Abstract
Tooth development involves the coordinated transcriptional regulation of extracellular matrix proteins produced by ameloblasts and odontoblasts. In this study, whole-genome ChIP-seq analysis was applied to identify the transcriptional regulatory gene targets of Sp6 in mesenchymal cells of the developing tooth. Bioinformatic analysis of a pool of Sp6 target peaks identified the consensus nine nucleotide binding DNA motif CTg/aTAATTA. Consistent with these findings, a number of enamel and dentin matrix genes including amelogenin (Amelx), ameloblastin (Ambn), enamelin (Enam) and dental sialophosphoprotein (Dspp), were identified to contain Sp6 target sequences. Sp6 peaks were also found in other important tooth genes including transcription factors (Dlx2, Dlx3, Dlx4, Dlx5, Sp6, Sp7, Pitx2, and Msx2) and extracellular matrix-related proteins (Col1a2, Col11a2, Halpn1). Unsupervised UMAP clustering of tooth single cell RNA-seq data confirmed the presence of Sp6 transcripts co-expressed with many of the identified target genes within ameloblasts and odontoblasts. Lastly, transcriptional reporter assays using promoter fragments from the Hapln1 and Sp6 gene itself revealed that Sp6 co-expression enhanced gene transcriptional activity. Taken together these results highlight that Sp6 is a major regulator of multiple extracellular matrix genes in the developing tooth.
Keywords: Tooth, Odontogenesis, Sp6, Sp7, ChIP-seq, scRNA-seq, Promoter, Transcriptional regulation, Matrix, Ameloblastin, Amelogenin, Hapln1, Col1a2
1. Introduction
During tooth formation, a complex series of spatial and temporal events is initiated by interactions between undifferentiated epithelium and mesenchyme leading to specialized ameloblasts and odontoblasts, coordinating the synthesis of enamel and dentin components [1]. The most abundant tooth protein is amelogenin, which comprises 80–90% of the proteinaceous material produced during the secretory stage [2]. Amelogenin has a unique ability to self-assemble and promote apatite nanocrystals leading to enamel mineralization [2,3]. Two minor proteins, ameloblastin [4] and enamelin, and trace amounts of Mmp-20 protease [5] are made by ameloblasts. During the maturation phase, ameloblasts produce the Klk4 protease that remodels the matrix scaffold by protein proteolysis required for the mineralization process to proceed [6]. Odontoblasts also produce collagens, including Col1a2 and Col11a2, small amounts of amelogenin and a number of proteoglycans [7,8]. In odontogenesis, the timing and spatial complexity of matrix protein synthesis, removal and control of mineralization involves more than 1000 genes [9,10]. Proteomic studies [11–13] and gene expression analysis [14–16] have demonstrated a number of structural and regulatory proteins involved in developing teeth. Importantly, extracellular matrix molecules such as collagens and proteoglycans provide scaffolds for organizing the complex tissue required for proper enamel and dentin assembly.
Despite the identification of key tooth proteins, little is known about the transcriptional regulation of matrix proteins during development. Genetic and molecular studies have identified tooth-specific transcription factors including Sox2, Msx1, Msx2, and Dlx3 [17–20], however, the downstream targets of these transcription factors are poorly characterized. One transcription factor, Sp6, shows high levels of mRNA and protein expression in the developing inner dental epithelium and dental papilla [21]. Sp6 belongs to a family of transcription factors and is related to another member, Sp7/osterix, which regulates a number of osteoblast genes controlling bone formation [22]. Interestingly, murine Sp6 gene knockout studies have demonstrated profound defects in tooth development including severe reductions in the differentiation of dental epithelium into enamel matrix-producing ameloblasts [23,24]. However, little mechanistic information is known by which the loss of Sp6 contributes to tooth defects.
In this study, chromatin immunoprecipitation profiling DNA sequencing (ChIP-seq) of mouse molar tissue identified a Sp6 consensus DNA-binding motif, which was identified in a number of tooth-specific matrix proteins and transcription factors. Functional studies confirmed Sp6 increased transcription activity substantiating its role to regulate gene expression. Together these studies highlight the role Sp6 plays as a key transcriptional regulator of tooth formation.
2. Materials and methods
2.1. Sp6 chromatin immunoprecipitation-sequencing (ChIP-seq) and peak visualization
The Sp6 ChIP-seq was used to identify in vivo binding sites of genomic DNA from developing mouse tooth molar tissue. Briefly, dental mesenchyme and epithelium tissue from 30 newborn C57/BL6 P1 molars (Under approval from NIDCR Animal Care and Use Committee) were isolated and fixed with a 1% formaldehyde solution for 15 min and quenched with 0.125 M glycine. Additional processing of the tissue and ChIP-seq was performed by Active Motif Inc. (San Diego, California). Dental tissues were sonicated and precleared before antibody incubation. Rabbit polyclonal antibody against mouse Sp6 was generated using a synthetic peptide corresponding to mouse Sp6 amino acids 210–227. For each of the immunoprecipitation samples, approximately 40 million high quality reads were derived by Illumina sequencing. The full dataset of identified Sp6 peaks from both mouse molar epithelial and mesenchyme have been deposited with GEO accession GSE145909 (token mjszmskqvnublit).
