Abstract
Infection with Borrelia garinii outer surface protein (Osp) A serotype 4 strains has been correlated with the development of neuroborreliosis in Lyme borreliosis patients in Europe. OspA serotype 4 isolates have been recovered primarily from human cerebrospinal fluid, suggesting a tropism for this environment. Previous studies with monoclonal antibodies directed against OspA and OspC demonstrated that OspA serotype 4 strains are antigenically closely related. In view of the pronounced antigenic and genetic variability that has been noted in the Osps of other Borrelia isolates, we sought to determine if OspA serotype 4 strains represent a recently emerged clonal lineage of B. garinii. Toward this goal, a representative group of OspA serotype 4 strains was analyzed for traits that typically exhibit hypervariability among isolates that cause Lyme borreliosis. The following criteria were assessed: (i) ospC sequences, (ii) plasmid composition, (iii) genomic restriction fragment length polymorphism (RFLP) patterns, and (iv) the RFLP patterns of the upstream homology box (UHB) element which flanks members of the UHB gene family at their 5′ end. Collectively, these analyses demonstrate genetic homogeneity, suggesting that OspA serotype 4 strains are a recently emerged clonal lineage with an apparent tropism for the central nervous system.
The genus Borrelia contains several spirochete species that are causative agents of important human diseases such as Lyme borreliosis and relapsing fever (for a review, see reference 7). Lyme borreliosis is caused by pathogenic species of the Borrelia burgdorferi sensu lato complex. While as many as nine species have been delineated in the B. burgdorferi sensu lato complex, disease in humans is attributed primarily to B. burgdorferi, B. garinii, and B. afzelii (2, 17, 18, 21, 23, 28, 29, 37, 38, 51, 52, 57). Members of the genus exhibit several unique features which distinguish them from other eubacteria. One such feature is their genome, which is comprised of a linear chromosome and a variable series of linear and circular plasmids (6). Interestingly, the plasmids carry a large number of genes that appear to be completely unique to the genus Borrelia (15). The proteins encoded by these Borrelia-specific genes likely play central roles in defining the unique aspects of Borrelia biology and pathogenesis. The plasmids also carry the majority of outer surface protein (Osp) genes including the ospAB operon, ospC, ospD, and the upstream homology box (UHB) gene family (5, 12, 32, 33, 35, 40). Like the plasmids that harbor them, the genes encoding surface-exposed proteins exhibit significant degrees of inter- and intraspecies variation (20, 30, 33, 48, 51, 57–59).
Analyses of phenotypic diversity of the Lyme disease spirochetes have centered primarily on the Osps, which have been the focus of efforts to design Borrelia vaccines and diagnostic assays (43, 46). On the basis of variation in the Osps, isolates of the B. burgdorferi sensu lato complex can be antigenically subtyped (53, 56, 57). On the basis of immunoreactivity with monoclonal antibodies, seven OspA serotypes have been defined, and specific serotypes have been demonstrated to correlate with specific species of the B. burgdorferi sensu lato complex (57). B. burgdorferi strains are serotype 1, and B. afzelii strains are serotype 2. The OspA proteins of B. garinii are more antigenically diverse and are divided into five distinct OspA serotypes (serotypes 3 through 7). Another important Osp carried by species of the B. burgdorferi sensu lato complex is OspC (16). The ospC gene has been mapped to a highly stable 26-kb circular plasmid that is universally distributed among isolates (32, 40). The highly immunogenic OspC protein exhibits a high degree of inter- and intraspecies variation, with 16 distinct OspC types having been delineated (48, 56). OspE and OspF (22) are additional outer surface proteins that are genetically and antigenically diverse (34, 47). These immunogenic proteins are members of a large protein family called the UHB protein family (34, 47). UHB gene family members are flanked at their 5′ ends by a highly conserved upstream homology box (UHB) element and are carried by a series of closely related 32-kb circular plasmids (cp32s) (12). A striking feature of this gene family is that it appears to be evolutionarily unstable and to undergo recombinational and mutational events at a high frequency during infection (25). It has been postulated that rearrangements in these genes may allow for the continual generation of new antigenic variants which could aid in evasion of the humoral immune response (34, 47).
Serotyping studies of isolates from Europe revealed a striking correlation between neuroborreliosis and infection with B. garinii OspA serotype 4 strains (55). Specifically, the majority of B. garinii isolates that have been recovered and cultured from cerebrospinal fluid from patients in Germany, The Netherlands, Denmark, and Slovenia are OspA serotype 4 (54, 55, 57). It is interesting that while strains of this serotype have been recovered from Lyme borreliosis patients, they have not been cultivated directly from ticks. Regarding the association between OspA serotype 4 strains, neuroborreliosis, and central nervous system invasion, it is possible that isolates of this serotype have a higher pathogenic potential than isolates of other OspA serotypes. Support for this comes from the observation that OspA serotype 4 strains exhibit a greater degree of resistance to serum than do other B. garinii OspA serotypes, which are serum sensitive (49).
