Abstract
Upf1p, Nmd2p, and Upf3p regulate the degradation of yeast mRNAs that contain premature translation termination codons. These proteins also appear to regulate the fidelity of termination, allowing translational suppression in their absence. Here, we have devised a novel quantitative assay for translational suppression, based on a nonsense allele of the CAN1 gene (can1-100), and used it to determine the regulatory roles of the UPF/NMD gene products. Deletion of UPF1, NMD2, or UPF3 stabilized the can1-100 transcript and promoted can1-100 nonsense suppression. Changes in mRNA levels were not the basis of suppression, however, since deletion of DCP1 or XRN1 or high-copy-number can1-100 expression in wild-type cells caused an increase in mRNA abundance similar to that obtained in upf/nmd cells but did not result in comparable suppression. can1-100 suppression was highest in cells harboring a deletion of UPF1, and overexpression of UPF1 in cells with individual or multiple upf/nmd mutations lowered the level of nonsense suppression without affecting the abundance of the can1-100 mRNA. Our findings indicate that Nmd2p and Upf3p regulate Upf1p activity and that Upf1p plays a critical role in promoting termination fidelity that is independent of its role in regulating mRNA decay. Consistent with these relationships, Upf1p, Nmd2p, and Upf3p were shown to be present at 1,600, 160, and 80 molecules per cell, levels that underscored the importance of Upf1p but minimized the likelihood that these proteins were associated with all ribosomes or that they functioned as a stoichiometric complex.
The pathways of gene expression include intricate mechanisms that safeguard against the accumulation of aberrant transcripts and proteins (6, 13, 14, 19, 30, 62). In addition to their protective functions, these pathways also contribute additional regulatory facility and complexity (57). The phenomenon of nonsense-mediated mRNA decay (NMD) exemplifies such mechanisms. NMD minimizes the synthesis of truncated polypeptides by eliminating mRNAs containing premature nonsense codons within their protein coding regions (19, 29, 39, 45, 46, 49, 51). NMD also provides the cell with a pathway for the selective degradation of a subset of mRNAs whose coding regions could be considered “normal” (37, 57).
In the yeast Saccharomyces cerevisiae, the rapid degradation of nonsense-containing mRNAs proceeds from deadenylation-independent removal of the 5′ cap by the decapping enzyme Dcp1p to 5′→3′ digestion of the remainder of the mRNA by the exoribonuclease Xrn1p (4, 5, 17, 27, 33, 40). Three additional factors are also essential for NMD in yeast: Upf1p, Nmd2p (Upf2p), and Upf3p (7, 20, 22, 34, 35). Mutations in the UPF1, NMD2, or UPF3 genes lead to the stabilization of mRNAs containing premature nonsense codons without affecting the rates of decay of most wild-type mRNAs. Since single or multiple mutations within UPF1, NMD2, or UPF3 yield similar decay phenotypes, all three gene products have been considered to be functionally related and to act in a common pathway (22). Substantial support for this conclusion has been derived from protein-protein interaction analyses (11, 22).
A more detailed understanding of the functions of Upf1p, Nmd2p, and Upf3p has been sought in several ways. Consistent with their roles in responding to aberrant translation, all three proteins have been shown to localize to the cytoplasm and to associate with polyribosomes (3, 38, 46). Upf1p is a 109-kDa protein that contains two putative zinc finger domains near its amino terminus and harbors seven motifs characteristic of RNA-DNA helicase superfamily I (1, 31). In vitro studies demonstrated that purified Upf1p has the ability to bind nucleic acids and that its ATPase and helicase activities are dependent upon nucleic acid binding (10, 60). Upf1p interacts with the polypeptide release factors Sup35p and Sup45p (11) and utilizes the same N-terminal zinc finger region for Nmd2p interaction, intramolecular interaction, and homodimerization (F. He and A. Jacobson, unpublished data). Little is known about the biochemical activities of the 127-kDa Nmd2p and 45-kDa Upf3p polypeptides.
The involvement of the UPF/NMD genes in regulating the stability of mRNAs containing premature nonsense codons and the interactions of Upf1p with Nmd2p, Upf3p, Sup35p, and Sup45p suggest that UPF1, NMD2, and UPF3 may all be regulators of translation termination and/or fidelity. Consistent with this notion are experiments which indicate that deletion of these genes leads to nonsense suppression (36, 58), allosuppression (9), and enhancement of programmed ribosomal frameshifting (8, 52). To investigate further the possible regulatory roles of Upf1p, Nmd2p, and Upf3p, we devised an assay that quantitatively monitors the effects of upf/nmd mutations on suppression of the can1-100 nonsense allele. Deletion of the genes encoding each of these factors was found to stabilize the can1-100 transcript and promote nonsense suppression. Strains harboring a deletion of UPF1 showed the highest levels of suppression, and overexpression of UPF1 in upf/nmd strains lowered the levels of nonsense suppression significantly without altering the steady-state levels of the can1-100 mRNA. These data and determinations of the abundance of all three factors indicate that Upf1p plays a critical role in regulating the efficiency of translation termination and that Nmd2p and Upf3p, in turn, regulate Upf1p activity.
MATERIALS AND METHODS
Strains, plasmids, and general methods.
The isogenic yeast strains used in this study are listed in Table 1. Preparation of standard yeast media and cell culturing were done as described by Rose et al. (50). Transformation of yeast strains was done by the rapid method described by Soni et al. (55). DNA manipulations were performed according to standard techniques (53). All PCR amplifications were performed with Taq DNA polymerase (61) and confirmed, where appropriate, by DNA sequencing using the method described by Sanger et al. (54). The can1-100 allele (28), characterized in this study by DNA sequencing (see Results), was recreated in a YEp24-CAN1 high-copy-number plasmid and a pRIP-CAN1 single-copy plasmid by PCR mutagenesis. CAN1 containing sequences that comprised a 3′ triple-hemagglutinin (HA) epitope tag was obtained from Duane Jenness. The 3′-HA-tagged can1-100 allele was constructed by inserting a SalI-EagI HA-containing fragment into the YEp24-can1-100 plasmid digested with the same enzymes. Plasmid DNAs were prepared from Escherichia coli DH5α
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| HFY1200 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 | 20 |
| HFY870 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::HIS3 NMD2 UPF3 | 22 |
| HFY1300 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 nmd2::HIS3 UPF3 | 20 |
| HFY861 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 upf3::HIS3 | 22 |
| HFY3000 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::URA3 nmd2::HIS3 UPF3 | 20 |
| HFY872 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::URA3 NMD2 upf3::HIS3 | 22 |
| HFY874 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 nmd2::URA3 upf3::HIS3 | 22 |
| HFY883 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::LEU2 nmd2::URA3 upf3::HIS3 | 22 |
RNA extraction and Northern blot analysis.
RNA was isolated using the hot phenol method as described by Herrick et al. (24). Aliquots (20 μg) of each RNA sample were analyzed by Northern blotting using radiolabeled probes prepared by random priming (12). mRNA steady-state levels were determined by quantitating Northern blots with a Bio-Rad Molecular Imager. The DNA probes used to detect specific transcripts included CYH2 (a 600-bp EcoRI-HindIII fragment from pGEM4Z-CYH2 which hybridizes to both the pre-mRNA and the mRNA) (24), CAN1 (a 1-kb EcoRI-SalI fragment from YEp24-CAN1), and SCR1 (a 400-bp fragment amplified from yeast genomic DNA using oligonucleotides SCR1-1 [5′-AGGCTGTAATGGCTTTCTGGTGGGATGGGA-3′] and SCR1-2 [5′-GATATGTGCTATCCCGGCCGCCTCCATCAC-3′]). Immunoprecipitation of capped mRNAs was performed as described by Muhlrad et al. (40) using polyclonal anti-m7G antibodies generously provided by Elsebet Lund.
Protein gels, Western blots, and antibodies.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described by Laemmli (32). Gels were electroblotted to Immobilon-P membranes (Millipore) under conditions recommended by the manufacturer. The binding conditions used for antibodies were as described by Harlow and Lane (18). Detection was enhanced by chemiluminescence with an ECL kit from Amersham Corp. Western blots were quantitated by densitometry or by Fluor-S (Bio-Rad) scanning of films exposed for different lengths of time. The anti-HA antibody (12CA5) used for Western blotting was obtained from Boehringer Mannheim Biochemicals.
