Abstract
Amyloid diseases are characterized by the aggregation of various proteins to form insoluble β-sheet rich fibrils leading to cell death. Vibrational spectroscopies have emerged as attractive methods to study this process due to the rich structural information that can be extracted without large, perturbative probes. Importantly, specific vibrations such as the amide-I band directly report on secondary structure changes, which are key features of amyloid formation. Beyond intrinsic vibrations, the incorporation of unnatural vibrational probes can improve sensitivity for secondary structure determination (e.g. isotopic labeling), provide residue-specific information of the surrounding polarity (e.g. unnatural amino acid), and are translatable into cellular studies. Here, we review the latest studies that have leveraged tools from chemical biology for the incorporation of novel vibrational probes into amyloidogenic proteins for both mechanistic and cellular studies.
1. Introduction
Amyloid diseases, including Alzheimer’s (AD), Parkinson’s (PD), Huntington’s, and type-II diabetes are a collection of protein misfolding diseases defined by the aggregation of various proteins into amyloid fibrils [1,2]. Amyloids are unbranched, filamentous assemblies rich in β-sheet secondary structure. A universal characteristic is the perpendicular alignment of the individual β-strands with respect to the fibril axis, known as the cross-β conformation, where the dominant interaction is hydrogen-bonding of backbone carbonyls (C=O) and amides (N–H) between monomeric units [3,4]. Additional side chain interactions such as steric zippers [5], salt bridges [6], and π-π stacking [7] can contribute to stabilizing amyloid structure formation.
The propagation of amyloid structures through self-templating is intrinsic to their formation and has been implicated in cell-to-cell transmission in some amyloid diseases [8–10]. Amyloid formation is an inherently stochastic process, making it highly sensitive to molecular perturbations. Solution conditions (e.g. pH, ionic strength, and temperature), post-translational modifications (e.g. acetylation, phosphorylation, glycosylation), and disease-related missense mutations have been demonstrated to dramatically alter not only the rate of aggregation but also the final fibril structure [11–14].
Fibril structural complexity and polymorphism have attracted interest in the application of vibrational spectroscopies to their studies [15]. In the context of protein structure, vibrational spectroscopies are particularly sensitive to changes in the amide-I band, which is composed primarily of the backbone carbonyl (C=O) stretching mode. Backbone interactions give rise to vibrational coupling between carbonyls, leading to amide-I frequencies that are characteristic for secondary structural motifs such as β-sheets. Amyloid fibrils are particularly amenable to vibrational spectroscopies due to the strong coupling of the carbonyls in the cross-β structure enhancing the intensity of the amide-I band [16–18]. Fourier-transform infrared spectroscopy (FTIR) has been extensively employed to characterize β-sheet structure in numerous amyloids in vitro [19], while Raman studies have been more limited by comparison.
Given the sensitivity of amyloid structures to solution conditions, it would be pertinent to perform secondary structural analysis in cells as it is not yet well understood how different cellular compartments and environments impact fibril structure and propagation. Case in point, recent structures of tau fibrils determined by cryoelectron microscopy of patient samples reveal that tau adopts unique structures in AD, chronic traumatic encephalopathy, Pick’s disease, and corticobasal degeneration, suggesting an intimate relationship between amyloid structure and disease phenotype [20]. More recently, α-synuclein (α-syn) fibril structures derived from patient samples of multiple system atrophy (MSA) and PD are also distinct [21,22]. In principle, vibrational techniques are capable of detecting β-sheet formation even in the early stages of aggregation, and assignment to specific residues is possible [15,19]. However, there are technical challenges to overcome due to spectral interference from endogenous biomolecular vibrations and, in the case of IR, strong water absorbance near the amide-I band.
In our own work, we have used Raman spectroscopy to study fibril formation of α-syn [11,23–25], which is the main amyloidogenic protein involved in PD, MSA, and dementia with Lewy bodies [26]. Notably, we have worked on the development of Raman probes towards fibril characterization in cells by Raman spectral imaging [27,28], where we have coupled a spectrometer to an inverted microscope to permit the acquisition of Raman spectra at multiple spatial locations. This is an important endeavor as there is substantial interest in observing where protein aggregation is initiated in the cell as well as to discern whether structural differences exist between aggregates in distinct cellular compartments. As part of this effort, we have utilized both isotopic labeling and unnatural amino acids (UAA) to facilitate these studies [27–29].
In this Review, we highlight work from the last three years leveraging chemical biological approaches such as expressed-protein ligation, metabolic labeling, and biosynthetic incorporation to install different vibrational probes in amyloidogenic proteins in vitro and in cells. Two major classes of vibrational probes are discussed—isotopic labeling and UAA incorporation. Isotopic labels offer the advantage of minimal perturbation and can enable site-specific examination of secondary structure, while UAAs can be used to introduce a wide variety of chemically and spectrally unique groups. The application of chemical biology to incorporate vibrational probes has significant potential to enable spectroscopic interrogations of protein misfolding and aggregation in cellular systems, which will contribute towards the mechanistic understanding of amyloid fibrils and their behaviors in diseases.