To determine if OspA serotype 4 strains represent a genetically and antigenically homogeneous group of neurotropic organisms, we have conducted a comparative analysis of a representative group of strains. As a means for assessing homogeneity, we have characterized traits of B. burgdorferi sensu lato isolates that have been demonstrated to be highly variable among isolates. These analyses demonstrate that B. garinii OspA type-4 serotype strains are significantly more genetically homogeneous than any other defined group of isolates analyzed to date. The data suggest that B. garinii OspA serotype 4 strains are a recently emerged clonal lineage that appear to have a strong potential to disseminate and a higher tropism for the central nervous system than strains of other OspA serotypes.
MATERIALS AND METHODS
Bacterial strains and DNA isolation.
The B. garinii OspA serotype 4 strains investigated in this study are described in Table 1. All strains except those specifically indicated in Table 1 were isolated at the Max von Pettenkofer-Institut, Munich, Germany. Strain AO1 was provided by A. van Dam, and strain DK6 was provided by K. Hansen. All bacterial isolates were cultivated in modified Kelly medium as described previously (59), and total genomic DNA was extracted as described previously (27).
TABLE 1.
B. garinii OspA serotype 4 strains analyzed in this study
| B. garinii isolate | Biological source | Geographic origin |
|---|---|---|
| A01 | Cerebrospinal fluid | The Netherlands |
| DK6 | Cerebrospinal fluid | Denmark |
| PBaeII, PBi, PFin, PFlk, PHoe | Cerebrospinal fluid | Germany |
| PMue, PScf, PSh, PWa | Cerebrospinal fluid | Germany |
| PRab | Synovia | Germany |
| PTrob | Cerebrospinal fluid | Slovenia |
Amplification and sequence analysis of the ospC gene.
The ospC genes were amplified from isolated genomic DNA by PCR with the OspC1 (5′-GAG GGA TCC ATC ATG AAA AAG AAT ACA TTA AGT GCG) and OspC3 (5′-GAG CTG CAG TTA AGG TTT TTT TGG ACT TTC TGC) primers. Cycling conditions and PCR conditions and reagents were as described previously (20, 50). The ospC amplicons were cloned by using the Original TA Cloning Kit as described by the manufacturer (Invitrogen). The ospC sequences were determined by the dideoxy chain termination method using a variety of oligonucleotide primers that target ospC. The sequences that were determined were translated and were aligned with the OspC sequences of three B. garinii OspA serotype 4 strains published previously (strain PBi, EMBL accession no. X69595; strain PTrob, EMBL accession no. X83554; and strain DK6, EMBL accession no. X73626).
Pulsed-field gel electrophoresis of borrelial DNA.
Borrelial strains were embedded in agarose plugs, and the DNA was released from the cells as described previously (9). To analyze the plasmid profiles, pulse times of 0.5 to 3.0 s for 30 h were used. For restriction fragment length polymorphism (RFLP) analyses, the embedded bacterial DNA was digested with ApaI, BssHII, SmaI, and MluI under the conditions recommended by the supplier (Boehringer Mannheim). The products obtained from the ApaI and SmaI digestions were separated for 30 h with pulse times of 1 to 20 and 1 to 30 s, respectively. The products obtained from the BssHII and MluI digestions were separated for 30 h with pulse times of 1 to 40 s.
Hybridization analyses.
To assess the UHB RFLP patterns, the isolated DNA was digested to completion with HaeIII, fractionated in a 0.8% GTG agarose gel, vacuum blotted with the VacuGene system (as described by the manufacturer; Pharmacia) onto a Hybond-N membrane (Amersham), and fixed to the membrane by UV cross-linking (Bio-Rad Gene Linker). Prehybridization and hybridization were conducted in hybridization buffer consisting of 0.2% (wt/vol) bovine serum albumin, 0.2% (wt/vol) polyvinylpyrrolidone (molecular weight, 40,000), 50 mM Tris-HCl (pH 7.5), 0.1% (wt/vol) sodium dodecyl sulfate (SDS), 10% (wt/vol) dextran sulfate, 100 μg of herring sperm DNA ml−1, and 1 M NaCl. The uhb(+) oligonucleotide (5′-GTT GGT TAA AAT TAC ATT TGC G) was radiolabeled with [γ-32P]ATP (6,000 Ci mmol−1; DuPont-NEN) by using polynucleotide kinase under standard conditions. Hybridizations were conducted in a Hybaid hybridization oven at 37°C overnight. Two 10-min washes with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS and a 1-h wash with 0.2× SSC–0.1% SDS were performed, followed by a final 5-min wash with 0.2× SSC–0.1% SDS at room temperature with vigorous shaking.