Purification of recombinant GST-Upf1p and GST-Nmd2p.
Extraction steps were carried out at between 0 and 4°C. All buffers included 0.1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, and the protease inhibitors bestatin (0.35 μg/ml), pepstatin (0.4 μg/ml), leupeptin (0.5 μg/ml), and benzamidine (20 μg/ml). Cell pellets were resuspended in 4 volumes of T(50) buffer (30 mM Tris-HCl [pH 7.9] 2 mM EDTA, 5% glycerol, 10 mM MgCl2, 50 mM KCl) per g of cell wet weight and lysed with a French press (cell pressure, 20,000 lb/in2). Lysates were cleared by centrifugation at 30,000 × g. The pellet was resuspended in denaturing buffer (6 M urea, 50 mM Tris-HCl [pH 7.9], 1 mM EDTA, 8 mM DTT), vortexed vigorously, homogenized with a B pestle, and centrifuged at 30,000 × g. Chromatography steps were carried out at room temperature. The supernatant was dialyzed against a buffer containing 50 mM Tris-HCl [pH 7.9], 1 mM EDTA, 1 mM DTT, and 20% glycerol. Extracts were bound in batches to glutathione-agarose (Sigma) previously equilibrated in T(50) buffer. After binding for 10 min on a platform shaker, the resin was washed three times with the same buffer. The resin was then transferred to a small column, and the protein was eluted with 10 column volumes of T(50) buffer containing 10 mM glutathione (Sigma). The purity of the protein was assessed by sodium dodecyl sulfate-polyacrylamide electrophoresis and staining with Coomassie blue R-250. Glutathione S-transferase (GST)–Upf1p (residues 876 to 971) was greater than 99% pure and was the only protein detected, while GST-Nmd2p was greater than 90% pure, with the majority of the contamination coming from proteolysis.
Quantitation of mRNA decay factors.
The relative abundance of Upf1p, Nmd2p, and Upf3p was determined by comparing the Western blot band intensities of the factors present in crude cell extracts to those of specific standards. For Upf1p, purified recombinant GST-Upf1p (residues 876 to 971) was used as a standard; for Nmd2p, purified recombinant Nmd2p was used as a standard; and for Upf3p, cells bearing an HA-NMD2 allele were used as a standard. For Western blotting, aliquots of crude cell extracts equivalent to 1 ml of cells at an optical density at 600 nm (OD600) of 0.2 were loaded onto polyacrylamide gels. The number of cells in each aliquot was determined by serially diluting and plating the cultures.
can1-100 nonsense suppression assay.
Multiple independent isolates of yeast strains to be assayed were grown in selective liquid media to mid-log phase (OD600 = 0.5 to 0.7). Samples from these cultures were serially diluted (1:10) four times, and aliquots (10 μl) of the four dilutions were spotted on SC-arg plates containing 0 to 500 μg of canavanine per ml. The final aliquots, used as the principal indicators of canavanine sensitivity, each contained approximately 100 cells. Growth on plates, monitored after incubation at 30°C for 2 days, yielded reproducible results for each strain.
Arginine uptake assay.
The arginine uptake assay was adopted from that previously described by Opekarova and Kubin (43). Yeast cultures were grown to mid-log phase (OD600 = 0.5 to 0.7) at 30°C in SC-arg medium and then supplemented with 50 mM l-arginine containing 5 μCi of l-[3H]arginine (Amersham). Aliquots of the cultures were then removed at specific intervals, diluted in 2 ml of 100 mM LiCl, filtered on GF/C filters (Whatman), and washed with 2 ml of water. Radioactive arginine associated with each filter was determined by scintillation counting.
RESULTS
The can1-100 transcript is a substrate for NMD.
To address the roles of UPF1, NMD2, and UPF3 in translation termination, we devised a quantitative assay for nonsense suppression, i.e., readthrough of a premature termination codon. This assay exploited the yeast CAN1 gene, which encodes a high-affinity permease (Can1p) responsible for the transport of arginine into cells (26). Previous studies indicated that a can1 allele, can1-100, was attributable to a nonsense mutation because it could be suppressed in strains containing an ochre suppressor tRNA (28). We confirmed this conclusion by sequence analysis of the can1-100 allele, identifying a single A-to-T mutation that results in the substitution of a lysine codon at position 47 of the CAN1 coding region with a UAA codon (data not shown).
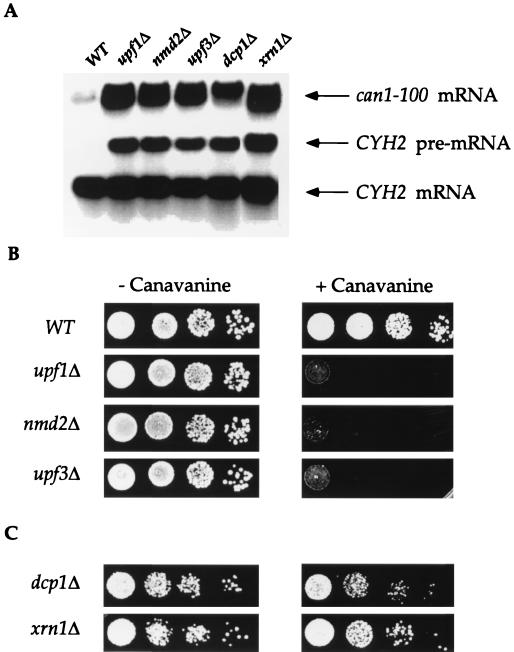
The occurrence of a premature termination codon in the can1-100 mRNA led us to predict that it would be a substrate for NMD. To test this possibility, single deletions of UPF1, NMD2, or UPF3 were constructed in yeast strains that harbored the can1-100 allele, and the effects of these mutations on the abundance of the can1-100 transcript were examined. Northern analyses of mRNA steady-state levels demonstrated that the can1-100 transcript was approximately fourfold more abundant in upf/nmd cells than in the isogenic UPF/NMD strain (Fig. 1A). Likewise, deletion of genes encoding general factors involved in mRNA decay (i.e., DCP1 and XRN1) also promoted a fourfold increase in can1-100 transcript abundance (Fig. 1A). These differences in mRNA abundance were consistent with the respective differences in the rates of decay of the CAN1 and can1-100 mRNAs in wild-type cells (half-lives of 8 and 2 min, respectively; data not shown). As a control for the experiments shown in Fig. 1A, the abundance of an endogenous substrate of the NMD pathway (19) was monitored. As expected, this experiment showed that the CYH2 pre-mRNA was barely detectable in wild-type cells and was abundant in all of the mutants. These results indicate that the can1-100 mRNA requires Upf1p, Nmd2p, Upf3p, Dcp1p, and Xrn1p for its degradation and that it is thus a typical substrate for NMD.
FIG. 1.
Deletion of UPF1, NMD2, or UPF3 stabilizes the can1-100 transcript and promotes nonsense suppression. (A) Deletion mutants that inactivate NMD stabilize the can1-100 transcript. Total RNA isolated from yeast strains with the indicated UPF/NMD genotypes was analyzed by Northern blotting with DNA probes that detected the can1-100 and CYH2 transcripts. WT, wild type. (B) Deletion of UPF1, NMD2, or UPF3 leads to a canavanine-sensitive phenotype. Aliquots (10 μl) of each of four 1:10 dilutions of liquid cultures of each yeast strain were spotted on SC-arg plates containing either 0 or 100 μg of canavanine per ml (− Canavanine or + Canavanine, respectively) and grown at 30°C for 2 days. (C) Deletion of DCP1 or XRN1 does not suppress the can1-100 mutation. Aliquots of serial 1:10 dilutions of each yeast strain were spotted on plates without or with canavanine as in panel B. Because these two mutants had slow doubling times, growth comparable to that of wild-type cells was obtained by maintaining the xm1Δ strain at 30°C for 3 days and the dcp1Δ strain at 30°C for 4 days.
Quantitative assay for nonsense suppression.
Mutations in the UPF1, NMD2, or UPF3 genes have been found to lead not only to increased abundance of substrate mRNAs but also to suppression of certain nonsense alleles, including leu2-2 and tyr7-1 (36, 58). To investigate nonsense suppression of the can1-100 allele, we took advantage of the observation that canavanine, a toxic arginine analog, is also transported into cells via Can1p (15). can1-100 cells are thus phenotypically canavanine resistant, and sensitivity to canavanine is indicative of can1-100 suppression.
