Summary
It can be challenging to maintain tissue integrity using established histology protocols. Here, we describe a protocol composed of Hartman’s fixation, window technique, microwave-based tissue processing, optimized depigmentation, and antigen retrieval pretreatment. This is followed by the ViewRNA single-molecule fluorescence in situ hybridization and immunofluorescence techniques to optimize routine histological staining and molecular histology multiplexing assays. Our protocol is highly reproducible in any laboratory and may decrease animal usage and lab resource expenditure.
For complete details on the use and execution of this protocol, please refer to Pang et al. (2021).
Subject areas: Antibody, In Situ Hybridization
Graphical abstract
Highlights
-
•
Step-by-step protocol to improve ocular histology
-
•
Tips to make multiple windows (notches) on a mouse eyeball
-
•
Multiplexing smFISH with immunofluorescence on an ocular paraffin section
It can be challenging to maintain tissue integrity using established histology protocols. Here, we describe a protocol composed of Hartman’s fixation, window technique, microwave-based tissue processing, optimized depigmentation, and antigen retrieval pretreatment. This is followed by the ViewRNA single-molecule fluorescence in situ hybridization and immunofluorescence techniques to optimize routine histological staining and molecular histology multiplexing assays. Our protocol is highly reproducible in any laboratory and may decrease animal usage and lab resource expenditure.
Before you begin
The purpose of developing this protocol is to preserve the structural integrity of mouse eyeball while subjecting it to paraffin processing, as well as to maintain the normal morphology of eye sections on a glass slide for molecular assays multiplexing after heat-induced antigen retrieval, H2O2 based de-pigmentation and/or protease pretreatment. Here, the mRNA and protein of connexin 43 (Cx43) are chosen as the example of molecular targets. Cx43, also called gap junction alpha-1 protein (Gja1), is one of the reported gap junction proteins expressed in the ciliary body (CB) of mouse, rat, and rabbit (Calera et al., 2009; Wolosin et al., 1997). Cx43 in the ciliary body is involved in aqueous humor secretion, maintaining intraocular pressure (IOP), and is required for maintaining the vitreous (Calera et al., 2006, 2009; Pang et al., 2021). Several human ocular diseases are reported to be associated with Cx43 mutation (Gabriel et al., 2011; Musa et al., 2009; Paznekas et al., 2003). Understanding the expression and cellular localization of Cx43 can be important in uncovering how defective Cx43 expression and localization contribute to the pathology of human eye diseases. Here we employ ViewRNA single molecular fluorescence in situ hybridization (smFISH) technology to detect Cx43 mRNA multiplexing with immunofluorescence assay to detect Cx43 protein on the same section. Of note, the protocol detailed below is a generic protocol that may work for other mRNA or protein targets after some minor modifications or optimizations.
Note: This protocol is a modified method of our previous research article (Pang et al., 2021) with additional multiplexing steps added. All mice used in this study were housed and cared according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and guidelines of the Institutional Animal Care and Use Committees at the Stowers Institute for Medical Research.
RNAse free handling
Timing: 30 min
CRITICAL: Throughout the protocol, make sure to work on a clean bench surface, use RNAse/DNase-free reagents, pipette tips, and Eppendorf tubes to avoid mRNA degradation until the RNA probe hybridization step is completed.
Be sure to apply RNAse-free practices for sample collection, processing, and handling when performing an RNA in situ hybridization assay.
-
1.
Wear a mask and full personal protective equipment (PPE) when working near or with the samples and/or slides.
-
2.
Wipe down all the outside working surfaces and tools with RNAse AWAY followed by 200 proof pure ethanol and then let everything air dry.
-
3.
Use RNAse-free water (0.1% v/v diethyl pyrocarbonate-treated and 20 min autoclaved water) to prepare any aqueous solutions and to fill the water bath for paraffin sectioning.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal Anti-Cx43 (1:200 when using) | Thermo Fisher Scientific | Cat# 35-5000; RRID: AB_2533207 |
| Mouse monoclonal Anti-Rhodopsin (1:250 when using) | EMD Millipore | Cat# MAB5356; RRID: AB_2178961 |
| Rabbit polyclonal Anti-Calbindin (1:250 when using) | Swant | Cat# CB38a; RRID: AB_10000340 |
| Goat polyclonal Anti-Calretinin (1:500 when using) | Chemicon | Cat# AB1550; RRID: AB_90764 |
| CF488A Donkey anti-mouse IgG (H+L) (1:300 when using) |
Biotium | Cat# 20014; RRID: AB_10561327 |
| Alexa Fluor 647 Donkey anti-mouse IgG (H+L) (1:300 when using) |
Invitrogen | Cat# A31571; RRID: AB_162542 |
| CF568 Donkey anti-rabbit IgG (H+L) (1:300 when using) |
Biotium | Cat# 20098; RRID: AB_10557118 |
| CF488A Donkey anti-goat IgG (H+L) (1:300 when using) |
Biotium | Cat# 20016; RRID: AB_10563028 |
| Chemicals, peptides, and recombinant proteins | ||
| Hartman’s Fixative | Sigma-Aldrich | Cat#H0290-500ML |
| Paraformaldehyde, 16% (Aqueous) | TED PELLA | Cat#18505 |
| Thermo Scientific™ Shandon Tissue Marking Dyes | Themo Fisher Scientific | Cat#NC9635094 |
| Diethyl pyrocarbonate (DEPC) | Sigma | Cat#D-5758 |
| RNAse AWAY | Thermo Fisher Scientific | Cat#7003 |
| Sodium Chloride | Fisher Scientific | Cat#BP358-212 |
| Potassium Chloride | Calbiochem | Cat#7300 |
| Sodium Biphosphate | JTBaker | Cat#3828-01 |
| Potassium Dihydrogen Phosphate | Macron | Cat#7100-12 |
| 200 proof pure ethanol | VWR | Cat#V1001TP |
| Paraffin (Richard Allen Scientific Type 9) | VWR | Cat#72050-030 |
| Isopropanol | VWR | Cat#BDH1133-4LP |
| Xylene | VWR | Cat#89370-088 |
| Sodium Citrate, Dihydrate | JTBaker 3646-05 | Cat#3646-05 |
| Citric Acid, Anhydrous | Sigma-Aldrich | Cat#C2404 |
| Hydrogen Peroxide (30%) | Sigma-Aldrich | Cat#216763-500ML |
| Formamide, deionized | VWR | Cat#0606-100ML |
| Protease QF | Thermo Fisher Scientific | Cat#QVT0512 |
| TritonX-100 | Fisher bioreagents | Cat#BP151-500 |
| SuperBlock (1×) | Thermo Fisher Scientific | Cat#37580 |
| 4′,6-diamidino-2-phenylindole (DAPI, 5 mg/mL) | BioLegend | Cat#422801 |