2. Isotopic Labeling
One approach to extracting residue-specific information is the use of isotopic labeling to shift vibrational modes of interest into a unique spectral region. Replacement of any nuclei involved in a vibrational mode, such as 13C-labeling of a backbone carbonyl shifts that vibrational mode to lower wavenumbers due to the change in reduced mass. Isotopic labeling has long been an attractive strategy in large part due to the minimally perturbative nature of these modifications. In its simplest form, isotopic labeling can be easily accomplished by expressing the protein of interest in isotope-enriched media or through use of isotopically-labeled amino acids in solid-phase peptide synthesis (SPPS). Okada et al. recently exemplified this approach in a study of amyloid-β (Aβ), which is the amyloidogenic protein involved in AD [30]. The authors incorporated 13C-carbonyls at specific sites using SPPS and measured carbonyl coupling in the β-sheet structure of these aggregates by FTIR. Using curve-fitting analysis, it was concluded that Aβ aggregates on GM1 lipid clusters are composed of unusual mixed parallel/antiparallel β-sheets, which are not observed for Aβ fibrils formed in solution.
Spectral resolution can be further enhanced by double isotopic labeling, such as by 13C18O-carbonyl labeling as demonstrated in prior work [31–33]. Zanni and coworkers have made extensive use of 13C18O-labeling in their two-dimensional IR (2D-IR) experiments with human islet amyloid polypeptide (hIAPP), which forms extracellular amyloid plaques in type-II diabetes [34,35]. For a full discussion on 2D-IR, we refer the reader to the many reviews on the topic [36–39]. Briefly, separate pump and probe lasers are used to produce a correlation map of vibrational coupling between functional groups. Interactions between residues appear as off-diagonal peaks in a manner analogous to two-dimensional NMR experiments, though 2D-IR is sensitive to events on much faster timescales. In addition to enhanced structural information, this method also offers significant improvements in sensitivity. 2D-IR has been used to identify amyloid in human cataracts and a novel hIAPP polymorph in human serum as well as in the design of a biosensor of protein structure [40–42]. More recently, Maj et al. demonstrated 2D-IR methods to study interactions between specific residue pairs by double-labeling hIAPP with 13C18O, revealing the existence of transient helical structure preceding oligomer formation [43]. Isotopic labeling can also be employed to probe side chain interactions, such as DeGrado and coworkers’ recent demonstration of Gln ladder formation in Tau306–311 and a yeast prion-derived peptide using a 13C18O-labeled Gln sidechain [44].
2.1. Raman study of segmentally 13C-labeled α-synuclein
Through the use of segmental 13C-labeling, we extended this approach to the study of specific polypeptide regions in α-syn [29]. Intein-mediated ligation was used to prepare α-syn uniformly labeled with 13C in the first 86 residues and 12C in the remaining 54 C-terminal residues (Fig. 1A). To accomplish this, α-syn was expressed in two segments in E. coli. This approach is analogous to a previous 2D-IR study by Moran et al. on the aggregation mechanism of segmentally 13C-labeled γD-crystallin [45]. Briefly, residues 1–86 were expressed with a C-terminal intein-tag followed by a chitin binding domain in 13C-enriched media, while residues 87–140 with a Ser-to-Cys mutation at position 87 were expressed in natural isotope abundance media. Autocleavage of the intein in the presence of a thiol produces an C-terminal thioester that reacts with the mutant Cys residue to generate the ligated S87C-α-syn with residues 1–86 and 87–140 uniformly labeled with 13C and 12C, respectively. This ligation reaction was performed as a one-pot synthesis, where the C-terminal fragment was supplemented in the cleavage buffer as the protein is released from the chitin column. Due to the intrinsically disordered nature of α-syn, the coupling was facile.
Figure 1.
