Abstract
Different SARS-CoV-2 vaccines are approved in various countries, but few direct comparisons of the antibody responses they stimulate have been reported. We collected plasma specimens in July 2021 from 196 Mongolian participants fully vaccinated with one of four COVID-19 vaccines: Pfizer/BioNTech, AstraZeneca, Sputnik V, and Sinopharm. Functional antibody testing with a panel of nine SARS-CoV-2 viral variant receptor binding domain (RBD) proteins revealed marked differences in vaccine responses, with low antibody levels and RBD-ACE2 blocking activity stimulated by the Sinopharm and Sputnik V vaccines in comparison to the AstraZeneca or Pfizer/BioNTech vaccines. The Alpha variant caused 97% of infections in Mongolia in June and early July 2021. Individuals who recover from SARS-CoV-2 infection after vaccination achieve high antibody titers in most cases. These data suggest that public health interventions such as vaccine boosting, potentially with more potent vaccine types, may be needed to control COVID-19 in Mongolia and worldwide.
Keywords: SARS-CoV-2, COVID-19, serology, viral variants, Mongolia, Pfizer/BioNTech, Sputnik V, Sinopharm, vaccine
Graphical abstract
Dashdorj et al. examined antibody responses in COVID-19-vaccinated Mongolian participants. Antibodies blocking ACE2-RBD binding across SARS-CoV-2 variants were highest among Pfizer/BioNTech vaccinees, followed by AstraZeneca, Sputnik V, and then Sinopharm vaccinees. Breakthrough infections in June through July of 2021 were predominantly the Alpha variant and induced higher blocking antibodies across vaccination groups.
Main text
Several different vaccines for SARS-CoV-2 have been approved for use in various countries and are being actively deployed in an effort to combat the COVID-19 pandemic, but few direct comparisons of the antibody responses they stimulate have been reported. Many viral variants have arisen since the initial months of the pandemic, and are in circulation with different geographical distributions and susceptibility to antibody responses elicited to Wuhan-Hu-1 antigens (Garcia-Beltran et al., 2021; Hoffmann et al., 2021; Muik et al., 2021; Planas et al., 2021a; Röltgen et al., 2021; Supasa et al., 2021). Breakthrough infections with SARS-CoV-2 in previously vaccinated individuals, together with data from the clinical trials supporting regulatory approval of the vaccines, indicate that there are disparities in the amount of protection against infection that they provide (AlQahtani et al., 2021; Khoury et al., 2021; Wall et al., 2021). Beginning on February 23, 2021, Mongolia carried out a vigorous campaign of vaccination of its citizens and achieved a high rate of 61.4% of the total population fully vaccinated, with an additional 6.3% having received a single dose, as reported in official Mongolian state news agency data (https://montsame.mn/en). The adult population has primarily been vaccinated with the Sinopharm vaccine (89.2% of vaccinated adults). In the summer of 2021, widespread outbreaks of SARS-CoV-2 infection were reported in Mongolia, with 86 cases per 100,000 in mid-June, decreasing to approximately 40 cases per 100,000 at the end of July; these cases included many vaccinated individuals. The viral variants responsible for these infections are currently unknown.
We collected plasma specimens in a five-day period from July 3, 2021 to July 7, 2021 from Mongolian participants who had been fully vaccinated with one of four COVID vaccines: Pfizer/BioNTech (BNT162b2), AstraZeneca (ChAdOx1-S), Sputnik V (Gam-COVID-Vac), and Sinopharm (BBIBP-CorV). Participants were recruited by public announcement, and volunteers were enrolled after signing the consent form approved by the Ethics Review Board at the Ministry of Health of Mongolia. Antibodies were analyzed in the plasmas of 196 participants divided between the vaccine groups (47, 50, 45, and 54 individuals for Pfizer/BioNTech, AstraZeneca, Sputnik V, and Sinopharm, respectively) and selected to balance age, sex, and time after second vaccine dose (Figure S1A). We measured antibody blocking of angiotensin-converting enzyme 2 (ACE2) host receptor protein binding to SARS-CoV-2 spike receptor binding domains (RBDs) from nine viral variants of concern or interest, according to CDC and WHO definitions, using an electrochemiluminescence (ECL) assay platform from Meso Scale Diagnostics. The RBDs that were tested were (with RBD amino acid changes from Wuhan-Hu-1 in parentheses): Alpha (N501Y), Beta (K417N, E484K, N501Y), Gamma (K417T, E484K, N501Y), Delta (L452R, T478K), Epsilon (L452R), Eta/Iota/Zeta (E484K), Kappa (L452R, E484Q), B.1.526.2 (S477N), and P.3 (E484K, N501Y) as well as Wuhan-Hu-1. Antibody blocking of ACE2 binding to each RBD for each vaccine type is shown in Figure 1 A. Wuhan-Hu-1 RBD-ACE2 blocking results are also displayed as a function of time of sample collection after vaccination (Figure S1B). RBD-ACE2 blocking antibody results for participants of different ages (< 60 or ≥ 60 years) and sexes are shown in Figure 1B. We additionally measured the concentration of IgG antibodies binding to RBD, spike (S) and nucleocapsid (N) antigens of Wuhan-Hu-1 SARS-CoV-2 in all specimens (Figure S1C), using a plasma dilution of 1:5,000 to ensure that measured antibody concentrations were within the linear range of the ECL assay (Figure S1D). Results from RBD-ACE2 blocking antibody assays and anti-RBD IgG binding assays have been shown to be correlated with neutralizing antibody titers from pseudotyped viruses displaying SARS-CoV-2 spike and authentic SARS-CoV-2 virus (Feng et al., 2021; Gilbert et al., 2021; Röltgen et al., 2020). We reconfirmed these previously reported correlations between RBD-ACE2 blocking antibody results, anti-RBD IgG antibody concentration, and virus neutralization for a subset of samples with a Wuhan-Hu-1 spike pseudovirus neutralization assay (Figure S1E). We further found that the ACE2 blocking and anti-RBD IgG assays used in this study are correlated with authentic SARS-CoV-2 virus neutralization results, in plasma specimens from Pfizer-BioNTech vaccine recipients from our recent publication (Arunachalam et al., 2021) (Figure S1F). High RBD-ACE2 blocking antibody values are associated with detectable neutralizing titers in the authentic SARS-CoV-2 virus neutralization assay.