Nucleotide sequence accession numbers.
The B. garinii ospC sequences determined in this report have been deposited in the EMBL database under the following accession numbers: AJ132793 through AJ132798 for isolates PBaeII, PWa, PHoe, PFin, PFlk, and PMue, respectively, and AJ236907 and AJ236908 for isolates PSh and PScf, respectively.
RESULTS AND DISCUSSION
Analysis of ospC genes in OspA type 4 strains.
To assess possible genetic variation among the ospC genes of B. garinii OspA serotype 4 strains, the genes were amplified with primers that target conserved segments of ospC. All isolates yielded ospC PCR amplicons that were approximately 650 bp in length, indicating size conservation among isolates. The ospC amplicons from B. garinii isolates PFin, PBae, PFlk, PMue, PSh, PScf, PWa, and PHoe were sequenced in their entirety (data not shown), and surprisingly, all were identical at the nucleotide level. Comparative analyses were facilitated by the extensive number of ospC sequences in the databases. Alignment of the sequences determined here with the ospC sequences available in the databases revealed that they are nearly identical to the ospC genes of B. garinii PBi, PTrob, and DK6. These isolates are serotype 4 strains, and both PBi and DK6 were isolated from cerebrospinal fluid (55). The only difference among the sequences was in that of the ospC gene of B. garinii PBi, which has a single nucleotide difference from the other ospC gene sequences. This single point mutation results in the replacement of a Gln residue by a Glu residue. The observed high degree of ospC sequence identity among strains is extremely unusual since ospC genes typically exhibit a high degree of sequence variation (20, 48, 55). In addition, diversity among ospC sequences is further driven by the lateral transfer of the circular plasmid carrying the ospC gene (20, 24). However, the conservation of the ospC sequences suggests that lateral transfer of the ospC plasmid between OspA serotype 4 strains and other OspA serotypes has not occurred to any significant extent. This does not, however, rule out the possibility that transfer has occurred among OspA serotype 4 isolates.
Plasmid and RFLP pattern analyses.
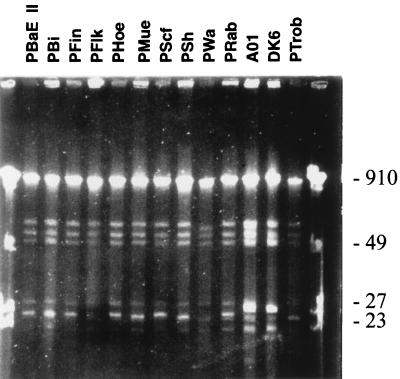
In recent years, the plasmid profiles of hundreds of B. burgdorferi sensu lato complex isolates have been determined and have been found to exhibit extensive variation. To compare the plasmid profiles of the OspA serotype 4 isolates, undigested genomic DNA was fractionated by pulsed-field gel electrophoresis (Fig. 1). Overall, the plasmid profiles were found to be strikingly similar. Previous studies of the plasmid profiles of Lyme disease spirochetes obtained from a defined geographic region have demonstrated that these patterns are rather variable (42). Three large plasmids of approximately 66, 57.5, and 50 kb were observed in each isolate. In addition, three to four plasmids between 20 and 30 kb were also noted. The similar plasmid profiles demonstrate an unusual degree of conservation of this component of the genome among B. garinii OspA serotype 4 strains.
FIG. 1.
Plasmid profile analysis of B. garinii OspA serotype 4 strains. Cells were embedded in agarose, treated to liberate the DNA, and electrophoresed as described in the text. The DNA was visualized by ethidium bromide staining. Size standards (in kilobases) are indicated on the right.
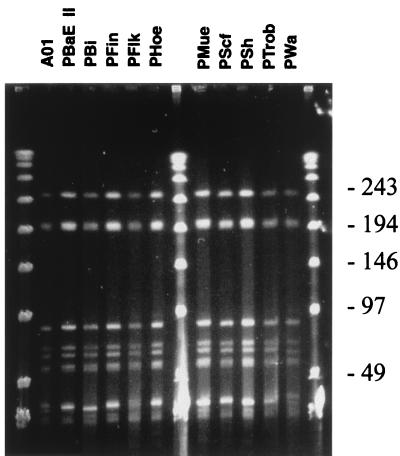
To further assess the genetic relatedness among OspA serotype 4 strains, a variety of RFLP analyses were conducted. DNA in agarose plugs was digested with ApaI, BssHII, MluI, or SmaI. The digested DNA was then fractionated by pulsed-field gel electrophoresis. Although some minor variations in RFLP patterns were noted, the patterns exhibited high degrees of conservation among OspA serotype 4 strains, further demonstrating an unusual level of genetic homogeneity and sequence conservation among the plasmids of these isolates. The results obtained for the BssHII digestion are shown in Fig. 2.
