Abstract
Introduction
The mouse placenta accumulates and possibly produces serotonin (5-hydroxytryptamine; 5-HT) in parietal trophoblast giant cells (pTGC) located at the interface between the placenta and maternal deciduum. However, the roles of 5-HT in placental function are unclear. This lack of information is unfortunate, given that selective serotonin-reuptake inhibitors are commonly used to combat depression in pregnant women. The high affinity 5-HT transporter SLC6A4 (also known as SERT) is the target of such drugs and likely controls much of 5-HT uptake into pTGC and other placental cells. We hypothesized that ablation of the Slc6a4 gene would result in morphological changes correlated with placental gene expression changes, especially for those involved in nutrient acquisition and metabolism, and thereby, provide insights into 5-HT placental function.
Methods
Placentas were collected at embryonic age (E) 12.5 from Slc6a4 knockout (KO) and wild-type (WT) conceptuses. Histological, RNAseq, qPCR, and integrative correlation analyses were performed.
Results
Slc6a4 KO placentas had a considerable increased pTGC to spongiotrophoblast area ratio relative to WT placentas and significantly elevated expression of genes associated with intestinal functions, including nutrient sensing, uptake, and catabolism, and blood clotting. Integrative correlation analyses revealed upregulation of many of these genes was correlated with pTGC layer expansion. One other key gene was dopa decarboxylase (Ddc), which catalyzes conversion of L-5-hydroxytryptophan to 5-HT.
Discussion
Our studies possibly suggest a new paradigm relating to how 5-HT operates in the placenta, namely as a factor regulating metabolic functions and blood coagulation. We further suggest that pTGC might be functional analogs of enterochromaffin 5-HT-positive cells of the intestinal mucosa, which regulate similar activities within the gut. Further work, including proteomics and metabolomic studies, are needed to buttress our hypothesis.
Keywords: Neurotransmitter, Trophoblast, RNAseq, Gestation, Conceptus, Nutrient metabolism, Nutrient acquisition, Serotonin
1. Introduction
The presence of 5-HT in the human placenta was first reported in 1965 [1] and has been an active topic of interest since then because of its role in fetal brain development and likely involvement in autism spectrum disorders (ASD) [2, 3] and fetal growth restriction (FGR) [4–7]. Accordingly, a disruption in the provision of 5-HT to the fetus likely leads to later behavioral abnormalities. Whether it is synthesized in significant amounts by human placental TB remains controversial [8], although the machinery for 5-HT biosynthesis and catabolism and a capacity for its uptake have been demonstrated in certain human TB cell types [9–12].
Disagreements about the source of placental 5-HT also exist for the mouse [13]. 5-HT immunoreactivity is visible in the ectoplacental cone of the developing mouse placenta. During emergence of the junctional zone (embryonic age-E9–12), it is localized strongly in pTGC, particularly those nearest decidual blood vessels [14]. Additionally, embryos collected during the E9–12 period can accumulate 5-HT from culture medium, with uptake sensitive to fluoxetine, a 5-HT uptake inhibitor. An apparent concentration gradient from the pTGC through the spongioTB has been proposed as a potential route for provisioning the fetus with 5-HT. In this regard, there appears to be a progressive switch during brain development from an early placental source of 5-HT, which is vital for proper forebrain development, to a later endogenous source from the fetal brain itself [14, 15]. That early placental source has been proposed to be either 5-HT accrued from the mother [14, 16, 17] or synthesized in the placenta at the expense of maternal tryptophan [10, 13, 15, 18, 19]. Comparison of E7.5 to 9.5 mouse fetal placentas indicate that the genetic machinery to produce 5-HT is upregulated during this window of gestation [18].
Whether 5-HT is important in placental function has clinical relevance as approximately 8–10% of pregnant women are prescribed selective serotonin-reuptake inhibitors (SSRI) to combat depression [20, 21]. Such drugs act by binding to SLC6A4/SERT within the intracellular membrane, which in the central nervous system prevents the presynaptic cells from taking up 5-HT. Inhibition of SLC6A4/SERT results in increased concentrations of 5-HT in the synaptic space. It seems likely that such drugs also inhibit SLC6A4/SERT within placental TB. In vitro studies reveal SSRI disrupt various structural and hormonal properties of placental cell lines [22–24]. In rats, in utero exposure to venlafaxine reduces fetal placental weight [25]. Pregnant mothers using SSRI have been reported to deliver lower birthweight infants and exhibit higher rates of placental-fetal vascular malperfusion than controls [26].
