Abstract
For over 20 years, peptide materials in their hydrogel or soluble fibril form have been used for biomedical applications such as drug delivery, cell culture, vaccines, and tissue regeneration. To facilitate the translation of these materials, key areas of research still need to be addressed. Their structural characterization lags compared to amyloid proteins. Many of the structural features designed to guide materials formation are primarily being characterized by their observation in atomic resolution structures of amyloid assemblies. Herein, these motifs are examined in relation to peptide designs identifying common interactions that drive assembly and provide structural specificity. Current efforts to design complex structures, as reviewed here, highlight the need to extend the structural revolution of amyloid proteins to peptide assemblies to validate design principles. With respect to clinical applications, the fundamental interactions and responses of proteins, cells, and the immune system to peptide materials are still not well understood. Only a few trends are just now emerging for peptide materials interactions with biological systems. Understanding how peptide material properties influence these interactions will enable the translation of materials towards current and emerging applications.
Introduction
Decades of research in the structure, formation, function, and biological effects of amyloid proteins have uncovered the main factors driving their assembly into supramolecular structures [1-4]. Many of the emerging structural features observed in amyloid proteins validate general design concepts that have historically been used in peptide materials, a field in which high-resolution structure determination is rare. For example, the same non-covalent interactions such as the hydrophobic effect, van der Waals forces, and hydrogen bonding drive assembly and provide structural specificity in protein and peptide assemblies. Several early classes of self-assembling peptide systems, from which much has been learned, take advantage of these overarching biophysical forces, including aromatic dipeptides, such as diphenylalanine (FF) and its Fmoc derivatives, and 10 - 20 residue amphiphilic or ionic self-complementary oligopeptides [5, 6]. However, beyond these general biophysical concepts, emerging amyloid structures provide insight into distinct inter-residue and main-chain interactions, and this, coupled with the diversity of natural and non-natural amino acid building blocks, should inspire creative new assembly designs.
Peptides prone to assembly can do so when initially dissolved in an aqueous solution or can be designed to form fibrils when triggered by different stimuli such as pH, temperature, and concentration. Depending on the concentration, peptide fibrils can either stay in solution or, at higher concentration, entangle to form physically crosslinked three-dimensional networks called hydrogels. β-sheet peptide materials bear attractive properties for biomedical applications either in their soluble fibril form or as hydrogels. The design of these peptides at the monomer level directly translates into the materials’ structure and properties, facilitating the incorporation of chemical diversity, bioactive cues, and motifs for further functionalization. Peptide materials are typically biocompatible and do not elicit adverse immune response unless designed to do so. Also, they are easily biodegradable, and their fibrous structure can be designed to resemble the natural extracellular matrix environment [7]. These properties facilitated the use of peptide materials in numerous applications, including vehicles for localized and controlled delivery of protein and nucleic acid therapeutics, small molecule drugs, and cell delivery [8-12], scaffolds for 2D and 3D cell culture for in vitro models [13, 14], vaccine adjuvants [15, 16], as surgical hemostatic or anastomotic agents [17-20], and materials for promoting wound healing and tissue regeneration [21, 22] (Figure 1).
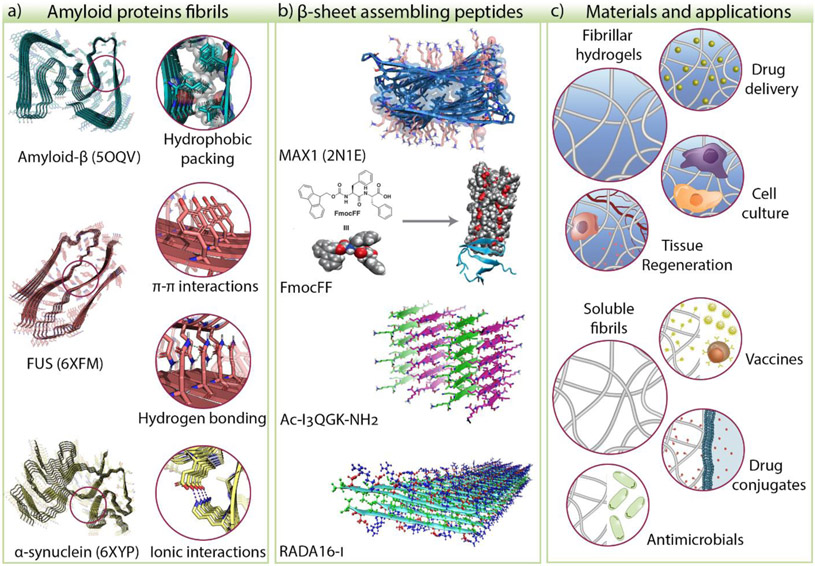
Figure 1.
a) The common structural features of amyloid proteins such as hydrophobic packing, π-π, hydrogen bonding, and ionic interactions have been used to develop β-sheet assembling peptides. b) Examples of peptide systems that rely on these features are MAX1, diphenylalanine derivatives, Ac-I3QGK-NH2, and RADA16-I, to name a few. c) Self-assembling peptides form materials such as fibrillar hydrogels and soluble fibrils used in biomedical applications. The Protein Data Bank (PDB) ID of amyloid-β [32], FUS [57], α-synuclein [39], and MAX1 [53] are shown in parenthesis. The other peptide structures are models or representations of proposed structures. FmocFF model adapted from [58]. © 2019, Ji et al. under CC BY license. Ac-I3QGK-NH2 adapted from [59] © Wang et al. under CC BY 4.0 license. RADA16-I structure image adapted from [54] © 2013, American Chemical Society.
To date, much effort has been dedicated to designing peptides capable of assembly, establishing the rules governing self-assembly, and developing applications for the resulting materials, mostly in drug delivery. However, our knowledge of the underlying structure of these systems at the molecular and atomic level greatly lags in contrast with the recent surge of amyloidogenic protein structure determination. Detailed structural knowledge of the diverse β-sheet peptide materials being developed may uncover new self-assembly principles and improve our ability to control the formation of well-defined nanostructured materials. Furthermore, there is still a void in our mechanistic understanding of the assembly process and the influence that the fibril structure and chemical composition has on the biological behavior. The ability to predict the cellular or immune responses to these fibrillar materials will inform the design of new materials and aid in the selection of existing materials for targeted applications. Despite an extensive exploration of peptide biomaterials, only a few self-assembling systems have achieved commercialization as scaffolds for cell culture and 3D printing, and only one has been cleared by the FDA for clinical use as a medical device for hemostasis and wound healing [23]. This review discusses the recent advances and trends in the design of biomaterials composed of β-sheet peptides directly derived from amyloid proteins or de novo designs. We focus on applications and observations on the cellular and immune responses to these systems and highlight areas in need of study using a few peptide materials as examples. The reader can find other excellent reviews that comprehensively cover the plethora of published peptide-material systems [6, 24-28].
Structural insights of β-sheet self-assembling peptide nanofibers
There has been a recent explosion of work elucidating the structure of amyloidogenic proteins and unraveling amyloid formation mechanisms, which can potentially uncover therapeutic liabilities of amyloid-associated diseases. Multiple structures from amyloid-beta (Aβ) [29-37], α-synuclein [38-40], Tau [41-43], FUS [44], among other proteins [45-48], have been obtained thanks to advances in crystallography, Cryo-EM, NMR, and computation. These studies revealed several common structural features that define amyloid proteins. First, they adopt a cross-β structure in which β-strands align perpendicular to the fibril growth axis while forming hydrogen bonds along the peptide backbone. Amyloid fibrils also form the so-called “steric zipper” composed of buried, mostly hydrophobic residues that drive aggregation by the hydrophobic effect, see amyloid-β in Figure 1a. These general features (main-chain H-bonding and hydrophobic collapse) have been inferred for many years based on biophysical studies, modeling, scattering, and microscopy. Although these proteins can form polymorphic fibrils depending on environmental conditions, these underlying structural features are always conserved. However, the exact β-fold of the main chain and the highly repetitive aromatic and polar interactions between residue side chains are illuminating. Inter-residue interactions are typically local in globular proteins, but in amyloid assemblies, they become repetitive long-range interaction highways that permeate the entire length of a given fibril. Great examples are the π-π aromatic interactions and H-bonding made between the tyrosine side chains in FUS and the salt bridge network found in α-synuclein, Figure 1a. These polar interactions often provide folding/assembly specificity. In addition to these contributions from the structural biology community, the field of crystal engineering has characterized similar intermolecular interactions that effect crystal packing and can be utilized in materials design [49, 50].
Amyloid and other natural proteins have been an inspiration for the development of peptide materials. Assembling sequences can be derived directly from protein motifs or created de novo utilizing the common structural features of amyloid protein. Zhang et al. provided one of the first examples by discovering ionic complementary self-assembling peptides derived from Zuotin protein [50]. An extensive body of work studying this system led to RADA-16-I, the only FDA-approved peptide gel, Figure 1b. Since then, various peptide designs have been reported that rely on hydrophobic packing, π-π interactions, ionic and polar interactions to control assembly and fiber formation. Most of these systems' structural characterization is limited to a few biophysical techniques that show the expected β-sheet fibrillar structure, such as circular dichroism, infrared spectroscopy, and nominal resolution TEM. High-resolution structures cannot be determined by these techniques, so in most cases, only schemes of predicted or desired models are provided. Although there has been a recent effort in determining the structures of model β-sheet forming peptides [51, 52], to our knowledge, only two structures have been determined for peptide fibrils, namely MAX1 [53] and RADA16-I [54]. In general, high resolution structural characterization of more systems is required to confirm existing design principles and uncover new ones leading to new materials, including ones with dynamic structures and function. To this end, although solid-state NMR continues to be extremely useful for self-assembled systems, recent advances in using cryo-EM to elucidate fibrillar structures should catalyze materials design. Further, computation can be a powerful tool in designing peptide and protein materials especially when couple with high-resolution structure determination as was demonstrated for the generation of filaments comprised of self-assembled helical protein [55]. Although computation has recently been applied to sheet-containing systems [56], to our knowledge no high-structures have been solved.