| ProLong™ Glass Antifade Mountant | Thermo Fisher Scientific | Cat#P36982 |
| Critical commercial assays | ||
| ST Infinity H&E Staining System | VWR | Cat#10015-090 |
| Surgipath Micromount Mounting Media | VWR | Cat#10014-976 |
| ViewRNA™ Cell Plus Assay Kit | Thermo Fisher Scientific | Cat#88-19000-99 |
| Trueview AF Quenching Kit | VECTOR LABORATORIES | Cat#SP-8400 |
| ViewRNA probe sets for mouse Gja1, 647 channel | Thermo Fisher Scientific | Cat#VX-06_VB6-17027-VCP |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6J | LASF of Stowers Institute for Medical Research | JAX: 000664. Wild type mice aging 1 week (for developing) and between 6-10 months (for adult) are used, both males and females. |
| Software and algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/download.html | ImageJ |
| Fiji-win64 |
Schindelin et al., 2012a Schneider et al., 2012b |
https://imagej.nih.gov/ij/https://imagej.net/Fiji |
| Other | ||
| 1 mL Slip-Tip Syringes with Precisionglide™ Subcutaneous Needles | BD Biosciences | Cat#309597 |
| Dissecting Forceps, non-serrated, fine tip, curved. | VWR | Cat#82027-406 |
| Vannas Scissors | World Precision Instruments | Cat#14003 |
| Titramax lab shaker | Heidolph Instruments | Model 101 |
| Lens Cleaning Tissues | VWR | Cat#52846-001 |
| Histoplex tissue processing cassettes | VWR | Cat#15001-138 |
| Ultrarapid microwave tissue processor | Milestone | PathosDelta |
| Automated rotary microtome | Leica Biosystems | RM2255 |
| Sure Bond Charged White Slides | Avantik | Cat#SL6332-1 |
| Surgipath stainless steel base molds - paraffin samples | VWR | Cat#75809-384 |
| Hydrophobic pen | VECTOR Laboratories | Cat#H-4000 |
| HybEZ™ II oven | Advanced Cell Diagnostics Inc | Cat#PN321710/321720 |
| HybEZ Humidity control Tray | Advanced Cell Diagnostics Inc | Cat#PN310012 |
| ACD HybEZ slide Rack | Advanced Cell Diagnostics Inc | Cat#310014 |
| Kimwipes | Kimberly Clark | Cat#34155 |
| 20 mL scintillation glass vials with attached foil lined cap | VWR | Cat#66022-060 |
| Pipettes | Gilson | Cat#FA10001M/FA10003M/FA10005M/FA10006M |
| Pipette tips (DNAse/RNAse free) | Ambion | Cat#12650/12660 |
| Antigen retrieval microwave | Biogenex | Model MW014-MO |
| Stereoscope | Leica | Model M80HD |
| Olympus Slide Scanner Automated Widefield Microscope | Olympus | Model VS120 |
| Spinning Disk Confocal System | Nikon | Eclipse Ti2 with CSU-W1 Spinning Disk |
Materials and equipment
10× Phosphate buffered saline (pH=7.4 when diluted to 1×) (PBS)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Chloride | 1.4 mol/L | 80 g |
| Potassium Chloride | 27 mmol/L | 2 g |
| Sodium Biphosphate | 100 mmol/L | 14.4 g |
| Potassium Dihydrogen Phosphate | 18 mmol/L | 2.4 g |
| RNAse-free ddH2O | n/a | Fill up to 1000 mL |
| Total | n/a | 1000 mL |
This solution is stable for 12 months when stored at 15°C–30°C, sterile.
Note: 1000mL 1× PBS (pH=7.4) can be obtained by mixing 100 mL 10× PBS and 900 mL RNAse-free ddH2O.
4% Paraformaldehyde (4% PFA)
| Reagent | Final concentration | Amount |
|---|---|---|
| RNAse-free ddH2O | n/a | 26 mL |
| PBS (10×) | 1× | 4 mL |
| Paraformaldehyde (16%) | 4% | 10 mL |
| Total | n/a | 40 mL |
This solution is stable for 1 month when stored at 2°C–8°C, sterile.
CRITICAL: Paraformaldehyde is flammable, toxic, and an irritant. Please use in a Labconco chemical safety fume hood and handle with full personal protective equipment.
0.1 mol/L Sodium Citrate
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Citrate, Dihydrate | 0.1 mol/L | 29.41 g |
| RNAse-free ddH2O | n/a | Fill up to 1000 mL |
| Total | n/a | 1000 mL |
This solution is stable for 3 months when stored at 2°C–8°C, sterile.
0.1 mol/L Citric Acid
| Reagent | Final concentration | Amount |
|---|---|---|
| Citric acid | 0.1 mol/L | 19.22 g |
| RNAse-free ddH2O | n/a | Fill up to 1000 mL |
| Total | n/a | 1000 mL |
This solution is stable for 3 months when stored at 2°C–8°C, sterile.
20× Sodium Chloride-Sodium Citrate buffer (pH=7.0) (SSC)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Chloride | 3 mol/L | 175.3 g |
| Sodium Citrate, Dihydrate | 0.3 mol/L | 88.2 g |
| RNAse-free ddH2O | n/a | Fill up to 1000 mL |
| Total | n/a | 1000 mL |
This solution is stable for 6 months when stored at 2°C–8°C, sterile.
Note: Adjust pH to 7.0 by 0.1 mol/L citric acid.
1× Citrate buffer (pH=6.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Citrate (0.1 mol/L) | 8.2 mmol/L | 82 mL |
| Citric Acid (0.1 mol/L) | 1.8 mmol/L | 18 mL |
| RNAse-free ddH2O | n/a | 900 mL |
| Total | n/a | 1000 mL |
This solution is stable for 3 months when stored at 2°C–8°C, sterile.
Note: Adjust pH to 6.0 by citric acid.
Bleaching solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 20× SSC | 0.5× | 1 mL |
| RNAse-free ddH2O | n/a | 31 mL |
| Formamide, deionized | 10% | 4 mL |
| Hydrogen Peroxide (30%) | 3% | 4 mL |
| Total | n/a | 40 mL |
This solution should be prepared right before use.
CRITICAL: Formamide is flammable, toxic, and an irritant. Hydrogen peroxide is an oxidate and irritant. Please use in hood and handle with full personal protective equipment. When making this mixture, do not directly add 30% hydrogen peroxide to formamide or vice versa. Add ddH2O in between these chemicals as a dilutant.
Protease QF working solution
| Reagent | Final concentration | Amount |
|---|---|---|
| RNAse free PBS (1×) | 1× | 1 mL |
| Protease QF | 1:500 | 2 μL |
| Total | n/a | 1 mL |
This solution should be prepared right before use.
PBSTx (Phosphate buffered saline/Triton-X)
| Reagent | Final concentration | Amount |
|---|---|---|
| RNAse-free ddH2O | n/a | 45 mL |
| PBS (10×) | 1× | 5 mL |
| Triton-X | 0.1% | 50 μL |
| Total | n/a | 50 mL |
This solution is stable for 3 months when stored at 15°C–30°C, sterile.
SSCTx (Sodium Chloride-Sodium Citrate/Triton-X)
| Reagent | Final concentration | Amount |
|---|---|---|
| RNAse-free ddH2O | n/a | 974 mL |
| SSC (20×) | 0.5× | 25 mL |
| Triton-X | 0.1% | 1 mL |
| Total | n/a | 1000 mL |
This solution is stable for 3 months when stored at 15°C–30°C, sterile.
Primary antibody working solution
| Reagent | Final concentration | Amount |
|---|---|---|
| SuperBlock (1×) | n/a | 100 μL |
| Mouse monoclonal anti-Cx43 (250 μg/mL) | 1.25 μg/mL (1:200) | 0.5 μL |
| Total | n/a | 100.5 μL |
This solution should be prepared for same day use only.