A dramatic decrease in placental serotonin (5-HT) concentrations has been noted following bisphenol A (BPA) or bisphenol S (BPS) exposure [27]. Immunohistochemistry revealed that the main group of cells displaying 5-HT immunoreactivity (ir) were pTGC [27], as had been inferred by others [14]. In addition, placentas from BPA/BPS exposed conceptuses had a reduced spongioTB to pTGC area [27], suggesting that 5-HT from the pTGC might be necessary for proper spongioTB development. Placenta of Slc64a4/SERT KO mice at E18 are reduced in weight compared to controls and have increased apoptosis and areas of necrosis, hemorrhage, and fibrosis in the spongioTB region [12]. The placental phenotype of tryptophan hydroxylase 1 (Tph1)−/− whole body KO mouse is less severe, possibly due to some conversion of tryptophan to 5-HT via tryptophan hydroxylase 2 (TPH2) but is otherwise similar to that of the Slc64a4/SERT KO.
Past studies thus suggest that whether 5-HT is synthesized by the placenta or acquired from the mother this neurotransmitter is important for placental function. To further examine the role of placental signaling in the placenta, we examined the histopathological changes in the placenta of WT and Slc6a4 KO mice at embryonic age (E) 12.5, a stage when the placenta appears to have attained functional maturity, to determine whether the morphological phenotype of this mouse was consistent with that reported for Slc6a4 KO mice at a later stage in pregnancy [12]. Our hypothesis was that any histopathological changes relating to interference with 5-HT transport would be causatively linked to gene expression changes identified by RNAseq analysis.
2. Material and methods
2.1. Animals, breedings, and placental collections
All animal experiments were approved by the University of Missouri Animal Care and Use Committee (Protocol #9590) and conformed to the NIH Guidelines for the Care and Use of Laboratory Animals. To establish a Slc6a4/SERT KO colony, five founder pairs of heterozygous female and male mice for Slc6a4 tm1Kpl were purchased from Jackson Laboratory (JAX stock #008355, Bar Harbor, ME) and bred together. These mice were originally generated by Dr. Dennis Murphy [28, 29] and were backcrossed to C57BL/6J mice for at least 24–25 generations prior to arrival at this facility. At the Jackson Laboratory, the mice were bred with C57BL/6J for an additional generation. To determine the genotype of the resulting pups, PCR analysis was performed as described previously (https://www.jax.org/Protocol?stockNumber=008355&protocolID=28217). Unrelated heterozygous pairs were bred until sufficient animals were available to permit all needed placental collections. This proved to be extremely difficult. One issue in interbreeding these heterozygous mice is that the newborn pups have to be fostered at birth to avoid them being cannibalized. As noted on the Jackson Laboratory page, the heterozygous dams demonstrate a high anxiety phenotype that renders them poor mothers; thereby necessitating such cross-fostering approaches. For example, in attempts to expand numbers, we used CF1 females bred to CF1 males as surrogates (all mice purchased from Envigo, Madison, WI). These CF1 females had been paired at the same time as the Slc6a4 tm1Kpl heterozygotes. Vaginal plugs were examined daily in both colonies, and day of the vaginal plug was considered embryonic age (E) 0.5. If no vaginal plug was observed in the morning, males and females were separated and placed together again that evening. Expanding the numbers of Slc6a4 tm1Kpl heterozygotes in the colonies required over one year. Of 27 litters born, only 10 litters provided pups who survived through weaning. Within these litters, only 28 pups (16 males and 12 females) were the correct genotype and interbred for the subsequent placental collection studies.
In those Slc6a4 tm1Kpl heterozygote pairs bred for placental collections, the breeder female was humanely euthanized on E12.5. Fetal tissue was collected for PCR sexing. Approximately half of the fetal placental tissue was frozen in liquid nitrogen after the underlying uterine tissue had been dissected away, while the other half with underlying uterine tissue attached was fixed in 4 % (w/v) paraformaldehyde (PFA) for histological analyses. The fetal and placental collections and preservation were performed as described previously [27, 30]. Out of six pregnant females sacrificed, only four females were pregnant with fetuses. Out of these four dams, a total of 23 fetuses and placenta samples were collected. Three were female wild type, five were male wild type, three were male homozygotes and twelve were heterozygotes (7 females and 5 males). There were no female homozygotes in any of the collections, even though there were often female fetuses within the litters. At present have no reason to believe that Slc6a4 −/− in female conceptuses is lethal as pup numbers were so low. Accordingly, just the three Slc6a4 tm1Kpl male homozygote placentas and three control male placenta in the same collection of fetuses were selected and used.
2.2. Fetal PCR sexing
To determine conceptus sex, DNA from the fetus was isolated with the DNeasy Blood & Tissue Kit (Catalogue #69504; Qiagen, Gaithersburg, MD) from fetal tissue. Polymerase Chain Reaction (PCR) amplification was then performed for Sry (Y-chromosome specific gene, forward primer: 5’TCATGAGACTGCCAACCACAG3’; reverse primer: 5’CATGACCACCACCACCACCAA3’) and myogenin (Myog- autosomal control gene; forward primer: 5’TTACGTCCATCGTGGACAGC3’; reverse primer: 5’TGGGCTGGGTGTTAGTCTTA3’), as detailed in [31]. For this study, we focused on examining the placenta of male conceptuses.