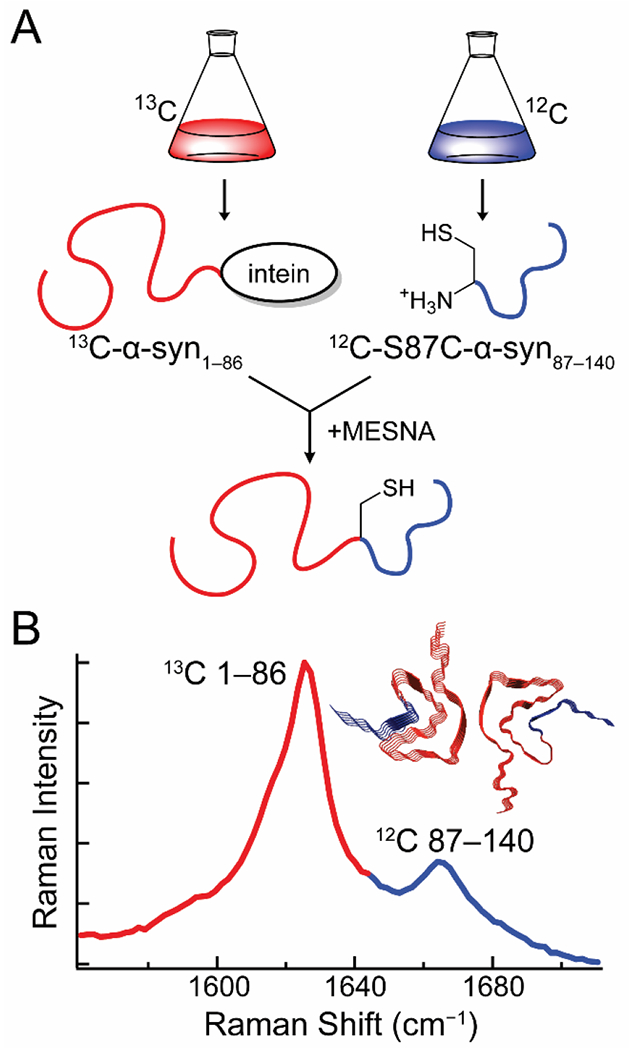
Expressed-protein ligation enables the creation of S87C-α-syn segmentally labeled with 13C and 12C at residues 1–86 and 87–140, respectively. (A) α-Syn1–86 is expressed with a C-terminal intein-tag in 13C-media, while S87C-α-syn is expressed in natural isotope abundance media. A one-pot reaction is carried out where autocleavage of the intein-tag in the presence of 2-mercaptoethanesulfonate (MESNA) leads to covalent ligation of the N-and C-terminal segments of the protein. (B) The 13C-and 12C-labeled segments of the fibrils produce two amide-I bands, enabling independent analysis of their secondary structures. (Inset) Fibril structure of α-syn with the 13C-and 12C-labeled fragments colored red and blue, respectively. Data originally published in [29].
The ligated protein produces two discrete amide-I bands in the Raman spectrum, permitting simultaneous secondary structural analysis of both regions (Fig. 1B). Raman spectral imaging enabled examination of early aggregates, which revealed the existence of a kinetic intermediate that displays a sharp 13C-amide-I peak, but a broad 12C-amide-I band. This suggests that early aggregation of α-syn involves the formation of β-sheet structure in the N-terminal region that later propagates into the C-terminal region of the protein. This work underscores the strength of coupling the chemical biological approach of chemical ligation with vibrational spectroscopy which provides unique information on secondary structure.
2.2. Raman spectral imaging of 13C2H15N-labeled α-synuclein fibrils in cells
Recently, we reported a different approach using uniformly 13C2H15N-labeled α-syn. The advantage here is the capability of resolving α-syn from endogenous proteins in a cellular environment due to the presence of the 13C-2H stretching bands, which appear in the center of the cellular quiet spectral region (Fig. 2A). The cellular quiet region is a region of the vibrational spectrum from ~1800 to 2400 cm −1 where no naturally-occurring biomolecules are vibrationally active.
Figure 2.
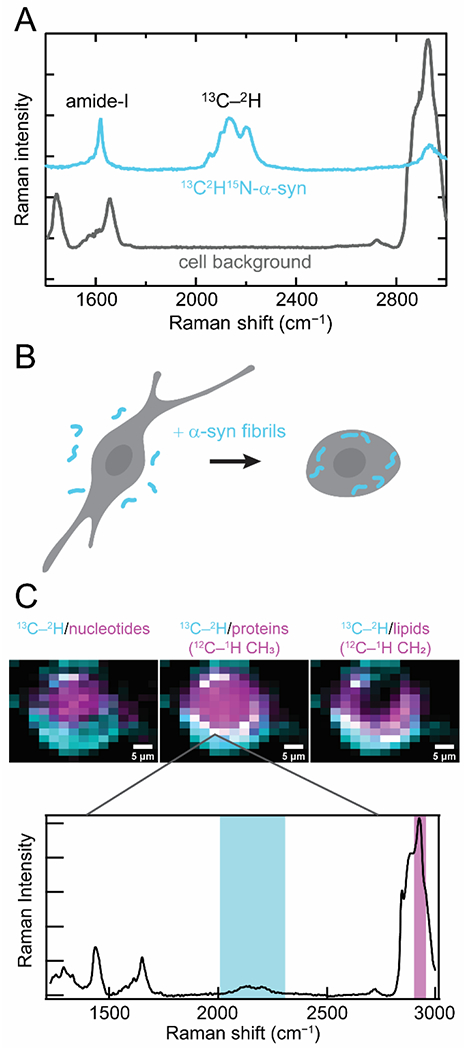
Raman spectral imaging of internalized 13C2H15N-α-syn fibrils in cells. (A) 13C2H15N-α-syn (cyan) possesses several unique Raman features that distinguish it against a cellular background (gray). The amide-I band is shifted to lower energy relative to endogenous amide-I and lipid C=C stretching bands, and the 13C–2H stretching bands appear in the cellular quiet region. (B) Schematic representation of our experimental conditions where 13C2H15N-α-syn fibrils formed in vitro (cyan) are added in media and readily uptaken by cultured mammalian cells. (C) Raman maps of internalized fibrils (cyan) and comparison to the localization of cellular components (magenta) by integrating over spectral regions characteristic of nucleotides, proteins, and lipids in human SK-MEL-28 cells (Top). Colocalization appears as white pixels. (Bottom) An example Raman spectrum measured at a pixel where α-syn fibrils are colocalized with endogenous proteins. Integrated spectral regions are indicated. Data originally published in [28].