Secondary antibody working solution
| Reagent | Final concentration | Amount |
|---|---|---|
| SuperBlock (1×) | n/a | 100 μL |
| CF488A Donkey anti-mouse IgG (H+L) | 1:300 | 0.3 μL |
| Total | n/a | 100.3 μL |
This solution should be prepared for same day use only.
DAPI working solution (4′,6-Diamidino-2-Phenylindole)
| Reagent | Final concentration | Amount |
|---|---|---|
| PBSTx (1×) | n/a | 1 mL |
| DAPI stock solution (5 mg/mL) | 10 μg/mL | 2 μL |
| Total | n/a | 1 mL |
This solution should be prepared for same day use only.
Autofluorescence blocking solution (Vector® TrueVIEW® Autofluorescence Quenching Kit)
| Reagent | Final concentration | Amount |
|---|---|---|
| Reagent A | 1:6 | 170 μL |
| Reagent B | 1:6 | 170 μL |
| Reagent C | 1:6 | 170 μL |
| PBS (1×) | n/a | 500 μL |
| Total | n/a | 1 mL |
This solution should be prepared right before use.
CRITICAL: Mix equal volumes of Reagent A and Reagent B, then add Reagent C to the mixture. Mix well and add PBS. The order of mixing is important. This formula is developed based on the user guide of Vector® TrueVIEW® Autofluorescence Quenching Kit:
https://vectorlabs.com/trueview-autofluorescence-quenching-kit.html#catalog.custom.product.faqs
Step-by-step method details
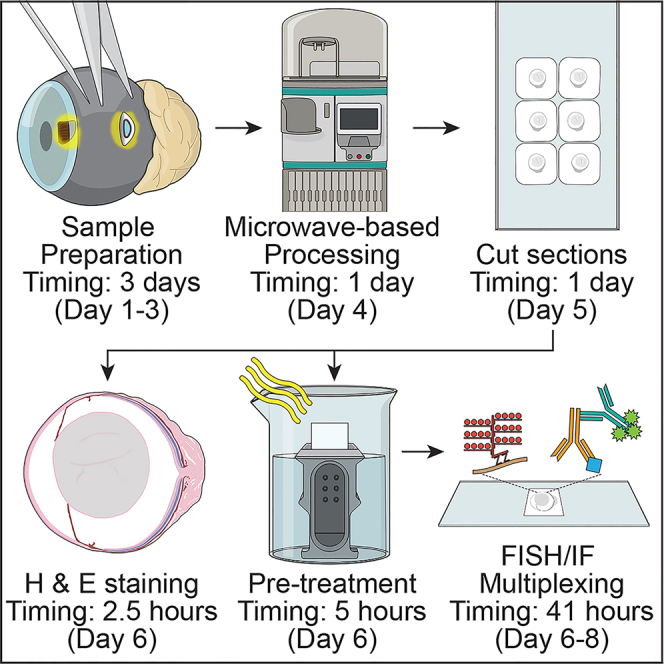
Sample preparation
Timing: 3 days, Day 1–3 for step 1
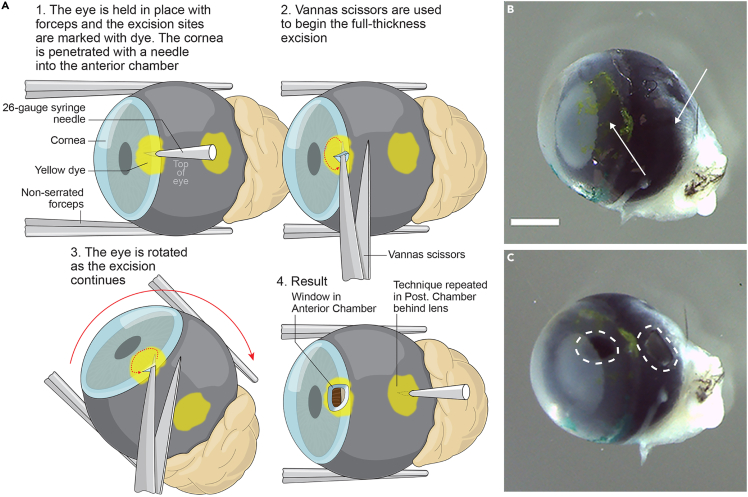
This section describes the protocol from the dissection of the mouse eye sample to tissue processing. The ‘Window Technique’ (making notches into the eye to allow optimal penetration of fluids and to balance the pressure) is essential for morphology preservation.
-
1.
Dissect eyeballs from the euthanized mice, as previously described by Sondereker, K.B. et al. (Sondereker et al., 2018)
Try to avoid scratching and hard extrusion on the eyeball tissue.
Note: Tissue marking dyes can be used to mark the eye sample for orientation purposes.
-
2.
Immerse the eye sample in a 20 mL disposable scintillation vial of Hartman’s fixative (commercially available).
Note: A minimum of 20:1 ratio of fixative volume to sample volume is recommended. In our observation, the size of an aqueous droplet of 0.5ml fixative was bigger than the size of an eyeball. Therefore, we dropped 2 eyeballs into 20ml fixative solution for tissue fixation.
-
3.
Place the vial with sample in fixative on a shaker with gentle agitation. Fix the eyeball at ambient temperature (20°C–25°C) for 20 h.
-
4.
Remove the Hartman’s fixative and fill the 20 mL sample vial with 4% PFA.
-
5.
Fix the eyeball sample in 4% PFA for another 20 h at ambient temperature.
-
6.
Remove the 4% PFA and rinse the eye sample with RNAse-free 1× PBS for 4 × 15min.
-
7.
Make windows on the eyeball wall as illustrated in Figure 1A. Individual eyeball is submerged in RNAse-free PBS and gently secured with non-serrated forceps. Use a 26-gauge syringe needle to penetrate and create a slight incision at one side (for this paper, yellow Thermo Scientific™ Shandon Tissue Marking Dye marked side) of the anterior chamber. Carefully insert a Vannas scissors into the incision to snip the tissue from (away from the central cornea) the incision, which expands the incision into a small window. Remove any air bubbles inside the sample by gently rolling the eye back and forth while submerged in the RNAse- free PBS.
CRITICAL: We recommend that the windows are created at the spots that will be trimmed away at the sectioning step. Do not allow the needle to penetrate too deeply into the eye because that may damage other parts of the eyeball tissue. Keep the eyeball in RNAse-free PBS during this step.
-
8.
Continue to make a window on the posterior part of the eyeball wall which is in line with the window developed at step 7. Create another window at the opposite side of the posterior eyeball wall and remove any air bubbles from the inside of eyeballs as needed.
CRITICAL: Steps 7 and 8 are defined as ‘window technique’ in this protocol. Window technique is essential for eye samples from mice older than 3 days to maintain the overall morphology, the size of the anterior chamber, and the vitreous during further processing.
CRITICAL: Windows at the anterior and posterior chamber are expected to facilitate reagent exchange and balance the pressure inside and outside the eye chambers during the paraffin processing steps.
-
9.
Rinse the eye sample with 1× PBS for 2 × 10 min.
-
10.