Trends in β-sheet peptide materials design
A current interest is to create materials with complex structures, hierarchical architectures, multiple functionalities, and dynamic properties. Typically, peptide materials are homogenous systems composed of a single self-assembling peptide, which provides a well-defined chemical composition, ease in design and preparation, and controlled physicochemical properties. One emerging strategy to increase structural and functional complexity involves designing multi-component systems [60] where different peptides and/or other molecules are mixed and allowed to form co-assembled [61, 62] or self-sorted structures [63] (Figure 2a). In co-assembled systems, individual fibers contain all the components available for assembly. Co-assembly can be directed by charge complementarity [62], hydrophobic interactions [58, 64], or stereocomplexation [65-68]. Co-assembly generates materials with enhanced or different properties than their single-component counterpart [69, 70]. For instance, Fmoc-protected diphenylalanine (Fmoc-FF) can undergo a structural transition from β-sheet nanoribbons to helical twisted fibrils and have responsiveness to light and redox conditions when co-assembled with bipyridine derivatives [58, 64]. Co-assembly can also be used to enhance peptide hydrogel stiffness either by stereocomplexation [66, 71] or inducing crosslink formation in a purely supramolecular manner [72]. In particular, stereocomplexation can be a predictive and useful tool, especially in the formation of rippled β-sheet assemblies as recently reviewed [73]. In self-sorting systems, individual components self-assemble independently, creating a mixture of coexisting but separate structures. For example, Webber et al. showed that a mixture of DWDW and KWKW tetrapeptides formed DWDW-based fibrillar gel containing KWKW-based spherical nanoparticles [63]. Self-sorting can also be achieved by mixing peptides with other molecules such as lipids [74], polysaccharides, and polymers [75, 76]. The development of these more complex systems should evolve together with biophysical and structural analysis of the resulting materials and strategies to favor the formation of selective structures [62, 77, 78].
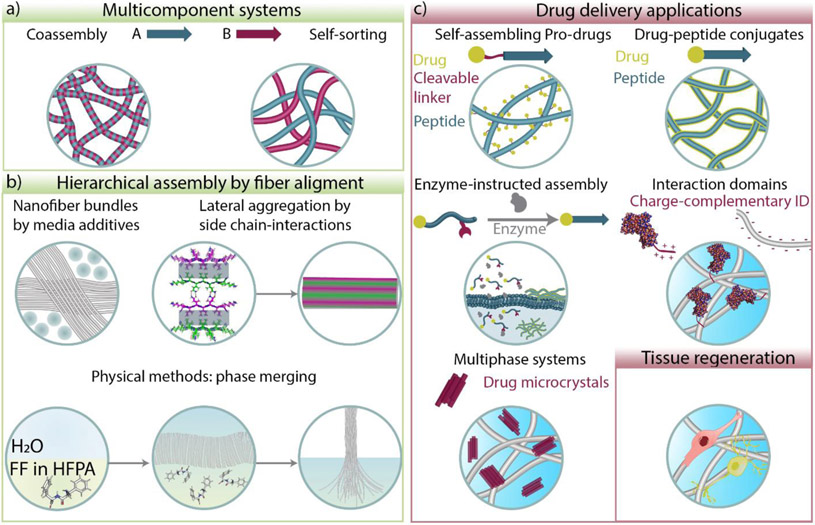
Figure 2.
Trends in β-sheet peptide materials design and emerging applications in drug delivery and tissue regeneration. a) Multicomponent peptide systems can coassemble or self-sort into fibrillar structures. b) Strategies to create hierarchical assembly and fiber alignment by molecular crowding, polar interactions, and phase merging. c) Different strategies used for controlled drug delivery of small molecules, nucleic acids, and proteins. Peptide materials are also used to promote tissue regeneration. Structure of Ac-I3QGK-NH2 representing lateral aggregation by side chain interactions was adapted from [59] © Wang et al. under CC BY 4.0 license.
Another strategy to introduce molecular complexity and structural diversity is creating hierarchical structures by inducing fiber alignment (Figure 2b). In nature, it is common to see complex and hierarchical structures, for example, collagen bundles within the extracellular matrix. The hierarchical assembly of collagen that results from post-translational modification of collagen peptides, including glycosylation and enzymatic crosslinking, is required to provide mechanical integrity and guide tissue formation [79]. Although peptide materials have not yet achieved the hierarchal complexity of tissues, progress has been made in inducing fiber-fiber interactions leading to hierarchical assembly. For example, the bundling of fibrils formed by the glycosylated peptide Q11 (QQKFQFQFEQQ) can be enhanced by inducing molecular crowding with polymeric additives, Figure 2b [80]. Xu et al. showed that fibril lamination could be facilitated by the incorporation of uncharged polar amino acids. Adding glutamine, serine, or asparagine to the self-assembling peptide Ac-I3XGK-NH2 (X = Q, S, R) favors the formation of wide ribbons comprised of laminated fibers, suggesting that favorable polar interactions are formed between individual fibers in the final laminated structure [59]. This phenomenon is also observed in heterochiral ld or dl FKFEFKFE peptides, which form wide helical tapes instead of typical nanofibers [81]. These examples highlight the promise of hierarchical peptide assembly and the opportunity to discover the structural principles guiding their formation and function. Lastly, aligned nanofiber bundles can be fabricated by physical methods (Figure 2b). Self-assembly of the dipeptide FF at the interface of hexafluoroisopropanol and water leads to the formation of fibers that can be physically pulled from the interface affording micrometer-long bundles that have electromechanical properties [82]. Further exploration of strategies to create hierarchical structures could ultimately result in materials that mimic anisotropic and highly ordered tissues.
Emerging biomedical applications of fibrous peptide materials
Peptide materials have been used extensively as scaffolds for the controlled delivery of proteins, nucleic acids, and small-molecule drugs. Generally, two main mechanisms dictate the release rate of a drug from a peptide gel network. The first is simple diffusion which depends on the size of the drug and the network's mesh size. As the size of the drug approaches the mesh size, steric interactions impact its release. The second factor involves the reversible formation of specific or non-specific interactions between the drug and the fibril network. Depending on the identity of the drug and the gel network, one or both factors can be at play. A current focus involves designing alternative strategies that enhance control over delivery rates, offer regiospecific control over drug action, and create generic hydrogel materials capable of delivering a broad range of therapeutics, eliminating the need to design custom gels for individual therapies.
Self-assembling prodrugs are simply peptides that contain a drug ligated via a cleavable linker (Figure 2c). The peptide portion is responsible for the assembly and formation of fibrils that display drugs from their surface. Drug release is dependent on linker degradation, typically the result of enzymatic action. These systems offer localized and sustained release [83]. Drug-peptide conjugates are a similar class of assembler [84-89]. Here, drugs are also appended to the peptide but can participate in the self-assembly process and become incorporated into the fibril's core architecture [84-86, 90]. For example, Hartgerink et al. designed a nanofibrous hydrogel called L-NIL-MDP, where the lysine residues of the self-assembling peptide K2(SL)6K2 were substituted by the small-molecule drug N6-(1-iminoethyl)-L-lysine (L-NIL). Gels prepared from this peptide retained native L-NIL bioactivity, inhibiting the production of inducible nitric oxide synthase in vitro and in vivo [84].
Peptide-drug conjugates can also be utilized to selectively deliver a therapeutic agent either at the cell surface or intracellularly by a process called enzyme-instructed self-assembly pioneered by Xu et al. [91-93] (Figure 2c). An enzymatically cleavable moiety is incorporated into the peptide to endear solubility, thus inhibiting self-assembly until a specific enzyme is encountered. Enzyme-mediated removal of the water-solubilizing moiety triggers peptide assembly in a regiospecific manner [87, 88, 94-96]. These enzyme-susceptible peptides can be functionalized with a drug to concentrate the therapy at desired cellular organelles through enzyme-triggered fibrillization. This strategy was used by Zhong et al. to deliver Lonidamine (LND) and inhibit glycolysis in the mitochondria of cancer cells [88]. In general, the use of fiber-forming peptides conjugated with drugs, whether self-assembling prodrugs or peptide-drug conjugates, provides several unique advantages. First, fiber formation can be triggered by environmental factors, such as pH, redox conditions, or enzymatic activity, to name a few, allowing regiospecific material formation. The resulting fibers can display the drug at high density, increasing its local concentration. Appending ligands to the self-assembling peptide to target unique cell surface receptors can deliver therapeutic material to select cells, potentially reducing side effects [97, 98]. The drug release profiles of these materials depend on their environment, for example, the presence of enzyme or a change in pH, affecting on-demand delivery. Lastly, some can form injectable hydrogels facilitating implantation [84] and be designed to co-deliver multiple components for combination therapy [86, 99].
Other recent strategies aim to produce general peptide-based delivery platforms that can be used for an entire class of cargo, eliminating the need to custom design a new hydrogel for each drug one wishes to deliver. One example involves protein therapeutics whose delivery can be challenging due to the possibility of protein-material adsorption, which can cause protein denaturation. Schneider et al. developed a protein delivery platform that limits direct contact between globular protein domains and the hydrogel fibrillar network (Figure 2c). Their approach uses complementary electrostatic interactions made between a suite of designed interaction domains (IDs), ligated to the terminus of a protein of interest, and a negatively charged hydrogel network. The gel network repels the globular domain of like-charged proteins with only the ID binding to the fibrillar network. Desorption energies can be modulated by the choice of ID to control protein release kinetics [100].