This isotopically labeled protein was used to examine fibril endocytosis, which is thought to be a key component of disease progression in synucleinopathies [46]. In this process, healthy cells uptake fibrils which seed amyloid formation of the endogenous soluble α-syn, leading to fibril propagation and cytotoxicity. Preformed 13C2H15N-labeled fibrils were fed to cultured mammalian cells and mapped by Raman spectral imaging to examine the fate of these endocytosed fibrils (Fig. 2B) [28]. Our data demonstrate that the 13C2H15N-amide-I band can be resolved from other cellular vibrational signals and that the strong 13C-2H stretching bands enable background-free mapping of exogenous α-syn in cells (Fig. 2C). Comparison of the amide-I band to the 13C-2H stretching modes, which are relatively insensitive to structural changes indicate that there is no appreciable loss of β-sheet structure upon endocytosis within our experimental timeframe of 24 h. In addition, we observed that the internalized fibrils largely accumulate at the cellular periphery, colocalizing with proteins and lipids. Interestingly, we observed lipid accumulation throughout the cytoplasm. This work demonstrates unambiguous localization of internalized fibrils and direct observation of β-sheet structure of amyloid fibrils in cells.
2.3. Stimulated-Raman imaging and selective deuteration of Gln residues of polyQ aggregates in cells
Isotopic labeling has recently been demonstrated for imaging aggregates formed from endogenous proteins. PolyQ diseases, including Huntington’s disease are characterized by translation of long chains of glutamine residues at the C-terminus of proteins such as Huntingtin [47]. These polyQ repeats trigger intracellular amyloid formation and subsequent cell death. Miao and Wei took advantage of the enrichment of Gin in these aggregates by introducing deuterium dab el ed Gin into the growth media of cells expressing these proteins [48]. Cells were imaged using stimulated Raman scattering (SRS), a two-photon approach which offers significantly enhanced intensity for a targeted vibrational mode compared to spontaneous Raman events [49]. Notably, it was suggested that comparable contrast is achieved to that of GFP fluorescence or fluorophore staining. Since Raman imaging is quantitative, it is superior to fluorescence methods, which can be complicated in very high density aggregates due to self-quenching or impermeability to exogenous fluorophores. The structure of intracellular aggregates was further interrogated by SRS analysis of the amide-I band. Interestingly, a blue-shift associated with β-sheet enrichment was not detectable, in agreement with other recent work [50]. Although this approach is not widely applicable as most amyloids are not so highly enriched in a single amino acid, the sensitivity of this technique stands out from other vibrational imaging methods in cells.
3. Unnatural Amino Acids
Although isotopic labeling offers the advantage of being inherently nonperturbative, the choice of labels and methods for site-specific incorporation are quite limited, especially in vivo. An alternative approach is UAA incorporation, which can be used to introduce a wide variety of chemically and spectroscopically unique functionalities into proteins. UAAs have been incorporated into a variety of peptides and proteins both by SPPS and chemical biology methods. UAAs such as 4-cyano-L-phenylalanine and azidohomoalanine are well studied and successful vibrational probes of protein folding [51–53]. In the context of amyloids, the use of UAAs as infrared probes has been largely limited to synthetic peptides [54–56]. One recent study, Pazos et al. employed two UAAs to examine Aβ16–22 hydrogen-bonding dynamics [57]. They introduced an ester group via an Asp derivative, L-aspartic acid 4-methyl ester (DM) and a second UAA, Lys(Nvoc) to improve fibril homogeneity. The authors used the unique vibrational band of DM to demonstrate that this position is exposed to two distinct chemical environments, one which is buried and the other solvent-exposed. Using the structural constraints from the FTIR data, a structural model of two antiparallel β-sheets packed parallel to one another was proposed. In a demonstration of the exquisite time resolution of vibrational spectroscopies, the authors also examined 2D-IR picosecond spectral diffusion dynamics to detect the DM carbonyl switching between H-bonds with individual hydrogens on a Lys amino-group.
3.1. Homopropargylglycine, a terminal alkyne Raman probe for cellular studies
In cells, the most straightforward method of unnatural amino acid incorporation is biosynthetic incorporation, which we employed in our own work with the alkyne-containing amino acid homopropargylglycine (HPG). HPG is a methionine analogue and was first introduced into proteins by Tirrell and coworkers, although they used it for bioorthogonal fluorophore labeling [58–60]. Others have used it as a metabolic label of the proteome in stimulated Raman imaging [61]. Here, HPG was introduced into α-syn biosynthetically in E. coli [27]. Because cells cannot distinguish HPG from methionine, Met residues can be readily replaced with HPG in a Met-auxotrophic strain of E. coli (Fig. 3A). The stretching vibrational mode of the alkynyl functional group appears at ~2110 cm−1 in the middle of the cellular quiet region (Fig. 3B). The alkyne moiety is sensitive to the polarity of its local surroundings, shifting to lower energies in more hydrophobic environments.