Figure 1B illustrates the canavanine resistance of can1-100 cells and demonstrates that deletion of UPF1, NMD2, or UPF3 results in a canavanine-sensitive phenotype when these cells are grown on media containing 100 μg of canavanine per ml. Although deletion of DCP1 and XRN1 led to can1-100 mRNA stabilization comparable to that seen in upf1Δ, nmd2Δ, or upf3Δ mutants (Fig. 1A), strains with the former deletions did not exhibit canavanine sensitivity (Fig. 1C). These results indicate that deletion of any of the UPF/NMD genes allows for suppression of the can1-100 nonsense mutation and that increased mRNA abundance alone is not sufficient to promote suppression (see below).
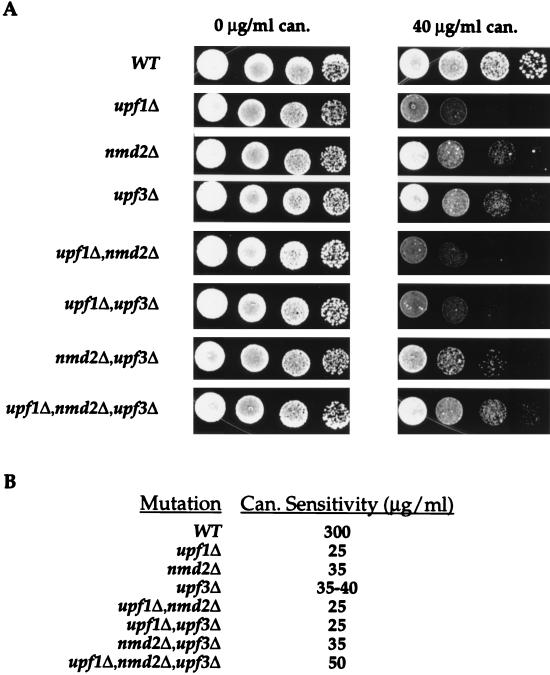
To quantitate the extent of nonsense suppression in the different mutant strains, they were grown on plates containing increasing amounts of canavanine, and the concentration at which each strain exhibited a canavanine-sensitive phenotype was determined. In this assay, canavanine sensitivity is defined as the minimum concentration of canavanine required to kill all cells at the end point of a serial dilution, i.e., approximately 100 cells. These experiments demonstrated that deletion of UPF1, NMD2, or UPF3 promoted different extents of can1-100 suppression. For example, Fig. 2A shows that 40 μg of canavanine per ml was sufficient to kill upf1Δ cells but was only partially toxic to comparable numbers of nmd2Δ or upf3Δ cells. Similar assays consistently demonstrated that the highest levels of nonsense suppression occurred in upf1Δ cells, which exhibited 12-fold greater sensitivity to canavanine than the isogenic wild-type strain (Fig. 2B). Suppression was found to be lower in nmd2Δ and upf3Δ cells, which exhibited 1.5-fold less sensitivity than upf1Δ cells (Fig. 2). Although the canavanine sensitivities of the nmd2Δ and upf3Δ strains were almost identical, subtle differences were detected which indicated that the nmd2Δ mutation was a slightly more effective suppressor than the upf3Δ mutation (Fig. 2).
FIG. 2.
Deletion of UPF1 promotes higher levels of can1-100 nonsense suppression than deletion of NMD2 or UPF3. (A) Growth of yeast strains with different UPF/NMD genotypes on SC-arg plates containing either 0 or 40 μg of canavanine (can.) per ml. Cells were grown for 2 days at 30°C. WT, wild type. (B) Canavanine sensitivities of different yeast strains. Suppression assays analogous to those shown in panel A were used to determine the minimum concentration of canavanine required to kill approximately 100 cells of the respective yeast strains (Can. Sensitivity) after 2 days of growth at 30°C.
Accumulation of functional Can1p correlates with nonsense suppression of can1-100.
To ensure that the respective differences in canavanine sensitivity reflected comparable changes in the extent of synthesis of functional Can1p, arginine permease activities were determined by monitoring the rate of uptake of [3H]arginine in wild-type and mutant cells. Consistent with the suppression assays of Fig. 1 and 2, these experiments demonstrated that deletion of UPF1, NMD2, or UPF3 allowed for enhanced transport of arginine (Fig. 3A).
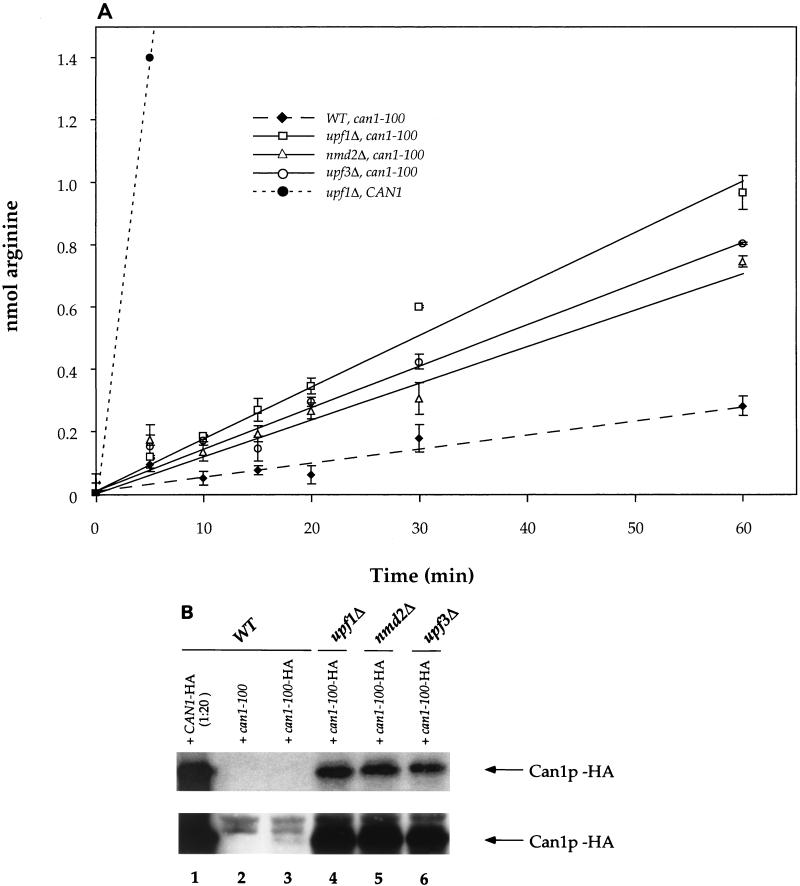
FIG. 3.
Accumulation of functional Can1p correlates with nonsense suppression of can1-100. (A) 3H-labeled arginine uptake in yeast strains with the indicated UPF/NMD and CAN1 genotypes. The control yeast strain harboring the CAN1 allele is PLY148 (36). WT, wild type. Error bars indicate standard deviations. (B) Western analysis of Can1p levels. Lysates of yeast strains with the indicated UPF/NMD genotypes and bearing either CAN1 or can1-100 plasmids were analyzed by Western blotting with HA-specific antibodies. The lower panel is a longer exposure of the same blot shown in the upper panel.
To test whether increased suppression and transport activity reflected enhanced synthesis of full-length Can1p, the expression of an HA epitope-tagged allele of can1-100 was monitored by Western blotting. As a control, we showed that all strains containing the can1-100–HA plasmid exhibited suppression phenotypes identical to those of strains containing the same plasmid lacking the triple-HA tag (data not shown). Figure 3B shows that Can1p-HA was barely detectable in wild-type cells (lower panel, lane 3) but increased approximately 10-fold in abundance in upf1Δ, nmd2Δ, and upf3Δ cells (compare lane 3 to lanes 4 to 6). Suppression of can1-100 yielded Can1p levels that were approximately 20-fold lower than those obtained from expression of the wild-type CAN1 gene, a result consistent with the high rate of arginine transport in CAN1 cells (Fig. 3A) and the sensitivity of the same cells to 0.7 μg of canavanine per ml (data not shown). Quantitation of the blot shown in Fig. 3B also provided an estimate of the reduction in CAN1 expression caused by the premature termination codon. Since the levels of Can1p in lanes 1 and 3 of Fig. 3B differ by approximately 10-fold and the sample in lane 1 is a 20-fold dilution, premature termination of CAN1 translation caused a 200-fold reduction in Can1p synthesis. The data in Fig. 3B also demonstrate that Can1p accumulation and the results of the plate assay for canavanine sensitivity approximate a linear relationship. This conclusion is drawn from the observations that wild-type cells harboring the CAN1 gene, wild-type cells harboring can1-100, and upf1Δ cells harboring can1-100 are sensitive to 0.7, 300, and 25 μg of canavanine per ml, respectively, and accumulate 200-, 1-, and 10-fold relative units of Can1p (Fig. 2B and 3B and data not shown).
can1-100 nonsense suppression by mutations in UPF1, NMD2, or UPF3 is only partially attributable to increases in mRNA abundance.