A general hydrogel has also been designed for the delivery of hydrophilic small molecules. For example, Tofacitinib, an immunosuppressant drug, is notoriously difficult to deliver because of its hydrophilicity. The drug quickly diffuses from most materials resulting in burst delivery. However, Tofacitinib forms microcrystals when dissolved together with hydrogel-forming peptides [101]. Triggered peptide assembly leads to the formation of hydrogel containing the soluble drug. Tofacitinib then crystallizes in the nanofibrous network forming a multiphase hydrogel where the two components coexist independently. Importantly, since the gel is non-interacting, the drug's release rate depends on crystal dissolution [101] (Figure 2c). In general, the approaches described in this section demonstrate the potential of finely controlled drug release. Future research might focus on achieving complete control over the on-demand drug release in vivo, where materials are exposed to the immune system and susceptible to fouling and degradation. Understanding in vivo response to these systems is important because it will ultimately affect these materials' efficacy and the resulting therapeutic effect, as will be described in the following sections.
Effects of peptide-based materials on cell responses
Peptide-based coatings and hydrogels have been widely used for two-dimensional and three-dimensional cell culture and as vehicles for exogenous cell delivery, commonly demonstrating high cytocompatibility [6, 9, 13, 25, 102]. Material cytocompatibility is assessed by quantifying viable cells and evaluating their metabolic activity, attachment (for adherent cells), and proliferation over time. Although experiments reported across the literature are performed with different cell types, highly diverse experimental conditions, and varying peptide designs, there are a few emerging trends in the cell response to this class of materials (Figure 3). For example, the hydrogel's charge state seems to be an important factor in determining cell behavior. Hydrogels prepared from negatively charged peptides seem to support cell viability. However, adherent cells do not adopt their native spread morphology and instead clump together. This phenomenon has been observed for human dermal fibroblasts cultured on gels formed from the β-hairpin peptide Ac-VES3 [9] (Figure 3a) and 3T3 fibroblasts seeded on glass coated with the decapeptide E1Y9 (Ac-EYEYKYEYKY) [103]. Similarly, viable fibroblasts do not spread homogenously and clump when cultured on coatings prepared from the zwitterionic self-assembling peptide QQKFQFQFEQQ (Q11) [80] or FEFEFKFK hydrogel [104] (Figure 3h). However, the incorporation of integrin-binding motifs or doping with other natural proteins facilitates cell adhesion on these peptide gels and coatings.
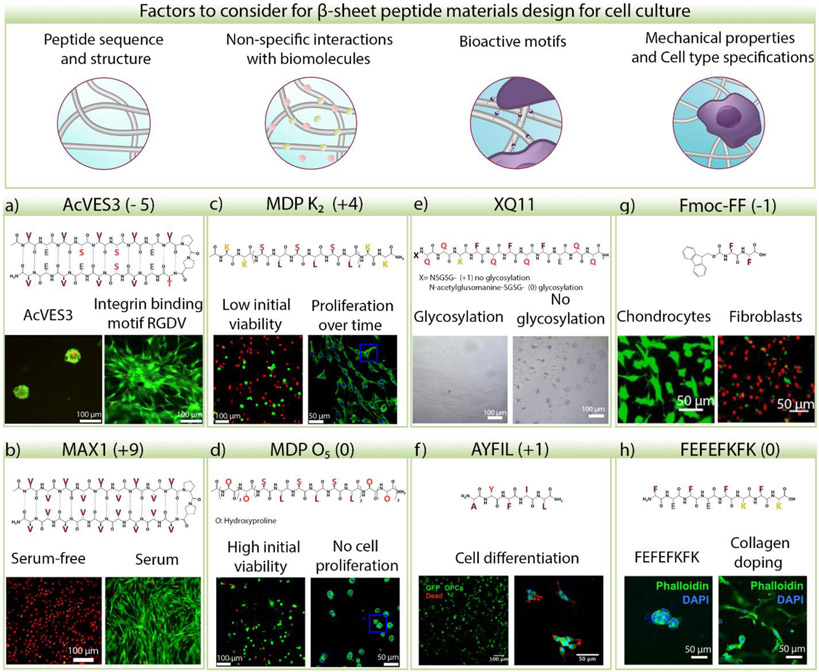
Figure 3.
Factors that play a role in cellular responses to β-sheet-rich peptide materials. a) Human Dermal Fibroblast (HDF) 2D cultured on the surface of 0.5 wt% AcVES3 and AcVES-GGGGRGDV for four days in medium without serum. Cells do not grow and spread on the negatively charged peptide gel unless an integrin-binding motif is incorporated. Green: calcein AM, red: ethidium homodimer I. Adapted with permission from [9]. © 2019, American Chemical Society. b) 2D cultured HDF cells on 0.5 wt% MAX1 gel with serum-free or serum-supplemented media for four days. Green: calcein AM, red: ethidium homodimer I. Adapted with permission from [105]. © 2021, American Chemical Society. c) 3D cultured NIH 3T3 fibroblasts on positively charged multidomain peptide (MDP) K2 gel, which is cytotoxic initially, but viable cells proliferate over time or d) on neutral MDP O5 gel, cells are highly viable but do not proliferate. Green: calcein AM or Phalloidin, red: ethidium homodimer I. Adapted with permission from [106]. © 2019, American Chemical Society. e) Adhesion of 2D cultured NIH 3T3 fibroblasts on glycosylated and non-glycosylated Q11 coatings in the presence of serum. Cells do not adhere to glycosylated Q11, but a few adhere and clump on the non-glycosylated material. Adapted with permission from [80]. © 2019, Restuccia et al. under CC BY 4.0 license. f) Oligodendrocyte precursor cells cultured on AYFIL gel for four days remain viable, and some neural processes are observed. Adapted with permission from [107]. © 2019, American Chemical Society. g) 2D culture of chondrocytes and NIH 3T3 fibroblasts on Fmoc-FF hydrogel for two days. High cell death is observed in fibroblasts in contrast to chondrocytes indicating that different cell types react differently to the same material. Green: calcein AM, red: ethidium homodimer I. Adapted with permission from [108]. © 2009 Acta Materialia Inc. Published by Elsevier Ltd. All rights reserved. h) 3D cultured human mammary fibroblast on FEFEFKFK gels with and without collagen. Cells spread and attach when collagen is present. Adapted with permission from [104]. © 2019, Ashworth et al. under CC BY license. The peptide charge at pH 7 is indicated in the parenthesis for each example.
In contrast, materials prepared from positively charged peptides sometimes present higher cytotoxicity, as has been observed for gels prepared from β-hairpin peptides MAX1 (Figure 3b), MAX8, and HLT2 [105], the multidomain peptide K2(SL)6K2 [106] (Figure 3c), pentapeptide KYFIL [107] among others. However, the cytotoxicity of these systems is highly dependent on the culture conditions, and alternative hydrogel preparation methods can be used to enhance cell survival. For example, the Schneider group demonstrated that the inherent cytotoxicity displayed by highly positively charged peptide hydrogels could be significantly reduced by priming the hydrogels with serum, a common element in cell culture media, or even single protein additives. They demonstrated that positively charged gels adsorb serum proteins via electrostatic interactions that passivate the surface charge and mediate cell signaling, resulting in the activation of both the PI3/Akt and MAPK/ERK pathways. These pathways are known to facilitate resistance to stress-induced apoptosis and overall cell survival [105] (Figure 3b). Thus, for both positive and negatively charged gels, protein adsorption seems to play an important role, but the specifics are not yet clear.
The case for materials prepared from neutral and non-ionic peptides is even less understood and divergent responses have been observed (Figure 3d-g). For example, in some systems such as SFFSF [72] and AYFIL (Figure 3f) [107], cells grow, proliferate, and differentiate; other peptides such as Fmoc-FF (Figure 3g) do not support fibroblast cell viability [8, 108]; and others such as glycosylated Q11 (Figure 3e) [80], and O5(SL)6O5 (O represents hydroxyproline, Figure 3d) [106, 109] do not induce cell attachment or proliferation when cells are encapsulated or seeded on the surface of the peptide hydrogels or peptide-coated glass. These observations suggest that other material attributes beyond charge are certainly at play.
Self-assembly pathways leading to fibrillization may also influence cell behavior, as is the case in amyloid formation. Early on, many thought that amyloid deposits were responsible for degenerative pathologies, but now there is strong evidence that oligomeric species formed during fibril formation are the toxic culprits [110]. In fact, several research groups are developing methods to catalyze the conversion of oligomers to non-toxic fibrils as a potential therapy [67, 111-113]. Oligomers might also form during the preparation of peptide materials. Using model peptides derived from prion proteins, Ventura et al. showed that cytotoxic oligomers were formed during the fibrillization process, which took seven days. The authors suggested that oligomer formation should also be assessed during the preparation of peptide-based materials [114]. With that said, these studies were performed under conditions that allowed slow fibrillization and concomitantly long persistence times for the oligomers. In contrast, most de novo designed material-forming peptides assemble quickly once triggered, where the persistence time of any oligomer would be short-lived if they are on-pathway to fibril formation. Certainly, it will be important in future studies to investigate the ramifications of self-assembly pathways and how material preparation methods influence cell behavior. Advancements in techniques such as liquid-cell TEM [115], solid-state NMR [116], and Cryo-EM will improve these efforts. For instance, Gelain and Weingarth utilized solid-state NMR to characterize the structure and assembly of several material-forming peptides for neural stem cell culture [116]. They observed a correlation between the degree of material formation, mechanical stiffness, and homogeneity on stem cell behavior, primarily cell differentiation. Heterogenous hydrogels were defined as those having a mixture of fibrillar networks and soluble "small assemblies". Heterogenous gels were characterized by low mechanical rigidities and poor cell differentiation potential. In contrast, homogenous gels, comprised mostly of fibrils, were mechanically stiff and better potentiated differentiation [116, 117]. These examples highlight that more mechanistic work is warranted and that the structural analysis of peptide materials can help define material attributes that influence cell behavior.