Dehydrate the eye sample with 20 mL 30% ethanol for 1 h.
-
11.
Replace the 30% ethanol with 50% ethanol and dehydrate samples for 1 h.
-
12.
Replace the 50% ethanol with 70% ethanol and further dehydrate samples for 1 h.
Pause point: Up to this point, eyeball samples can be stored in 70% ethanol in 4°C for up to 2 weeks.
Figure 1.
Window technique
(A) An illustration demonstrating how windows are created on the eye wall (A). This operation applies to all three windows although the sample illustration is made on the anterior wall.
(B) is eye sample before applying the window technique. White arrow points to the region for the 2 windows.
(C) is the eye sample after making windows on an eyeball. Dashed white polygons highlight the 2 windows. The third window is not shown. Scale bar: 1000 μm.
Microwave-based paraffin processing
Timing: 1 day, Day 4 for step 13
This section describes the protocol of paraffin infiltration and embedding. With the aid of a microwave-based automatic tissue processor, samples are dehydrated, cleared, and then infiltrated with paraffin.
-
13.
Carefully wrap eyeball sample in a piece of lens cleaning tissue.
-
14.
Put each sample into an individually pre-labeled tissue cassette.
Note: Immerse all cassettes with samples in 70% ethanol until loading onto the tissue processor.
-
15.
Load cassettes onto the tissue processor and select the appropriate program.
CRITICAL: The different development stages of mouse eyeball may require different tissue processing procedures. Program 1 is for eye samples from embryo or mouse at or younger than 3 days old. Program 2 is for eye samples from mouse older than 3 days. To reduce the shrinking artifact that results in larger eyes, heat and pressure have been eliminated or reduced in program 2 and thus longer incubation times are needed to counteract this change. Processing times are approximate since some added time is needed to heat the fluids. In the table, mBar = millibar.
| Program 1 | Program 2 | |
|---|---|---|
| Reagent | Condition | Condition |
| Ethanol | 10 min @ 65°C | 30 min @ 37°C |
| Isopropanol | 45 min @ 68°C | 2 h @ 45°C |
| Paraffin 1 | 10 min @ 70°C, 500 mBar | 30 min @ 62°C |
| Paraffin 2 | 10 min @ 70°C, 400 mBar | 30 min @ 62°C |
| Paraffin 3 | 2 min @ 70°C, 300 mBar | 30 min @ 62°C |
| Paraffin 4 | 2 min @ 70°C, 200 mBar | 30 min @ 62°C |
| Paraffin 5 | 2 min @ 70°C, 150 mBar | 30 min @ 62°C |
| Paraffin 6 | 27 min @ 65°C, 100 mBar | 30 min @ 62°C |
| Approximate total time | 1 h:48 min | 5 h:30 min |
-
16.
When processing is complete, embed each eye into a separate mold.
Note: To get sections of transverse plane, orient the eyeball to ensure that the central part of the cornea and the optic nerve are on the same focal plane before embedding.
Pause point: The paraffin blocks can be stored at room temperature for up to 1 year.
Sections cutting
Timing: 1 day, Day 5
This section describes the protocol from a paraffin block with an eye sample in it until it is trimmed and cut into transverse cross sections, which can be stored or processed with histological staining.
-
17.
Set thickness setting for the microtome and water bath temperature setting as desired.
Note: For this paper, section thickness is set at 5 μm and temperature of water bath is set at 42°C.
-
18.
Attach paraffin block containing eye sample to microtome.
-
19.
Slowly trim into the block until the sample is located.
-
20.
Switch from a trimming mode to a sectioning mode and begin to create paraffin ribbons to float on the water bath. A single paraffin section is created by one full turn of the microtome wheel and continual revolutions of the wheel create a paraffin ribbon.
Note: Wheel speed should not be very fast when sectioning eyes. A slow and steady revolution will lessen the chances of a shattered lens appearance. A shattered lens appearance can also be caused by dryness of the lens. Soaking the lens in an icy slurry (created by allowing a tray of ice to partially melt) before making sections or adding moisture to the paraffin block by dabbing with a moist tissue soaked with ice-cold water between each section helps to eliminate this shattered artifact.
-
21.
Lift the paraffin ribbon from the microtome plate using forceps or a small brush. Carefully lay the ribbon onto the water bath and allow to rest for 15–30 s.
-
22.
Guide the floating paraffin ribbons onto a test slide.
-
23.
Check the sections microscopically for quality and proper location within the eyeball.
Optional: To get transverse sections near the central plane, trim further into the block to the point where both the iris and the retinal pigmented epithelium (RPE) begin to break open.
Note: This can be checked under a stereoscope using the brightfield setting, as the dark line of the pigmented iris and RPE will show up against the white background. If the eye is oriented correctly, a break in the anterior dark line (the iris) will appear first, which locates the pupil. Further sections will reveal a break in the posterior dark line, the RPE, opposite the pupil. This opening will be the optic nerve head. Once both openings are observed, begin saving these ribbons or sections onto slides for further testing. Both the anterior opening (the pupil) and the posterior opening (the optic nerve head) should be visible in the same section for the central plane of the eye.
-
24.
Once the slide is complete, stand it upright to drain while continuing to make the next slide.
-
25.
Allow the collection of slides to dry overnight while still in a vertical position.
Note: Slides should have no visible water on them before they are ready for further staining.
Pause point: Slides can be stored at this point for up to 1 year at ambient temperature (20°C–25°C) or used for a staining procedure (Hematoxylin & Eosin (H&E), In situ hybridization (ISH), Immunofluorescence (IF) and/or immunohistochemistry (IHC).
Deparaffinization
Timing: 2 h, Day 6
This section describes the protocol from a paraffin preserved eye sample section until the paraffin is removed and the tissue is exposed to solutions and ready for the staining procedure (H&E, ISH, IF, and/or IHC).
-
26.
Heat the slides in a 60°C oven for 1 h to melt away the paraffin.
-
27.
Immerse the slides in xylene at ambient temperature 3 times, 5 min each time.
-
28.
Immerse the slides in 100% ethanol at ambient temperature 3 times, 3 min each time.
-
29.
Air dry the slides at ambient temperature for 15 min. The sections are then ready for H&E staining (steps 30–43) for morphology check. The sections are also ready for tissue pre-treatment (steps 44–54) with single molecule mRNA Fluorescent in situ Hybridization (smFISH) staining (steps 55–79) and/or Immuno-Fluorescence (IF) staining (steps 80–93) as follows.
Hematoxylin and eosin staining
Timing: 20 min, Day 6
This section describes the protocol from a de-paraffinized and untreated eye section until the eye section is stained by Hematoxylin (H) for nuclei and Eosin (E) for cytoplasm. The H&E-stained sections are typical for morphology observation of the tissue at a histological level.
-
30.
Following step 29, load the slides on the Leica Autostainer XL and run the pre-set eye staining program which utilizes reagents and dyes from the St Infinity H&E kit, supplied by Leica Biosystems. The following steps, steps 31–42, are done automatically and times are as follows:
-
31.
Rinse the sections in tap water for 1 min.
-
32.
Incubate the sections in Hemalast for 30 s.
-
33.
Incubate the sections in Hematoxylin for 45 s.
-
34.
Rinse the sections in tap water for 2 min.
-
35.