Evaluation of the dataset using Model-based Analysis of ChIP-seq (MACs) with a cutoff p value of 10−7, identified 11,436 peaks. Some of the peaks (n = 2727) were not close enough to be assigned to a gene and were therefore excluded from the Excel files in GEO (GSE145909). The false discovery rate by MACs analysis was estimated to be <1%. Sp6-enriched immunoprecipitation peak sequences were compiled as binary analysis results (BAR) files and analyzed using the Integrated Genome Browser (https://bioviz.org).
2.2. Multiple expectation maximization for motif elicitation (MEME) for defining the Sp6 consensus binding motif
To determine the Sp6 consensus binding motif, 1400 DNA sequences were randomly subjected to MEME analysis [25]. WebLogo was used for visualization of the DNA consensus sequence. To identify genes from the ChIP-seq enriched in the tooth with the consensus motif, the ChIP-seq browser extensible MACs peaks in Excel format (GSM4339188) and the Pemberton et al. data set of 100 tooth-enriched genes [14] were joined by employing the Excel power query option. To elicit potential DNA motifs in tooth-enriched genes, 40 bp on either side of the MAX peak of the 37/100 shared genes (i.e., MACs peak and tooth-enriched) were analyzed by MEME.
2.3. Single-cell RNA sequencing (scRNA-seq) and UMAP analysis
scRNA-seq from mouse P7 incisor was described in detail previously [16] and only a brief description is presented. Here, scRNA was processed through the 10X Genomics workflow, cDNA libraries were prepared using a Nextera XT kit (Illumina) and sequencing performed on a NextSeq500 sequencer (Illumina). After Cell Ranger processing, removing low quality cell reads (cells with less than 700 identified genes and containing a mean value of greater than 9% mitochondrial genes) 6260 cells with 7356 median UMIs per cell were included in the analysis. Unsupervised clustering of cells populations was performed using Uniform Manifold Projection Approximation (UMAP) analysis from the Seurat R package [26].
2.4. Sp6 transcriptional regulation of the Hapln1 and Sp6 gene promoters
The 0.9 kb promoter region of the Hapln1 gene [27] was used to create a construct, pGL3-pHapln1, in the pGL3 promoterless firefly luciferase reporter plasmid (Promega). A mutant of the Hapln1 promoter (pGL3-pHapln1–4bp) was also constructed containing a 4 bp substitution in the core TAATTA sequence located at −219 to −218 and was changed to TAGCCG. Previous studies elucidated an important regulatory element in the Hapln1 promoter which was designated HLBS [28] and in this study, found to overlap the Sp6-enriched sequence. To study the transcriptional activity of this sequence, four direct repeats of this Sp6 target sequence (HLBS; 5’-GTTAGGCTGTAATTAGAGGA) were utilized driving a minimal promoter in pGL4.23 firefly luciferase reporter plasmid and was designated pGL4–4X-HLBS [28]. A series of Sp6 target mutants were constructed in a similar fashion in pGL4.23 including pGL4–4X-HLBS-Mut1 (5’- ACCGAGCTGTAATTAGAGGA), pGL4–4X-HLBS-Mut2 (5’-GTTAGATCAC AATTAGAGGA), pGL4–4X-HLBS-Mut3 (5’-GTTAGGCTGTGGCCGGAGGA), and pGL4–4X-HLBS-Mut4 (5’ GTTAGGCTGTAATTAAGAAG). A plasmid containing 53 bp (5’-CTCACCTTATAATTTATGCAAATCAACTCTGTAATTAACTA-TAATTTGGAGGC) from the Sp6 mouse gene itself located at −2015 was also subcloned in a plasmid designated pSp6–53bp. For analysis, HEK-293T cells were transfected with 1 μg of either empty vector (pGL4.23), wild type pGL4.23–4X HLBS, or the corresponding mutant targets along with 25 ng of pCMV-Sp, 25 ng of pCMV-Sp6-VP16 (a construct for enhanced cofactor recruitment), and 25 ng of CMV-Renilla luciferase. Transfections were performed in duplicate, and 48 h post-transfection, the cells were lysed in passive lysis buffer (Promega). Transcriptional activity normalized to transfection efficiency by sequentially assaying for firefly and Renilla luciferase activity with a luminometer (Berthold LB 940). Data was plotted comparing the pGL4.23 basal promoter co-transfected with a CMV expression vector without the gene insert.
3. Results
3.1. ChIP-seq analysis identifies Sp6 regulatory sites
To understand the gene networks controlled by the Sp6 in tooth development, genome-wide ChIP-seq analysis was utilized to identify in vivo genomic targets. Mouse mesenchymal and epithelial molar tissues were separately dissected, formaldehyde crosslinked, sonicated and used in subsequent ChIP analysis. Either input or affinity-purified rabbit anti-Sp6 antibody immunoprecipitated DNA pools were utilized, in which recovered DNA pools were sequenced to a depth of approximately 40 million reads per sample. Bioinformatic analysis was used to remove redundant sequences and identity candidate peaks in target genes based on the relative frequency of enriched genomic sequences. The ChIP-seq dataset derived from tooth mesenchyme, accession (GSM4339188), was utilized in this report.