Figure 3.
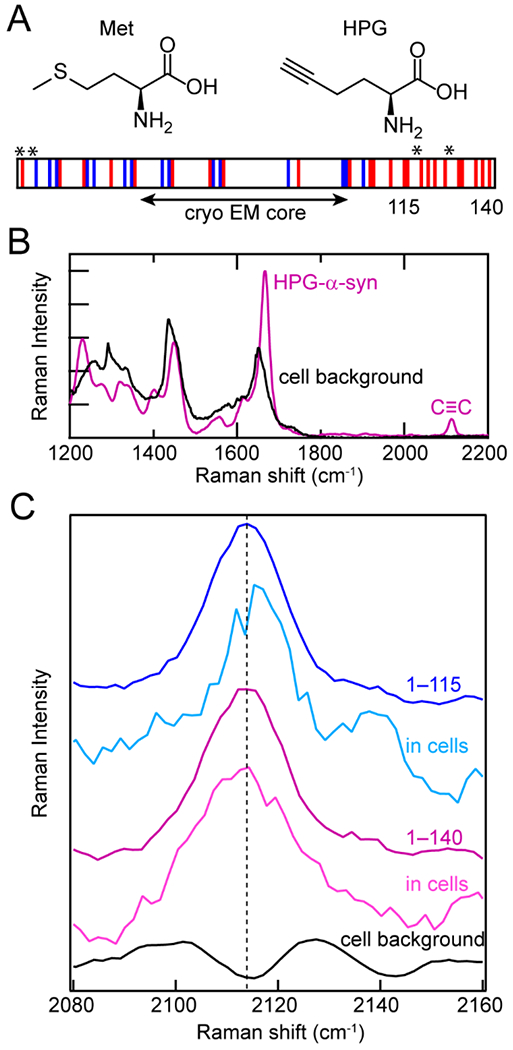
Biosynthetic incorporation of HPG, a Met analogue, into α-syn as site-specific Raman probes. (A) Structures of Met and HPG. HPG is readily incorporated into α-syn by Met-auxotrophic E. coli. Schematic of α-syn sequence showing acidic (red) and basic (blue) residues, as well as the fibrillar core determined by cryo-EM and the locations of Met residues replaced with HPG (asterisks). The C-terminal truncation site (115) is also indicated. (B) In the Raman spectrum, the alkyne stretching mode (C≡C) of HPG-α-syn (purple) appears in the quiet region of the cellular background (black). (C) The alkyne stretching band of HPG-α-syn1–115 in cells (cyan) is narrower and blue-shifted relative to full-length HPG-α-syn1–140 fibrils in vitro (purple) and in cells (magenta) as well as HPG-α-syn1–115 in vitro (blue).The cell background (black) is shown for comparison. Data originally published in [27].
Since α-syn contains four native Met residues at positions 1, 5, 116 and 127, the alkyne stretching band of HPG-labeled α-syn simultaneously reports on the local environments of the N-and C–termini. To examine these ends independently, a C-terminally truncated variant comprising residues 1–115 was also studied. Whereas the alkyne stretching bands of HPG-α-syn and HPG-α-syn1–115 fibrils were similar in vitro, the peak narrowed and blue-shifted for HPG-α-syn1–115 when measured in cultured N27 rat dopaminergic cells (Fig. 3C). This suggests that the N- and C-termini experience very different local environments in cells, despite recent cryo-EM structures indicating that neither region is well-structured in the fibrillar state (Fig. 3A) [24,62,63].
By coupling HPG- and 13C-labeling of α-syn, it was also possible to analyze protein secondary structure in the cellular environment, which suggested that although removal of the C-terminus did not significantly affect β-sheet formation, it did lead to reduced lipid accumulation. To our knowledge, this is the first Raman study of protein-lipid interactions and amyloid formation using HPG. Furthermore, it is also the first reported direct observation of β-sheet secondary structure and region-specific differences in α-syn fibrils in cellular environments.
4. Conclusions and Outlook
The vibrational spectroscopic studies reviewed here exemplify the utility and versatility of the probes and methods available to study amyloids from the atomic to cellular scale. Vibrational probes can be very small and minimally perturbative, making them particularly attractive in amyloid formation studies. Though a variety of vibrational probes have been incorporated into amyloidogenic proteins by SPPS, the work reviewed here demonstrates the efforts to extend this approach to a wider array of amyloids. Despite their potential, vibrational studies in cells are underexplored, highlighting a tremendous opportunity for further advances in this area. As amyloid research moves towards understanding fibril formation by larger, more complex proteins and in cellular environments, new methods of vibrational probe incorporation are needed. Perhaps the most notable absence among these methods is UAA incorporation using evolved aminoacyl-tRNA/tRNA pairs (21st-pair technology). This approach has been employed to incorporate a variety of vibrational probes for protein folding studies, but remains an untapped resource in amyloid research. It has been demonstrated in prokaryotic and eukaryotic expression systems, as well as cultured mammalian cells, making it an attractive and vetted method for both in vitro and in cellulo studies [64]. As this field continues to develop, we expect that chemical biology will play an increasingly prominent role in vibrational spectroscopic studies of amyloid formation.