Incubate the sections in Differentiator for 45 s.
-
36.
Rinse the sections in tap water for 1 min.
-
37.
Incubate the sections in Bluing for 1 min.
-
38.
Rinse the sections in tap water for 1 min.
-
39.
Rinse the sections in 80% ethanol for 1 min.
-
40.
Incubate the sections in Eosin for 2 min.
Note: Eosin solution can be diluted by 100% ethanol if the sections are overstained. In this protocol, we purposefully did a lighter eosin stain (1% vol/vol) to give an enhanced contrast for the nuclei hematoxylin stain.
-
41.
Rinse the sections in 100% ethanol 2 times, 1 min each time.
-
42.
Rinse the sections in xylene 4 times, 1 min each time.
-
43.
Mount the slides with Surgipath Micromount Mounting Media.
Pause point: Slides can be stored at this point for up to 1 year, microscopically observed, or digitally imaged. For this paper, H&E-stained slides are imaged by Olympus Slide Scanner Automated Widefield Microscope.
Tissue pre-treatment
Timing: 3 h, Day 6
This section describes the protocol from a de-paraffinized and untreated eye section until the eye section is pre-treated and ready for further molecular assays such as single molecule mRNA FISH and immunofluorescence multiplexing, which provide spatial information on both the transcription level of mRNA targets and expression level of protein targets.
-
44.
Heat the citrate buffer for 10 min using a BioGenex EZ-Retriever microwave (set a temperature at 95°C).
CRITICAL: This step can remove most of the dissolved air in the solution and thus decrease the number of air bubbles generated at the tissue, which can cause tissue lifting during step 45.
-
45.
Immerse the deparaffinized slides (following step 29) into the heated citrate buffer for 10 min. Make sure the temperature is maintained between 95°C to 100°C.
Note: If you don’t need immunofluorescence, but need only mRNA FISH, lower temperature (70°C for 30min) works better to avoid lifting of sections. For immunofluorescence, 95°C is required depending on targeted proteins.
-
46.
Remove the slides from the hot citrate buffer and immediately rinse them in RNAse free ddH2O at ambient for 1 min.
-
47.
Immerse the slides in 100% ethanol for 2 × 2 min each time. Then air dry the slides for 15 min.
-
48.
Draw a hydrophobic barrier using an ImmEdge Pen on the glass slide to surround tissue sections and air dry.
Note: It might take 30–60 min for the hydrophobic barrier to dry out.
-
49.
Bleach the pigment of eyeball tissue by applying bleaching solution to sections and illuminate by direct cool light for 20 min (see our previously published method (Jordan et al., 2019)).
Note: If the pigment bleaching is not required, apply 3% H2O2 for 15 min to inactivate the endogenous peroxidase enzyme activity (the direct light illumination is unnecessary).
-
50.
Rinse the sections in RNAse free ddH2O for 2 × 1 min each time.
-
51.
Prepare protease QF working solution by diluting protease QF in RNAse free PBS at 1:500 as indicated in materials and equipment. Apply 200 μL protease QF working solution to the sections and incubate for 2 min at room temperature.
Note: This step requires optimization to fit the fixation and tissue type used in the experiment. If tissue is lifting or missing during optimization, the concentration of protease QF or the treatment time can be decreased. If the detected number of mRNA particles is lower than expected or the signal to noise ratio is too low, then the concentration of protease QF or the treatment time needs to be increased. Such a pre-treatment step is unnecessary for immunofluorescence assay. If FISH experiment is not required, this step for the immunostaining experiment can be skipped. Also, this step becomes optional if bleaching solution is applied on the sections at step 49, and the protein targets can be better preserved without protease treatment. Solution volume needs to be adjusted depending on the size of hydrophobic barrier. 200 μL of Protease QF working solution is recommended for 2 cm × 1.5 cm area.
-
52.
Wash the slides with RNAse free ddH2O for 3 × 3 min each time.
-
53.
Apply PBSTx to the section and incubate for 15 min at room temperature.
-
54.
Wash the slides with 1X PBS for 2 × 10 min each time. Sections are then ready for single molecule mRNA FISH and/or Immunofluorescence assay.
Single molecule mRNA fluorescent in situ hybridization (FISH)
Timing: 21 h, Day 6–7
This section describes the protocol from a pre-treated eye section until the eye section is processed with the ViewRNA kit. A HybEZ™ II oven, HybEZ Humidity control Tray and ACD HybEZ slide Rack are used for FISH assay. The following steps (steps 55–79) are developed based on the manuals and protocols of ViewRNA™ Cell Plus Assay Kit:
https://www.thermofisher.com/order/catalog/product/88-19000-99#/88-19000-99
Note: Any incubator or oven is acceptable provided the temperature can be kept at 40°C. Also, any slide rack, chamber or container is acceptable for incubation of slides if is maintained inside the container to avoid drying out of slides.
-
55.
Switch on HybEZ™ II oven and prewarm probe set diluent at 40°C for 10 min.
-
56.
Apply 200 μL of 1X PBS to the slide to cover the section. Put the slides in humidity control tray with humidified tissue paper and incubate for 20 min at 40°C.
Note: Volume of 1X PBS needs to be adjusted to cover the sections depending on the size of hydrophobic barrier.
-
57.
Prepare probe mix working solution by diluting the ViewRNA Probe in pre-warmed probe set diluent at 1:100 as indicated in materials and equipment.
Note: Volume of probe mix working solution need to be adjusted depending on the size of hydrophobic barrier. 100 μL of probe mix working solution is recommended for 2cm × 1.5 cm area.
-
58.
Remove PBS and apply 100 μL of probe mix working solution to the sections. Incubate in humidity control tray overnight (approximately 16 h) at 40°C.
Note: Volume of probe mix working solution needs to be adjusted depending on the size of hydrophobic barrier. 100 μL of probe mix working solution is recommended for 2cm × 1.5 cm area. Overnight incubation is not essential, but over 5 h incubation time is recommended for the best result.
-
59.
Prewarm Amplifier diluent at 40°C for 10 min.
-
60.
Prepare 10 mL of ViewRNA wash buffer and 100 μL of PreAmplifier working solution as indicated in materials and equipment.
Note: Required volume depends on the size of hydrophobic barrier. 200 μL of viewRNA wash buffer is recommended for 2cm × 1.5 cm area. 100 μL of PreAmplifier working solution is recommended for 2cm × 1.5 cm area.
-
61.
Aspirate probe mix working solution and rinse with 200 μL of viewRNA wash buffer for 2 × 2 min each time.
-
62.
Rinse the sections in 25mL of SSCTx for 2 × 5 min.
-
63.
Rinse the sections with 200 μL of viewRNA wash buffer for 2 × 2 min each time.
-
64.
Apply 100 μL of PreAmplifier working solution to the sections and incubate in the humidity control tray for 1 h at 40°C.
Note: Volume of PreAmplifier mix working solution needs to be adjusted depending on the size of hydrophobic barrier. 100 μL of PreAmplifier working solution is recommended for 2cm × 1.5 cm area.
-
65.
Prewarm Amplifier diluent at 40°C for 10 min.
-
66.
Prepare 100 μL of Amplifier working solution by diluting Amplifier mix in Amplifier diluent at 1:25 dilution as indicated in materials and equipment.