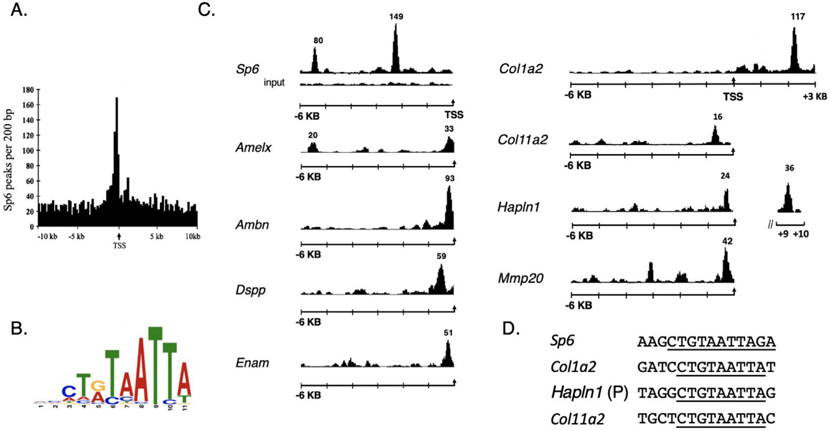
From Sp6 peak evaluation from the tooth mesenchyme using MACs with a cutoff p-value of 10−7, identified 11,436 enriched peaks. Approximately 29% of the 11,436 peaks were located within 10 Kb of the transcriptional start sites of the target genes, in which a large cluster of the Sp6 sites were often found directly preceding the transcriptional start sites (Fig. 1A). From unbiased, bioinformatic analysis of the nucleotide sequences within the Sp6 peak binding regions of the 1400 target genes, a consensus sequence comprising the nine-nucleotide binding sequence of C-T-A/G-T-A-A-T-T-A was identified with high probability (4.0e-219, Fig. 1B). Evaluation of the dataset using the IGB genome browser revealed that many tooth genes contained Sp6 peaks in their gene promoters including amelogenin, (Amelx), ameloblastin (Ambn), enamelin (Enam), dental sialophosphoprotein (Dspp), membrane-type matrix metalloprotease-20 (Mmp20) and several matrix genes (Col1a2, Col11a2 and Hapln1 (Fig. 1C). Peaks for Ambn, Enam, Mmp20, and Dspp were all located within 300 bp upstream of the start of transcription, but for Amelx, the peak was located at 5 kb upstream (Fig.1C). No prominent peaks were observed in these genes or other evaluated Sp6 target genes using control input sequence (only the input tracing for Sp6 is shown, as well, see Supplementary Fig. 2). MEME analysis also defined the core sequence motif in Sp6, Hapln1, and Col11a2 (Fig. 1D, and Supplementary Fig. 1).
Fig. 1.
ChIP-Seq identifies Sp6 promoter binding sites and a consensus binding motif. In total, 11,436 high confidence Sp6-enriched peaks were identified from tooth mesenchymal tissue. A. 3305 ChIP-seq peaks were identified within 10 Kb of the transcriptional start sites of the target genes. B. MEME bioinformatic analysis of the ChIP-seq data set identified a Sp6 consensus binding motif. C. ChIP-Seq peaks are shown for Sp6 binding sites identified near the transcription start sites (arrow) for the following genes: Sp6, Amelx, Ambn, DSPP, Enam, Col1a2, Col11a2, Hapln1, Mmp20. The black tracing for each gene and the numbers above it represents the Sp6-enriched peak. For this figure, the Sp6 and Col1a2 enriched peak heights are not drawn to scale. D. Extracted peaks from MEME analysis shows CTGTAATTA at SP6, Hapln1, Col11a2, (Col1a2, a major tooth matrix protein did not occur in the Pendleton dataset was evaluated visually).
To evaluate the extent of Sp6 interactions in tooth-specific genes in an unbiased fashion, Sp6 ChIP peak data in browser extensible data (BED) format was correlated to 100 genes known to be highly expressed in the developing tooth determined by microarray analysis [14]. Comparison of the data files showed that 37 genes of the 100 tooth-enriched genes harbored Sp6 binding sites near their promoter or enhancer (Table 1).
Table 1:
Sp6 tooth-enriched genes.
| Gene Symbol | Gene Name | Sp6 Peak Locationa |
|---|---|---|
| Ambn | Ameloblastin | −172 |
| Amelx | Amelogenin | −5499 |
| Asxl3 | Sex combs like | −4873 |
| Bmp7 | Bone morphogenetic protein 7 | + 1314 |
| Cldn1 | Claudin 1 | +3125 |
| Col11a2 | Collagen 11a2 | −446 |
| Cxadr | Coxsackie adeno receptor | −1520 |
| Dlx2 | Dlx homeobox 2 | −1538 |
| Dlx3 | Dlx homeobox 3 | +8935 |
| Dlx4 | Dlx homeobox 4 | +425 |
| Dlx5 | Dlx homeobox 5 | −124 |
| Dspp | Dentin sialo phospho protein | −339 |
| Dlx6os1 | Dlx6 opposite strand | −562 |
| Edg3 | G-protein-receptor 3 | +971 |
| Enam | Enamelin | −168 |
| Fras1 | Fraser syndrome 1 homolog | +2274 |
| Gal3st4 | Galactose-3-O-sulfotransferase 4 | −6722 |
| Hapln1 | Hyaluronan and proteoglycan binding link protein 1 | +9622 |
| Ibsp | Integrin binding sialoprotein | −221 |
| Klk4 | Kallikrein 4 | −838 |
| Maf | Avian fibrosarcoma homolog | −886 |
| Mmp14 | Matrix metallopeptidase 14 | −264 |
| Mmp20 | matrix metallopeptidase 20 | −135 |
| Msx2 | Msx2 homeo box | −203 |
| Nedd4 | Neural down-regulated gene 4 | +4854 |
| Papln | Sulfated glycoprotein | +749 |
| Phex | Phosphate regulating gene | −270 |
| Pitx2 | Paired-like homeodomain | +460 |
| Rnd3 | Rho family GTPase 3 | +23 |
| Shh | Sonic hedgehog | +28 |
| Slc39a8 | solute carrier 39 | +2372 |
| Sox4 | SRY-box4 | −17 |
| Sp6 | Sp6 | −2115 |
| Sp7 | Sp7 | −84 |
| Trp63 | Transformation related protein 63 | −28 |
| Vangl2 | Van gogh like-2 | −1097 |
| Wnt6 | wingless-6 | −7650 |
Numbers denote the location of the Sp6 target peak in either the promoter (negative number) or the first intron (positive number).