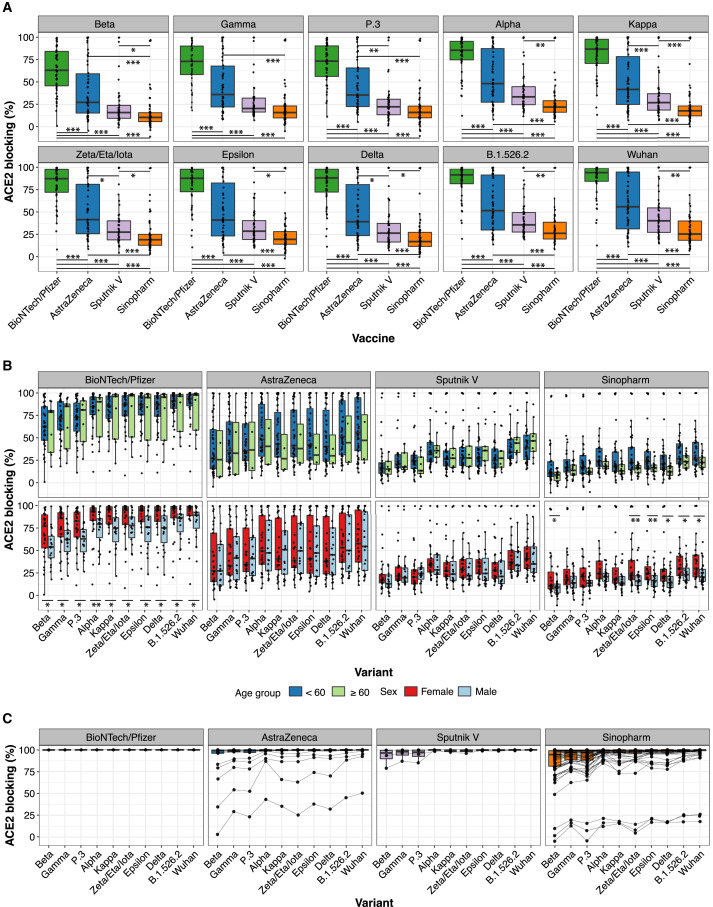
Figure 1.
Vaccine-induced antibody blocking of RBD-ACE2 binding for different viral variants
(A) Percentage blocking of ACE2 binding to RBD of specified viral variants by plasma antibodies of recipients of Pfizer/BioNTech, AstraZeneca, Sputnik V, and Sinopharm vaccines is shown. Significance of differences between pairwise combination of vaccine groups was calculated by Wilcoxon test with Bonferroni correction to adjust for multiple hypothesis correction (∗, ∗∗, and ∗∗∗ indicate p < 0.05, p < 0.01, and p < 0.001 respectively).
(B) Blocking antibody responses stratified by participant age (< 60 years, or ≥ 60 years) and sex. Significance between two groups (age groups and male versus female) was calculated by Wilcoxon test (∗ and ∗∗ indicate p < 0.05 and p < 0.01 respectively).
(C) RBD-ACE2 blocking antibody responses for 99 participants with confirmed SARS-CoV-2 infection post-vaccination with the indicated vaccines. Data points for samples from the same individual are connected with a line.