Note: Required volume depends on the size of hydrophobic barrier. 100 μL of Amplifier working solution is recommended for 2cm × 1.5 cm area.
-
67.
Aspirate PreAmplifier working solution and rinse with 200 μL of viewRNA wash buffer for 2 × 2 min each time.
-
68.
Rinse the sections in 25mL of SSCTx for 2 × 5 min each time.
-
69.
Rinse the sections with 200 μL of viewRNA wash buffer for 2 × 2 min each time.
-
70.
Apply 100 μL of Amplifier working solution to the sections and incubate in the humidity control tray for 1 h at 40°C.
Note: Solution volume needs to be adjusted depending on the size of hydrophobic barrier. 100 μL of Amplifier working solution is recommended for 2 cm × 1.5 cm area.
-
71.
Prewarm Label probe diluent at 40°C for 10 min.
-
72.
Prepare 100 μL of Label probe working solution by diluting Label probe mix in Label probe diluent at 1:25 dilution as indicated in materials and equipment.
Note: Required volume depends on the size of hydrophobic barrier. 100 μL of Amplifier working solution is recommended for 2cm × 1.5 cm area.
-
73.
Aspirate Amplifier working solution and rinse with 200 μL of viewRNA wash buffer for 2 × 2 min each time.
-
74.
Rinse the sections in SSCTx for 2 × 5 min each time.
-
75.
Rinse the sections with 200 μL of viewRNA wash buffer for 2 × 2 min each time.
-
76.
Apply 100 μL of Label probe working solution to the sections and incubate in the humidity control tray for 1 h at 40°C.
Note: Solution volume needs to be adjusted depending on the size of hydrophobic barrier. 100 μL of Label probe working solution is recommended for 2cm × 1.5 cm area. Incubation must be performed under dark conditions because Label probe working solution contains fluorescent dyes. Avoid direct light after this step to protect fluorophore from photobleaching.
-
77.
Aspirate Label probe working solution and rinse with 200 μL of viewRNA wash buffer for 2 × 5 min each time.
-
78.
Rinse the sections with SSCTx for 2 × 5 min each time.
-
79.
Place the sections in 1× PBS for 2 × 5 min each time. The sections are then ready for Immunofluorescence assay (steps 80–93) as follows.
Note: If only smFISH staining is required and IF is not necessary, steps 80–86 can be skipped.
Immunofluorescence
Timing: 20 h, Day 7–8
This section describes the protocol of the immunofluorescence procedure that is performed on pre-treated sections with or without FISH assay.
-
80.
Apply 100 μL of SuperBlock to the section and incubate for 1 h in humid chamber.
Note: Any commercial blocking reagent for immunofluorescence may be used but requires optimization to ensure the correct signal-to-noise ratio.
-
81.
Prepare primary antibodies working solution by diluting primary antibodies in SuperBlock solution at the desired concentration.
Note: The concentration of primary antibody requires optimization according to different protein expression level, pretreatment conditions, and upstream assays such as FISH.
-
82.
Drain the slide without rinsing and apply 100 μL primary antibodies working solution to the section and incubate at 4°C overnight (about 16 h) in humid chamber.
-
83.
Rinse the sections with PBSTx for 3 × 15 min each time.
-
84.
Prepare secondary antibodies working solution by diluting secondary antibodies in 1× SuperBlock solution at the desired concentration.
Note: The concentration of secondary antibody requires optimization as well. DAPI can be diluted at 10ug/ml into secondary antibody working solution.
-
85.
Apply 100 μL secondary antibodies working solution to the section and incubate for 1 h at room temperature in humid chamber.
Note: For some lowly expressed proteins, Tyramide Signal Amplification based signal detection method is recommended for FISH/IF multiplexing assay. Again, the condition requires optimization.
-
86.
Rinse the sections with PBSTx for 3 × 15 min each time.
-
87.
Incubate the sections in 200 μL of PBST containing DAPI (10 μg/mL) for 15 min.
Note: This step can be skipped if DAPI was applied at step 84.
-
88.
Rinse the sections with PBSTx for 3 × 5 min each time.
-
89.
Rinse the sections with 1× PBS for 3 min.
-
90.
Prepare Autofluorescence blocking solution as indicated in materials and equipment.
Note: Solution volume needs to be adjusted depending on the size of hydrophobic barrier. 300 μL of working solution is recommended for 2cm × 1.5 cm area.
-
91.
Apply 300 μL working solution of autofluorescence blocking reagent to the sections and incubate for 5 min at room temperature under dark conditions.
-
92.
Rinse the section with 1× PBS for 3 min.
-
93.
Coverslip sections with two drops of ProLong™ Glass antifade mounting media.
Pause point: Slides can be stored at this point for up to 4 weeks in 4°C without experiencing noticeable fluorescence signal loss. For this paper, all the fluorescent stained slides are imaged by a Nikon Spinning Disk Confocal System.
Expected outcomes
The common artifacts (see Figure S1) of ocular specimens are collapse, shrinkage, deformation, misalignment of the ciliary body or lens while sectioning, shattered looking sections or lenses, and retinal separation. In addition, heat-induced antigen retrieval, depigmentation, or enzymatic digestion pretreatments before immunofluorescence and in situ hybridization assays, often cause detachment of tissue sections from glass slides. In particular, the cornea, the iris, the ciliary body, the lens, and the retinal pigmented epithelium are more susceptible. In this protocol, we have provided a strategy of applying a window technique on Hartman’s fixative-fixed samples, microwave-based tissue processing, de-pigmentation with H2O2 and formamide, and other pretreatments for immunofluorescence and in situ hybridization assays. Our protocol has been proven to be easily reproduced by different individuals in our Stowers histology group and the Xie lab, and thus would be expected to be reproducible in any histology laboratory.
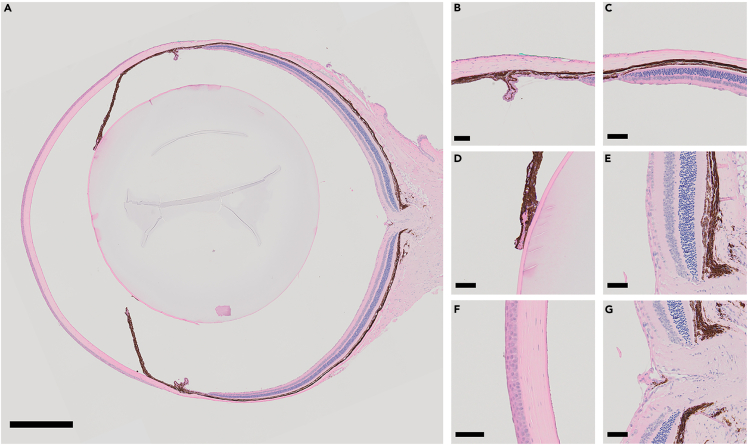
Figure 2 is an example of anticipated results for the morphological study of the mouse eyeball using paraffin sections. The window technique combined with microwave-based paraffin processing helps to avoid common artifacts such as eyeball collapse, shrinkage, retina separation, etc. On those tissue sections, the round shape of the mouse eyeball, anterior and posterior chambers (Figure 2A), delicate structures such as the ciliary body (CB) and the trabecular meshwork (TM) region (Figure 2B), peripheral retina (Figure 2C), iris and anterior lens (Figure 2D), near-central retina (Figure 2E), central cornea (Figure 2F) and optic nerve head (Figure 2G) are well-preserved.