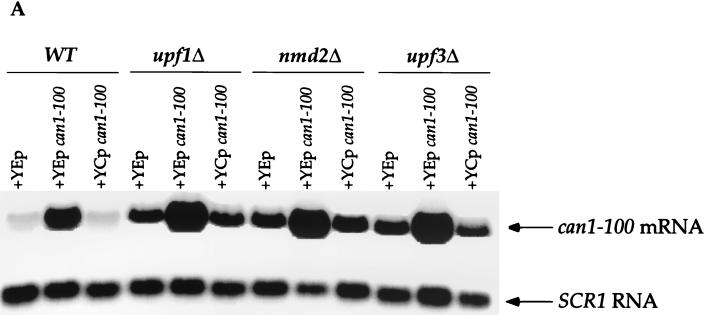
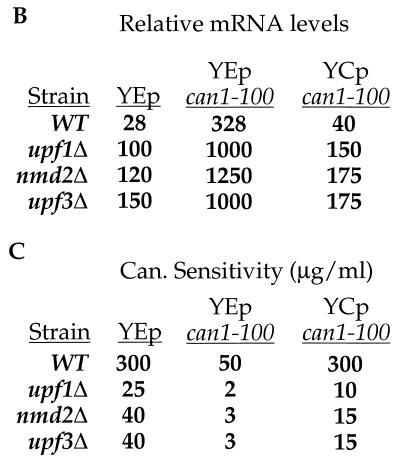
Since the can1-100 mRNA was stabilized in upf1Δ, nmd2Δ, and upf3Δ mutants (Fig. 1), suppression might be attributable to a constant but low rate of “leaky” termination that becomes functionally significant as mRNA levels increase. To directly address the contribution of mRNA abundance to the suppression phenotypes, the can1-100 allele was subcloned into single-copy and high-copy-number plasmids that were then introduced into cells that were wild type for NMD and already harbored a genomic copy of the can1-100 allele. Levels of the can1-100 mRNA were then measured by Northern analysis (Fig. 4A and B), and the respective suppression phenotypes (i.e., canavanine sensitivities) of the different strains were determined (Fig. 4C). Wild-type cells expressing an additional copy of can1-100 (YCp can1-100) showed a slight (1.4-fold) increase in can1-100 mRNA levels (Fig. 4A and B), but this increase did not alter the suppression phenotype of wild-type cells containing either single-copy or high-copy-number vectors without inserts (Fig. 4C, compare WT–YCp can1-100 with WT–YEp; also, data not shown). Wild-type cells transformed with the high-copy-number plasmid containing the can1-100 allele showed a 12-fold increase in can1-100 mRNA abundance compared to the same cells containing only the vector (Fig. 4A and B, compare WT–YEp can1-100 with WT–YEp). Accompanying this increase in mRNA levels was a sixfold increase in sensitivity to canavanine (Fig. 4C).
FIG. 4.
can1-100 nonsense suppression is only partially attributable to increased mRNA abundance. (A) Northern analysis of can1-100 mRNA levels. RNA isolated from yeast strains with the indicated genotypes was analyzed by Northern blotting with probes specific for can1-100 mRNA and SCR1 RNA (the latter to serve as an internal loading control). Each of the indicated strains contained either a high-copy-number can1-100 plasmid (YEp can1-100), a single-copy can1-100 plasmid (YCp can1-100), or an empty vector as a control (YEp). WT, wild type. (B) can1-100 steady-state mRNA levels. Data from the blot in panel A were quantitated by phosphorimaging, standardized to SCR1 RNA levels, and normalized to data for the upf1Δ strain. (C) Canavanine sensitivities of strains harboring single-copy or high-copy-number plasmids. Suppression assays analogous to those shown in Fig. 2 were used to define the canavanine (Can.) sensitivities of cells with different UPF/NMD genotypes.
The same phenomena were exhibited when this experiment was repeated with upf1Δ, nmd2Δ, and upf3Δ mutants. All strains expressing an additional copy of the can1-100 allele exhibited modest increases in can1-100 mRNA levels (15 to 50%; Fig. 4A and B) but showed approximately threefold increases in their respective levels of suppression (Fig. 4C, compare YCp can1-100 with YEp for all three mutants). When the can1-100 allele was expressed in these mutants from the high-copy-number plasmid, there was a 10-fold increase in the abundance of its mRNA (Fig. 4A and B) and a comparable increase in the level of nonsense suppression (Fig. 4C). These results indicate that increased mRNA abundance contributes to nonsense suppression but is not its sole determinant. This conclusion is illustrated further by direct comparisons of mRNA levels and extents of suppression in mutant and wild-type cells. For example, UPF/NMD wild-type cells overexpressing can1-100 (WT–YEp can1-100) had two- to threefold higher levels of can1-100 mRNA than any of the upf/nmd mutant cells (Fig. 4A and B), yet the level of suppression in the WT–YEp can1-100 strain was still lower than that in any of the mutants (Fig. 4C).
Additional support for the notion that increased mRNA abundance is not sufficient for can1-100 nonsense suppression is the finding that single deletions of UPF1, NMD2, UPF3, DCP1, or XRN1 were found to stabilize can1-100 mRNA to comparable levels (approximately fourfold; Fig. 1A), yet there were substantial differences in the canavanine sensitivities of the respective strains (Fig. 1B and C and 2B). Collectively, the data in Fig. 1 to 4 provide strong support for the notion that the UPF/NMD genes regulate not only the rates of decay of nonsense-containing mRNAs but also their efficiencies of translation.
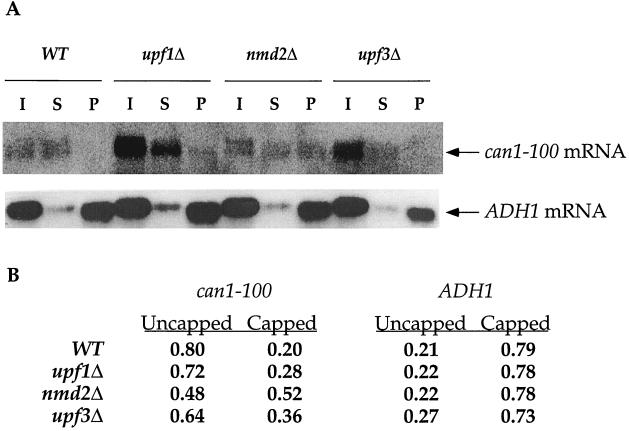
Different efficiencies of suppression are not attributable to changes in the fraction of capped can1-100 mRNA.
Recent experiments have indicated that deletions of UPF1, NMD2, or UPF3 inhibit the decay of nonsense-containing mRNAs prior to the decapping step; i.e., such deletions increase the steady-state ratio of capped to uncapped mRNAs (He and Jacobson, unpublished). Since the upf/nmd mutations affected the efficiency of translational suppression (see above), we considered the possibility that this effect, in turn, reflected substantial alterations in the relative percentages of capped can1-100 mRNA in wild-type and mutant cells. Immunoprecipitation experiments with anti-cap antibodies were used to examine the 5′ cap status of the can1-100 mRNA and a control (ADH1) mRNA in wild-type, upf1Δ, nmd2Δ, and upf3Δ strains. In both wild-type and mutant cells, the ADH1 mRNA was predominantly capped (Fig. 5). However, the can1-100 mRNA was predominantly uncapped in wild-type cells, and deletion of UPF1, NMD2, or UPF3 led to a slight increase in the percentage of capped molecules (Fig. 5). These changes in the ratios of capped to uncapped can1-100 mRNAs do not correlate with the suppression data of Fig. 1 to 4 and indicate that variations in suppression efficiencies must reflect events unrelated to mRNA cap status. This conclusion is underscored by experiments indicating that dcp1Δ and xm1Δ, two mutations that have negligible effects on can1-100 suppression (Fig. 1), lead to the accumulation of mRNAs that are predominantly capped or uncapped, respectively (4, 27, 40; also data not shown).