Besides major tooth matrix proteins, two tooth proteases were identified, kallikren4 (Klk4) and membrane-type matrix metalloprotease-14 (Mmp14), both of which are involved in enamel formation and dentinogenesis [6,29]. A large number of established tooth-specific transcription factors controlling dental tissue patterning and gene expression harbored Sp6 binding sites in their promoter or first intron regions including Dlx2, Dlx3, Dlx4, Dlx5, Sp6, Sp7, Pitx2, and Msx2 [18–20]. Additionally, genes for growth factors (Bmp7, Wnt6), and matrix proteins (Hapln1 (hyaluronan and proteoglycan binding link protein 1)) and Col11a2 had Sp6 binding sites in their promoters or first introns. The matrix protein Col11a2 was previously reported in tooth odontogenic cells [8]. Additional Sp6 ChIP-seq gene mapping data are found in Supplementary Table I and the Excel datasheets in GEO:GSE145909.
3.2. Evaluation of scRNA-seq from the developing tooth
Based on the finding that numerous tooth-specific genes had in vivo Sp6 binding sites near their transcriptional start sites, we investigated the mRNA expression of Sp6 by scRNA-seq from developing mouse incisor using the dataset (GSE146855). UMAP employing Seurat R-based package 3.0 was utilized. UMAP analysis showed Sp6 transcripts throughout multiple cell types of the incisor, including pre-ameloblast and ameloblast cell populations that co-expressed transcripts for Amelx, Ambn, DSPP, and Enam. Sp6 transcripts were also identified in a previously defined odontogenic cell population containing transcripts for the bone and tooth mineralization factor sphingomyelin phosphodiesterase 3 (Smpd3, [30]). Transcripts for Col1a2, Col11a2, Hapln1 (Fig. 2) and additional matrix proteins such as versican (not shown) were identified in the odontogenic cell populations.
Fig. 2.
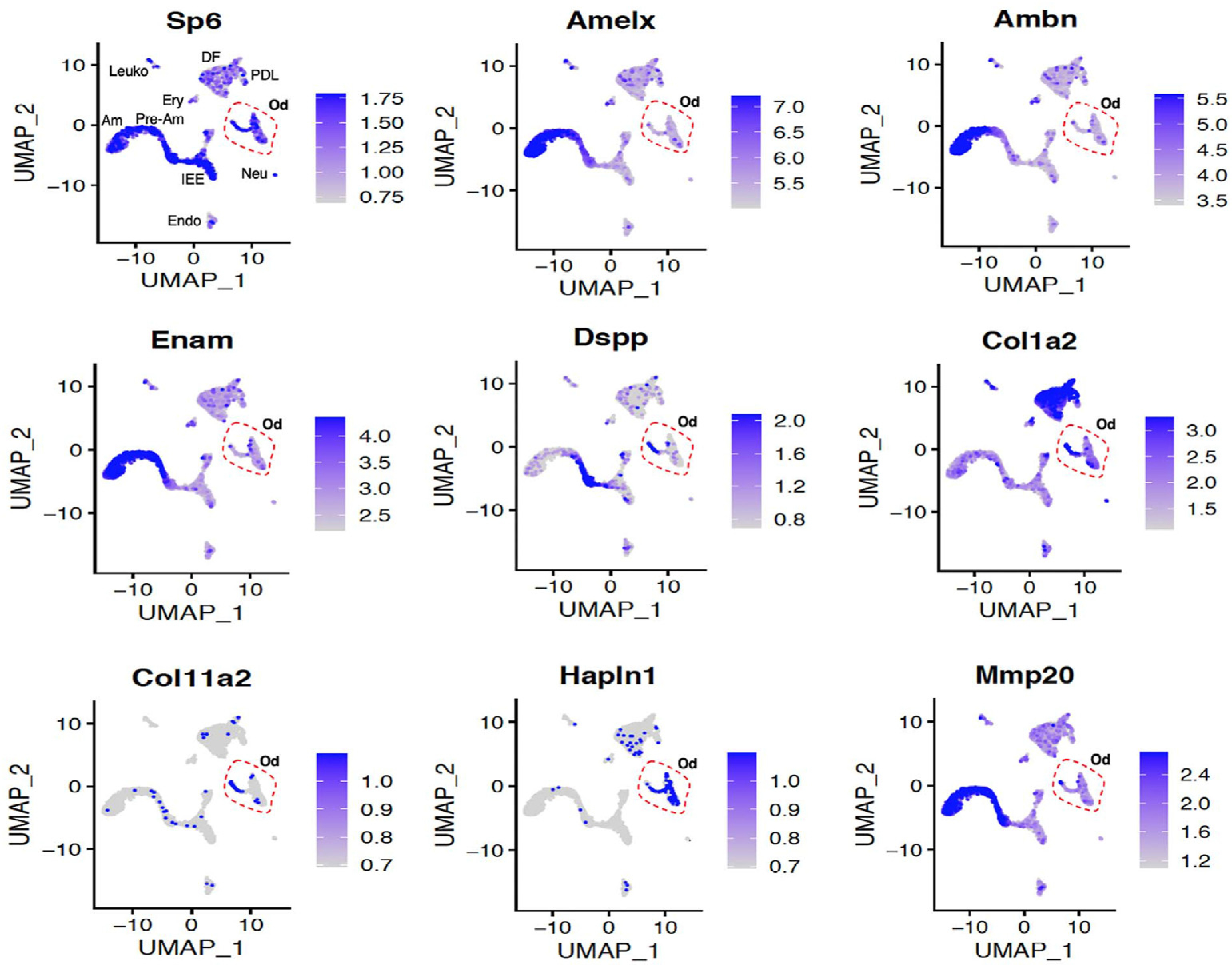
A. Differential expression analysis of Sp6, Amelx, Ambn, Enam, MMP20, DSPP, Cola11a2, Hapln1, Col1a2 by UMAP analysis from mouse P7 incisor dataset (GSE146855). Gene signatures and inferred functions were previously described: IEE, inner enamel epithelium; Pre-Am, pre-ameloblast; Am, ameloblast; SI/SR/OEE, stratum intermedium, stellate reticulum, and outer enamel epithelium; DF, dental follicle; Od, odontoblast; PDL, periodontal ligament; Ery, erythrocyte; Leuko, leukocyte; Endo, endothelium; Neu, neural clusters [30].
