis of human prostate cancer cells through suppression of KLF5, ICAM-1 and Stat5b, and upregulation of Bax/Bcl-2 ratio: an in vitro and in vivo study. Oncol. Rep. 2015;34:2953–2960. doi: 10.3892/or.2015.4293. [DOI] [PubMed] [Google Scholar]
- 121.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y., Gan B., Liu D., Paik J.H. FoxO family members in cancer. Cancer Biol. Ther. 2011;12:253–259. doi: 10.4161/cbt.12.4.15954. [DOI] [PubMed] [Google Scholar]
- 123.Lu W.Y. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16:589–590. doi: 10.1080/15384101.2017.1278935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang C., Tao W., Ni S., Chen Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco Targets Ther. 2019;12:8085–8094. doi: 10.2147/OTT.S215823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Polacheck W.J., Zervantonakis I.K., Kamm R.D. Tumor cell migration in complex microenvironments. Cell. Mol. Life Sci. 2013;70:1335–1356. doi: 10.1007/s00018-012-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 127.Zhang J., Fan X.S., Wang C.X., Liu B., Li Q., Zhou X.J. Up-regulation of Ago2 expression in gastric carcinoma. Med. Oncol. 2013;30:628. doi: 10.1007/s12032-013-0628-2. [DOI] [PubMed] [Google Scholar]
- 128.Papachristou D.J., Korpetinou A., Giannopoulou E., Antonacopoulou A.G., Papadaki H., Grivas P., Scopa C.D., Kalofonos H.P. Expression of the ribonucleases drosha, dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011;459:431–440. doi: 10.1007/s00428-011-1119-5. [DOI] [PubMed] [Google Scholar]
- 129.Qu H., Zheng L., Song H., Jiao W., Li D., Fang E., Wang X., Mei H., Pu J., Huang K., et al. microRNA-558 facilitates the expression of hypoxia-inducible factor 2 alpha through binding to 5’-untranslated region in neuroblastoma. Oncotarget. 2016;7:40657–40673. doi: 10.18632/oncotarget.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Y., Wang B., Chen X., Li W., Dong P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget. 2017;8:54735–54746. doi: 10.18632/oncotarget.18047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ao M., Williams K., Bhowmick N.A., Hayward S.W. Transforming growth factor-beta promotes invasion in tumorigenic but not in nontumorigenic human prostatic epithelial cells. Cancer Res. 2006;66:8007–8016. doi: 10.1158/0008-5472.CAN-05-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin T.H., Lee S.O., Niu Y., Xu D., Liang L., Li L., Yeh S.D., Fujimoto N., Yeh S., Chang C. Differential androgen deprivation therapies with anti-androgens casodex/bicalutamide or MDV3100/Enzalutamide versus anti-androgen receptor ASC-J9(R) lead to promotion versus suppression of prostate cancer metastasis. J. Biol. Chem. 2013;288:19359–19369. doi: 10.1074/jbc.M113.477216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barrett C.S., Millena A.C., Khan S.A. TGF-β effects on prostate cancer cell migration and invasion require FosB. Prostate. 2017;77:72–81. doi: 10.1002/pros.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang L., Wu H., Wang L., Zhang H., Lu J., Liang Z., Liu T. Asporin promotes pancreatic cancer cell invasion and migration by regulating the epithelial-to-mesenchymal transition (EMT) through both autocrine and paracrine mechanisms. Cancer Lett. 2017;398:24–36. doi: 10.1016/j.canlet.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 136.Araki K., Shimura T., Suzuki H., Tsutsumi S., Wada W., Yajima T., Kobayahi T., Kubo N., Kuwano H. E/N-cadherin switch mediates cancer progression via TGF-β-induced epithelial-to-mesenchymal transition in extrahepatic cholangiocarcinoma. Br. J. Cancer. 2011;105:1885–1893. doi: 10.1038/bjc.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cornford P., Bellmunt J., Bolla M., Briers E., De Santis M., Gross T. Henry AM., Joniau S., Lam TB., Mason MD, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur. Urol. 2017;71:630–642. doi: 10.1016/j.eururo.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 138.Walsh P.C. Immediate versus deferred treatment for advanced prostatic cancer: initial results of the Medical Research Council trial. The Medical Research Council Prostate Cancer Working Party Investigators Group. J. Urol. 1997;158:1623–1624. [PubMed] [Google Scholar]
- 139.Galsky M.D., Vogelzang N.J. Docetaxel-based combination therapy for castration-resistant prostate cancer. Ann. Oncol. 2010;21:2135–2144. doi: 10.1093/annonc/mdq050. [DOI] [PubMed] [Google Scholar]