FIG. 5.
Suppression phenotypes are not a consequence of changes in the relative fractions of capped can1-100 mRNA. (A) Northern analysis of mRNAs fractionated by 5′-cap immunoprecipitation. Total RNA from yeast strains with the indicated UPF/NMD genotypes was separated into capped and uncapped fractions by use of polyclonal anti-m7G antibodies and analyzed by Northern blotting with DNA probes for either the ADH1 mRNA or the can1-100 mRNA. I, input RNA; S, RNA in the supernatant fraction (represents the uncapped fraction); P, RNA in the pellet fraction (represents the capped fraction). WT, wild type. (B) Relative amounts of capped and uncapped can1-100 and ADH1 transcripts. RNA in the S and P fractions of panel A was quantitated by phosphorimaging, and the relative percentages of capped and uncapped transcripts were determined by calculating the fraction each sample represented of its respective total (S + P).
The relative distributions of capped and uncapped can1-100 mRNA species differed not only from that observed for the ADH1 mRNA but also from that seen with nonsense-containing PGK1, MER2, and CYH2 transcripts (41; He and Jacobson, unpublished). This finding was unexpected and may reflect the possibility that, for some mRNAs, decapping is not immediately followed by exonucleolytic digestion. This conclusion is supported by experiments showing that at least one other NMD substrate, the his4-38 mRNA, behaves similarly (He and Jacobson, unpublished) and that uncapped mRNAs accumulate in a temperature-sensitive eukaryotic initiation factor 5A mutant (63).
Epistatic relationships of Upf1p, Nmd2p, and Upf3p in nonsense suppression.
Since the different upf/nmd mutations showed small but highly reproducible differences in the extents of can1-100 suppression that they promoted (Fig. 2B), we were able to exploit those differences to determine epistatic relationships of Upf1p, Nmd2p, and Upf3p. To resolve epistatic relationships, mutants containing double deletions of the UPF1, NMD2, or UPF3 genes were constructed and assayed for their sensitivity to canavanine. Analyses of these mutants demonstrated that any strain harboring a deletion of UPF1 exhibited the highest levels of suppression (i.e., sensitivity to 25-μg/ml of canavanine) and, conversely, that strains harboring a wild-type UPF1 gene showed lower levels of suppression (i.e., sensitivity to 35 μg of canavanine per ml). As shown in Fig. 2, double deletion of UPF1 and either NMD2 or UPF3 resulted in a suppression phenotype identical to that caused by upf1Δ alone. This result indicates that combining an nmd2Δ or upf3Δ mutation with upf1Δ does not have an additive effect on nonsense suppression and that the upf1Δ phenotype supersedes the nmd2Δ and upf3Δ phenotypes. Of the double mutants, the nmd2Δ upf3Δ mutant showed the lowest level of suppression, displaying a phenotype like that of an nmd2Δ strain (compare nmd2Δ upf3Δ to nmd2Δ in Fig. 2A). These results indicate that Upf1p is epistatic to Nmd2p and Upf3p and suggest a role for Upf1p in affecting the efficiency of premature translation termination.
While the suppression phenotypes of the double mutants suggested relatively straightforward epistatic relationships, the phenotype of the triple mutant, lacking UPF1, NMD2, and UPF3, was somewhat surprising. This mutant showed a lower level of suppression than any of the upf/nmd mutants tested (sensitivity to 50 μg of canavanine per ml; Fig. 2), demonstrating that the efficiency of translation termination is greater in the absence of all three UPF/NMD gene products than in the presence of any one of them. This result suggests either the existence of an alternate mechanism of termination fidelity that functions in the absence of the UPF/NMD gene products or that the presence of one of the UPF/NMD factors without the other two acts dominantly to prevent proper termination.
Overexpression of UPF1 decreases the efficiency of nonsense suppression without altering can1-100 mRNA levels.
As an additional approach to characterizing the functional relationships of Upf1p, Nmd2p, and Upf3p, these gene products were overexpressed in all of the upf/nmd mutant backgrounds, and the resulting effects on nonsense suppression were examined. Overexpression was accomplished by cloning UPF1, NMD2, or UPF3 under the control of the strong ADH1 promoter on a high-copy-number plasmid (22). Expression of the UPF/NMD genes from these constructs was found to increase the accumulation of the respective proteins at least 10-fold (data not shown). As controls for these experiments, we utilized mutant strains transformed with only the high-copy-number vector. The presence of this plasmid did not alter the suppression phenotypes of any of the mutant strains (compare Table 2 [YEp column] with Fig. 2B).
TABLE 2.
Overexpression of UPF1, NMD2, or UPF3 in wild-type and upf/nmd strainsa
| Strain | Canavanine sensitivity (μg/ml)
|
|||
|---|---|---|---|---|
| YEp | YEp-UPF1 | YEp-NMD2 | YEp-UPF3 | |
| Wild type | 300 | 300 | 300 | 300 |
| upf1Δ | 25 | 300 | 25 | 25 |
| nmd2Δ | 35 | 75 | 300 | 35 |
| upf3Δ | 35–40 | 75 | 25 | 300 |
| upf1Δ nmd2Δ | 25 | 75 | 25 | 25 |
| upf1Δ upf3Δ | 25 | 75 | 25 | 25 |
| nmd2Δ upf3Δ | 35 | 75 | 35 | 35 |
| upf1Δ nmd2Δ upf3Δ | 50 | 125 | 50 | 50 |
Suppression assays are summarized, with the concentration of canavanine at which each strain began to exhibit the canavanine-sensitive phenotype indicated (canavanine sensitivity). Yeast strains of the indicated genotypes were transformed with the high-copy-number vector alone (YEp; control) or high-copy-number plasmids expressing UPF1, NMD2, and UPF3 (YEp-UPF1, YEp-NMD2, and YEp-UPF3, respectively).
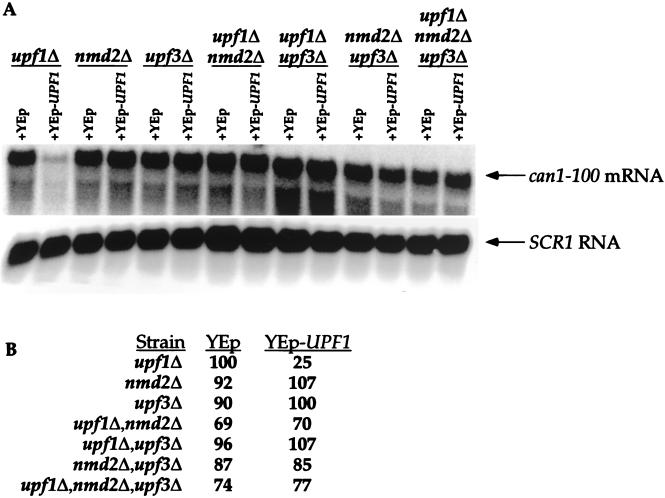
Overexpression of UPF1 in all of the single, double, and triple mutant strains (not including the upf1Δ control) was found to lower suppression levels two- to threefold (Table 2, compare YEp and YEp-UPF1 columns). These results are consistent with the notion that Upf1p can, by itself, enhance termination fidelity and also implicate a regulatory role for Nmd2p and Upf3p, since Upf1p can lower suppression in the absence of either of the other proteins. Overexpressing UPF3 complemented its own deletion, had no effect on any other single or double mutation, and did not change the phenotype of the triple mutant. The latter phenomenon, however, could be considered to reflect a modest increase in canavanine resistance over that observed in a upf1Δ nmd2Δ strain (Table 2). Overexpression of NMD2 had comparable effects, except that, in upf3Δ cells, it also enhanced suppression to a level comparable to that obtained in upf1Δ cells (Table 2). This result suggests that Nmd2p may be a negative regulator of the activity of Upf1p or is capable of simply titrating available Upf1p.