RBD-ACE2 blocking antibody activity was strikingly different between the vaccines tested. Within each vaccine group, differences were also observed in antibody activity for the different viral variant antigens, although these were smaller than the differences between the vaccine groups. The Pfizer/BioNTech vaccine elicited the strongest RBD-ACE2 blocking antibody activity, followed by the AstraZeneca vaccine, then Sputnik V, with the lowest levels from Sinopharm (Figure 1A). Differences between the vaccine responses were highly significant for most viral variant antigens, although differences between the Sputnik V and AstraZeneca did not always reach significance. RBD-ACE2 blocking antibody activity for RBD antigens of viral variants of concern or interest showed a consistent hierarchy of decreased blocking, with the greatest decrease for the Beta, Gamma, and P.3 variants and more modest decreases for the other variants (Figure 1B), similar to previously reported results (Garcia-Beltran et al., 2021; Röltgen et al., 2021). Median times of sampling after second vaccination were between 2 and 3 months for all vaccines (Figure S1A); we note that some Sputnik V recipient plasmas were collected at later time points after the second vaccine dose compared to the other vaccines, but plotting RBD-ACE2 blocking as a function of time after the booster dose indicated that this had little effect on the results (Figure S1B). Anti-RBD and anti-spike binding assay data were similar to RBD-ACE2 blocking antibody results, with decreasing antibody concentrations from Pfizer/BioNTech to AstraZeneca to Sputnik V to Sinopharm (Figure S1C). The age of vaccine recipients and proportions of males and females in each group were comparable (Figure S1A). RBD-ACE2 blocking antibody median values were lower for males than females for the Pfizer/BioNTech and Sinopharm vaccines, but not for the AstraZeneca and Sputnik V vaccines (Figure 1B).
Testing for antibodies to SARS-CoV-2 N antigen assessed for evidence of prior infection, since the Pfizer/BioNTech mRNA vaccine and the AstraZeneca and Sputnik V adenoviral vectored vaccines do not contain or produce N antigen. Most recipients of the Sinopharm vaccine, which contains inactivated SARS-CoV-2 viruses, showed the expected increased levels of anti-N antibodies compared to other vaccine recipients (Figure S1C), although most were below the cutoff for seroconversion in this assay. The anti-N IgG assay results also identified some participants with evidence of prior infection among the other vaccine recipients who had no reported history of infection (5, 8, and 4 individuals for Pfizer/BioNTech, AstraZeneca, and Sputnik V, respectively). Very high amounts of anti-N IgG well above the cutoff for seroconversion were observed in three Sinopharm individuals. Pre-vaccination specimens were not available for participants to further evaluate evidence of infection prior to vaccination, but the Sinopharm and Sputnik V recipients with the highest anti-N antibodies had significantly higher ACE2-blocking antibody activity than others in their vaccination groups, suggesting that these individuals had a combination of infection and vaccination. To further evaluate the serological effects of combined SARS-CoV-2 infection and vaccination, an additional cohort of 99 participants who had been vaccinated outside of this study and then had documented SARS-CoV-2 infections were recruited (1, 21, 4, and 73 recipients of Pfizer/BioNTech, AstraZeneca, Sputnik V, and Sinopharm vaccines, respectively) (Figure S1G) and analyzed with the RBD-ACE2 blocking antibody assay (Figure 1C). RBD-ACE2 blocking antibody activity was very high in almost all of these individuals, with a median of > 99% blocking against all variants except the Beta, Gamma, and P.3 variants. While most AstraZeneca and Sinopharm recipients showed high levels of RBD-ACE2 blocking activity, a few showed lower blocking activity against most variants after testing positive for infection post-vaccination (Figure 1C).
It is important to note that RBD-ACE2 blocking antibody assay results are only a surrogate for potential protection of the individual from infection by SARS-CoV-2, as are the results from other assays such as viral neutralization assays. Initial attempts to evaluate decreases in risk of SARS-CoV-2 infection following vaccination suggest that the results of surrogate assays, such as anti-RBD or anti-spike IgG measurement, and neutralization assays are well correlated with protection against symptomatic infection (Feng et al., 2021; Gilbert et al., 2021).
The serological data from recipients of the four vaccines tested suggested that Sinopharm recipients, who then comprised 89.2% of vaccinated adults in Mongolia, as well as the smaller number of individuals vaccinated with Sputnik V or AstraZeneca vaccines could be particularly susceptible to breakthrough infections. To assess whether viral variants that are more resistant to antibodies elicited by Wuhan-Hu-1 antigens are responsible for the ongoing wave of breakthrough infections in Mongolia, we carried out viral genotyping with spike N501Y, E484K, and L452R mutation-specific RT-qPCR on 182 nasopharyngeal swabs collected between June 18, 2021 and July 5, 2021 from individuals with Sinopharm post-vaccination SARS-CoV-2 infection. Viral genotyping showed that these infections were dominated by Alpha variants with the N501Y mutation, accounting for 97.3% (177/182) of cases tested. The other samples were comprised of two samples with the L452R mutation, consistent with several lineages including the Delta variant, and three samples without N501Y, E484K, or L452R mutations, unlikely to represent current variants of concern. Samples containing N501Y were further tested for spike del69-70 by RT-qPCR; this characteristic deletion was detected in all (177/177) of these samples, confirming that Sinopharm post-vaccination cases were caused primarily by the Alpha variant. The Alpha variant has minimal evasion of antibody responses elicited by Wuhan-Hu-1 antigens (Muik et al., 2021; Figure 1A), suggesting that the breakthrough infections in Mongolia between June 18, 2021 and July 5, 2021 were related to the overall low antibody levels to all variants in the Sinopharm-vaccinated population rather than being driven by highly immune-evasive viral variants.