2.3. Placental processing and histology
Prior to histological embedding, placenta tissue was marked with tattoo ink to aid in orientation. Sections of placenta for histology were fixed overnight in a 4% PFA solution at 4 °C. They were then subjected to three washes in 1X phosphate buffered saline (PBS) before being placed in histology cassettes (Fisher Scientific, St. Louis. MO) and stored in 70% ethanol until processed. Histological sections (4–5 μm thick) were cut at the IDEXX Co. Histology Laboratory (Columbia, MO) and stained with PAS-Hematoxylin (PAS-H). Histological slides were viewed, and images photographed under a Leica DM 5500B (Wetzlar, Germany) upright microscope with Leica DFC290 color digital camera at the University of Missouri Cytology Core Facility. Images taken from same tissue sections were stitched together by using Leica Application Suite software (V4.12, Leica). Morphometric analyses on the obtained images were performed with the Adobe Photoshop program (v2020, San Jose, CA). The labyrinth (LA), spongiotrophoblast (spongioTB), and parietal trophoblast giant cell (pTGC) areas were measured within each placental section and compared relative to each other to control for differences in size across sections examined.
2.4. Placental RNA isolation
RNA was isolated from each of the selected placental samples with the Qiagen AllPrep DNA/RNA/miRNA Universal Kit (Catalogue #80224; Qiagen, Germantown, MD). The quantity and quality of the RNA was determined with a Nanodrop ND1000 spectrophotometer (Nanodrop Products, Wilmington, DE). Results were further confirmed by analyzing RNA with the Fragment Analyzer (Agilent Technologies, Inc.). Only those samples that had an RNA quality number (RQN) score above 7 were selected for RNA sequencing (RNAseq).
2.5. Illumina TruSeq RNA library preparation and sequencing
Libraries were constructed per the manufacturer’s protocol with reagents supplied in Illumina’s TruSeq mRNA Stranded Library Preparation Kit and sequenced at the University of Missouri Genomics Technology Core Facility. In brief, poly-A containing mRNA was purified from total RNA (500 ng) on poly-T oligo beads, mRNA fragmented, and double-stranded cDNA generated from cleaved RNA by using random hexamers. The cDNA then underwent end repair and adapter ligation followed by PCR amplification. Unique dual indexing was used to mitigate index hopping events known to be an issue on patterned flow cells with exclusion amplification chemistry.
The size and purity of the final library was determined with the Fragment Analyzer (Agilent Technologies, Inc.) and quantified with the Qubit fluorometer by means of the Qubit dsDNA HS Assay kit (Invitrogen). Libraries were pooled and run on an Illumina NovaSeq 6000 sequencer by using a paired end 50 base read format on a S1 flow cell to generate ~35 million paired reads per sample. The actual number of reads and mapped reads obtained for each sample is listed in Supplementary Table 1.
2.6. RNAseq data processing
The reads were trimmed with cutadapt (Version 1.18), and reference: DOI:10.14806/ej.17.1.200 for Illumina adapters, for ambiguous nucleotides (N’s), and for artificial poly-G [32] for reads whose 3’ ends overlap with the adapter for a minimum of 3 bases with 90% identity. After trimming, reads with fewer than ten bases were discarded. The filtered trimmed reads were aligned to the reference mouse genome (GRCm38) by HISAT2 version 2.0.5 to achieve a high overall alignment (~94%) [33]. The aligned reads were further filtered to remove reads that mapped to the mitochondrial genome and those that aligned to pseudochromosomes. The number of reads that aligned to each protein-coding gene were counted with the featureCount tool from the Subread software suite Version 2.0.0 [34].
2.7. Differential gene expression analysis (DGEA): DESeq2
The raw read counts were used to carry out differential gene expression analysis (DGEA) by means of DESeq2 used previously to study the effects of maternal oxycodone exposure on the placental transcriptome [35]. Those sequences with an average of less than 5 read counts in at least one group were filtered out before carrying out DGEA. Genes were considered upregulated if they had an absolute fold-change ≥ 1.5 and adjusted p-value ≤ 0.05.
2.8. Tissue-specific gene enrichment analysis
Tissue-specific gene enrichment analysis was determined by TissueEnrich [36]. We used the mouse ENCODE [37] dataset to carry out the enrichment analysis with default settings. Enrichments were considered significant if p was ≤ 0.01 and fold-change ≥ 2.