- 140.Sekino Y., Oue N., Koike Y., Shigematsu Y., Sakamoto N., Sentani K., Teishima J., Shiota M., Matsubara A., Yasui W. KIFC1 inhibitor CW069 induces apoptosis and reverses resistance to docetaxel in prostate cancer. J. Clin. Med. 2019;8:225. doi: 10.3390/jcm8020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lai J., Yang H., Zhu Y., Ruan M., Huang Y., Zhang Q. MiR-7-5p-mediated downregulation of PARP1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer. 2019;19:602. doi: 10.1186/s12885-019-5798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jia B., Liu W., Gu J., Wang J., Lv W., Zhang W., Hao Q., Pang Z., Mu N., Zhang W., et al. MiR-7-5p suppresses stemness and enhances temozolomide sensitivity of drug-resistant glioblastoma cells by targeting Yin Yang 1. Exp. Cell Res. 2019;375:73–81. doi: 10.1016/j.yexcr.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 143.Hu H., Yang L., Li L., Zeng C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem. Biophys. Res. Commun. 2018;503:2400–2406. doi: 10.1016/j.bbrc.2018.06.168. [DOI] [PubMed] [Google Scholar]
- 144.Mellado B., Codony J., Ribal M.J., Visa L., Gascón P. Molecular biology of androgen-independent prostate cancer: the role of the androgen receptor pathway. Clin. Transl. Oncol. 2009;11:5–10. doi: 10.1007/s12094-009-0304-3. [DOI] [PubMed] [Google Scholar]
- 145.Tran C., Ouk S., Clegg N.J., Chen Y., Watson P.A., Arora V., Wongvipat J., Smith-Jones P.M., Yoo D., Kwon A., et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ramadan W.H., Kabbara W.K., Al Basiouni Al Masri H.S. Enzalutamide for patients with metastatic castration-resistant prostate cancer. Onco Targets Ther. 2015;8:871–876. doi: 10.2147/OTT.S80488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Massard C., Fizazi K. Targeting continued androgen receptor signaling in prostate cancer. Clin. Cancer Res. 2011;17:3876–3883. doi: 10.1158/1078-0432.CCR-10-2815. [DOI] [PubMed] [Google Scholar]
- 148.Palapattu G.S. Commentary on "AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer." Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J, Division of urologic oncology, department of urology, university of Michigan, MI. N Engl J Med 2014; 371(11):1028-1038. Urol. Oncol. 2016;34:520. doi: 10.1016/j.urolonc.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 149.Antonarakis E.S., Lu C., Luber B., Wang H., Chen Y., Nakazawa M., Nadal R., Paller C.J., Denmeade S.R., Carducci M.A., et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015;1:582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Onstenk W., Sieuwerts A.M., Kraan J., Van M., Nieuweboer A.J., Mathijssen R.H., Hamberg P., Meulenbeld H.J., De Laere B., Dirix L.Y., et al. Efficacy of cabazitaxel in castration-resistant prostate cancer is independent of the presence of AR-V7 in circulating tumor cells. Eur. Urol. 2015;68:939–945. doi: 10.1016/j.eururo.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 151.Sita T.L., Petras K.G., Wafford Q.E., Berendsen M.A., Kruser T.J. Radiotherapy for cranial and brain metastases from prostate cancer: a systematic review. J. Neurooncol. 2017;133:531–538. doi: 10.1007/s11060-017-2460-6. [DOI] [PubMed] [Google Scholar]
- 152.Picardi C., Perret I., Miralbell R., Zilli T. Hypofractionated radiotherapy for prostate cancer in the postoperative setting: what is the evidence so far? Cancer Treat. Rev. 2018;62:91–96. doi: 10.1016/j.ctrv.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 153.Kamran S.C., D'Amico A.V. Radiation therapy for prostate cancer. Hematol. Oncol. Clin. North Am. 2020;34:45–69. doi: 10.1016/j.hoc.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 154.Chang L., Graham P.H., Hao J., Bucci J., Cozzi P.J., Kearsley J.H., Li Y. Emerging roles of radioresistance in prostate cancer metastasis and radiation therapy. Cancer Metastasis Rev. 2014;33:469–496. doi: 10.1007/s10555-014-9493-5. [DOI] [PubMed] [Google Scholar]
- 155.Chen L., Zhou H., Guan Z. CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR-9/platelet-derived growth factor receptor B axis. Biochem. Biophys. Res. Commun. 2019;512:786–792. doi: 10.1016/j.bbrc.2019.03.126. [DOI] [PubMed] [Google Scholar]
- 156.Yang W., Liu Y., Gao R., Xiu Z., Sun T. Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell Signal. 2019;60:122–135. doi: 10.1016/j.cellsig.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 157.Zhao M., Xu J., Zhong S., Liu Y., Xiao H., Geng L., Liu H. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Oncol. Rep. 2019;41:1893–1900. doi: 10.3892/or.2019.6972. [DOI] [PubMed] [Google Scholar]
- 158.Wang G., Zhao D., Spring D.J., DePinho R.A. Genetics and biology of prostate cancer. Genes Dev. 2018;32:1105–1140. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stamey T.A., Yang N., Hay A.R., McNeal J.E., Freiha F.S., Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 1987;317:909–916. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- 160.Moyer V.A. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2012;157:120–134. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 161.Hugosson J., Carlsson S. Overdetection in screening for prostate cancer. Curr. Opin. Urol. 2014;24:256–263. doi: 10.1097/MOU.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 162.Grossman D.C., Curry S.J., Owens D.K., Bibbins-Domingo K., Caughey A.B., Davidson K.W., Doubeni C.A., Ebell M., Epling J.W., Jr.., Kemper A.R., et al. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. JAMA. 2018;319:1901–1913. doi: 10.1001/jama.2018.3710. [DOI] [PubMed] [Google Scholar]