This direct comparison of vaccine-elicited functional antibody responses to a panel of nine SARS-CoV-2 viral variant RBDs indicates that there are marked differences in the serological responses generated by each vaccine, with relatively low antibody concentrations and RBD-ACE2 blocking activity stimulated by the Sinopharm and Sputnik V vaccines, intermediate levels for the AstraZeneca vaccine, and the highest values for the Pfizer/BioNTech vaccine. The reasons for the differences in the serological responses between these vaccine types are the subject of intense research, but they are likely to include factors such as the antigen doses provided or expressed by the recipient’s cells, the anatomical distribution of antigen, adjuvant effects and the degree of stimulation of innate immune mechanisms, and the timing and nature of the priming and boost vaccinations, among other possibilities.
Most individuals who recover from infection with SARS-CoV-2 after receiving any of the vaccines studied show elevated ACE2 blocking antibody activity comparable to that seen in uninfected recipients of the Pfizer/BioNTech vaccine. Breakthrough infections in Mongolia in June and July 2021 are largely attributable to the more infectious, but not highly immune-evasive, Alpha variant (Davies et al., 2021; Planas et al., 2021b). Our data concerning the breakthrough infections in recipients of the different vaccines are not controlled for the degree of exposure to virus after vaccination, time since vaccination, or other clinical variables, and they cannot be used to infer vaccine efficacy values for the four vaccines. However, a recent preprint analyzing health care records in Bahrain to identify breakthrough infections for the same four vaccines we evaluated found that, while all vaccines decreased infections and mortality, Sinopharm vaccine recipients had a higher risk of post-vaccination infections, hospitalizations, ICU admissions, and deaths than Pfizer/BioNTech recipients (AlQahtani et al., 2021); other recent studies have highlighted the value of antibody measurements as correlates of vaccine-mediated protection (Feng et al., 2021; Gilbert et al., 2021).
Limitations of this study include its retrospective observational design without randomized assignment of individuals to vaccine groups, the lack of pre-vaccination plasma samples, and the lack of plasma specimens following vaccination but before infection in the 99 individuals with documented breakthrough infections. While samples from breakthrough infection cases were collected continuously as they were reported, the limited number of samples precludes evaluation of vaccine efficacy in this study. Our serological analysis focused on RBD-binding antibodies and RBD-ACE2 blocking antibodies and does not evaluate other potential antibody-mediated immunological mechanisms or T cell responses that may play a role in vaccine efficacy. We observed lower median values of RBD-ACE2 blocking in males than females for the Pfizer and Sinopharm vaccines. Although there are significant sex-related differences in health outcomes in Mongolia, including a 9.6 year shorter life expectancy at birth in males than females, per recent reports from the World Bank (https://documents1.worldbank.org/curated/en/153011608186666637) and the Mongolian Center for Health Development (http://hdc.gov.mn/media/uploads/2021-05/Health_Indicator_2019_ENG.pdf), definitive evaluation of sex-related differences in vaccine responses in this population will require further study with trials designed to address this topic.
In summary, our data indicate that there are major differences in the magnitude of functional antibody responses stimulated by the four vaccines studied and suggest that additional public health interventions such as booster vaccine doses, potentially with the more potent vaccine types, may be needed to further control the COVID-19 pandemic in Mongolia and worldwide. However, faced with the public health crisis of increasing SARS-CoV-2 infections and limited supply or distribution of the most effective vaccines, widespread vaccination with a lower-efficacy vaccine may still represent a route to decreasing infections, hospitalizations, and mortality.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Plasma samples from 196 vaccinated individuals | ND Dashdorj, Ulaanbaatar, Mongolia | http://www.onomfoundation.org |
| Plasma samples from 99 individuals with breakthrough infections | ND Dashdorj, Ulaanbaatar, Mongolia | http://www.onomfoundation.org |
| Nasopharyngeal swab specimens from 182 individuals with breakthrough infections | ND Dashdorj, Ulaanbaatar, Mongolia | http://www.onomfoundation.org |
| Chemicals, peptides, and recombinant proteins | ||
| Calcium Chloride | Sigma-Aldrich | Cat# C4901 |
| HEPES | Sigma-Aldrich | Cat# H3375 |
| Sodium Chloride | Sigma-Aldrich | Cat# S9888 |
| Sodium Phosphate Dihydrate | Sigma-Aldrich | Cat# 71505 |
| Polybrene Transfection Reagent | Milipore Sigma | Cat# TR-1003-G |
| BriteLite Plus Reporte Gene Assay System | Perkin Elmer | Cat# 6066761 |
| Dulbecco’s Modification of Eagle’s Medium (DMEM) | Cytiva | Cat# SH30243.01 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat# 12306C |