3.3. Sp6 activation of the Hapln1 gene promoter
Based on the co-expression of SP6 with a number of tooth-specific target genes, we tested whether one such target gene, Hapln1, was regulated by Sp6. For these studies, cell transcription reporter assays were utilized employing a 900 bp Hapln1 genomic promoter fragment driving luciferase reporter activity. In Hapln1, the Sp6 consensus sequence is located at −220 bp in the promoter and overlapped a previously characterized HLBS target sequence [28]. Transfection of Sp6 or the Sp6-Vp16 chimeric activator expression vectors in HEK-293 cells activated the basal promoter activity of pGL3 control vector (Fig. 3A). Sp6 co-transfection with the pGL-Hapln1 promoter also demonstrated a 2-fold activation, and the Sp6-Vp16 chimeric construct produced 5-fold transcriptional activation (Fig. 3A). Importantly, a pGL-Hapln1 reporter with a 4bp substitution (i.e., from ATTA to GCCG) within the core Sp6 binding motif in the promoter demonstrated no significant activation by either Sp6 or the Sp6-Vp16 constructs (Fig. 3A). Lastly, a 53 bp fragment from the mouse Sp6 promoter region containing the Sp6 binding motif was also activated 3-fold by SP6 expression and increased over 30-fold by Sp6-Vp16 construct (Fig. 3A). These results demonstrate that the consensus Sp6 regulatory motif found in the Hapln1 and Sp6 promoters are functionally active in vivo and regulatable by Sp6.
Fig. 3.
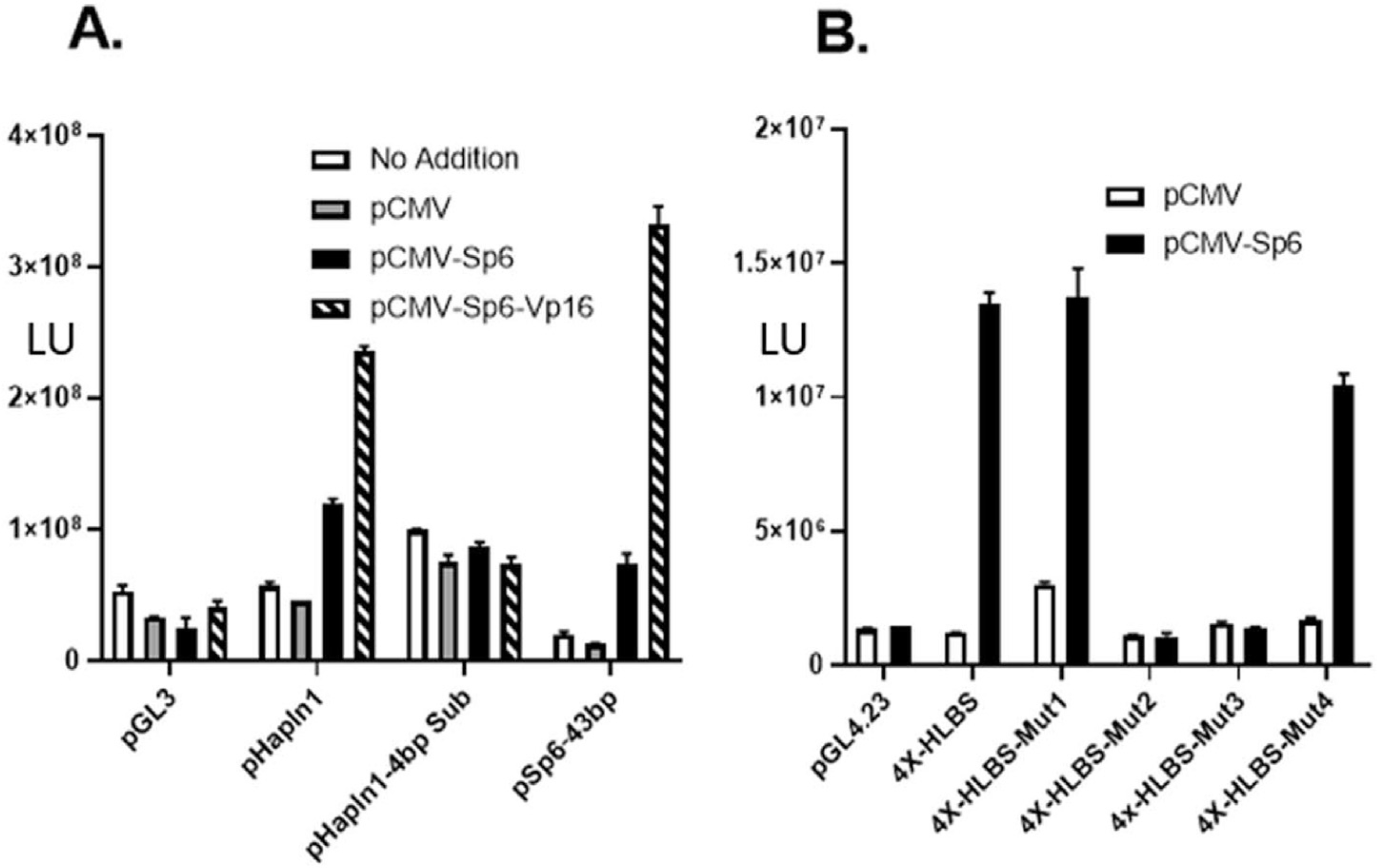
Hapln1 and Sp6 gene promoters are transactivated by Sp6. A. Transfection experiment in HEK-293 cells were performed with luciferase reporter constructs for the promoterless pGL3 vector, 0.9 Kb promoter (pHapln1), a promoter with a 4 bp substitution (pHapln1-4bpSub) in the core AT-rich Sp6 binding sequence and with pSp6–53 bp, a construct containing 53bp of the rat Sp6 gene containing Sp6 target sequence. In these experiments, no addition, pCMV empty vector, pCMV-Sp6 and pCMV-Sp6-Vp16 expression vectors were utilized and following transfection, luciferase reporter activity was assessed. B. As described in the Material and Methods, transient transfection experiments were performed using pGL4.23 (a basal promoter luciferase reporter vector) and corresponding 4X wild type and mutant 4X HLBS reporter constructs.