FIG. 2.
RFLP pattern analysis of BssHII-digested DNA from B. garinii OspA serotype 4 strains. The DNA embedded in agarose plugs was digested with BssHI and was then fractionated by pulsed-field gel electrophoresis as described in the text. The DNA was visualized by staining with ethidium bromide. Size standards (in kilobases; lanes 1, 8, and 14) are indicated on the right.
Hybridization analysis of UHB element.
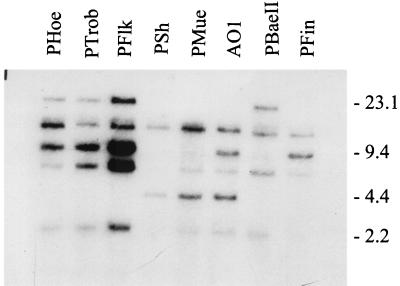
The UHB gene family is a large group of lipoprotein-encoding genes, all of which are flanked at their 5′ end by a highly conserved, upstream, promoter-carrying sequence called the UHB element (34, 47). Although the sequence of the UHB element itself is highly conserved, previous analyses of the UHB-flanked genes have revealed that their coding sequences exhibit variability among isolates, and the RFLP patterns of the UHB element have been demonstrated to be hypervariable among isolates of the B. burgdorferi sensu lato complex even at the intraspecies level (34, 47). It has been hypothesized that the pronounced variation in the RFLP patterns of UHB elements among isolates has resulted from recent molecular rearrangements in the UHB-flanked genes and the plasmids that carry them (34, 47), and recent data suggest that immune pressures drive or select for rearrangements (25). To assess the degree of variability in the RFLP patterns of UHB elements of OspA serotype 4 strains, DNA isolated from each strain was digested with HaeIII, fractionated, and probed with the uhb(+) oligonucleotide. In contrast to that noted in other B. burgdorferi sensu lato complex isolates (34, 47), the RFLP patterns of the OspA serotype 4 strains differed from one another only in the number of hybridizing bands detected (from two to five) rather than in variations in the sizes of individual bands (Fig. 3). The nature of these differences is important because the observed different numbers of hybridizing bands suggest that differences in the RFLP patterns of UHB elements are due primarily to the loss of some UHB element-carrying plasmids among these isolates rather than to molecular rearrangement events. The absence of detectable recombination events in the UHB-flanked genes is consistent with the possibility that OspA serotype 4 strains represent a recently emerged clonal lineage.
FIG. 3.
RFLP pattern analysis of the UHB element. DNA isolated from each strain was digested with HaeIII and was fractionated in a 0.8% GTG agarose gel. The DNA was then transferred to Hybond-N membranes and was hybridized with the 5′-end-labeled uhb(+) oligonucleotide probe. All procedures are described in the text. Size standards (in kilobases) are indicated on the right.
Conclusions.
It has been demonstrated that infection with particular species or subspecies (OspA types) of the B. burgdorferi sensu lato complex correlates with the development of certain clinical manifestations of Lyme borreliosis (1, 9, 10, 58, 60, 61). This has been demonstrated for the dermatological manifestation acrodermatitis chronica atrophicans, which is associated with B. afzelii. B. afzelii is OspA serotype 2. In contrast, the causative agents of neuroborreliosis and arthritis in Europe are heterogeneous, and the development of these manifestations cannot be attributed to infection with a specific species or OspA serotype. The diversity of the isolates associated with neuroborreliosis and arthritis is reflective of the diversity of isolates recovered from ticks in Europe. While OspA serotype 4 is not the only OspA serotype associated with neuroborreliosis, it is striking that isolates of this serotype have been recovered almost exclusively from human cerebrospinal fluid (8, 13, 14, 50, 58, 60, 61). The goal of this study was to determine if OspA serotype 4 strains represent a recently emerged clonal lineage. If this is in fact the case, then one would expect genetic homogeneity in traits that have been widely demonstrated to exhibit variability among isolates. To assess this we analyzed features or components of the Borrelia genome that are known to be highly variable among isolates. These comparative analyses focused on four areas: (i) ospC gene sequences, (ii) plasmid profiles, (iii) RFLP patterns obtained with infrequently cutting enzymes, and (iv) the RFLP patterns of the UHB element. In summary, all four of these areas were found to be conserved among the OspA serotype 4 strains investigated in this study, suggesting that they represent a recently emerged clonal lineage.