| Penicillin/Streptomycin/Glutamine | Life Technologies | Cat# 10367-016 |
| 0.05% Trypsin-EDTA | GIBCO | Cat# 25300054 |
| Dulbecco’s Phosphate-Buffered Saline, without Calcium and Magnesium | Cytiva | Cat# SH30028.03 |
| Critical commercial assays | ||
| V-PLEX SARS-CoV-2 Panel 11 (ACE2) kit | Meso Scale Discovery | Cat# K15458U-2 |
| V-PLEX Coronavirus Panel 4 (IgG) kit | Meso Scale Discovery | Custom plate |
| Chemagic Viral DNA/RNA 300 Kit | Perkin-Elmer, Waltham, MA | Cat# CMG-1033-S |
| SuperScript™ III Platinum™ One-Step qRT-PCR Kit | Invitrogen, Carlsbad | Cat# 11732088 |
| Deposited Data | ||
| Demographic data and electrochemiluminescence data | This paper | Mendeley Data:http://dx.doi.org/10.17632/hy3zm69f57.1 |
| Experimental models: Cell lines | ||
| HEK293T | Laboratory of William Weis | ATCC CRL-3216 |
| HeLa expressing human ACE2 | Laboratory of Dennis Burton (Provided by Laboratory of Peter Kim) | ATCC CRM-CCL-2 |
| Oligonucleotides | ||
| N501Y_FWD GTTTTAATTGTTACTTT CCTTTACAATC; final conc. 360 nM |
Wang et al., 2021 | PMID: 34037430 |
| N501Y_REV CTTTTTAGGTCCACAA ACAGT; final conc. 360 nM |
Wang et al., 2021 | PMID: 34037430 |
| N501Y_MT_FAM TTTCCAACCCACT TATGGT; final conc. 80 nM; 5' Mod: FAM; 3' Mod: BHQ-1 |
Wang et al., 2021 | PMID: 34037430 |
| del69_70_FWD CATTAAATGGTAGG ACAGGGTTA; final conc. 360 nM |
Wang et al., 2021 | PMID: 34037430 |
| del69_70_REV ACATTCAACTCAGG ACTTGTT; final conc. 360 nM |
Wang et al., 2021 | PMID: 34037430 |
| del69_70_MT_HEX TTGGTCCCAGA GATAGCATG; final conc. 80 nM; 5' Mod: HEX; 3' Mod: BHQ-1 |
Wang et al., 2021 | PMID: 34037430 |
| P681H_FWD CAGGTATATGCGCTA GTTATCAG; final conc. 360 nM |
Wang et al., 2021 | PMID: 34037430 |
| P681H_REV CACCAAGTGACATAG TGTAGG; final conc. 360 nM |
Wang et al., 2021 | PMID: 34037430 |
| P681H_MT_Cy5 CAGACTAATTCTC ATCGGCG; final conc. 80 nM; 5' Mod: Cy5; 3' Mod: BHQ-2 |
Wang et al., 2021 | PMID: 34037430 |
| Recombinant DNA | ||
| Backbone lentiviral plasmid (pHAGE_Luc2_IRES_ZsGreen-W) | Laboratory of Jesse Bloom lab (Provided by Laboratory of Peter Kim); Crawford et al., 2020 | BEI Cat# NR-52516 |
| SARS-CoV-2 Wuhan-Hu1 Spike plasmid (pHDM-IDTSpike_fixK) | Laboratory of Jesse Bloom lab (Provided by Laboratory of Peter Kim); Crawford et al., 2020 | BEI Cat# NR-51514 |
| Gag-Pol plasmid (pHDM-Hgpm2) | Laboratory of Jesse Bloom lab (Provided by Laboratory of Peter Kim); Crawford et al., 2020 | BEI Cat# NR-52517 |
| Tat plasmid (pHDM-Tat1b) | Laboratory of Jesse Bloom lab (Provided by Laboratory of Peter Kim); Crawford et al., 2020 | BEI Cat# NR-52518 |
| Rev plasmid (pRC-CMV_Rev1b) | Laboratory of Jesse Bloom lab (Provided by Laboratory of Peter Kim); Crawford et al., 2020 | BEI Cat# NR-52519 |
| Software and algorithms | ||
| R base packages (for statistical analysis) | R Foundation for Statistical Computing, Vienna, Austria; RStudio Team (2020). RStudio: Integrated Development for R. RStudio, PBC, Boston, MA | https://www.rstudio.com/products/rstudio/download/ |
| ggplot2 package (for graphs) | Wickham, H. (2009) ggplot2: elegant graphics for data analysis. Springer New York. | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Other | ||
| Syringe filter unit, 0.45 μm, polyethersulfone | Milipore Sigma | Cat# SLHP033RS |
| BD Luer-lock general use syringe | Fisher Scientific | Cat# 22-124-969 |
| 96-well white-walled, clear bottom plates (Grenier Bio-One CellStar Microplate) | Fisher Scientific | Cat# 07-000-167 |
| 96-well U-bottom clear plates (Corning Costar Assay plate) | Corning | Cat# 3788 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to, and if reasonable will be fulfilled by, the lead contact, Dr. Scott Boyd (sboyd1@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Recruitment and informed consent of research participants
Research participants were recruited by public announcement. Informed consent for research participation and collection of blood and nasopharyngeal swab specimens was obtained, with research volunteers signing a consent form approved by the Ethics Review Board at the Ministry of Health of Mongolia. Blood specimens were collected in a five-day period from July 3 to 7, 2021. Of the initial 794 participants enrolled in the study, 196 participants (111 females and 85 males, aged 20-85) balanced according to age, sex and time post-second vaccine dose were selected for serological analysis, with 47, 50, 45 and 54 recipients of the Pfizer/BioNTech, AstraZeneca, Sputnik V and Sinopharm vaccines, respectively. Research participants were healthy at the time of sample collection and had no known history of immunodeficiency. No other procedures or medications were recorded. An additional 99 participants (67 females and 32 males, aged 20-72) who had a history of SARS-CoV-2 infection after vaccination (1, 21, 4 and 73 recipients of the Pfizer/BioNTech, AstraZeneca, Sputnik V and Sinopharm vaccines, respectively) confirmed by real-time PCR or rapid diagnostic testing (RDT), were also studied with serological assays. Average time between infection and sample collection was 18 days (range from 7 to 63 days). Demographic data for this study are available at Mendeley Data: https://doi.org/10.17632/hy3zm69f57.1.