The different peptide-based materials developed over the years present distinct amino acid compositions, charge, self-assembling pathways, physical and mechanical properties, comprise numerous fiber morphologies, and often contain additional bioactive motifs to increase their cytocompatibility [8, 9, 109, 118]. Further, the effects of cell-induced peptide modifications or enzymatic degradation products have yet to be explored. Determining if and how each of these factors affects cell behavior is a daunting but necessary task. The result will be principles that globally unite the myriad of published observations and improve future material designs.
Immune response to β-sheet peptide materials: a relevant factor and target for materials design
Advances in immunology and immunoengineering have emphasized the immune system’s role in determining the overall response to materials and how material properties can be designed to modulate and induce specific immune responses [119-121]. Most of these studies used natural or synthetic polymers. With respect to β-sheet peptide materials, there has been incredible progress in their design, biophysical and mechanical characterization, and their assessment in vitro. In vivo studies have largely been carried out to evaluate their performance in affecting an intended function, for example, drug delivery or tissue engineering. However, the immune response to the materials was often not studied in any depth. In fact, efforts to define the immune response to peptide materials significantly lag behind those dedicated to commonly used polymers.
Early in the development of peptide materials, many outside the field believed that materials prepared from peptides would, a priori, elicit adverse immune responses precluding their biomedical use. Unfortunately, our community had not yet collected enough data to refute those concerns. Gratifyingly, over the last several years the field has begun to examine immune responses in earnest. As expected, responses vary and depend on the exact material, but adverse reactions in animal models have been rare.
Several independent observations of tissue responses to implanted peptide hydrogels show that these materials can be infiltrated by cells with different profiles depending on the peptide sequence and stereochemistry, hydrogel cargo such as cells or drugs, and the tissue type [21, 22, 84, 100, 101, 106, 109, 122] (Figure 4a). For example, gels prepared from the positively charged β-hairpin peptide MAX8, when injected subcutaneously in mice, were mildly infiltrated initially by neutrophils that gave way to macrophages after a few days. The gel persists for over a month but is ultimately degraded by macrophage-dependent phagocytosis and remodeled to native tissue [101]. A separate study suggested that stereochemistry may influence the immune response. Gels prepared from the negatively charged β-hairpin peptide AcVES3 had high macrophage infiltration after a few days in stark contrast to a gel prepared from its enantiomer d-AcVES3, which had significantly fewer infiltrating cells [100]. The basis for this stereochemical response is not yet known. Concerning the hairpin gels studied to date, neutrophils and macrophages largely represent the infiltrating milieu, with little representation of B- or T-cells. However, these cells can be recruited if certain cytokines are encapsulated in the gel (unpublished). Conversely, the global cellular response can be dampened by adding immunosuppressants to the hydrogel [101].
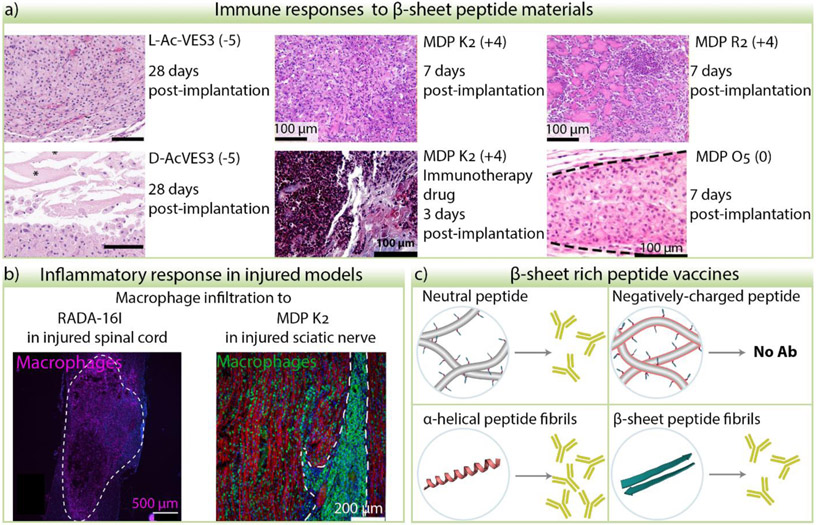
Figure 4.
Immune and inflammatory responses to β-sheet peptide materials. a) Examples of peptide hydrogel sections from subcutaneous injection of materials stained with Hematoxylin and Eosin. Cell nuclei are stained purple, cell cytoplasm, collagen, and hydrogel are stained in different shades of pink. Distinct cell morphology, infiltration degree, and cell distribution is observed depending on the peptide sequence, stereochemistry, immunotherapy drug cargo, and time. L- and D- AcVES3 images were adapted from [100] with permission from the American Chemical Society. Images of MDP K2 and R2 were adapted from [122] and MDP K2 with immunotherapy drug from [123]. © 2020 and 2019 Elsevier Ltd. All rights reserved. Image from MDP O5 was adapted from [106]. © 2019, American Chemical Society. The peptide charge at pH 7 is indicated in the parenthesis. b) When peptide hydrogels are implanted in injured nerve tissue such as spinal cord or sciatic nerve, macrophage infiltration is observed. RADA16-I image adapted from [21]. © 2020 Acta Materialia Inc. Published by Elsevier Ltd. All rights reserved. MDP K2 nerve image was adapted from [109]. © 2020 Elsevier Ltd. All rights reserved. c) Effects of peptide charge and structure in the efficacy of antibody production of epitope-bearing peptide fibers.
Hartgerink et al. compared the host immune response to the β-sheet peptide gels X2(SL)6X2, where X can be lysine, arginine, glutamic acid, or aspartic acid residues [122] (Figure 4). They found that the X-residue identity and the charge dictated the immune response to those materials. The negatively charged gels were infiltrated by tissue-resident macrophages that do not induce proinflammation and cleared the material within ten days. In contrast, the positively charged peptide gels provoked higher cellular infiltration of inflammatory monocytes, macrophages, and granulocytes and presented higher levels of pro-inflammatory cytokines, particularly the arginine-based peptide. Further, the clearance was much slower for the positively charged gels. The immune response to the lysine-based peptide can be modified when immunotherapy drugs are loaded into the gel [123]. The neutral peptide from this family containing hydroxyproline in the flanking domains is also infiltrated only by macrophages and is easily degraded within two weeks [106] (Figure 4a). In injury models, hydrogels prepared from zwitterionic RADA16-I or positively charged MDP K2 elicit robust macrophage responses in nerve tissues (Figure 4b) [21, 109].
Although the immune response varies depending on the exact peptide material, nearly all the materials reported to date are eventually biodegraded and replaced with native tissue [100, 101, 106, 122]. Similar to the discussion earlier on in vitro cellular responses to peptide materials, the factors influencing the immune response are not completely understood. The chemical composition, charge, shape, size, mechanical properties, and tissue type could all influence the response to these materials.
The most comprehensive work in analyzing immune responses to peptide nanofibers (as opposed to hydrogels) has been done in vaccine development. Peptide nanofibers bearing different immunogenic epitopes have been used to induce strong humoral immune responses without the use of adjuvants. The efficacy of these materials is due to their ability to present a high density of epitopes on the nanofiber surface, which can be easily customized during peptide synthesis and self-assembly [16, 124-126]. It is important to note that the fibers, by themselves, are non-immunogenic and require the addition of the epitope to elicit an adaptive immune response to specific targets. However, the peptide sequence does influence the vaccines’ effectiveness, although it is not understood exactly how. Collier and coworkers compared the antibody responses against OVA, an ovalbumin epitope, when presented on soluble nanofibers prepared from negatively versus positively-charged peptides derived from the parent peptide Q11 [127]. T-cell and B-cell responses were raised by OVA-Q11 and lysine modified-Q11 (KnSGSG-Q11) but not by the nanofibers containing negative residues (EnSGSG-Q11) (Figure 4c). When comparing the efficacy of peptide vaccines composed of the β-sheet-rich Q11 fibrils versus an α-helical peptide fiber, formed from the peptide Coil29, Wu et al. observed that the α-helical fibers elicited a greater antibody response suggesting that fibril morphology also influences activity [128] (Figure 4c). The immune and inflammatory responses to β-sheet peptide fibers and hydrogels represent the foundation of how these materials interact with living systems. However, it is still unclear what factors and material properties are responsible for evoking specific immune responses. To better guide efforts in the clinical translation of peptide materials, there is a need to better understand how their individual attributes (e.g. sequence composition, charge, biophysical/mechanical properties, etc.) influence the immune system. Further, a significant opportunity exists in the immunoengineering of these materials to control and inform the immune system.
Conclusion
Over the last 20 years, research on β-sheet peptide fibrils, coatings, and gels has evolved from understanding and controlling the mechanisms and forces driving assembly to their use in a broad range of biomedical applications. Novel designs shift from single to multiple component peptide systems and hierarchical assembly to incorporate structural and functional diversity into the fibrillar materials. The fabrication of more complex systems requires a deeper understanding of their structure to enhance our ability to favor the formation of specific and well-defined fibril morphologies and resulting materials. Among the most explored applications of peptide materials is their use as vehicles or scaffolds for drug delivery, cell culture, regenerative medicine, and vaccines. Significant advances have been made with the design of self-assembling prodrugs, peptide-drug conjugates, and hydrogels for controlled drug delivery, which represents a new frontier in therapeutics. The development of these materials can potentially impact the challenges associated with drug stability, localization, and side effects. Furthermore, β-sheet peptide materials display promising properties for delivering cell therapy and promoting tissue regeneration and wound healing. However, there is still a lack of fundamental understanding of the peptide material interactions with cells and organisms, particularly the immune system. Exploring the influence of material properties on biological systems will complement our ability to design new systems and guide the selection of existing materials for a particular application. Interdisciplinary collaboration could expedite efforts to establish rules that connect peptide composition, sequence, and structure with material biophysical/mechanical attributes, and cellular/immune responses in vivo. We intended to highlight needed research efforts throughout the review that should help catalyze the translation of academic discoveries through preclinical evaluation and into clinical use.