Comparative analyses of the OspC amino acid sequences revealed sequence identities of greater than 99%. ospC sequence conservation of this extent has not been reported for any other OspA serotypes. In fact, comparative analyses of OspC amino acid sequences from isolates of different B. burgdorferi sensu lato species have revealed extensive divergence, with identity values dropping to as low as 62% (20). At the intraspecies level, OspC identities were found to be as low as 71, 76, and 68% in B. burgdorferi, B. afzelii, and B. garinii, respectively. When OspC sequences from isolates of OspA serotype 6 were analyzed, sequence identities were as low as 68% (20). Hence, the ospC sequences determined here provide a clear indication of the genetic homogeneity at this typically variable locus in OspA serotype 4 strains.
Pulsed-field gel electrophoresis fractionation of genomic DNA and analysis of the plasmid profiles revealed strong conservation of the plasmid composition among the OspA serotype 4 strains. As discussed above, although the plasmids are thought to be essential for survival, as inferred from their ubiquitous distribution among B. burgdorferi sensu lato complex isolates, it has been clearly demonstrated that plasmid composition can vary widely among B. burgdorferi sensu lato complex strains (3, 11, 26, 31, 42, 45). Mechanisms that contribute to plasmid heterogeneity include plasmid loss (35, 41), lateral transfer (20, 33), recombination (30, 33, 39, 47), and dimer formation (19, 26). The presence of repeated sequences and gene families on plasmids may allow for interplasmid homologous recombination (4, 12, 34, 36, 44, 62).
To further analyze possible genetic homogeneity, RFLP patterns were assessed with the enzymes BssHII, MluI, ApaI, and SmaI. The RFLP patterns exhibited an unusual degree of conservation, with only minor differences noted. This observation indicates that both the plasmids and the chromosomes of OspA serotype 4 strains have not undergone significant molecular rearrangement events. Similarly, analysis of the UHB-element RFLP patterns of the serotype 4 strains revealed that they are also highly conserved. Previous analyses of UHB-element RFLP patterns have demonstrated that they are variable even among closely related isolates of the same species (47). In the OspA serotype 4 strains, the differences in the UHB-element RFLP patterns were primarily due to the absence of some hybridizing bands from some isolates. This is in contrast to the case for other B. burgdorferi sensu lato isolates, which differ not only in the numbers of fragments but also in the sizes of the hybridizing fragments. Hence, it appears that in the OspA serotype 4 strains, differences in the UHB-element RFLP patterns are due to the loss of some of the UHB-element-carrying plasmids rather than to rearrangements among plasmids.
From the analyses presented here it can be concluded that OspA serotype 4 strains are a genetically homogeneous group of B. burgdorferi sensu lato complex strains. The observed genetic homogeneity indicates that this serotype is a recently emerged clonal variant of B. garinii. It is interesting that strains of this serotype have not been recovered from ticks but have been recovered only from humans. The basis for this observation is undefined and warrants further study. Furthermore, it remains to be determined why this particular variant has established an apparent tropism for the central nervous system. Comparative analysis of type 4 strains with those of other OspA serotypes may allow the identification of specific factors that facilitate invasion of the central nervous system and persistence in cerebrospinal fluid. Since serum resistance promotes dissemination and, therefore, by extension, may allow OspA serotype 4 strains to have greater pathogenic potential than strains of other OspA serotypes, comparative studies of the dissemination characteristics of OspA serotypes in ticks and mammals are warranted.
ACKNOWLEDGMENTS
R. T. Marconi and C. P. LaVoie are supported in part by grants from the Jeffress Trust and the National Institutes of Health.