Care and maintenance of cell lines
Human embryonic kidney (HEK293T) and human ACE2-expressing HeLa cells were cultured at 37 °C, 5% CO2, in DMEM (Cytiva) supplemented with 10% FBS (Sigma-Aldrich), 10 mM HEPES (Sigma-Aldrich), and Penicillin-Streptomycin-Glutamine (Life Technologies). At 90% confluence, cells were washed with Dulbecco’s Phosphate-Buffered Saline, without calcium and magnesium (Cytiva), and chemically dissociated by 0.05% trypsin-EDTA with Phenol Red (GIBCO). Cells were gently pipetted and sub-cultured at a 1:5-1:10 ratio.
Method details
Electrochemiluminescence (ECL) IgG binding assay
Vaccine recipient plasma samples were heat-inactivated at 56 °C for 30 min. IgG antibodies targeting the Wuhan-Hu-1 SARS-CoV-2 nucleocapsid (N), spike (S) and spike receptor binding domain (RBD) were detected using MSD V-PLEX Coronavirus Panel 4 (IgG) kits (Meso Scale Discovery) according to the manufacturer protocols. The kits measure samples in a 96-well plate by multiplexed indirect serology using patterned arrays of the target antigens and electrochemiluminescence (ECL) detection. Plasma samples were analyzed in duplicate at a 1:5,000 dilution, detected with anti-human IgG antibodies labeled with an ECL label (SULFO-TAG), and quantified with an MSD MESO QuickPlex SQ 120 instrument. Sample dilution of 1:5000 ensured that measured antibody concentrations were within the linear range of the ECL assay (Figure S3A). In addition to test samples, each plate contained a blank well, three positive control samples, and a 7-point calibration curve in duplicate generated by serial dilution of a reference standard. The calibration sample data for each antigen was fit to a 4-parameter logistic (4-PL) model using 1/Y2 weighting. Antibody unit concentrations (MSD AU/mL) for samples were calculated by backfitting ECL signals to the models.
ACE2-variant RBD antibody blocking assays
Antibodies blocking the binding of ACE2 protein to SARS-CoV-2 RBD for viral variants Alpha (N501Y), Beta (K417N, E484K, N501Y), Gamma (K417T, E484K, N501Y), Delta (L452R, T478K), Epsilon (L452R), Eta/Iota/Zeta (E484K), Kappa (L452R, E484Q), B.1.526.2 (S477N) and P.3 (E484K, N501Y), and Wuhan-Hu-1were detected with MSD V-PLEX SARS-CoV-2 Panel 11 (ACE2) kits according to the manufacturer’s protocols. Heat inactivated plasma samples from vaccinees were analyzed in duplicate at a dilution of 1:10. Samples were added to wells of 96-well plates presenting arrays of the different RBDs and incubated to allow antibodies in the samples to bind. Human ACE2 protein conjugated with the SULFO-TAG label was then added to the wells. After incubating to let labeled ACE2 bind to ACE2-binding sites that were not blocked by antibodies, the plates were read with a MESO QuickPlex SQ 120 instrument. In addition to the test samples, each plate contained a 7-point calibration curve in duplicate generated by serial dilution of a reference standard and a blank well. Results are reported as percent inhibition calculated based on the equation ((1 – Average Sample ECL Signal / Average ECL signal of blank well) x 100).
Production of SARS-CoV-2 pseudotyped lentivirus
As previously described by Crawford et al. (Crawford et al., 2020), HEK293T cells were transfected via calcium phosphate with a five-plasmid 3rd generation lentiviral system composed of a lentiviral packaging vector (pHAGE_Luc2_IRES_ZsGreen-W), SARS-CoV-2 Wuhan-Hu-1 Spike plasmid (pHDM-IDTSpike_fixK), and 3 Helper plasmids containing Gag-Pol (pHDM-Hgpm2), Tat (pHDM-Tat1b), and Rev (pRC-CMV_Rev1b). Briefly, HEK293T cells were plated in 10 cm tissue culture dishes and grown to 70% confluence in DMEM supplemented with 10% FBS, Penicillin/Streptomycin/Glutamine, and 10 mM HEPES. Plasmids were combined in 500 μL of water in the following quantities: 10 μg lentiviral packaging vector, 3.4 μg Spike plasmid, 2.2 μg each Helper plasmid. 500 μL 2X HEPES-Buffered Saline was added dropwise to the DNA mixture followed by 100 μL 2.5 M Calcium Chloride dropwise while agitating the mixture to avoid clumping the DNA. After a 20 min incubation at room temperature, the transfection reaction was added dropwise to the cells with gentle swirling. After 24 h incubation at 37°C, 5% CO2, media was gently aspirated from cells, spun down (300 x g, 5 min) to remove cellular debris, then passed through a 0.45 μm filter, aliquoted, and stored at −80°C.