- 163.Nadler R.B., Humphrey P.A., Smith D.S., Catalona W.J., Ratliff T.L. Effect of inflammation and benign prostatic hyperplasia on elevated serum prostate specific antigen levels. J. Urol. 1995;154:407–413. doi: 10.1097/00005392-199508000-00023. [DOI] [PubMed] [Google Scholar]
- 164.Hu J.C., Williams S.B., Carter S.C., Eggener S.E., Prasad S., Chamie K., Trinh Q.D., Sun M., Nguyen P.L., Lipsitz S.R. Population-based assessment of prostate-specific antigen testing for prostate cancer in the elderly. Urol. Oncol. 2015;33 doi: 10.1016/j.urolonc.2014.06.003. 69.e29-34. [DOI] [PubMed] [Google Scholar]
- 165.Borghesi M., Ahmed H., Nam R., Schaeffer E., Schiavina R., Taneja S., Weidner W., Loeb S. Complications after systematic, random, and image-guided prostate biopsy. Eur. Urol. 2017;71:353–365. doi: 10.1016/j.eururo.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 166.Lim M.C.J., Baird A.M., Aird J., Greene J., Kapoor D., Gray S.G., McDermott R., Finn S.P. RNAs as candidate diagnostic and prognostic markers of prostate cancer-from cell line models to liquid biopsies. Diagnostics (Basel) 2018;8:60. doi: 10.3390/diagnostics8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kolling M., Haddad G., Wegmann U., Kistler A., Bosakova A., Seeger H., Hubel K., Haller H., Mueller T., Wuthrich R.P., et al. Circular RNAs in urine of kidney transplant patients with acute T cell-mediated allograft rejection. Clin. Chem. 2019;65:1287–1294. doi: 10.1373/clinchem.2019.305854. [DOI] [PubMed] [Google Scholar]
- 169.Luo J., Li Y., Zheng W., Xie N., Shi Y., Long Z., Xie L., Fazli L., Zhang D., Gleave M., et al. Characterization of a prostate- and prostate cancer-specific circular RNA encoded by the androgen receptor gene. Mol. Ther. Nucleic Acids. 2019;18:916–926. doi: 10.1016/j.omtn.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Huang E., Chen X., Yuan Y. Downregulated circular RNA itchy E3 ubiquitin protein ligase correlates with advanced pathologic T stage, high lymph node metastasis risk and poor survivals in prostate cancer patients. Cancer Biomark. 2019;26:41–50. doi: 10.3233/CBM-182111. [DOI] [PubMed] [Google Scholar]
- 171.Wang S., Su W., Zhong C., Yang T., Chen W., Chen G., Liu Z., Wu K., Zhong W., Li B., et al. An eight-CircRNA assessment model for predicting biochemical recurrence in prostate cancer. Front. Cell Dev. Biol. 2020;8:599494. doi: 10.3389/fcell.2020.599494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang M., Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 2018;11:21. doi: 10.1186/s13045-018-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rochow H., Franz A., Jung M., Weickmann S., Ralla B., Kilic E., Stephan C., Fendler A., Jung K. Instability of circular RNAs in clinical tissue samples impairs their reliable expression analysis using RT-qPCR: from the myth of their advantage as biomarkers to reality. Theranostics. 2020;10:9268–9279. doi: 10.7150/thno.46341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bossuyt P.M., Reitsma J.B., Bruns D.E., Gatsonis C.A., Glasziou P.P., Irwig L., Lijmer J.G., Moher D., Rennie D., de Vet H.C., et al. StardTARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin. Chem. 2015;61:1446–1452. doi: 10.1373/clinchem.2015.246280. [DOI] [PubMed] [Google Scholar]
- 175.Ling H. Non-coding RNAs: therapeutic strategies and delivery systems. Adv. Exp. Med. Biol. 2016;937:229–237. doi: 10.1007/978-3-319-42059-2_12. [DOI] [PubMed] [Google Scholar]
- 176.Yingling Y.G., Shapiro B.A. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett. 2007;7:2328–2334. doi: 10.1021/nl070984r. [DOI] [PubMed] [Google Scholar]
- 177.Afonin K.A., Bindewald E., Yaghoubian A.J., Voss N., Jacovetty E., Shapiro B.A., Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Westhof E., Masquida B., Jaeger L. RNA tectonics: towards RNA design. Fold Des. 1996;1:R78–R88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 179.Guo S., Piao X., Li H., Guo P. Methods for construction and characterization of simple or special multifunctional RNA nanoparticles based on the 3WJ of phi29 DNA packaging motor. Methods. 2018;143:121–133. doi: 10.1016/j.ymeth.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jasinski D., Haque F., Binzel D.W., Guo P. Advancement of the emerging field of RNA nanotechnology. ACS Nano. 2017;11:1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]