REFERENCES
- 1.Assous M V, Postic D, Nevot P, Paul G, Baranton G. Western blot analysis of sera from Lyme borreliosis patients according to the genomic species of the Borrelia strains used as antigens. Eur J Clin Microbiol Infect Dis. 1993;12:261–268. doi: 10.1007/BF01967256. [DOI] [PubMed] [Google Scholar]
- 2.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J-C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 3.Barbour A G. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J Clin Microbiol. 1988;26:475–478. doi: 10.1128/jcm.26.3.475-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour A G, Carter C J, Bundoc V, Hinnebusch J. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J Bacteriol. 1996;178:6635–6639. doi: 10.1128/jb.178.22.6635-6639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour A G, Garon C F. The genes encoding the major surface proteins of Borrelia burgdorferi are located on a plasmid. Ann N Y Acad Sci. 1988;539:144–153. doi: 10.1111/j.1749-6632.1988.tb31847.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbour A G, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 7.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busch U, Hizo-Teufel C, Boehmer R, Fingerle V, Nitschko H, Wilske B, Preac-Mursic V. Three species of Borrelia burgdorferi sensu lato (B. burgdorferi sensu lato, B. afzelii, and B. garinii) identified from cerebrospinal fluid isolates by pulsed field gel electrophoresis and PCR. J Clin Microbiol. 1996;34:1072–1078. doi: 10.1128/jcm.34.5.1072-1078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busch U, Hizo-Teufel C, Bohmer R, Fingerle V, Robler D, Wilske B, Preac-Mursic V. Borrelia burgdorferi sensu lato strains isolated from cutaneous Lyme borreliosis biopsies differentiated by pulsed-field gel electrophoresis. Scand J Infect Dis. 1996;28:583–589. doi: 10.3109/00365549609037965. [DOI] [PubMed] [Google Scholar]
- 10.Canica M M, Nato F, du Merle L, Mazie J C, Baranton G, Postic D. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand J Infect Dis. 1993;25:441–448. doi: 10.3109/00365549309008525. [DOI] [PubMed] [Google Scholar]
- 11.Carlyon J A, LaVoie C, Sung S Y, Marconi R T. Analysis of the organization of multicopy linear- and circular-plasmid-carried open reading frames in Borrelia burgdorferi sensu lato isolates. Infect Immun. 1998;66:1149–1158. doi: 10.1128/iai.66.3.1149-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiffert H, Karsten A, Thomssen R, Christen H-J. Characterization of Borrelia burgdorferi strains in Lyme arthritis. Scand J Infect Dis. 1998;24:437–439. doi: 10.1080/00365549850160918. [DOI] [PubMed] [Google Scholar]
- 14.Eiffert H, Ohlenbusch A, Christen H-J, Thomssen R, Spielman A, Matuschka F-R. Nondifferentiation between Lyme disease spirochetes from vector ticks and human cerebrospinal fluids. J Infect Dis. 1995;171:476–479. doi: 10.1093/infdis/171.2.476. [DOI] [PubMed] [Google Scholar]
- 15.Fraser C, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischman R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Cotton F C M C, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs R, Jauris S, Lottspeich F, Preac-Mursic V, Wilske B, Soutschek E. Molecular analysis and expression of a Borrelia burgdorferi gene encoding a 22 kDa protein (pC) in Escherichia coli. Mol Microbiol. 1992;6:503–509. doi: 10.1111/j.1365-2958.1992.tb01495.x. [DOI] [PubMed] [Google Scholar]
- 17.Fukunaga M, Hamase A, Okada K, Nakao M. Borrelia tanuki sp. nov. and Borrelia turdae sp. nov. found from Ixodid ticks in Japan: rapid species identification by 16S rRNA gene-targeted PCR analysis. Microbiol Immunol. 1996;40:877–881. doi: 10.1111/j.1348-0421.1996.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 18.Fukunaga M, Takahashi Y, Tsuruta Y, Matsushita O, Ralph D, McClelland M, Nakao M. Genetic and phenotypic analysis of Borrelia miyamotoi sp. nov., isolates from the Ixodid tick Ixodes persulcatus, the vector for Lyme disease in Japan. Int J Syst Bacteriol. 1995;45:804–810. doi: 10.1099/00207713-45-4-804. [DOI] [PubMed] [Google Scholar]
- 19.Hyde F W, Johnson R C. Characterization of a circular plasmid from Borrelia burgdorferi, etiologic agent of Lyme disease. J Clin Microbiol. 1988;26:2203–2205. doi: 10.1128/jcm.26.10.2203-2205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jauris-Heipke S, Liegl G, Preac-Mursic V, Robler D, Schwab E, Soutschek E, Will G, Wilske B. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J Clin Microbiol. 1995;33:1860–1866. doi: 10.1128/jcm.33.7.1860-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 22.Lam T T, Nguyen T-P K, Montgomery R R, Kantor F S, Fikrig E, Flavell R A. Outer surface protein E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Fleche A, Postic D, Girardet K, Peter O, Baranton G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int J Syst Bacteriol. 1997;47:921–925. doi: 10.1099/00207713-47-4-921. [DOI] [PubMed] [Google Scholar]
- 24.Livey I, Gibbs C P, Schuster R, Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol. 1995;18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 25.Marconi, R. T. Unpublished data.