Acknowledgements
This work was supported by the National Institutes of Health / National Cancer Institute.
Footnotes
Declaration of competing interest
The authors declare that they have no competing financial or personal interest that could have influence this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- [1].Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE, A new era for understanding amyloid structures and disease, Nature Reviews Molecular Cell Biology 19(12) (2018) 755–773. [DOI] [PubMed] [Google Scholar]
- [2].Ke PC, Zhou R, Serpell LC, Riek R, Knowles TPJ, Lashuel HA, Gazit E, Hamley IW, Davis TP, Fändrich M, Otzen DE, Chapman MR, Dobson CM, Eisenberg DS, Mezzenga R, Half a century of amyloids: past, present and future, Chemical Society Reviews 49(15) (2020) 5473–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Otzen D, Riek R, Functional Amyloids, Cold Spring Harbor Perspectives in Biology 11(12) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lee EY, Srinivasan Y, de Anda J, Nicastro LK, Tükel Ç, Wong GCL, Functional Reciprocity of Amyloids and Antimicrobial Peptides: Rethinking the Role of Supramolecular Assembly in Host Defense, Immune Activation, and Inflammation, Frontiers in Immunology 11 (2020) 1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wei G, Su Z, Reynolds NP, Arosio P, Hamley IW, Gazit E, Mezzenga R, Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology, Chemical Society Reviews 46(15) (2017) 4661–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Du X, Zhou J, Shi J, Xu B, Supramolecular Hydrogelators and Hydrogels: From Soft Matter to Molecular Biomaterials, Chemical Reviews 115(24) (2015) 13165–13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prince E, Kumacheva E, Design and applications of man-made biomimetic fibrillar hydrogels, Nature Reviews Materials 4(2) (2019) 99–115. [Google Scholar]
- [8].Wang Y, He X, Bruggeman KF, Gayen B, Tricoli A, Lee WM, Williams RJ, Nisbet DR, Peptide Programmed Hydrogels as Safe Sanctuary Microenvironments for Cell Transplantation, Advanced Functional Materials 30(9) (2020) 1900390. [Google Scholar]
- [9].Yamada Y, Patel NL, Kalen JD, Schneider JP, Design of a Peptide-Based Electronegative Hydrogel for the Direct Encapsulation, 3D Culturing, in Vivo Syringe-Based Delivery, and Long-Term Tissue Engraftment of Cells, ACS Applied Materials & Interfaces 11(38) (2019) 34688–34697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Koch F, Ekat K, Kilian D, Hettich T, Germershaus O, Lang H, Peters K, Kreikemeyer B, A Versatile Biocompatible Antibiotic Delivery System Based on Self-Assembling Peptides with Antimicrobial and Regenerative Potential, Advanced Healthcare Materials 8(13) (2019) 1900167. [DOI] [PubMed] [Google Scholar]
- [11].Karavasili C, Andreadis DA, Katsamenis OL, Panteris E, Anastasiadou P, Kakazanis Z, Zoumpourlis V, Markopoulou CK, Koutsopoulos S, Vizirianakis IS, Fatouros DG, Synergistic Antitumor Potency of a Self-Assembling Peptide Hydrogel for the Local Co-delivery of Doxorubicin and Curcumin in the Treatment of Head and Neck Cancer, Molecular Pharmaceutics 16(6) (2019) 2326–2341. [DOI] [PubMed] [Google Scholar]
- [12].Liu S, Zhao M, Zhou Y, Li L, Wang C, Yuan Y, Li L, Liao G, Bresette W, Chen Y, Cheng J, Lu Y, Liu J, A self-assembling peptide hydrogel-based drug co-delivery platform to improve tissue repair after ischemia-reperfusion injury, Acta Biomaterialia 103 (2020) 102–114. [DOI] [PubMed] [Google Scholar]
- [13].Hainline KM, Gu F, Handley JF, Tian YF, Wu Y, de Wet L, Vander Griend DJ, Collier JH, Self-Assembling Peptide Gels for 3D Prostate Cancer Spheroid Culture, Macromolecular Bioscience 19(1) (2019) 1800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Faroni A, Workman VL, Saiani A, Reid AJ, Self-Assembling Peptide Hydrogel Matrices Improve the Neurotrophic Potential of Human Adipose-Derived Stem Cells, Advanced Healthcare Materials 8(17) (2019) 1900410. [DOI] [PubMed] [Google Scholar]
- [15].Shores LS, Kelly SH, Hainline KM, Suwanpradid J, MacLeod AS, Collier JH, Multifactorial Design of a Supramolecular Peptide Anti-IL-17 Vaccine Toward the Treatment of Psoriasis, Frontiers in Immunology 11 (2020) 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kelly SH, Wu Y, Varadhan AK, Curvino EJ, Chong AS, Collier JH, Enabling sublingual peptide immunization with molecular self-assemblies, Biomaterials 241 (2020) 119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Smith DJ, Brat GA, Medina SH, Tong D, Huang Y, Grahammer J, Furtmüller GJ, Oh BC, Nagy-Smith KJ, Walczak P, Brandacher G, Schneider JP, A multiphase transitioning peptide hydrogel for suturing ultrasmall vessels, Nat Nanotechnol 11(1) (2016) 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen C, Zhang Y, Fei R, Cao C, Wang M, Wang J, Bai J, Cox H, Waigh T, Lu JR, Xu H, Hydrogelation of the Short Self-Assembling Peptide I3QGK Regulated by Transglutaminase and Use for Rapid Hemostasis, ACS Applied Materials & Interfaces 8(28) (2016) 17833–17841. [DOI] [PubMed] [Google Scholar]
- [19].Wu M, Ye Z, Zhu H, Zhao X, Self-Assembling Peptide Nanofibrous Hydrogel on Immediate Hemostasis and Accelerative Osteosis, Biomacromolecules 16(10) (2015) 3112–3118. [DOI] [PubMed] [Google Scholar]
- [20].Luo Z, Wang S, Zhang S, Fabrication of self-assembling D-form peptide nanofiber scaffold d-EAK16 for rapid hemostasis, Biomaterials 32(8) (2011) 2013–2020. [DOI] [PubMed] [Google Scholar]
- [21].Tran KA, Partyka PP, Jin Y, Bouyer J, Fischer I, Galie PA, Vascularization of self-assembled peptide scaffolds for spinal cord injury repair, Acta Biomaterialia 104 (2020) 76–84. [DOI] [PubMed] [Google Scholar]
- [22].Feng Z, Su Q, Zhang C, Huang P, Song H, Dong A, Kong D, Wang W, Bioinspired Nanofibrous Glycopeptide Hydrogel Dressing for Accelerating Wound Healing: A Cytokine-Free, M2-Type Macrophage Polarization Approach, Advanced Functional Materials n/a(n/a) (2020) 2006454. [Google Scholar]
- [23].Hainline KM, Fries CN, Collier JH, Progress Toward the Clinical Translation of Bioinspired Peptide and Protein Assemblies, Advanced Healthcare Materials 7(5) (2018) 1700930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gelain F, Luo Z, Zhang S, Self-Assembling Peptide EAK16 and RADA16 Nanofiber Scaffold Hydrogel, Chemical Reviews 120(24) (2020) 13434–13460. [DOI] [PubMed] [Google Scholar]
- [25].Moore AN, Hartgerink JD, Self-Assembling Multidomain Peptide Nanofibers for Delivery of Bioactive Molecules and Tissue Regeneration, Accounts of Chemical Research 50(4) (2017) 714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rughani RV, Schneider JP, Molecular Design of beta-Hairpin Peptides for Material Construction, MRS Bull 33(5) (2008) 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gazit E, Reductionist Approach in Peptide-Based Nanotechnology, Annual Review of Biochemistry 87(1) (2018) 533–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Levin A, Hakala TA, Schnaider L, Bernardes GJL, Gazit E, Knowles TPJ, Biomimetic peptide self-assembly for functional materials, Nature Reviews Chemistry 4(11) (2020) 615–634. [Google Scholar]
- [29].Lu J-X, Qiang W, Yau W-M, Charles D Schwieters, Stephen C. Meredith, R. Tycko, Molecular Structure of β-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue, Cell 154(6) (2013) 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH, Atomic-Resolution Three-Dimensional Structure of Amyloid β Fibrils Bearing the Osaka Mutation, Angewandte Chemie International Edition 54(1) (2015) 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, Griffin RG, Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils, Journal of the American Chemical Society 138(30) (2016) 9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gremer L, Schölzel D, Schenk C, Reinartz E, Labahn J, Ravelli RBG, Tusche M, Lopez-Iglesias C, Hoyer W, Heise H, Willbold D, Schröder GF, Fibril structure of amyloid-β(1–42) by cryo–electron microscopy, Science 358(6359) (2017) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R, Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril, Proceedings of the National Academy of Sciences 113(34) (2016) E4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y, Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease, Nature Structural & Molecular Biology 22(6) (2015) 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R, 3D structure of Alzheimer's amyloid-β(1–42) fibrils, Proc Natl Acad Sci U S A 102(48) (2005) 17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kollmer M, Close W, Funk L, Rasmussen J, Bsoul A, Schierhorn A, Schmidt M, Sigurdson CJ, Jucker M, Fändrich M, Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue, Nature Communications 10(1) (2019) 4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ghosh U, Thurber KR, Yau W-M, Tycko R, Molecular structure of a prevalent amyloid-β fibril polymorph from Alzheimer's disease brain tissue, Proceedings of the National Academy of Sciences 118(4) (2021) e2023089118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li B, Ge P, Murray KA, Sheth P, Zhang M, Nair G, Sawaya MR, Shin WS, Boyer DR, Ye S, Eisenberg DS, Zhou ZH, Jiang L, Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel, Nature Communications 9(1) (2018) 3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schweighauser M, Shi Y, Tarutani A, Kametani F, Murzin AG, Ghetti B, Matsubara T, Tomita T, Ando T, Hasegawa K, Murayama S, Yoshida M, Hasegawa M, Scheres SHW, Goedert M, Structures of α-synuclein filaments from multiple system atrophy, Nature 585(7825) (2020) 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Guerrero-Ferreira R, Taylor NMI, Arteni A-A, Kumari P, Mona D, Ringler P, Britschgi M, Lauer ME, Makky A, Verasdonck J, Riek R, Melki R, Meier BH, Böckmann A, Bousset L, Stahlberg H, Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy, eLife 8 (2019) e48907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW, Cryo-EM structures of tau filaments from Alzheimer’s disease, Nature 547(7662) (2017) 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, Goedert M, Structures of filaments from Pick’s disease reveal a novel tau protein fold, Nature 561(7721) (2018) 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, Scheres SHW, Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules, Nature 568(7752) (2019) 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R, Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains, Cell 171(3) (2017) 615–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lu J, Cao Q, Hughes MP, Sawaya MR, Boyer DR, Cascio D, Eisenberg DS, CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid, Nature Communications 11(1) (2020) 4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS, Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils, Nature Structural & Molecular Biology 27(7) (2020) 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS, Cryo-EM structures of four polymorphic TDP-43 amyloid cores, Nature Structural & Molecular Biology 26(7) (2019) 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Iadanza MG, Silvers R, Boardman J, Smith HI, Karamanos TK, Debelouchina GT, Su Y, Griffin RG, Ranson NA, Radford SE, The structure of a β2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism, Nature Communications 9(1) (2018) 4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Desiraju GR, Crystal Engineering: From Molecule to Crystal, Journal of the American Chemical Society 135(27) (2013) 9952–9967. [DOI] [PubMed] [Google Scholar]