Figure 2.
Representative images of paraffin section with H&E stain
The overall morphology (A), fine structure of multiple tissues such as the ciliary body (CB) and the trabecular meshwork (TM) region (B), peripheral retina (C), iris and anterior lens (D), near-central retina (E), central cornea (F) and optic nerve head (G) are well-preserved. Scale bar: 500 μm for (A) and 50 μm for (B–G).
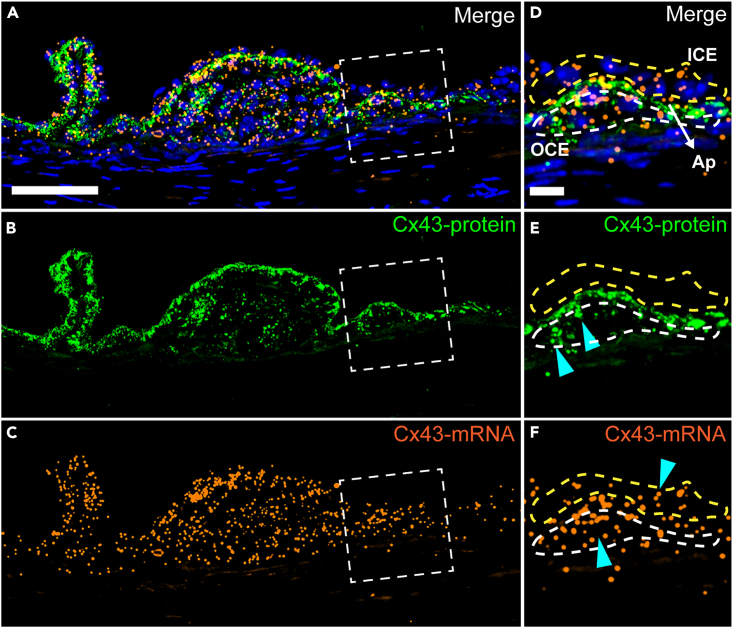
The expected results from smFISH with immunofluorescence multiplexing assay are shown in Figure 3. Cx43 mRNA molecules are detected in the apical and the lateral-basal sides of both inner ciliary epithelia (ICE) and outer ciliary epithelia (OCE), which is consistent with previous publications (Calera et al., 2009; Pang et al., 2021). The distribution of Cx43 mRNA is more diffused in the entire cells in ICE and OCE (Figure 3C). However, the Cx43 protein that is detected by immunofluorescence assay demonstrates a cellular localization predominately on the apical side of both ICE and OCE cells, and the lateral-basal side of OCE cells, but with almost no detectable expression at the lateral-basal side of the ICE (Figure 3B, E). This result is also consistent with findings from previous publications (Calera et al., 2006, 2009; Pang et al., 2021), which indicates that Cx43 undergoes different posttranscriptional regulations and cellular localizations in the ICE and OCE.
Figure 3.
Expected result of Cx43 smFISH and IF multiplexing assay
(A), the merged image of IF signal of Cx43 protein expression (green, B), smFISH fluorescence signal (orange, C), and DAPI (blue). (D–F) images of the enlarged region highlighted by the dashed white square in (A–C). The yellow dashed polygon in (D–F) shows the lateral-basal side of inner ciliary epithelium (ICE). The white dashed polygon in (D–F) shows the lateral-basal side of outer ciliary epithelium (OCE). The gap between the 2 polygons is the apical side of ICE and OCE. Cyan arrow heads point to the region with positive signal at the lateral-basal side of ICE and OCE in E and F. Scale bar: 50 μm for (A–C) and 15 μm for (D–F).
Limitations
Our methodology overcomes the morphology preservation and section detachment issues associated with ocular histology, but the success rate is not one hundred percent. About a quarter of adult mouse eyeballs will still show some morphology artifacts such as collapse or shrinkage, or some part of tissue sections will be lifted from glass slides after pretreatment. In addition, we have only optimized the procedures for developing and adult mouse eyeballs. Our protocol may need more optimization for lens fixation. Although the antigen retrieval and enzymatic digestion pretreatments may be similar for different tissues, it requires more optimization in fixation and tissue processing for larger eyeballs such as human and monkey. It is worth noting that this protocol is based on paraffin processing, which is expected to better maintain the morphology of the tissue. However, some antibodies may not work on paraffin sections. In such cases, assays based on those antibodies should use sections from frozen tissues instead.
Troubleshooting
Problem 1
Severe collapse appears at the anterior chamber and the vitreous body after step 16 if steps 7 and 8 were not followed
Potential solution
Window technique (steps 7 and 8) is important to maintain the shape of the anterior chamber and the vitreous body. Therefore, it is highly recommended that this technique should not be skipped for the paraffin processing of the eyeball samples. We recommend one window toward the anterior chamber and two windows at the opposite region toward the vitreous. Figure 1B shows the eye sample before applying the window technique and the white arrows point to the region for the windows to be made, and Figure 1C shows the two windows that were highlighted by the dashed white polygons (The one at the opposite side is not shown in both figures). The window technique is critical for eye samples from mouse that is older than 3 days. Figure S1B shows the result of an eye sample from a 1-week-old mouse without window technique. The eye sample deforms severely with the anterior chamber and the vitreous largely lost, compared with the result shown in Figure S1A, which comes from an eye sample processed exactly with our protocol. The possible reason could be that during the paraffin processing, the windows help to facilitate the fluid exchange and avoid pressure alteration between inside and outside of eyeballs, which helps to maintain the overall eyeball morphology. If this issue persists when window technique is applied, try larger windows to facilitate fluid exchange during tissue processing.
Problem 2
At the completion of step 43, multiple artifacts such as cracks, shredded lenses, shrinkage, and separation between different tissue layers are present on paraffin sections if 4% PFA was used in step 2. (Figure S1C).
Potential solution
The Hartman’s fixative is critical for fixation of the eyeball samples. 4% PFA cannot take the place of the Hartman’s fixative in terms of morphology. The possible reason is that the delayed fixation for the inner part of eyeball tissue may cause those artifacts. It is known that Hartman’s fixative outperforms 4% PFA in terms of maintaining the morphology of the eye tissues (Isam Eltoum and Grizzle, 2001). Part of the reason may be due to the solvent (ethanol) used in Hartman’s fixative, which can permeabilize the cell membrane while fixation takes place. Therefore, it defeats the delayed fixation effect that is common for PFA immersion fixation (Figure S1C). Interestingly, we tried window techniques with 4% PFA-fixed eyeballs before, after, and in the middle of fixation, but none of those conditions prevented artifacts on paraffin sections.
Problem 3
A bubble is found within the eye after slightly trimming into the sample (step 19).
Potential solution
A skilled technician performing the sectioning will decide if it is possible to obtain a section even with a bubble present, but time and effort may be saved by melting the eye sample and re-embedding. With a portion of the eye already trimmed off during the initial paraffin trimming, it is easy to float the bubble out of the interior chamber while immersed in liquid paraffin. Melting the paraffin block after trimming into the sample will also allow a chance to observe the optic nerve and the iris opening to see if they are in parallel alignment with the trimmed area. If so, the optimal plane and true center of the eyeball will be reached upon deeper sectioning.