- 26.Marconi R T, Casjens S, Munderloh U G, Samuels D S. Analysis of linear plasmid dimers in Borrelia burgdorferi sensu lato isolates: implications concerning the potential mechanism of linear plasmid replication. J Bacteriol. 1996;178:3357–3361. doi: 10.1128/jb.178.11.3357-3361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marconi R T, Garon C F. Development of polymerase chain reaction primer sets for diagnosis of Lyme disease and for species-specific identification of Lyme disease isolates by 16S rRNA signature nucleotide analysis. J Clin Microbiol. 1992;30:2830–2834. doi: 10.1128/jcm.30.11.2830-2834.1992. . (Erratum, 31:1026, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marconi R T, Garon C F. Identification of a third genomic group of Borrelia burgdorferi through signature nucleotide analysis and 16S rRNA sequence determination. J Gen Microbiol. 1992;138:533–536. doi: 10.1099/00221287-138-3-533. [DOI] [PubMed] [Google Scholar]
- 29.Marconi R T, Garon C F. Phylogenetic analysis of the genus Borrelia: a comparison of North American and European isolates of B. burgdorferi. J Bacteriol. 1992;174:241–244. doi: 10.1128/jb.174.1.241-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marconi R T, Konkel M E, Garon C F. Variability of osp genes and gene products among species of Lyme disease spirochetes. Infect Immun. 1993;61:2611–2617. doi: 10.1128/iai.61.6.2611-2617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marconi R T, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marconi R T, Samuels D S, Landry R K, Garon C F. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marconi R T, Sung S Y, Hughes C N, Carlyon J A. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J Bacteriol. 1996;178:5615–5626. doi: 10.1128/jb.178.19.5615-5626.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porcella S F, Popova T G, Akins D R, Li M, Radolf J R, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multicopy open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postic D, Belfazia J, Isogai E, Saint Girons I, Grimont P A D, Baranton G. A new genomic species in Borrelia burgdorferi sensu lato isolated from Japanese ticks. Res Microbiol. 1993;144:467–473. doi: 10.1016/0923-2508(93)90054-6. [DOI] [PubMed] [Google Scholar]
- 38.Postic D, Edlinger C, Richaud C, Grimont F, Dufresne Y, Perolat P, Baranton G, Grimont P A D. Two genomic species in Borrelia burgdorferi. Res Microbiol. 1990;141:465–475. doi: 10.1016/0923-2508(90)90072-x. [DOI] [PubMed] [Google Scholar]
- 39.Rosa P A, Schwan T, Hogan D. Recombination between genes encoding major outer surface proteins A and B of Borrelia burgdorferi. Mol Microbiol. 1992;6:3031–3040. doi: 10.1111/j.1365-2958.1992.tb01761.x. [DOI] [PubMed] [Google Scholar]
- 40.Sadziene A, Wilske B, Ferdows M S, Barbour A G. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwan T G, Schrumpf M E, Karstens R H, Clover J R, Wong J, Daugherty M, Struthers M, Rosa P A. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J Clin Microbiol. 1993;31:3096–3108. doi: 10.1128/jcm.31.12.3096-3108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigal L H, Zahradnik J M, Lavin P, Patella S J, Bryant G, Haselby R, Hilton E, Kunkel M, Adler-Klein D, Doherty T, Evans J, Malawista S. A vaccine consisting of recombinant Borrelia burgdorferi outer surface protein A to prevent Lyme disease. N Engl J Med. 1998;339:216–222. doi: 10.1056/NEJM199807233390402. [DOI] [PubMed] [Google Scholar]
- 44.Simpson W J, Garon C F, Schwan T G. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]
- 45.Stålhammar-Carlemalm M, Jenny E, Gern L, Aeschlimann A, Meyer J. Plasmid analysis and restriction fragment length polymorphisms of chromosomal DNA allow a distinction between Borrelia burgdorferi strains. Zentbl Bakteriol Parasitenkd Infectkrankh Hyg Abt 1 Orig. 1990;274:28–39. doi: 10.1016/s0934-8840(11)80972-2. [DOI] [PubMed] [Google Scholar]
- 46.Steere A C, Sikand V K, Meurice F, Parenti D L, Fikrig E, Schoen R T, Nowakowski J, Schmid C H, Laukamp S, Buscarino C, Krause D S. Vaccination against Lyme disease with recombinant Borrelia burgdorferi outer surface lipoprotein A with adjuvant. N Engl J Med. 1998;339:209–215. doi: 10.1056/NEJM199807233390401. [DOI] [PubMed] [Google Scholar]