Since the overexpression of UPF1 altered the suppression phenotypes of all of the mutants, we investigated whether these effects might be caused by restoration of the rapid rate of decay of the can1-100 transcript. To this end, steady-state levels of the can1-100 mRNA were examined in upf/nmd mutants overexpressing UPF1. As expected, overexpression of UPF1 in the upf1Δ strain restored NMD to wild-type levels, resulting in a fourfold decrease in can1-100 mRNA levels (Fig. 6, compare upf1Δ–YEp to upf1Δ–YEp-UPF1). Accompanying this restoration of decay function was the restoration of the wild-type suppression phenotype (Table 2). In all of the other upf/nmd mutant strains, UPF1 overexpression did not significantly alter can1-100 mRNA levels compared to those seen in the starting mutant strains that contained the vector only (Fig. 6). Additionally, overexpression of NMD2 or UPF3 in any of the mutant backgrounds had no effect on steady-state can1-100 mRNA levels, other than those involving direct complementation of the respective single deletions (data not shown). These results demonstrate that changes in the suppression phenotype caused by overexpression of UPF1 are not attributable to changes in can1-100 mRNA levels. These observations are consistent with the proposed role of Upf1p in controlling the efficiency of translation termination, provide further support for a regulatory function for Nmd2p and Upf3p, and comprise additional evidence for the separation of the activities of Upf1p in mRNA decay and translation (11, 58, 59).
FIG. 6.
Overexpression of UPF1 in upf/nmd mutant strains does not affect can1-100 mRNA abundance. (A) Northern analysis of can1-100 mRNA levels. Total RNA isolated from yeast strains with the indicated genotypes was analyzed by Northern blotting as described in the legend to Fig. 4. Each of the mutant strains contained either a high-copy-number UPF1 plasmid (YEp-UPF1) or an empty vector as a control (YEp). (B) Quantitation of can1-100 steady-state mRNA levels. can1-100 mRNA levels were determined, standardized to SCR1 RNA, and normalized to data for the upf1Δ strain as described in the legend to Fig. 4.
Upf1p is considerably more abundant than Nmd2p or Upf3p but is not stoichiometric with ribosomes.
The suppression analyses described above indicated that Upf1p was a critical regulator of termination fidelity and that Nmd2p and Upf3p regulated the activity of Upf1p. These putative regulatory relationships are consistent with the results of previous protein-protein interaction analyses (11, 21, 22, 58) but raise the question as to whether these interactions occur as part of a stoichiometric complex or are more transient events. To address this issue further, we determined the cellular abundance of each of these factors. Western blotting was used to compare the amounts of epitope-tagged Upf1p and Nmd2p in a fixed number of cells with those present in purified samples of each of the two proteins. Relative levels of Nmd2p and Upf3p in crude extracts were determined by comparing the relative Western blot intensities of the two proteins when each harbored the same epitope tag. Using this approach, Upf1p was found to be the most abundant of the three factors, with approximately 1,600 molecules of Upf1p/cell (Table 3). Nmd2p was found to be 10-fold less abundant than Upf1p (160 molecules of Nmd2p/cell), and Upf3p was found to be the least abundant of the NMD factors (80 molecules of Upf3p/cell) (Table 3). These experiments indicate that the cellular concentrations of Upf1p, Nmd2p, and Upf3p differ greatly and do not approach the cellular levels of ribosomes, release factors, or the major cellular exonuclease, Xrn1p (23) (Table 3). These data are, however, consistent with the putative role of Upf1p as a regulator of termination fidelity, as well as the implied roles of Nmd2p and Upf3p as regulators of Upf1p.
TABLE 3.
Cellular levels of Upf1p, Nmd2p, and Upf3pa
| Protein or ribosomes | Protein
|
|
|---|---|---|
| No. of molecules per cell | % Soluble | |
| Upf1p | 1,600 | 0.007 |
| Nmd2p | 160 | 0.0007 |
| Upf3p | 80 | 0.00035 |
| Sup45p | 29,600b | 0.2b |
| Xrn1p | 29,000c | 0.2c |
| Ribosomes | 300,000d | 7 |
The relative abundance of Upf1p and Nmd2p was determined by comparing Western blot band intensities of the UPF/NMD factors present in crude cell extracts to those of the individual proteins. For Upf1p and Nmd2p, highly purified recombinant GST-Upf1p and GST-Nmd2p were used as standards, respectively. For Upf3p, cells bearing an HA-NMD2 allele were used as the standard. Calculations used to derive protein abundance are summarized in Materials and Methods.
Cellular Sup45p levels determined by Gygi et al. (16).
Cellular Xrn1p levels determined by Heyer et al. (25).
Cellular levels of ribosomes determined by Waldron and Lacroute (56).
DISCUSSION
Suppression of the can1-100 nonsense allele is enhanced by upf1Δ, nmd2Δ, and upf3Δ mutations.
The UPF1, NMD2, and UPF3 genes regulate NMD (7, 20, 22, 34–36, 45). Mutations in any of these genes generally promote the stabilization of nonsense-containing mRNAs by reducing the rate at which the recognition of a premature termination codon by the translation apparatus triggers mRNA decapping (He and Jacobson, unpublished). These effects of upf/nmd mutations on mRNA stability and parallel enhancing effects on nonsense suppression (9, 36, 58, 59) and programmed ribosomal frameshifting (8, 52) suggested a regulatory role in translation termination and/or fidelity for Upf1p, Nmd2p, and Upf3p. Strong support for this conclusion was obtained from experiments demonstrating interactions between Upf1p and the polypeptide release factors Sup35p and Sup45p (11).
To characterize further the roles of the UPF/NMD gene products in translation termination, we developed an assay that examined the effects of upf/nmd mutations on suppression of the can1-100 allele. A single A→T mutation in this allele leads to the synthesis of a transcript in which codon 47 has been changed to UAA. As a consequence, the can1-100 mRNA is a substrate for NMD. Mutations in UPF1, NMD2, or UPF3 not only stabilized the can1-100 transcript but also promoted its suppression. Quantitative measurement of the extent of can1-100 suppression by these mutations was achieved by varying the canavanine concentration of the growth media and determining the specific concentration that effectively killed diluted samples of the respective mutants. Since the degree of suppression (i.e., enhanced canavanine sensitivity) was found to correlate with the level and activity of Can1p in the cells, we conclude that the can1-100 system provides a reliable assay for nonsense suppression. Further support for the reliability of this assay was provided by experiments showing that the qualitative aspects of can1-100 suppression were comparable to those obtained in independent assays with the leu2-1 (UAA) and tyr7-1 (UAG) nonsense alleles (data not shown).
We initially investigated the effects of single deletions of UPF1, NMD2, and UPF3 on can1-100 nonsense suppression. Individual deletions of each of these genes were shown to have comparable stabilizing effects on the can1-100 mRNA but to produce differential effects on suppression. Strains harboring the upf1Δ mutation consistently showed a higher level of nonsense suppression than strains harboring either the nmd2Δ or the upf3Δ mutation. We inferred from this observation that Upf1p might play a more direct role in regulating the translation of nonsense-containing mRNAs than the other two factors; further experimentation appears to have substantiated this conclusion (see below).
can1-100 nonsense suppression: a loss in termination fidelity?
Mutations in the UPF/NMD genes have previously been shown to promote the suppression of leu2, tyr7, met8, and his4 nonsense alleles (7, 36, 58, 59). Since these mutations invariably led to increases in the levels of the corresponding mRNAs (35, 36) but failed to generate evidence for an effect on the readthrough of premature stop codons, it was initially concluded that suppression was due solely to the combination of enhanced mRNA abundance and an inherent rate of readthrough that was sufficient to generate the minimal amount of protein required for function of the respective genes (36, 47). However, the experiments of Weng et al. (58, 59) suggested that an alternative explanation was more likely. They generated a large set of upf1 alleles and identified several in which the effects on mRNA decay and translational suppression could be separated. More specifically, they identified two significant classes of upf1 alleles: (i) those which, when expressed at a high copy number, inactivated mRNA decay but failed to allow suppression (e.g., DE572AA) (58) and (ii) those which, when expressed in a single copy, promoted normal mRNA decay but allowed suppression to occur (e.g., C84S) (59). The phenotypes of these mutants indicated that suppression was unlikely to be caused solely by changes in mRNA levels and established the notion that Upf1p could regulate both mRNA decay and translation. Since upf1 mutations had no effect on polysome profiles (19, 35) and since Upf1p was known to be of relatively low abundance, it was considered likely that the translational effects were targeted not to general initiation or elongation but rather to the premature termination event.