Pseudovirus neutralization assay
HeLa cells expressing human ACE2 were plated on the inner 60 wells of a 96-well white-walled, flat clear bottom plates at a density of 5,000 cells per well (100 uL of a 50,000 cell/mL suspension). 200 μL of PBS was placed in the outer wells to reduce evaporation during incubation periods. After 24 h incubation at 37 °C, 5% CO2, media was replaced with 100 μL of pseudovirus/serum mixtures, with virus only wells serving as a positive control and media only wells as a negative control. To prepare pseudovirus/serum mixtures, serum was heat inactivated at 56 °C for 30 min, diluted 1:25 in media, then serial diluted by 2-fold. Pseudovirus was diluted 1:2 with media and supplemented with Polybrene at 1:1,000 to improve infection efficiency. Serum dilutions and pseudovirus was then combined 1:1 for a final starting dilution of 1:50. After 48 h incubation, plates were read by replacing 50 μL of pseudovirus/serum mixture with 50 μL Perkin Elmer BriteLite Plus Luciferase reagent. Plates were read by BioTek Synergy 2 plate reader.
SARS-CoV-2 variant genotyping
Total nucleic acids were extracted from 182 SARS-CoV-2 positive nasopharyngeal swab specimens (300 μL) using the Chemagic Viral DNA/RNA 300 Kit on the Chemagic 360 extraction instrument (both from Perkin-Elmer, Waltham, MA) according to the manufacturer’s instructions.
Purified nucleic acid eluates were genotyped using a multiplex, mutation-specific RT-qPCR targeting N501Y, E484K, and L452R, as previously described (Wang et al., 2021). A second confirmatory genotyping RT-qPCR assay was then used to identify the Alpha variant in N501Y mutation-positive samples (n = 177) from the first multiplex reaction. For this assay we designed primers and a dual-labeled hydrolysis probe targeting spike del69-70 (Table S2). The N501Y mutation was also included as a positive control, as this Alpha variant confirmation assay was run only on samples positive for N501Y in the first reaction. The del69-70 mutation was selected for Alpha variant confirmation given that 99.93% (929,411/930,076) of adequately covered [unidentified nucleotides (N) < 5%], full-length SARS-CoV-2 sequences in GISAID (as of June 22, 2021) with N501Y and del69-70 belonged to the Alpha variant B.1.1.7 lineage. Primers and probes for the spike P681H mutation were also included in the reaction, but this target was not utilized in this study.
Primer/probe mix (1 μL, final concentration 360 nM each primer, 80 nM each probe) was combined with a one-step RT-qPCR system (12.5 μL master mix + 0.5 μL Taq polymerase, SuperScript III Platinum One-Step qRT-PCR Kit, Invitrogen, Carlsbad), nuclease-free water (6 μL), and template (5 μL) in a 25 μL reaction. All experiments were conducted on a BioRad CFX96 real-time PCR instrument (BioRad, Hercules, CA, USA). One mutant control (pooled mutant ssDNA) and one wild-type control (whole-genome synthetic RNA from Twist Bioscience, South San Francisco, CA) were included in each RT-qPCR experiment. Cycling conditions were as follows: 52 °C for 15:00, 94 °C for 2:00, and then 45 cycles of 94 °C for 00:15, 59.0 °C for 00:40, and 68 °C for 00:20. Fluorescence thresholds were manually set at 500 for both N501Y-FAM and del69-70-HEX.
Quantification and statistical analysis
Statistical analysis of RBD-ACE2 blocking assay
To assess for differences between pairwise combination of vaccine groups, significance was calculated using Wilcoxon test with Bonferroni correction to adjust for multiple hypothesis correction. Significance between two groups (age groups and male versus female) was calculated by Wilcoxon test. Statistical analyses were performed in R using base packages for statistical analysis and the ggplot2 package for graphs. Statistical tests used are indicated in the respective figure legends.
Statistical analysis for pseudovirus neutralization assays
Luciferase readout values were normalized by the average positive and negative control values on each sample’s respective plate. IC50 values were calculated by 4-point non-linear regression with a constraint of 0% set to the bottom of the fit and 100% for the top of the fit.
Acknowledgments
The authors gratefully acknowledge the study participants for their contributions to this research. The authors thank the laboratories of Jesse Bloom and Peter Kim for providing the pseudovirus plasmid system and the laboratories of Dennis Burton and Peter Kim for providing HeLa cells expressing hACE2. We thank Maria Filsinger Interrante and Abigail Powell for guidance with the pseudovirus assay and Javaria Najeeb and Ana Rita Cardoso for helpful discussions.
This work was supported by NIH/NIAID HHSN272201700013C (G.B.S.); NIH/NIAID R01AI127877 (S.D.B.), NIH/NIAID R01AI130398 (S.D.B.), NIH/NCI 1U54CA260517 (S.D.B., B.A.P., K.C.N, T.J.), and an Early Postdoc.Mobility Fellowship Stipend to O.W. from the Swiss National Science Foundation (SNSF).