Additional experiments were designed to test whether the Sp6-binding motif from the Hapln1 promoter was itself necessary and sufficient for activation. Four copies of the Sp6 target sequence (designated HLBS) were placed in front of the basal promoter and tested in co-expression experiments. As shown in HLBS reporter constructs were activated by Sp6 resulting in an up-regulation of promoter activity by 30-fold in HEK293 cells (Fig. 3B). Co-transfection experiments with other transcription factors, including Sox9 and β-catenin, were unable to activate the HLBS reporter (data not shown). Mutations within the HLBS sequence showed markedly less activity inducible by Sp6 transactivation, in which changing the core sequence within the motif resulted in no promoter activity (Fig. 3B). Specifically, two mutants pGL4–4X-HLBS-Mut2 and pGL4–4X-HLBS-Mut3 disrupting the AT-rich region within the consensus motif were essentially transcriptionally inactive (Fig. 3B). Taken together, these results confirm that Sp6 is a regulator of both the Sp6 and the Hapln1 gene and likely other matrix genes.
4. Discussion
Tooth development is a highly complex process involving the expression of many different specialized matrix proteins. Evaluating a list of the 100 most tooth-enriched genes revealed that 37 tooth-specific genes has Sp6 binding sites identified by ChIP-seq. Sp6 motifs were found in ameloblastin, amelogenin, enamelin and mmp20, representing some of the most abundant tooth proteins. Further evidence for a key regulatory role of the Sp6 transcription factor in tooth morphogenesis was the finding that several other critical tooth-enriched transcription factors including Dlx2, Dlx3, Dlx4, Dlx5, and Msx2, contained Sp6 binding sites in their promoter regions. In many tooth-related genes, the Sp6 binding sites were enriched in close proximity to the start of gene transcription. These findings along with the observation that Sp6 gene itself had two Sp6 binding sites in its promoter suggests that it is a major coordinator acting at a central hub of this gene network during tooth development. The large number of gene targets identified provides an extensive road map for more detailed analyses of tooth development.