- [50].Saha S, Mishra MK, Reddy CM, Desiraju GR, From Molecules to Interactions to Crystal Engineering: Mechanical Properties of Organic Solids, Acc Chem Res 51(11) (2018) 2957–2967. [DOI] [PubMed] [Google Scholar]
- [51].Truex NL, Wang Y, Nowick JS, Assembly of Peptides Derived from β-Sheet Regions of β-Amyloid, Journal of the American Chemical Society 138(42) (2016) 13882–13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kreutzer AG, Nowick JS, Elucidating the Structures of Amyloid Oligomers with Macrocyclic beta-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases, Acc Chem Res 51(3) (2018) 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[53]. Nagy-Smith K, Moore E, Schneider J, Tycko R, Molecular structure of monomorphic peptide fibrils within a kinetically trapped hydrogel network, Proceedings of the National Academy of Sciences 112(32) (2015) 9816. "This work utilized solid-state NMR to determine the molecular structure of fibrils comprised of the self-assembling peptide MAX1. The kinetically trapped MAX1 fibrils are monomorphic and showed a double-layered cross-β structure in a Syn/Anti configuration. This is one of the few examples that characterize the molecular structure of peptides in their fibril state."
- [54].Cormier AR, Pang X, Zimmerman MI, Zhou H-X, Paravastu AK, Molecular Structure of RADA16-I Designer Self-Assembling Peptide Nanofibers, ACS Nano 7(9) (2013) 7562–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shen H, Fallas JA, Lynch E, Sheffler W, Parry B, Jannetty N, Decarreau J, Wagenbach M, Vicente JJ, Chen J, Wang L, Dowling Q, Oberdorfer G, Stewart L, Wordeman L, De Yoreo J, Jacobs-Wagner C, Kollman J, Baker D, De novo design of self-assembling helical protein filaments, Science 362(6415) (2018) 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jonnalagadda SVR, Kokotidou C, Orr AA, Fotopoulou E, Henderson KJ, Choi C-H, Lim WT, Choi SJ, Jeong H-K, Mitraki A, Tamamis P, Computational Design of Functional Amyloid Materials with Cesium Binding, Deposition, and Capture Properties, The Journal of Physical Chemistry B 122(30) (2018) 7555–7568. [DOI] [PubMed] [Google Scholar]
- [57].Lee M, Ghosh U, Thurber KR, Kato M, Tycko R, Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS, Nature Communications 11(1) (2020) 5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ji W, Yuan C, Chakraborty P, Gilead S, Yan X, Gazit E, Stoichiometry-controlled secondary structure transition of amyloid-derived supramolecular dipeptide co-assemblies, Communications Chemistry 2(1) (2019) 65. [Google Scholar]
- [59].Wang M, Wang J, Zhou P, Deng J, Zhao Y, Sun Y, Yang W, Wang D, Li Z, Hu X, King SM, Rogers SE, Cox H, Waigh TA, Yang J, Lu JR, Xu H, Nanoribbons self-assembled from short peptides demonstrate the formation of polar zippers between β-sheets, Nature Communications 9(1) (2018) 5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vantomme G, Meijer EW, The construction of supramolecular systems, Science 363(6434) (2019) 1396. [DOI] [PubMed] [Google Scholar]
- [61].Candreva J, Chau E, Aoraha E, Nanda V, Kim JR, Hetero-assembly of a dual β-amyloid variant peptide system, Chemical Communications 54(49) (2018) 6380–6383. [DOI] [PubMed] [Google Scholar]
- [62].Seroski DT, Dong X, Wong KM, Liu R, Shao Q, Paravastu AK, Hall CK, Hudalla GA, Charge guides pathway selection in β-sheet fibrillizing peptide co-assembly, Communications Chemistry 3(1) (2020) 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sahoo JK, VandenBerg MA, Ruiz Bello EE, Nazareth CD, Webber MJ, Electrostatic-driven self-sorting and nanostructure speciation in self-assembling tetrapeptides, Nanoscale 11(35) (2019) 16534–16543. [DOI] [PubMed] [Google Scholar]
- [64].Ji W, Yuan C, Chakraborty P, Makam P, Bera S, Rencus-Lazar S, Li J, Yan X, Gazit E, Coassembly-Induced Transformation of Dipeptide Amyloid-Like Structures into Stimuli-Responsive Supramolecular Materials, ACS Nano 14(6) (2020) 7181–7190. [DOI] [PubMed] [Google Scholar]
- •[65]. Nagy-Smith K, Beltramo PJ, Moore E, Tycko R, Furst EM, Schneider JP, Molecular, Local, and Network-Level Basis for the Enhanced Stiffness of Hydrogel Networks Formed from Coassembled Racemic Peptides: Predictions from Pauling and Corey, ACS Central Science 3(6) (2017) 586–597. " Nagy-smith et al. performed a series of structural and spectroscopy experiments to understand why an enantiomeric coassembled system of L-peptide MAX1 and D-peptide DMAX1 form a more rigid hydrogel in comparison with the individual peptide materials. Using TEM, ssNMR, small-angle neutron scattering, and other spectroscopy analyses, they found that the coassembled system forms an extended heterochiral rippled sheet as predicted in 1953 by Pauling and Corey. This structure allows for the formation of nested hydrophobic interactions that are not present in the enantiomeric pure peptide fibrils."
- [66].Swanekamp RJ, Welch JJ, Nilsson BL, Proteolytic stability of amphipathic peptide hydrogels composed of self-assembled pleated β-sheet or coassembled rippled β-sheet fibrils, Chemical Communications 50(70) (2014) 10133–10136. [DOI] [PubMed] [Google Scholar]
- [67].Dutta S, Foley AR, Warner CJA, Zhang X, Rolandi M, Abrams B, Raskatov JA, Suppression of Oligomer Formation and Formation of Non-Toxic Fibrils upon Addition of Mirror-Image Aβ42 to the Natural l-Enantiomer, Angewandte Chemie International Edition 56(38) (2017) 11506–11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Swanekamp RJ, DiMaio JTM, Bowerman CJ, Nilsson BL, Coassembly of Enantiomeric Amphipathic Peptides into Amyloid-Inspired Rippled β-Sheet Fibrils, Journal of the American Chemical Society 134(12) (2012) 5556–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chakraborty P, Tang Y, Guterman T, Arnon ZA, Yao Y, Wei G, Gazit E, Co-Assembly between Fmoc Diphenylalanine and Diphenylalanine within a 3D Fibrous Viscous Network Confers Atypical Curvature and Branching, Angewandte Chemie International Edition 59(52) (2020) 23731–23739. [DOI] [PubMed] [Google Scholar]
- [70].Marshall LR, Jayachandran M, Lengyel-Zhand Z, Rufo CM, Kriews A, Kim M-C, Korendovych IV, Synergistic Interactions Are Prevalent in Catalytic Amyloids, ChemBioChem 21(18) (2020) 2611–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Nagy KJ, Giano MC, Jin A, Pochan DJ, Schneider JP, Enhanced Mechanical Rigidity of Hydrogels Formed from Enantiomeric Peptide Assemblies, Journal of the American Chemical Society 133(38) (2011) 14975–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Scelsi A, Bochicchio B, Smith A, Workman VL, Castillo Diaz LA, Saiani A, Pepe A, Tuning of hydrogel stiffness using a two-component peptide system for mammalian cell culture, Journal of Biomedical Materials Research Part A 107(3) (2019) 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Raskatov JA, Schneider JP, Nilsson BL, Defining the Landscape of the Pauling-Corey Rippled Sheet: An Orphaned Motif Finding New Homes, Accounts of Chemical Research 54(10) (2021) 2488–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kubota R, Nagao K, Tanaka W, Matsumura R, Aoyama T, Urayama K, Hamachi I, Control of seed formation allows two distinct self-sorting patterns of supramolecular nanofibers, Nature Communications 11(1) (2020) 4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nowak BP, Ravoo BJ, Photoresponsive hybrid hydrogel with a dual network of agarose and a self-assembling peptide, Soft Matter 16(31) (2020) 7299–7304. [DOI] [PubMed] [Google Scholar]
- [76].Ishikawa S, Iijima K, Matsukuma D, Asawa Y, Hoshi K, Osawa S, Otsuka H, Interpenetrating Polymer Network Hydrogels via a One-Pot and in Situ Gelation System Based on Peptide Self-Assembly and Orthogonal Cross-Linking for Tissue Regeneration, Chemistry of Materials 32(6) (2020) 2353–2364. [Google Scholar]
- •[77]. Shao Q, Wong KM, Seroski DT, Wang Y, Liu R, Paravastu AK, Hudalla GA, Hall CK, Anatomy of a selectively coassembled β-sheet peptide nanofiber, Proceedings of the National Academy of Sciences 117(9) (2020) 4710. " Shao et al. characterized the molecular structure of coassemble ionic complementary peptides CATCH (+) and CATCH (−). They demonstrated that these peptides coassemble predominantly in an ABAB-type arrangement using molecular dynamics simulations and NMR techniques. Some deviations of AA and AB type arrangements are also detected. This work is among the firsts to support the assumed alternate structure of coassembly systems experimentally and is a guide to characterize peptide fibrils at a molecular level."