Author contributions
N.J.D. coordinated and oversaw the clinical study design and conduct and contributed to data analysis. O.F.W. and K.R. contributed to study design and had the main responsibility for acquisition, analysis, and interpretation of the data. E.H., A.S.B., M.S., H.W., J.A.M., D.S., M.K.S., A.S.L., M.M.S., and J.L. contributed to acquisition and analysis of the data. S.B., P.B-U., A.E., E.B., D.Z., B.O., T.K., G.D., N.B., U.B., N.A., O.O., A.S.B., Z.G., D.Y., and A.M. carried out sample and data collection. P.S.A. and B.P. shared and contributed to interpretation of the data. S.C. and K.C.N. shared specimens and contributed to interpretation of the data. B.A.P. and T.J. contributed to study design and oversaw collection and interpretation of the data. J.L.W., J.N.W., and G.B.S. contributed reagents, provided support for the study, and contributed to data analysis. S.D.B. contributed to study design, served as the principal investigator for the experimental methods, acquisition, analysis, and interpretation of the data, and supported the study. N.D.D. contributed to study design and served as the principal investigator for the clinical study. The manuscript was mainly written by S.D.B., O.F.W., and K.R. with contributions from B.A.P. and A.S.B. All authors reviewed and approved the manuscript.
Declaration of interests
S.D.B. has consulted for Regeneron, Sanofi, and Novartis on topics unrelated to this study and owns stock in AbCellera Biologics; B.P. and P.S.A. are inventors on a provisional patent application (no. 63/026,577) submitted by the Board of Trustees of the Leland Stanford Junior University, Stanford, CA, that covers the use of “Therapeutic Methods for Treating COVID-19 Infections”; B.P. serves on the External Immunology Network of GSK and on the Scientific Advisory Board of Medicago and Boehringer Ingelheim; and K.C.N. reports grants from National Institute of Allergy and Infectious Diseases (NIAID), Food Allergy Research & Education (FARE), End Allergies Together (EAT), National Heart, Lung, and Blood Institute (NHLBI), and National Institute of Environmental Health Sciences (NIEHS). K.C.N. is Director of FARE and World Allergy Organization (WAO) Center of Excellence at Stanford, Advisor at Cour Pharmaceuticals, Cofounder of Before Brands, Alladapt, Latitude, and IgGenix, National Scientific Committee member for the Immune Tolerance Network (ITN) of NIAID, recipient of a Research Sponsorship from Nestle, Consultant and Advisory Board Member at Before Brands, Alladapt, IgGenix, NHLBI, and ProBio, and Data and Safety Monitoring Board member at NHLBI; R.S. Chinthrajah receives grant support from CoFAR National Institute of Allergy and Infectious Diseases, Aimmune, DBV Technologies, Astellas, AnaptysBio, Novartis, and Regeneron and is an advisory board member for Alladapt Immunotherapeutics Inc., Novartis, and Genentech. J.L.W., J.N.W., and G.B.S. are employees of Meso Scale Diagnostics (MSD). The rest of the authors declare that they have no conflicts of interest relevant to this manuscript.
Published: November 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.11.004.
Supplemental information
Data and code availability
-
•
Demographic data and electrochemiluminescence data for this study are available in a file at Mendeley Data: https://doi.org/10.17632/hy3zm69f57.1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- AlQahtani M., Bhattacharyya S., Alawadi A., Mahmeed H.A., Sayed J.A., Justman J., El-Sadr W.M., Hidary J., Mukherjee S. Research Square; 2021. Morbidity and mortality from COVID-19 post-vaccination breakthrough infections in association with vaccines and the emergence of variants in Bahrain. [DOI] [Google Scholar]
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. CMMID COVID-19 Working Group. COVID-19 Genomics UK (COG-UK) Consortium Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. MedRxiv. 2021 doi: 10.1038/s41591-021-01540-1. 2021.06.21.21258528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A., Fong Y., Benkeser D.C., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune Correlates Analysis of the mRNA-1273 COVID-19 Vaccine Efficacy Trial. MedRxiv. 2021 doi: 10.1126/science.abm3425. 2021.08.09.21261290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. BioRxiv. 2021 2021.05.26.445838. [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Arunachalam P.S., Yang F., Hoh R.A., Wirz O.F., Lee A.S., Gao F., Mallajosyula V., Li C., et al. mRNA vaccination compared to infection elicits an IgG-predominant response with greater SARS-CoV-2 specificity and similar decrease in variant spike recognition. MedRxiv. 2021 2021.04.05.21254952. [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall E.C., Wu M., Harvey R., Kelly G., Warchal S., Sawyer C., Daniels R., Hobson P., Hatipoglu E., Ngai Y., et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Miller J.A., Verghese M., Sibai M., Solis D., Mfuh K.O., Jiang B., Iwai N., Mar M., Huang C., et al. Multiplex SARS-CoV-2 Genotyping Reverse Transcriptase PCR for Population-Level Variant Screening and Epidemiologic Surveillance. J. Clin. Microbiol. 2021;59:e0085921. doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Demographic data and electrochemiluminescence data for this study are available in a file at Mendeley Data: https://doi.org/10.17632/hy3zm69f57.1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.