Consistent with the study by Wang, X. et al., 2020 [30], a cell population with a gene expression odontogenic profile was identified. This cell population contained transcripts for matrix genes Col11a2, Col1a2, and Hapln1, which all harbor Sp6 binding elements occurred in the promoter or first intron. The finding of Hapln1 in the developing tooth is consistent with multiple other studies [13,14,31,32]. Here we further demonstrate that a 0.9 kb promoter region of the Hapln1 gene contains a single Sp6 site was regulated by Sp6 co-expression, and this regulation was lost when the conserved sequence in the core motif was mutated in the promoter. Sp6-dependent transcriptional activity was also substantiated when four copies of the Sp6 binding site from Halpn1 was placed in front of a basal promoter. An Sp6-binding site derived from the Sp6 gene itself was controlled by Sp6 expression highlighting a regulatory loop that may amplify matrix deposition during critical developmental stages. Although our in vitro analysis with these regulatory regions from Hapln1 and Sp6 suggest Sp6 acts as an activator, it is possible that the transcriptional milieu during in vivo development is much more complex, and Sp6 may function as either a repressor or an activator depending on the stoichiometry of other nuclear factors.
Taken together these studies shed light on the Sp6 transcription factor and its regulatory activity via the core motif containing the CTg/aTAATTA DNA sequence. Mechanistically, the Sp6 TAATTA identified contrasts to other well-characterized GC-rich binding sequences required for activation by Sp1-Sp4 clade members. The Sp6 consensus sequence identified in our study is nearly identical to the known binding motif reported for Sp7, a factor controlling osteoblast specification of bone formation [22]. Sp7 knockout mice exhibit skeletal defects in ossification through alterations in the regulation of mineralizing chondrocytes. Comparison of the Sp6 and Sp7 ChIP-seq datasets revealed multiple common matrix gene targets (e.g., Hapln1, Col1a1, Col11a2) found in both dental and bone cell types (Supplementary Fig. 2). These findings demonstrate that Sp6 and Sp7 members can share essentially identical in vivo DNA elements, but function in a tissue-specific fashion by expression in either odontoblast-like cells, or osteoblasts, respectively. Overall these observations provide potential mechanistic insight for the development of tooth and bone mineralized tissues.
Supplementary Material
Acknowledgements
An appreciation to Dr. Yoshihiko Yamada, he passed in December 2019. We appreciate Dr. Paul Labhart (Active Motif Inc.) for ChIP-seq assistance. We thank Drs. Marian Young and Kenneth Yamada for critical feedback on the manuscript. This work was supported by the intramural research program of the National Institute of Dental and Craniofacial Research and Combined Technical Research Core Facility (ZICDE000729–09). This work utilized the High-Performance Computation Biowulf Cluster (NIH, http://hpc.nih.gov).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.10.017.
References
- [1].Jussila M, Thesleff I, Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages, Cold Spring Harb Perspect Biol 4 (2012) a008425, 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bartlett JD, Dental enamel development: proteinases and their enamel matrix substrates, ISRN Dent 2013 (2013) 684607, 10.1155/2013/684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ruan Q, Moradian-Oldak J, Amelogenin and enamel biomimetics, J. Mater. Chem. B 3 (2015) 3112–3129, 10.1039/C5TB00163C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fan D, Iijima M, Bromley KM, Yang X, Mathew S, Moradian-Oldak J, The cooperation of enamelin and amelogenin in controlling octacalcium phosphate crystal morphology, Cells Tissues Organs 194 (2011) 194–198, 10.1159/000324208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, Wakida K, Simmer JP, Yamada Y, Bartlett JD, Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis, J. Dent. Res. 77 (1998) 1580–1588, 10.1177/00220345980770080501. [DOI] [PubMed] [Google Scholar]
- [6].Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP, Functions of KLK4 and MMP-20 in dental enamel formation, Biol. Chem. 389 (2008) 695–700, 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].He P, Zhang Y, Kim SO, Radlanski RJ, Butcher K, Schneider RA, DenBesten PK, Ameloblast differentiation in the human developing tooth: effects of extracellular matrices, Matrix Biol. 29 (2010) 411–419, 10.1016/j.matbio.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hamada Y, Sumiyoshi H, Matsuo N, Yun-Feng W, Nakashima M, Yanagisawa S, Yoshioka H, The pro-alpha2(XI) collagen gene is expressed in odontoblasts, Biochem. Biophys. Res. Commun. 392 (2010) 166–170, 10.1016/j.bbrc.2010.01.001. [DOI] [PubMed] [Google Scholar]
- [9].Sehic A, Risnes S, Khan QE, Khuu C, Osmundsen H, Gene expression and dental enamel structure in developing mouse incisor, Eur. J. Oral Sci. 118 (2010) 118–130, 10.1111/j.1600-0722.2010.00722.x. [DOI] [PubMed] [Google Scholar]
- [10].Lacruz RS, Smith CE, Chen YB, Hubbard MJ, Hacia JG, Paine ML, Gene-expression analysis of early- and late-maturation-stage rat enamel organ, Eur. J. Oral Sci. 119 (Suppl 1) (2011) 149–157, 10.1111/j.1600-0722.2011.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eckhardt A, Jagr M, Pataridis S, Miksik I, Proteomic analysis of human tooth pulp: proteomics of human tooth, J. Endod. 40 (2014) 1961–1966, 10.1016/j.joen.2014.07.001. [DOI] [PubMed] [Google Scholar]