Problem 4
Paraffin ribbon separates and expands when floating on the water bath too quickly or slowly. (step 21).
Potential solution
The proper temperature of the water bath will allow a paraffin ribbon to float on the water bath for a minute or two without signs of spreading apart. The proper temperature will also allow ease of separating the paraffin ribbon into smaller increments. Although 42°C works best in our lab for the water bath during paraffin sectioning of eye samples, this optimized temperature could vary in a different lab with a different brand of paraffin, water bath or the varied preference of different technicians. If a paraffin ribbon quickly begins to spread apart once placed on the water bath, the temperature is too warm. Lower the temperature and wait until the set temperature is reached. If a paraffin ribbon is difficult to separate into individual sections, the temperature may be too cool. Adjust the temperature to a warmer setting.
Problem 5
The windows appear as undesired breaks along the cornea or RPE when paraffin sections are viewed under the brightfield setting of the stereoscope (step 23).
Potential solution
Windows made during the window technique steps should not interfere with the section images if the eyeball is properly oriented during the embedding step. Because the two windows made to the posterior chamber of the eye are made on opposite sides, one window should be embedded down into the mold, thus the other window would then be at the top of the mold. The window that is embedded down will be cut away during sectioning and the window that is at the top of the mold will not be reached, since sectioning will end before that point. The window that is made in the cornea is in alignment with one of the posterior windows and embedded in the same plane so that it also doesn’t come into view. A deeper trimming or an adjustment of the angle of the microtone and the sample will help to get entire sections without the windows coming into view.
Problem 6
The heavy pigment in the tissue, like the ciliary body, affects the observation of the fluorescent signal from the molecular histology testing (Figure S2).
Potential solution
Although fluorescent background originating from other tissue components can be completely suppressed by the TrueView background suppressing kit (as indicated in steps 90 and 91), the autofluorescence from remaining pigment is still visible after applying the Trueview kit. The pigment can be totally removed by adjusting the concentration of H2O2 and the time and intensity of LED light illumination at step 49 (Figure S2A). The protease treatment can also be reduced or skipped after the bleaching treatment. However, we noticed that longer time of bleaching or higher concentration of H2O2 can affect downstream smFISH and/or immunofluorescence outcomes and cause tissue lifting.
Problem 7
Antigen retrieval causes tissue lifting (step 45).
Potential solution
The cross-linking caused by nearly two days of aldehyde fixation at ambient temperature requires antigen retrieval for achieving a successful immunofluorescence experiment. The temperature and the time of heat-induced antigen retrieval in a citrate buffer (pH 6.0) affects both the efficiency of epitope exposure and tissue attachment to a glass slide. The mild heating condition is beneficial for tissue attachment on glass slides, but it may be insufficient to expose epitopes for an antibody assay.
We recommend optimizing those conditions before the actual experiment to achieve the best immunofluorescence outcome for different protein targets. As an example, a comparison of the immunofluorescence results from sections treated with 70°C heating for 30 min to those sections treated with 95°C heating for 10 min is illustrated in Figure S3. Different protein targets responded to the antigen retrieval conditions differently.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yongfu Wang (yow@stowers.org).
Materials availability
This study does not generate new unique reagents.
Acknowledgments
We thank Dorothy Stanley (Stowers Institute) for editorial assistance, the Lab Animal Services Facility for animal husbandry services, Steven Hoffman and Zulin Yu (Stowers Institute) for imaging consultation, Seth Malloy and Karen Smith (Stowers Institute) for histological technical assistance, and the Xie laboratory members and Histology team members for proofreading. This work was supported by the National Eye Institute (EY027441, T.X.), the Stowers Institute for Medical Research (T.X.), and a scholarship by the China Scholarship Council (J.P.).
Author contributions
J.P., N.T, and D.T. performed the experiments and collected image data; J.P., N.T, D.T., and Y.W. wrote the manuscript; D.Y., T.P. T.X., and Y.W. supervised the work.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100879.
Contributor Information
Ting Xie, Email: tgx@stowers.org.
Yongfu Wang, Email: yow@stowers.org.
Supplemental information
Data and code availability
No datasets or codes were generated during this study. Primary data files can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1634).
References
- Calera M.R., Topley H.L., Liao Y., Duling B.R., Paul D.L., Goodenough D.A. Connexin43 is required for production of the aqueous humor in the murine eye. J. Cell Sci. 2006;119:4510–4519. doi: 10.1242/jcs.03202. [DOI] [PubMed] [Google Scholar]
- Calera M.R., Wang Z., Sanchez-Olea R., Paul D.L., Civan M.M., Goodenough D.A. Depression of intraocular pressure following inactivation of connexin43 in the nonpigmented epithelium of the ciliary body. Invest. Ophthalmol. Vis. Sci. 2009;50:2185–2193. doi: 10.1167/iovs.08-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel L.A., Sachdeva R., Marcotty A., Rockwood E.J., Traboulsi E.I. Oculodentodigital dysplasia: new ocular findings and a novel connexin 43 mutation. Arch. Ophthalmol. 2011;129:781–784. doi: 10.1001/archophthalmol.2011.113. [DOI] [PubMed] [Google Scholar]
- Isam Eltoum J.F., Myers R.B., Grizzle W.E. Introduction to the theory and practice of fixation of tissues. J. Histotechnol. 2001;24:23. [Google Scholar]
- Jordan T., Williams D., Criswell S., Wang Y. Comparison of bleaching protocols utilizing hematoxylin and eosin stain and immunohistochemical proliferation marker MCM3 in pigmented melanomas. J. Histotechnol. 2019;42:177–182. doi: 10.1080/01478885.2019.1649886. [DOI] [PubMed] [Google Scholar]
- Musa F.U., Ratajczak P., Sahu J., Pentlicky S., Fryer A., Richard G., Willoughby C.E. Ocular manifestations in oculodentodigital dysplasia resulting from a heterozygous missense mutation (L113P) in GJA1 (connexin 43) Eye. 2009;23:549–555. doi: 10.1038/eye.2008.77. [DOI] [PubMed] [Google Scholar]
- Pang J., Le L., Zhou Y., Tu R., Hou Q., Tsuchiya D., Thomas N., Wang Y., Yu Z., Alexander R., et al. NOTCH signaling controls ciliary body morphogenesis and secretion by directly regulating Nectin protein expression. Cell Rep. 2021;34:108603. doi: 10.1016/j.celrep.2020.108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paznekas W.A., Boyadjiev S.A., Shapiro R.E., Daniels O., Wollnik B., Keegan C.E., Innis J.W., Dinulos M.B., Christian C., Hannibal M.C., et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am. J. Hum. Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondereker K.B., Stabio M.E., Jamil J.R., Tarchick M.J., Renna J.M. Where you cut matters: a dissection and analysis guide for the spatial orientation of the mouse retina from ocular landmarks. J. Vis. Exp. 2018:57861. doi: 10.3791/57861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosin J.M., Schutte M., Chen S. Connexin distribution in the rabbit and rat ciliary body. A case for heterotypic epithelial gap junctions. Invest. Ophthalmol. Vis. Sci. 1997;38:341–348. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets or codes were generated during this study. Primary data files can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1634).