On the basis of the results of Weng et al. (58, 59), we anticipated that the suppression of can1-100 by upf/nmd mutations would also be attributable to more than simple increases in mRNA levels. This assumption was substantiated by several new lines of experimentation which demonstrated that (i) xrn1Δ- and dcp1Δ-mediated increases in can1-100 mRNA abundance, to levels comparable to those obtained in upf/nmd mutant cells, did not promote canavanine sensitivity; (ii) high-copy-number expression of the can1-100 allele in wild-type cells, leading to can1-100 mRNA levels which exceeded those obtained in upf/nmd mutant cells 2- to 3-fold, was less effective in promoting canavanine sensitivity than single upf1Δ, nmd2Δ, or upf3Δ mutations; (iii) when UPF1 was overexpressed, large changes in the extent of nonsense suppression could be attained without significant alterations in can1-100 mRNA abundance; (iv) in upf/nmd mutant cells harboring an additional copy of the can1-100 allele, 2- to 3-fold increases in canavanine sensitivity were obtained when levels of the corresponding mRNA increased only 50% or less; and (v) high-copy-number expression of the can1-100 allele led to 3- to 4-fold higher levels of the corresponding mRNA in upf/nmd mutant cells than in wild-type cells but to 16- to 25-fold higher levels of suppression. Interestingly, the observation that the canavanine sensitivity of wild-type cells increased at all in response to enhanced abundance of the can1-100 mRNA indicates that mRNA abundance contributes to suppression and that the premature termination codon in the can1-100 mRNA must be leaky. The latter conclusion is substantiated by the identification of small amounts of full-length Can1p in wild-type cells harboring the can1-100 allele (Fig. 3B).
Given that the premature termination codon in the can1-100 allele has an intrinsic, albeit low, rate of readthrough, two explanations for the mechanism of suppression appear plausible. In the first, translation initiation of the can1-100 mRNA is somehow increased, and in the second, the efficiency of the premature termination event is decreased. While there is no evidence supporting global effects on translation initiation by upf/nmd mutants (19, 35), inactivation of the NMD pathway has been shown to promote a modest increase in the translational efficiency of nonsense-containing mRNAs (42). Moreover, recent studies have demonstrated that the upf1Δ, nmd2Δ, and upf3Δ mutations alter the distributions of capped and uncapped transcripts (He and Jacobson, unpublished). Therefore, suppression by deletion of UPF1, NMD2, or UPF3 could, in principle, have been caused by subtle increases in the translational efficiency of can1-100 mRNA, possibly because of changes in its extent of capping. However, since deletion of UPF1, NMD2, or UPF3 produced differential effects on the amounts of capped and uncapped can1-100 mRNAs (Fig. 5) that did not correlate with their respective suppression phenotypes, it is unlikely that suppression is dependent on changes in the fraction of capped can1-100 mRNA. We therefore consider it likely that suppression caused by deletion of UPF1, NMD2, or UPF3 is due either to a loss in termination fidelity at the premature nonsense codon or to additional rounds of translational initiation on an mRNA with an inherent, low rate of leaky termination. The demonstration of interactions between Upf1p and the polypeptide release factors (11) suggests that the former model is more likely.
Upf1p plays a central role in regulating nonsense suppression.
The finding that deletion of UPF1 resulted in a greater extent of suppression than deletion of NMD2 or UPF3 either implicates Upf1p as the most critical of the three factors for the maintenance of termination fidelity or suggests that Upf1p and either Nmd2p (i.e., as in the upf3Δ strain) or Upf3p (i.e., as in the nmd2Δ strain) may enhance termination fidelity cooperatively. To distinguish between these possibilities, strains harboring double mutations were constructed, and nonsense suppression by these strains was monitored. Any double mutant harboring a deletion of UPF1 showed the highest levels of suppression and, alternatively, the nmd2Δ upf3Δ mutant (the only double mutant expressing UPF1) showed lower suppression levels. Therefore, the suppression phenotype mediated by the deletion of UPF1 supersedes an additional mutation of NMD2 or UPF3, suggesting that, of the three proteins, Upf1p is the central factor involved in regulating the translational efficiency of nonsense-containing mRNAs. This conclusion was significantly reinforced by analyses of the consequences of UPF1 overexpression (see below). Interestingly, since deletion of both NMD2 and UPF3 does not have an additive effect on suppression, it appears that Nmd2p and Upf3p may act in concert, as opposed to independently, to regulate Upf1p activity.
Deletion of all three UPF/NMD genes resulted in significantly lower levels of suppression than that seen in any of the other mutants tested. This result was surprising, since this mutant was expected to exhibit a phenotype characteristic of upf1Δ strains. Since deletion of the genes encoding all three factors enhances termination efficiency, either an alternate fidelity pathway may function in the absence of the UPF/NMD-mediated mechanism or any one of the UPF/NMD factors without the other two may act in a dominant-negative manner.
Overexpression of UPF1 restores termination fidelity without affecting mRNA decay.
As noted above, Weng et al. (58, 59) showed that specific upf1 alleles could separate the translation and turnover functions of Upf1p, i.e., some alleles resulted in normal mRNA decay but impaired termination fidelity, whereas others resulted in the opposite phenotype. Curiously, these phenotypes are not reproduced in the can1-100 system. Strains with upf1 alleles shown to result in normal decay but impaired fidelity (e.g., C84S) behaved like the wild-type strain in the can1-100 system, and strains with alleles resulting in inactive mRNA decay but functional fidelity (e.g., DE572AA) behaved just like upf1Δ strains (data not shown). However, we have been able to obtain independent evidence for the separation of the two putative functions of Upf1p. In analyses of the effects of overexpression of each of the UPF/NMD genes in the different mutant backgrounds, we observed that high-copy-number expression of UPF1 led to substantial decreases in can1-100 nonsense suppression without having any significant effect on can1-100 mRNA levels. This finding underscores the existence of a separate translational role for Upf1p, reinforces the notion of Upf1p as the preeminent of the three factors in regulating termination fidelity, and implies a regulatory function for Nmd2p and Upf3p (since the overexpression of Upf1p has the ability to enhance fidelity even in the absence of Nmd2p or Upf3p).
The overexpression of NMD2 in a upf3Δ strain enhanced nonsense suppression to an extent comparable to that observed in strains harboring only a UPF1 deletion. This result suggests that Nmd2p may negatively regulate the activity of Upf1p, such that an excess of this negative regulator renders Upf1p inactive. Alternatively, since Nmd2p and Upf1p interact (21, 22), the overexpression of NMD2 may simply sequester Upf1p molecules and prevent their proper functioning by hindering additional interactions. The latter hypothesis leaves open the possibility that Nmd2p and Upf3p are actually activators of Upf1p activity, a model consistent with the decreases in suppression observed when UPF1 was overexpressed. If Nmd2p and Upf3p are indeed such activators, then the results of their respective overexpression would indicate that high levels of either factor alone are not sufficient to promote such activation.
Cellular concentrations of Upf1p, Nmd2p, and Upf3p are consistent with their apparent regulatory interactions.
Earlier studies recognized that the UPF/NMD factors were relatively low in abundance (3, 37, 46), but their actual cellular concentrations were not determined previously. Here, using Western blotting of crude cell extracts and purified proteins as standards, we found approximately 1,600, 160, and 80 molecules of Upf1p, Nmd2p, and Upf3p per cell, respectively. The abundance of these factors is consistent with the proposed central role of Upf1p in regulating termination fidelity, as well as with the hypothesis that Nmd2p and Upf3p regulate the activity of Upf1p. Although Upf1p, Nmd2p, and Upf3p have all been shown to be interacting proteins that associate with polyribosomes (3, 20–22, 38, 46, 58, 59), these data make it unlikely that these proteins exist in a stable complex or that they associate with all ribosomes. Rather, their interactions and ribosome association must be transient, with the latter limited to ribosomes recognizing newly synthesized mRNAs or their termination codons. An association with ribosomes actively recognizing termination codons would be consistent with recent studies demonstrating that Upf1p interacts with the peptide release factors Sup35p and Sup45p (11).