- 47.Sung S-Y, LaVoie C, Carlyon J A, Marconi R T. Evolutionary instability of ospE related members of the UHB gene family in Borrelia burgdorferi sensu lato complex isolates. Infect Immun. 1998;66:4656–4668. doi: 10.1128/iai.66.10.4656-4668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theisen M, Frederiksen B, Lebech A-M, Vuust J, Hansen K. Polymorphism in ospC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implications for taxonomy and for use of OspC Protein as a diagnostic antigen. J Clin Microbiol. 1993;31:2570–2576. doi: 10.1128/jcm.31.10.2570-2576.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Dam A P, Oei A, Jaspars R, Fijen C, Wilske B, Spanjaard L, Dankert J. Complement-mediated serum sensitivity among spirochetes that cause Lyme disease. Infect Immun. 1997;65:1228–1230. doi: 10.1128/iai.65.4.1228-1236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasiliu V, Herzer P, Rossler D, Lehnert G, Wilske B. Heterogeneity of Borrelia burgdorferi sensu lato demonstrated by an ospA type specific PCR in synovial fluid from patients with Lyme arthritis. Med Microbiol Immunol. 1998;187:97–102. doi: 10.1007/s004300050079. [DOI] [PubMed] [Google Scholar]
- 51.Wallich R, Helmes C, Schaible U E, Lobet Y, Moter S E, Kramer M D, Simon M M. Evaluation of genetic divergence among Borrelia burgdorferi isolates by use of OspA, fla, HSP60, and HSP70 gene probes. Infect Immun. 1992;60:4856–4866. doi: 10.1128/iai.60.11.4856-4866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G, van Dam A P, Le Flecha A, Postic D, Peter O, Baranton G, de Boer R, Spanjaard L, Dankert J. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19) Int J Syst Bacteriol. 1997;47:926–932. doi: 10.1099/00207713-47-4-926. [DOI] [PubMed] [Google Scholar]
- 53.Will G, Jaruis-Heipke S, Schwab E, Busch U, Rossler D, Soutschek E, Wilske B, Preac-Mursic V. Sequence analysis of the ospA genes shows homogeneity within Borrelia burgdorferi sensu stricto and Borrelia afzelii strains but reveals major subgroups within the Borrelia garinii species. Med Microbiol Immunol. 1995;184:73–80. doi: 10.1007/BF00221390. [DOI] [PubMed] [Google Scholar]
- 54.Wilske B, Busch U, Eiffert H, Fingerle V, Pfister H-W, Rossler D, Will G. Diversity of OspA and OspC among cerebrospinal fluid isolates of Borrelia burgdorferi sensu lato from patients with neuroborreliosis in Germany. Med Microbiol Immunol. 1996;184:195–201. doi: 10.1007/BF02456135. [DOI] [PubMed] [Google Scholar]
- 55.Wilske B, Busch U, Fingerle V, Jauris-Heipke S, Preac-Mursic V, Robler D, Will G. Immunological and molecular variability of OspA and OspC: implications for Borrelia vaccine development. Infection. 1996;24:208–212. doi: 10.1007/BF01713341. [DOI] [PubMed] [Google Scholar]
- 56.Wilske B, Jauris-Heipke S, Lobentanzer R, Pradel I, Preac-Mursic V, Roessler D, Soutschek E, Johnson R C. Phenotypic analysis of the outer surface protein C (OspC) of Borrelia burgdorferi sensu lato by monoclonal antibodies. J Clin Microbiol. 1995;33:103–109. doi: 10.1128/jcm.33.1.103-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilske B, Preac-Mursic V, Gobel U B, Graf B, Jauris S, Soutschek E, Schwab E, Zumstein G. An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J Clin Microbiol. 1993;31:340–350. doi: 10.1128/jcm.31.2.340-350.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Schwab E, Will G, Wanner G. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61:2182–2191. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilske B, Preac-Mursic V, Schierz G, Busch K V. Immunochemical and immunological analysis of European Borrelia burgdorferi strains. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt 1 Orig Reihe A. 1986;263:92–102. doi: 10.1016/s0176-6724(86)80108-0. [DOI] [PubMed] [Google Scholar]
- 60.Wilske B, Preac-Mursic V, Schierz G, Gueye W, Herzer P, Weber K. Immunochemical analysis of the immune response in late manifestations of Lyme borreliosis. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt 1 Orig Reihe A. 1988;267:549–558. [PubMed] [Google Scholar]
- 61.Wilske B, Preac-Mursic V, Schierz G, Kühbeck R, Barbour A G, Kramer M. Antigenic variability of Borrelia burgdorferi. Ann N Y Acad Sci. 1988;539:126–143. doi: 10.1111/j.1749-6632.1988.tb31846.x. [DOI] [PubMed] [Google Scholar]
- 62.Zuckert W R, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]