- [78].Wong KM, Wang Y, Seroski DT, Larkin GE, Mehta AK, Hudalla GA, Hall CK, Paravastu AK, Molecular complementarity and structural heterogeneity within co-assembled peptide β-sheet nanofibers, Nanoscale 12(7) (2020) 4506–4518. [DOI] [PubMed] [Google Scholar]
- [79].Mouw JK, Ou G, Weaver VM, Extracellular matrix assembly: a multiscale deconstruction, Nature Reviews Molecular Cell Biology 15(12) (2014) 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Restuccia A, Seroski DT, Kelley KL, O’Bryan CS, Kurian JJ, Knox KR, Farhadi SA, Angelini TE, Hudalla GA, Hierarchical self-assembly and emergent function of densely glycosylated peptide nanofibers, Communications Chemistry 2(1) (2019) 53. [Google Scholar]
- [81].Clover TM, O’Neill CL, Appavu R, Lokhande G, Gaharwar AK, Posey AE, White MA, Rudra JS, Self-Assembly of Block Heterochiral Peptides into Helical Tapes, Journal of the American Chemical Society 142(47) (2020) 19809–19813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[82]. Chen J, Yan K, Xiong S, Wei T, Wu X, Chu PK, Controlled fiberization of dipeptide in merging phases leads to collagen-level strength and opto/electric mechanofunctionalities, Biomaterials 208 (2019) 1–7. " This work describes a method for forming self-assembled diphenylalanine long aligned fiber bundles using a phase merging process. In this process, diphenylalanine monomers are dissolved in hexafluoroisopropanol, and water is deposed on top of the organic layer. Slow mixing of both phases results in the controlled assembly of nanofibrils that can be pulled out of the solution while forming long aligned fiber bundles."
- [83].Schiapparelli P, Zhang P, Lara-Velazquez M, Guerrero-Cazares H, Lin R, Su H, Chakroun RW, Tusa M, Quiñones-Hinojosa A, Cui H, Self-assembling and self-formulating prodrug hydrogelator extends survival in a glioblastoma resection and recurrence model, Journal of Controlled Release 319 (2020) 311–321. [DOI] [PubMed] [Google Scholar]
- [84].Leach DG, Newton JM, Florez MA, Lopez-Silva TL, Jones AA, Young S, Sikora AG, Hartgerink JD, Drug-Mimicking Nanofibrous Peptide Hydrogel for Inhibition of Inducible Nitric Oxide Synthase, ACS Biomaterials Science & Engineering 5(12) (2019) 6755–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fu W, Farhadi Sabet Z, Liu J, You M, Zhou H, Wang Y, Gao Y, Li J, Ma X, Chen C, Metal ions modulation of the self-assembly of short peptide conjugated nonsteroidal anti-inflammatory drugs (NSAIDs), Nanoscale 12(14) (2020) 7960–7968. [DOI] [PubMed] [Google Scholar]
- [86].Mei L, He S, Liu Z, Xu K, Zhong W, Co-assembled supramolecular hydrogels of doxorubicin and indomethacin-derived peptide conjugates for synergistic inhibition of cancer cell growth, Chemical Communications 55(30) (2019) 4411–4414. [DOI] [PubMed] [Google Scholar]
- ••[87]. Feng Z, Wang H, Xu B, Instructed Assembly of Peptides for Intracellular Enzyme Sequestration, Journal of the American Chemical Society 140(48) (2018) 16433–16437. " Enzyme-instructed self-assembly was used to sequestrate enzymes intracellularly in the endoplasmic reticulum selectively. The nonsteroidal anti-inflammatory drug, Naproxen, was conjugated to a phosphorylated self-assembling peptide precursor. Naproxen serves as a binding moiety for cyclooxygenase-2, while the phosphorylated peptide is a substrate for phosphatases. This design results in the selective concentration of specific enzymes in the ER and has potential use in therapeutics.•
- [88].Wu C, Liu J, Tang X, Zhai Z, Xu K, Zhong W, An enzyme-assisted self-delivery system of lonidamine–peptide conjugates for selectively killing cancer cells, Chemical Communications 55(98) (2019) 14852–14855. [DOI] [PubMed] [Google Scholar]
- [89].Yang C, Hu F, Zhang X, Ren C, Huang F, Liu J, Zhang Y, Yang L, Gao Y, Liu B, Liu J, Combating bacterial infection by in situ self-assembly of AIEgen-peptide conjugate, Biomaterials 244 (2020) 119972. [DOI] [PubMed] [Google Scholar]
- [90].Guo Q, Liu Y, Mu G, Yang L, Wang W, Liu J, Liu J, A peptide-drug hydrogel to enhance the anti-cancer activity of chlorambucil, Biomater Sci 8(20) (2020) 5638–5646. [DOI] [PubMed] [Google Scholar]
- [91].Yang Z, Gu H, Fu D, Gao P, Lam JK, Xu B, Enzymatic Formation of Supramolecular Hydrogels, Advanced Materials 16(16) (2004) 1440–1444. [Google Scholar]
- [92].Yang Z, Liang G, Xu B, Enzymatic Hydrogelation of Small Molecules, Accounts of Chemical Research 41(2) (2008) 315–326. [DOI] [PubMed] [Google Scholar]
- [93].Zhou J, Du X, Yamagata N, Xu B, Enzyme-Instructed Self-Assembly of Small d-Peptides as a Multiple-Step Process for Selectively Killing Cancer Cells, Journal of the American Chemical Society 138(11) (2016) 3813–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ""[94]. Yang L, Peltier R, Zhang M, Song D, Huang H, Chen G, Chen Y, Zhou F, Hao Q, Bian L, He M-I, Wang Z, Hu Y, Sun H, Desuccinylation-Triggered Peptide Self-Assembly: Live Cell Imaging of SIRT5 Activity and Mitochondrial Activity Modulation, Journal of the American Chemical Society 142(42) (2020) 18150–18159. " This work utilized the enzyme instructed self-assembly strategy to program the self-assembly of di- and tri-phenylalanine peptide derivatives in the mitochondria of living cells. Peptides contained a succinyl group and a fluorescent module are uptaken by cells, and the mitochondria-localized enzyme, SIRT5, cleaves the succinyl group inducing assembly into nanofibers. This strategy can be used to study the activity of SIRT5 by imaging and boost cell response to anticancer drugs."