Possible functions of Upf1p, Nmd2p, and Upf3p in translation termination.
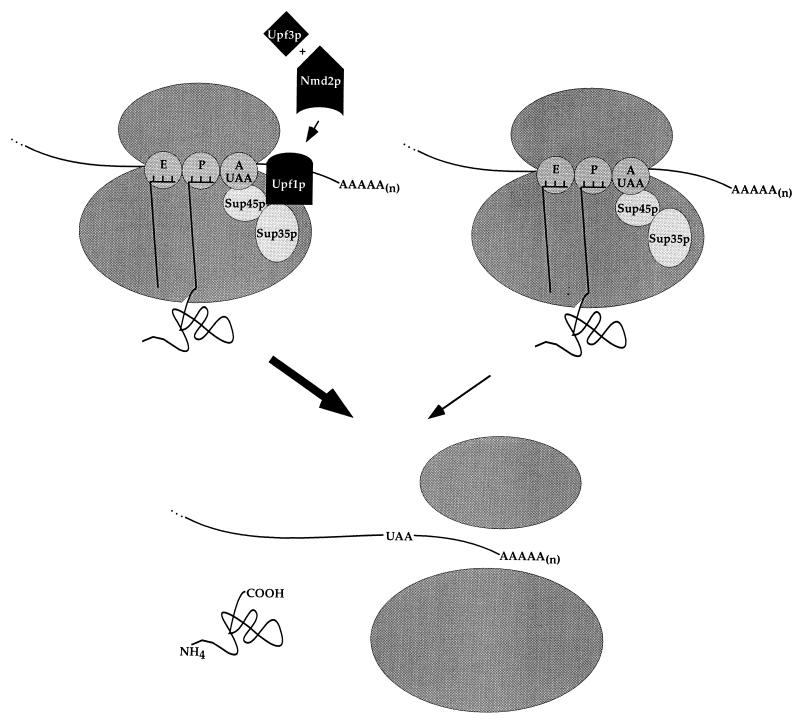
Taken together, the findings presented are consistent with Upf1p playing an important role in regulating the efficiency of translation termination, with Nmd2p and Upf3p serving as codependent activators of Upf1p function (Fig. 7). The importance of UPF1 is highlighted by the observations that deletion of UPF1 results in the highest levels of suppression, overexpression of UPF1 can restore termination fidelity, and Upf1p is the most abundant of the three proteins involved in NMD. Further, homologs of Upf1p have been identified in other organisms, including Caenorhabditis elegans (44, 49) and humans (2, 48), indicating evolutionary conservation of this factor. It is possible that the role of Upf1p in translation termination simply involves stimulation of the activity of the peptide release factors (K. Czaplinski et al., submitted for publication), such that efficient release allows for enhanced fidelity. Alternatively, Upf1p may play a more elaborate role in termination, including the regulation of ribosome release and recycling and the stimulation of decapping concurrent with premature nonsense codon recognition. Experiments to be described elsewhere suggest that these activities are also within the realm of Upf1p (R. Ganesan, F. He, and A. Jacobson, unpublished data; He and Jacobson, unpublished).
FIG. 7.
Model for functional relationships of Upf1p, Nmd2p, and Upf3p in translation termination. Upf1p is depicted as a positive regulator of the efficiency of translation termination mediated by Sup35p and Sup45p. The activity of Upf1p is postulated to be dependent on the function of both Nmd2p and Upf3p. Regulation of Upf1p by Upf3p and Nmd2p is postulated to occur as a consequence of either the combined or the sequential action of Upf3p and Nmd2p. The left and right complexes depict translation termination with and without nonsense decay factors, respectively, with the breadth of the large arrows indicating the relative efficiencies of the two events. E, P, and A represent the exit, peptidyl, and aminoacyl sites on the ribosome (dark gray ovals).
ACKNOWLEDGMENTS
This work was supported by a grant (GM27757) to A. J. from the National Institutes of Health, a predoctoral NRSA fellowship (GM18043) to A.B.M. from the National Institutes of Health, and a postdoctoral fellowship to D.A.M. from The Medical Foundation/Charles A. King Trust.
We thank Elsebet Lund for anti-cap antibodies, Duane Jenness for CAN1-HA and for teaching us about the potential value of a CAN1-based assay, and members of the Jacobson laboratory for their helpful editorial comments and occasional moral support.
REFERENCES
- 1.Altamura N, Groudinsky O, Dujardin G, Slonimski P P. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- 2.Applequist S E, Selg M, Roman C, Jack H M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and the Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 4.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 5.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin K, Pyle A M. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 8.Cui Y, Dinman J D, Peltz S W. Mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culbertson M R, Underbrink K M, Fink G R. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1p: a factor involved in mRNA turnover. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 13.Friest W, Sternbach H, Cramer F. Phenylalanyl-tRNA-synthetase from yeast and its discrimination of 19 amino acids in aminoacylation of tRNA(Phe)-C-C-A and tRNA(Phe)-C-C-A(3′NH2) Eur J Biochem. 1996;240:526–531. doi: 10.1111/j.1432-1033.1996.0526h.x. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman S, Wickner R, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie C, Fink G R, editors. Methods in enzymology: molecular biology of Saccharomyces cerevisiae. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 16.Gygi S P, Rochon Y, Franza B R, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 19.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc. Natl. Acad. Sci. USA . 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway using an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 21.He F, Brown A H, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]
- 22.He F, Brown A H, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hereford L M, Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977;10:453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- 24.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heyer W D, Johnson A W, Reinhart U, Kolodner R D. Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol Cell Biol. 1995;15:2728–2736. doi: 10.1128/mcb.15.5.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. A transmembrane protein without N-terminal hydrophobic signal sequence. J Biol Chem. 1985;260:11831–11837. [PubMed] [Google Scholar]
- 27.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurt D J, Wang S S, Lin Y H, Hopper A K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987;3:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 30.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koonin E V. A new group of putative RNA helicases. Trends Biochem Sci. 1992;17:495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.LaGrandeur T E, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 36.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lelivelt M J, Culbertson M R. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;10:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangus D A, Jacobson A. Linking turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 39.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 40.Muhlrad D, Decker C, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 41.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 42.Muhlrad D, Parker R. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol Biol Cell. 1999;10:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opekarova M, Kubin J. On the unidirectionality of arginine uptake in the yeast Saccharomyces cerevisiae. FEMS Microbiol Lett. 1997;152:261–267. doi: 10.1111/j.1574-6968.1997.tb10437.x. [DOI] [PubMed] [Google Scholar]
- 44.Page M F, Carr B, Anders K R, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 46.Peltz S W, Trotta C, He F, Brown A, Donahue J, Welch E, Jacobson A. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay. In: Brown A, Tuite M, McCarthy J, editors. Protein synthesis and targeting in yeast. Berlin, Germany: Springer-Verlag; 1993. pp. 1–10. [Google Scholar]
- 47.Peltz S W, He F, Welch E, Jacobson A. Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acids Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 48.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Mammalian orthologues of a yeast regulator of nonsense-transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 50.Rose M D, Winston F, Heiter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 51.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz-Echevarria M J, Yasenchak J M, Han X, Dinman J D, Peltz S W. The Upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc Natl Acad Sci USA. 1998;95:8721–8726. doi: 10.1073/pnas.95.15.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soni R, Carmichael J P, Murray J A H. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simple procedure. Curr Genet. 1993;24:455–459. doi: 10.1007/BF00351857. [DOI] [PubMed] [Google Scholar]
- 56.Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welch E M, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weng Y, Czaplinski K, Peltz S W. ATP is a cofactor of the Upf1p protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 61.White T J, Arnheim N, Erlich H A. The polymerase chain reaction. Trends Genet. 1989;5:185–189. doi: 10.1016/0168-9525(89)90073-5. [DOI] [PubMed] [Google Scholar]
- 62.Yarus M. Proofreading, NTPases and translation: successful increase in specificity. Trends Biochem Sci. 1992;17:171–174. doi: 10.1016/0968-0004(92)90257-a. [DOI] [PubMed] [Google Scholar]
- 63.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]