- [12].Jagr M, Ergang P, Pataridis S, Kolrosova M, Bartos M, Miksik I, Proteomic analysis of dentin-enamel junction and adjacent protein-containing enamel matrix layer of healthy human molar teeth, Eur. J. Oral Sci. 127 (2019) 112–121, 10.1111/eos.12594. [DOI] [PubMed] [Google Scholar]
- [13].Green DR, Schulte F, Lee KH, Pugach MK, Hardt M, Bidlack FB, Mapping the tooth enamel proteome and amelogenin phosphorylation onto mineralizing porcine tooth crowns, Front. Physiol. 10 (2019) 925, 10.3389/fphys.2019.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pemberton TJ, Li FY, Oka S, Mendoza-Fandino GA, Hsu YH, Bringas P Jr., Chai Y, Snead ML, Mehrian-Shai R, Patel PI, Identification of novel genes expressed during mouse tooth development by microarray gene expression analysis, Dev. Dynam. 236 (2007) 2245–2257, 10.1002/dvdy.21226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lacruz RS, Smith CE, Bringas P Jr., Chen YB, Smith SM, Snead ML, Kurtz I, Hacia JG, Hubbard MJ, Paine ML, Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling, J. Cell. Physiol. 227 (2012) 2264–2275, 10.1002/jcp.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chiba Y, Saito K, Martin D, Boger ET, Rhodes C, Yoshizaki K, Nakamura T, Yamada A, Morell RJ, Yamada Y, Fukumoto S, Single-cell RNA-sequencing from mouse incisor reveals dental epithelial cell-type specific genes, Front Cell Dev Biol 8 (2020) 841, 10.3389/fcell.2020.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhao M, Gupta V, Raj L, Roussel M, Bei M, A network of transcription factors operates during early tooth morphogenesis, Mol. Cell Biol. 33 (2013) 3099–3112, 10.1128/MCB.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramanathan A, Srijaya TC, Sukumaran P, Zain RB, Abu Kasim NH, Homeobox genes and tooth development: understanding the biological pathways and applications in regenerative dental science, Arch. Oral Biol. 85 (2018) 23–39, 10.1016/j.archoralbio.2017.09.033. [DOI] [PubMed] [Google Scholar]
- [19].Bei M, Molecular genetics of tooth development, Curr. Opin. Genet. Dev. 19 (2009) 504–510, 10.1016/j.gde.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ruspita I, Das P, Xia Y, Kelangi S, Miyoshi K, Noma T, Snead ML, D’Souza RN, Bei M, An Msx2-Sp6-Follistatin pathway operates during late stages of tooth development to control amelogenesis, Front. Physiol. 11 (2020) 582610, 10.3389/fphys.2020.582610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nakamura T, Unda F, de-Vega S, Vilaxa A, Fukumoto S, Yamada KM, Yamada Y, The Kruppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation, J. Biol. Chem. 279 (2004) 626–634, 10.1074/jbc.M307502200. [DOI] [PubMed] [Google Scholar]
- [22].Hojo H, Ohba S, He X, Lai LP, McMahon AP, Sp7/Osterix is restricted to bone-Forming vertebrates where it acts as a Dlx Co-factor in osteoblast specification, Dev. Cell 37 (2016) 238–253, 10.1016/j.devcel.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nakamura T, de Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y, Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number, J. Biol. Chem. 283 (2008) 4825–4833, 10.1074/jbc.M708388200. [DOI] [PubMed] [Google Scholar]
- [24].Hertveldt V, Louryan S, van Reeth T, Dreze P, van Vooren P, Szpirer J, Szpirer C, The development of several organs and appendages is impaired in mice lacking Sp6, Dev. Dynam. 237 (2008) 883–892, 10.1002/dvdy.21355. [DOI] [PubMed] [Google Scholar]
- [25].Bailey TL, Elkan C, Fitting a mixture model by expectation maximization to discover motifs in biopolymers, Proc. Int. Conf. Intell. Syst. Mol. Biol. 2 (1994) 28–36. [PubMed] [Google Scholar]
- [26].Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data, Nat. Biotechnol. 33 (2015) 495–502, 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rhodes C, Yamada Y, Characterization of a glucocorticoid responsive element and identification of an AT-rich element that regulate the link protein gene, Nucleic Acids Res. 23 (1995) 2305–2313, 10.1093/nar/23.12.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rhodes CS, Matsunobu T, Yamada Y, Analysis of a limb-specific regulatory element in the promoter of the link protein gene, Biochem. Biophys. Res. Commun. 518 (2019) 672–677, 10.1016/j.bbrc.2019.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu H, Snider TN, Wimer HF, Yamada SS, Yang T, Holmbeck K, Foster BL, Multiple essential MT1-MMP functions in tooth root formation, dentinogenesis, and tooth eruption, Matrix Biol. 52–54 (2016) 266–283, 10.1016/j.matbio.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang X, Chiba Y, Jia L, Yoshizaki K, Saito K, Yamada A, Qin M, Fukumoto S, Expression patterns of claudin family members during tooth development and the role of claudin-10 (Cldn10) in cytodifferentiation of stratum intermedium, Front Cell Dev Biol 8 (2020) 595593, 10.3389/fcell.2020.595593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yamauchi S, Cheng H, Neame P, Caterson B, Yamauchi M, Identification, partial characterization, and distribution of versican and link protein in bovine dental pulp, J. Dent. Res. 76 (1997) 1730–1736, 10.1177/00220345970760110301. [DOI] [PubMed] [Google Scholar]
- [32].Sato R, Yamamoto H, Kasai K, Yamauchi M, Distribution pattern of versican, link protein and hyaluronic acid in the rat periodontal ligament during experimental tooth movement, J. Periodontal. Res. 37 (2002) 15–22, 10.1034/j.1600-0765.2002.90770.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.