- [95].Li J, Shi K, Sabet ZF, Fu W, Zhou H, Xu S, Liu T, You M, Cao M, Xu M, Cui X, Hu B, Liu Y, Chen C, New power of self-assembling carbonic anhydrase inhibitor: Short peptide–constructed nanofibers inspire hypoxic cancer therapy, Science Advances 5(9) (2019) eaax0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].He H, Liu S, Wu D, Xu B, Enzymatically Formed Peptide Assemblies Sequestrate Proteins and Relocate Inhibitors to Selectively Kill Cancer Cells, Angewandte Chemie International Edition 59(38) (2020) 16445–16450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang L, Lv Y, Li C, Yang G, Fu B, Peng Q, Jian L, Hou D, Wang J, Zhao C, Yang P, Zhang K, Wang L, Wang Z, Wang H, Xu W, Transformable Dual-Inhibition System Effectively Suppresses Renal Cancer Metastasis through Blocking Endothelial Cells and Cancer Stem Cells, Small 16(40) (2020) 2004548. [DOI] [PubMed] [Google Scholar]
- [98].Xiao W-Y, Wang Y, An H-W, Hou D, Mamuti M, Wang M-D, Wang J, Xu W, Hu L, Wang H, Click Reaction-Assisted Peptide Immune Checkpoint Blockade for Solid Tumor Treatment, ACS Applied Materials & Interfaces 12(36) (2020) 40042–40051. [DOI] [PubMed] [Google Scholar]
- [99].Leach DG, Dharmaraj N, Lopez-Silva TL, Venzor JR, Pogostin BH, Sikora AG, Hartgerink JD, Young S, Biomaterial-Facilitated Immunotherapy for Established Oral Cancers, ACS Biomaterials Science & Engineering (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Miller SE, Yamada Y, Patel N, Suárez E, Andrews C, Tau S, Luke BT, Cachau RE, Schneider JP, Electrostatically Driven Guanidinium Interaction Domains that Control Hydrogel-Mediated Protein Delivery In Vivo, ACS Central Science 5(11) (2019) 1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Majumder P, Zhang Y, Iglesias M, Fan L, Kelley JA, Andrews C, Patel N, Stagno JR, Oh BC, Furtmüller GJ, Lai CC, Wang Y-X, Brandacher G, Raimondi G, Schneider JP, Multiphase Assembly of Small Molecule Microcrystalline Peptide Hydrogel Allows Immunomodulatory Combination Therapy for Long-Term Heart Transplant Survival, Small 16(38) (2020) 2002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Dou X-Q, Feng C-L, Amino Acids and Peptide-Based Supramolecular Hydrogels for Three-Dimensional Cell Culture, Advanced Materials 29(16) (2017) 1604062. [DOI] [PubMed] [Google Scholar]
- [103].Tsutsumi H, Matsubara D, Mihara H, Functionalization of self-assembling peptide materials using molecular recognition of supramolecular peptide nanofibers, Polymer Journal 52(8) (2020) 913–922. [Google Scholar]
- [104].Ashworth JC, Thompson JL, James JR, Slater CE, Pijuan-Galitó S, Lis-Slimak K, Holley RJ, Meade KA, Thompson A, Arkill KP, Tassieri M, Wright AJ, Farnie G, Merry CLR, Peptide gels of fully-defined composition and mechanics for probing cell-cell and cell-matrix interactions in vitro, Matrix Biology 85-86 (2020) 15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[105]. Yamada Y, Fichman G, Schneider JP, Serum Protein Adsorption Modulates the Toxicity of Highly Positively Charged Hydrogel Surfaces, ACS Applied Materials & Interfaces (2021). " The authors demonstrate that the cytotoxicity of highly positively charged peptide hydrogels is eliminated by the adsorption of proteins present in serum-containing media. This study recognizes the importance of protein adsorption in peptide materials."
- [106].Lopez-Silva TL, Leach DG, Li IC, Wang X, Hartgerink JD, Self-Assembling Multidomain Peptides: Design and Characterization of Neutral Peptide-Based Materials with pH and Ionic Strength Independent Self-Assembly, ACS Biomaterials Science & Engineering 5(2) (2019) 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Tang JD, Mura C, Lampe KJ, Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engineering, Journal of the American Chemical Society 141(12) (2019) 4886–4899. [DOI] [PubMed] [Google Scholar]
- [108].Jayawarna V, Richardson SM, Hirst AR, Hodson NW, Saiani A, Gough JE, Ulijn RV, Introducing chemical functionality in Fmoc-peptide gels for cell culture, Acta Biomaterialia 5(3) (2009) 934–943. [DOI] [PubMed] [Google Scholar]
- [109].Lopez-Silva TL, Cristobal CD, Edwin Lai CS, Leyva-Aranda V, Lee HK, Hartgerink JD, Self-assembling multidomain peptide hydrogels accelerate peripheral nerve regeneration after crush injury, Biomaterials 265 (2021) 120401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Choi ML, Gandhi S, Crucial role of protein oligomerization in the pathogenesis of Alzheimer's and Parkinson's diseases, The FEBS Journal 285(19) (2018) 3631–3644. [DOI] [PubMed] [Google Scholar]
- [111].Bieschke J, Herbst M, Wiglenda T, Friedrich RP, Boeddrich A, Schiele F, Kleckers D, Lopez del Amo JM, Grüning BA, Wang Q, Schmidt MR, Lurz R, Anwyl R, Schnoegl S, Fändrich M, Frank RF, Reif B, Günther S, Walsh DM, Wanker EE, Small-molecule conversion of toxic oligomers to nontoxic β-sheet–rich amyloid fibrils, Nature Chemical Biology 8(1) (2012) 93–101. [DOI] [PubMed] [Google Scholar]
- [112].Wobst HJ, Sharma A, Diamond MI, Wanker EE, Bieschke J, The green tea polyphenol (−)-epigallocatechin gallate prevents the aggregation of tau protein into toxic oligomers at substoichiometric ratios, FEBS Letters 589(1) (2015) 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sonzini S, Stanyon HF, Scherman OA, Decreasing amyloid toxicity through an increased rate of aggregation, Physical Chemistry Chemical Physics 19(2) (2017) 1458–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Díaz-Caballero M, Navarro S, Ventura S, Soluble Assemblies in the Fibrillation Pathway of Prion-Inspired Artificial Functional Amyloids are Highly Cytotoxic, Biomacromolecules 21(6) (2020) 2334–2345. [DOI] [PubMed] [Google Scholar]
- •[115]. Touve MA, Carlini AS, Gianneschi NC, Self-assembling peptides imaged by correlated liquid cell transmission electron microscopy and MALDI-imaging mass spectrometry, Nature Communications 10(1) (2019) 4837. 'This study combined electron microscopy (LCTEM) with MALDI-imaging mass spectrometry to observe the peptide self-assembly process induced by enzymatic activity or chemical reduction. The focus of this work is analyzing the damage that the electron beam can induce in the peptide species and optimize a methodology for accurate visualization of peptide assembly structures. The development of these techniques will facilitate the mechanistic study of assembling peptides."
- ••[116]. Jekhmane S, Prachar M, Pugliese R, Fontana F, Medeiros-Silva J, Gelain F, Weingarth M, Design Parameters of Tissue-Engineering Scaffolds at the Atomic Scale, Angewandte Chemie International Edition 58(47) (2019) 16943–16951. " Using solid-state NMR and Molecular dynamics simulations, this study analyzes mechanical properties' correlation to the homogeneity and self-assembly degree of several peptide hydrogels. NMR allowed the differentiation between peptides with mobile components that do not contribute to fiber formation and peptides that form only rigid fibrillar assemblies. The hydrogel's properties at the atomic level correlate with their performance as scaffolds for neural stem cells."
- [117].Gelain F, Silva D, Caprini A, Taraballi F, Natalello A, Villa O, Nam KT, Zuckermann RN, Doglia SM, Vescovi A, BMHP1-Derived Self-Assembling Peptides: Hierarchically Assembled Structures with Self-Healing Propensity and Potential for Tissue Engineering Applications, ACS Nano 5(3) (2011) 1845–1859. [DOI] [PubMed] [Google Scholar]
- [118].Yin H, Strunz F, Yan Z, Lu J, Brochhausen C, Kiderlen S, Clausen-Schaumann H, Wang X, Gomes ME, Alt V, Docheva D, Three-dimensional self-assembling nanofiber matrix rejuvenates aged/degenerative human tendon stem/progenitor cells, Biomaterials 236 (2020) 119802. [DOI] [PubMed] [Google Scholar]
- [119].Chung L, Maestas DR, Housseau F, Elisseeff JH, Key players in the immune response to biomaterial scaffolds for regenerative medicine, Advanced Drug Delivery Reviews 114 (2017) 184–192. [DOI] [PubMed] [Google Scholar]
- [120].Dellacherie MO, Seo BR, Mooney DJ, Macroscale biomaterials strategies for local immunomodulation, Nature Reviews Materials 4(6) (2019) 379–397. [Google Scholar]
- [121].Hotaling NA, Tang L, Irvine DJ, Babensee JE, Biomaterial Strategies for Immunomodulation, Annual Review of Biomedical Engineering 17(1) (2015) 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lopez-Silva TL, Leach DG, Azares A, Li IC, Woodside DG, Hartgerink JD, Chemical functionality of multidomain peptide hydrogels governs early host immune response, Biomaterials 231 (2020) 119667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Leach DG, Dharmaraj N, Piotrowski SL, Lopez-Silva TL, Lei YL, Sikora AG, Young S, Hartgerink JD, STINGel: Controlled release of a cyclic dinucleotide for enhanced cancer immunotherapy, Biomaterials 163 (2018) 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Fries CN, Curvino EJ, Chen J-L, Permar SR, Fouda GG, Collier JH, Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health, Nat Nanotechnol (2020). [DOI] [PubMed] [Google Scholar]
- •[125]. Si Y, Tian Q, Zhao F, Kelly SH, Shores LS, Camacho DF, Sperling AI, Andrade MS, Collier JH, Chong AS, Adjuvant-free nanofiber vaccine induces in situ lung dendritic cell activation and TH17 responses, Science Advances 6(32) (2020) eaba0995. " Self-assembling peptide Q11 bearing a pEα peptide antigen was used as an intranasal vaccine without exogenous adjuvants and minimal inflammation. The authors found that the peptide nanofiber length is relevant for a successful presentation by dendritic cells and promoting immune activation. This work expands on the use of β-sheet fibrils as vaccines and their enhanced antigen presentation mechanisms."
- [126].Yang P, Song H, Qin Y, Huang P, Zhang C, Kong D, Wang W, Engineering Dendritic-Cell-Based Vaccines and PD-1 Blockade in Self-Assembled Peptide Nanofibrous Hydrogel to Amplify Antitumor T-Cell Immunity, Nano Letters 18(7) (2018) 4377–4385. [DOI] [PubMed] [Google Scholar]
- [127].Wen Y, Waltman A, Han H, Collier JH, Switching the Immunogenicity of Peptide Assemblies Using Surface Properties, ACS Nano 10(10) (2016) 9274–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wu Y, Kelly SH, Sanchez-Perez L, Sampson JH, Collier JH, Comparative study of α-helical and β-sheet self-assembled peptide nanofiber vaccine platforms: influence of integrated T-cell epitopes, Biomaterials Science 8(12) (2020) 3522–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]