Acknowledgements.
MDW and JCL are supported by the Intramural Research Program at the National Institutes of Health, National Heart, Lung, and Blood Institute.
Abbreviations
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- FTIR
Fourier-transform infrared spectroscopy
- α-syn
α-synuclein
- MSA
multiple system atrophy
- UAA
unnatural amino acid
- SPPS
solid-phase peptide synthesis
- Aβ
amyloid-β
- 2D-IR
two-dimensional infrared spectroscopy
- hIAPP
human islet amyloid polypeptide
- MESNA
2-mercaptoethanesulfonate
- SRS
stimulated Raman scattering
- DM
L-aspartic acid 4-methyl ester
- HPG
homopropargylglycine
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Annotated References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
••of outstanding interest
- 1.Chiti F, Dobson CM: Protein misfolding, functional amyloid, and human disease. Amm Rev Biochem 2006, 75:333–366. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ: Folding proteins in fatal ways. Nature 2003, 426:900–904. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg DS, Sawaya MR: Structural studies of amyloid proteins at the molecular level. Amm Rev Biochem 2017, 86:69–95. [DOI] [PubMed] [Google Scholar]
- 4.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC: The common architecture of cross-β amyloid. J Mol Biol 2010, 395:717–727. [DOI] [PubMed] [Google Scholar]
- 5.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. : Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007, 447:453–457. [DOI] [PubMed] [Google Scholar]
- 6.Boyer DR, Li B, Sun C, Fan W, Zhou K, Hughes MP, Sawaya MR, Jiang L, Eisenberg DS: The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc Natl Acad Sci USA 2020, 117:3592–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC: Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci USA 2005, 102:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harper JD, Lansbury PT Jr.: Models of amyloid seeding in Alzheimer’s disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Amm Rev Biochem 1997, 66:385–407. [DOI] [PubMed] [Google Scholar]
- 9.Powers ET, Powers DL: Mechanisms of protein fibril formation: nucleated polymerization with competing off-pathway aggregation. Biophys J 2008, 94:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JL, Lee VM: Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat Med 2014,20:130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson MD, Lee JC: N-terminal acetylation affects α-synuclein fibril polymorphism. Biochemistry 2019, 58:3630–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sneideris T, Darguzis D, Botyriute A, Grigaliunas M, Winter R, Smirnovas V: pH-driven polymorphism of insulin amyloid-like fibrils. PLoS One 2015, 10:e0136602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero-Ferreira R, Kovacik L, Ni D, Stahlberg H: New insights on the structure of α-synuclein fibrils using cryo-electron microscopy. Carr Opin Neurobiol 2020, 61:89–95. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Liu C: Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat Chem Biol 2021, 10.1038/s41589-020-00708-z. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Lantz R, Du D: Vibrational approach to the dynamics and structure of protein amyloids. Molecules 2019, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarroukh R, Goormaghtigh E, Ruysschaert JM, Raussens V: ATR-FTIR: a “rejuvenated” tool to investigate amyloid proteins. Biochim Biophys Acta 2013, 1828:2328–2338. [DOI] [PubMed] [Google Scholar]
- 17.Lomont JP, Ostrander JS, Ho JJ, Petti MK, Zanni MT: Not all β-sheets are the same: Amyloid infrared spectra, transition dipole strengths, and couplings investigated by 2D IR spectroscopy. J Phys Chem B 2017, 121:8935–8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zandomeneghi G, Krebs MR, McCammon MG, Fandrich M: FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci 2004, 13:3314–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran SD, Zanni MT: How to get insight into amyloid structure and formation from infrared spectroscopy. J Phys Chem Lett 2014, 5:1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheres SH, Zhang W, Falcon B, Goedert M: Cryo-EM structures of tau filaments. Curr Opin Struct Biol 2020, 64:17–25. [DOI] [PubMed] [Google Scholar]
- 21.Schweighauser M, Shi Y, Tarutani A, Kametani F, Murzin AG, Ghetti B, Matsubara T, Tomita T, Ando T, Hasegawa K, et al. : Structures of α-synuclein filaments from multiple system atrophy. Nature 2020, 585:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahnawaz M, Mukherjee A, Pritzkow S, Mendez N, Rabadia P, Liu X, Hu B, Schmeichel A, Singer W, Wu G, et al. : Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 2020, 578:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn JD, McGlinchey RP, Walker RL 3rd, Lee JC: Structural features of α-synuclein amyloid fibrils revealed by Raman spectroscopy. J Biol Chem 2018, 293:767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni X, McGlinchey RP, Jiang J, Lee JC: Structural insights into α-synuclein fibril polymorphism: effects of Parkinson’s disease-related C-terminal truncations. J Mol Biol 2019, 431:3913–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flynn JD, Lee JC: Raman fingerprints of amyloid structures. Chem Commun (Camb) 2018, 54:6983–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees AJ, Hardy J, Revesz T: Parkinson’s disease. Lancet 2009, 373:2055–2066. [DOI] [PubMed] [Google Scholar]
- ••27.Flynn JD, Gimmen MY, Dean DN, Lacy SM, Lee JC: Terminal alkynes as Raman probes of α-synuclein in solution and in cells. ChemBioChem 2020, 21:1582–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a Met-auxotropic expression system, the unnatural amino acid HPG was biosynthetically incorporated into α-syn. Analysis of the alkyne stretching band by Raman spectral imaging revealed local sidechain differences in the cellularly internalized fibils.
- ••28.Watson MD, Flynn JD, Lee JC: Raman spectral imaging of (13)C(2)H(15)N-labeled a-synuclein amyloid fibrils in cells. Biophys Chem 2021, 269:106528. [DOI] [PMC free article] [PubMed] [Google Scholar]; Raman spectral imaging of cells treated with 13C2H15N-labeled α-syn fibrils demonstrated background-free detection and colocalization of β-sheet amyloid structure with endogenous biomolecules in a cellular environment.
- ••29.Flynn JD, Jiang Z, Lee JC: Segmental 13C-labeling and Raman microspectroscopy of α-synuclein amyloid formation. Atigew Chem Int Ed Engl 2018, 57:17069–17072. [DOI] [PMC free article] [PubMed] [Google Scholar]; A method for segmental 13C-labeling of α-syn using intein-mediated expressed-protein ligation was reported. Aggregation kinetics were monitored by the respective amide-I bands revealing region-specific β-sheet structural changes.
- •30.Okada Y, Okubo K, Ikeda K, Yano Y, Hoshino M, Hayashi Y, Kiso Y, Itoh-Watanabe H, Naito A, Matsuzaki K: Toxic amyloid tape: A novel mixed antiparallel/parallel β-sheet structure formed by amyloid β-protein on GM1 clusters. ACS Chem Nenrosci 2019, 10:563–572. [DOI] [PubMed] [Google Scholar]; The authors measured carbonyl coupling in 13C-labeled Aβ fibrils to demonstrate the formation of unusual mixed parallel/antiparallel β-sheet structures on GM1 lipid clusters.
- 31.Brewer SH, Song B, Raleigh DP, Dyer RB: Residue specific resolution of protein folding dynamics using isotope-edited infrared temperature jump spectroscopy. Biochemistry 2007, 46:3279–3285. [DOI] [PubMed] [Google Scholar]
- 32.Davis CM, Cooper AK, Dyer RB: Fast helix formation in the B domain of protein A revealed by site-specific infrared probes. Biochemistry 2015, 54:1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang C, Wang J, Charnley AK, Barber-Arm strong W, Smith Iii AB, Decatur SM, Hochstrasser RM: Two-dimensional infrared measurements of the coupling between amide modes of an α-helix. Chem Phys Lett 2003, 382:586–592. [Google Scholar]
- 34.Buchanan LE, Maj M, Dunkelberger EB, Cheng PN, Nowick JS, Zanni MT: Structural polymorphs suggest competing pathways for the formation of amyloid fibrils that diverge from a common intermediate species. Biochemistry 2018, 57:6470–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrano AL, Lomont JP, Tu LH, Raleigh DP, Zanni MT: A free energy barrier caused by the refolding of an oligomeric intermediate controls the lag time of amyloid formation by hIAPP. J Am Chem Soc 2017, 139:16748–16758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganim Z, Chung HS, Smith AW, Deflores LP, Jones KC, Tokmakoff A: Amide I two-dimensional infrared spectroscopy of proteins. Acc Chem Res 2008, 41:432–441. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh A, Ostrander JS, Zanni MT: Watching proteins wiggle: Mapping structures with two-dimensional infrared spectroscopy. Chem Rev 2017, 117:10726–10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraack JP, Hamm P: Surface-sensitive and surface-specific ultrafast two-dimensional vibrational spectroscopy. Chem Rev 2017, 117:10623–10664. [DOI] [PubMed] [Google Scholar]
- 39.Le Sueur AL, Horness RE, Thielges MC: Applications of two-dimensional infrared spectroscopy. Analyst 2015, 140:4336–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alperstein AM, Ostrander JS, Zhang TO, Zanni MT: Amyloid found in human cataracts with two-dimensional infrared spectroscopy. Proc Natl Acad Sci USA 2019, 116:6602–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fields CR, Dicke SS, Petti MK, Zanni MT, Lomont JP: A different hIAPP polymorph is observed in human serum than in aqueous buffer: demonstration of a new method for studying amyloid fibril structure using infrared spectroscopy. J Phys Chem Lett 2020, 11:6382–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostrander JS, Lomont JP, Rich KL, Saraswat V, Feingold BR, Petti MK, Birdsall ER, Arnold MS, Zanni MT: Monolayer sensitivity enables a 2D IR spectroscopic immuno-biosensor for studying protein structures: Application to amyloid polymorphs. J Phys Chem Lett 2019, 10:3836–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43.Maj M, Lomont JP, Rich KL, Alperstein AM, Zanni MT: Site-specific detection of protein secondary structure using 2D IR dihedral indexing: A proposed assembly mechanism of oligomeric hIAPP. Chem Sci 2018, 9:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vibrational coupling between pairs of 13C18O-labeled residues in hIAPP was measured by 2D-IR and used to identify transient helical structure in a prefibrillar species.
- •44.Wu H, Saltzberg DJ, Kratochvil HT, Jo H, Sali A, DeGrado WF: Glutamine side chain 13C–18O as a nonperturbative IR probe of amyloid fibril hydration and assembly. J Am Chem Soc 2019, 141:7320–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]; FTIR-based isotope dilution experiments using 13C18O-labeled Gin sidechains in peptides revealed the formation of Gln ladders in yeast prion and tau-derived peptides.
- 45.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM, Zanni MT: Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc Natl Acad Sci USA 2012, 109:3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez L, Marano MM, Tandon A: Import and export of misfolded α-synuclein. Front Nenrosci 2018, 12:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan HC, Ho LI, Chi CS, Chen SJ, Peng GS, Chan TM, Lin SZ, Harn HJ: Polyglutamine (PolyQ) diseases: Genetics to treatments. Cell Transplant 2014, 23:441–458. [DOI] [PubMed] [Google Scholar]
- ••48.Miao K, Wei L: Live-cell imaging and quantification of polyQ aggregates by stimulated Raman scattering of selective deuterium labeling. ACS Cent Sci 2020, 6:478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quantitative imaging of endogenous poly-Q aggregates in live cells was accomplished using stimulated Raman scattering by introduction of deuterated Gin into growth media.
- 49.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS: Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008, 322:1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warner JBt, Ruff KM, Tan PS, Lemke EA, Pappu RV, Lashuel HA: Monomeric huntingtin exon 1 has similar overall structural features for wild-type and pathological polyglutamine lengths. J Am Chem Soc 2017, 139:14456–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bagchi S, Boxer SG, Fayer MD: Ribonuclease S dynamics measured using a nitrile label with 2D IR vibrational echo spectroscopy. J Phys Chem B 2012, 116:4034–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung JK, Thielges MC, Fayer MD: Conformational dynamics and stability of HP35 studied with 2D IR vibrational echoes. J Am Chem Soc 2012, 134:12118–12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taskent-Sezgin H, Chung J, Banerjee PS, Nagarajan S, Dyer RB, Carrico I, Raleigh DP: Azidohomoalanine: A conformationally sensitive IR probe of protein folding, protein structure, and electrostatics. Angew Chem Int Ed Engl 2010, 49:7473–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia B, Sun Y, Yang L, Yu Y, Fan H, Ma G: A structural model of the hierarchical assembly of an amyloid nanosheet by an infrared probe technique. Phys Chem Chem Phys 2018, 20:27261–27271. [DOI] [PubMed] [Google Scholar]
- 55.Marek P, Mukherjee S, Zanni MT, Raleigh DP: Residue-specific, real-time characterization of lag-phase species and fibril growth during amyloid formation: A combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J Mol Biol 2010, 400:878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh KI, Lee JH, Joo C, Han H, Cho M: β-Azidoalanine as an IR probe: Application to amyloid Aβ(16–22) aggregation. J Phys Chem B 2008, 112:10352–10357. [DOI] [PubMed] [Google Scholar]
- •57.Pazos IM, Ma J, Mukheijee D, Gai F: Ultrafast hydrogen-bonding dynamics in amyloid fibrils. J Phys Chem B 2018, 122:11023–11029. [DOI] [PMC free article] [PubMed] [Google Scholar]; The ester moiety of a methylated Asp residue was used as a vibrational probe of intersheet hydrogen bonding in fibrils formed by an Aβ-derived peptide.
- 58.van Hest JCM, Kiick KL, Tirrell DA: Efficient incorporation of unsaturated methionine analogues into proteins in vivo. J Am Chem Soc 2000, 122:1282–1288. [Google Scholar]
- 59.Beatty KE, Tirrell DA: Two-color labeling of temporally defined protein populations in mammalian cells. Bioorg Med Chem Lett 2008, 18:5995–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beatty KE, Xie F, Wang Q, Tirrell DA: Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc 2005, 127:14150–14151. [DOI] [PubMed] [Google Scholar]
- 61.Wei L, Hu F, Shen Y, Chen Z, Yu Y, Lin CC, Wang MC, Min W: Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat Methods 2014, 11:410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerrero-Ferreira R, Taylor NM, Mona D, Ringler P, Lauer ME, Riek R, Britschgi M, Stahlberg H: Cryo-EM structure of α-synuclein fibrils. Elife 2018, 7:e36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B, Ge P, Murray KA, Sheth P, Zhang M, Nair G, Sawaya MR, Shin WS, Boyer DR, Ye S, et al. : Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat Commim 2018, 9:3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young DD, Schultz PG: Playing with the molecules of life. ACS Chem Biol 2018, 13:854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]