2.9. Functional enrichment analysis
Potential protein-protein interactions (PPI) for proteins encoded by DEG for the comparisons between Slc6a4 KO mice and control mice were determined with the STRING Database [38]. The PPI files generated with STRING were imported into the Cytohubba app [39] in Cytoscape [40] in order to identify so-called hub genes by using Maximal Clique Centrality (MCC) analysis [39]. Hub genes are inferred to have key roles in controlling unrelated biological pathways. For functional enrichment analysis, DEG were imported into the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) [41] and GO molecular biological terms were searched to determine which pathways might be affected.
2.10. qPCR validation
Ten DEG that were considered hub genes, as determined by Cytohubba app [39] in Cytoscape [40], were analyzed by qPCR analysis. Total RNA, which had been treated with DNase to remove any genomic DNA contamination, was reverse transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Catalogue #205310, Qiagen). The qPCR procedure was performed on The Bio-Rad CFX Connect Real-Time PCR Detection System (Bio-Rad). Primers were designed by using NCBI Primer-Blast online (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and purchased from IDT technologies (Coralville, IA). Primer sequences and efficiencies for the genes examined are listed in Supplementary Table 2. The Bio-Rad SYBR Green Master Mix (Catalogue # 1725121, Hercules, CA) was used according to the manufacture’s protocol. The cycling conditions for qPCR were 1) 95°C for 5 min for polymerase activation 2) 40 cycles of denaturation 15 sec at 95 °C, annealing and extension 30 sec at 56 °C, and 3) dissociation melt curve analysis of denaturation at 95°C, complete annealing at 56 °C, followed by a gradual increase in temperature up to 95°. Ubiquitin C (Ubc), Rpl7, and Gapdh genes were used as internal reference controls. Ubc has been shown to be an appropriate reference control for the mouse placenta [42]. The two additional reference controls Ubc and Gapdh, were included to check the strength and confidence of the comparisons.
2.11. Statistical analyses for placental histology and qPCR analyses
Data for dependent variables ratios between LA, spongioTB, and pTGC areas in placenta were analyzed for normality by using the Wilk-Shapiro test (V9.4; SAS Analytics, Cary, NC). These data were then analyzed by the PROC MIXED procedure of SAS v9.4. Sources of variation considered were treatment, sex, and treatment × sex interaction with an individual mouse serving as the experimental unit to determine treatment effects. For all data, a p value of ≤0.05 was considered significant. All data are presented as actual means . The error bars for all figures and reported data represent the standard error of the mean (SEM).
Gene expression data as determined by qPCR analyses were normalized by using combined average dCt values of the three housekeeping genes: Rpl7, Gapdh and Ubc and then analyzed based on treatment X sex interactions with the PROC GLM procedure of SAS. Graphs are based though on 2−ΔΔCt values relative to CTL values for each sex with the mean value for these CTL groups set to 1 for graphing purposes. For all data, a p-value of ≤ 0.05 was considered significant. All data are presented as means ± standard error of the means (SEM).
2.12. Weighted correlation network analysis (WGCNA)
WGCNA describes correlation patterns among genes based on their gene expression data. The WGCNA package in R [43] was used to find unsigned weighted gene co-expression modules. The blockwiseModules function was run with a soft thresholding power of 18 to indicate the gene cluster. In order to identify significant gene modules, correlation scores between genes and traits were used to rank the gene cluster, which utilized the eigengene network methodology through use of Pearson Correlation/Bicor. Trait data used for this analysis were LA to spongioTB, LA to pTGC ratio, and spongioTB to pTGC ratio. Genes within module eigengenes (ME) that were significant were further analyzed as detailed for functional enrichment analyses for DEG.
3. Results
3.1. Histopathological changes in Slc6a4 (SERT) KO placenta
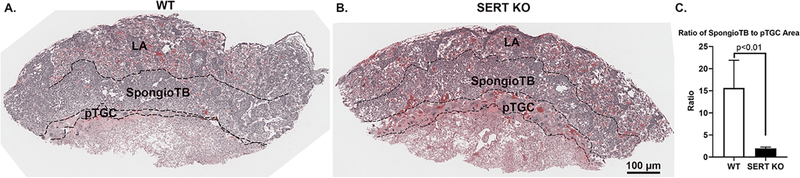
The relative areas (measured empirically as pixels) of pTGC, spongiotrophoblast, and labyrinth in E12.5 placentas are shown in Table 1. There was no significant difference in the relative areas of labyrinth and spongioTB between the Slc6a4 KO conceptuses and controls. However, the pTGC area of the Slc6a4 KO conceptuses was 3.6-fold greater than that in WT conceptuses (p = 0.002). The large variance in all area values is likely due to two factors: 1) the initial slicing of the placenta prior to freezing was not precisely symmetrical and 2) the orientation of the tissue in the block when the sections were cut was not precise. The ratios of the LA to spongioTB, LA to pTGC ratio, and spongioTB to pTGC were determined from these pixel values (Table 1; Fig. 1). The only ratio found to be significantly different between the two genotypes was that of spongioTB to pTGC area (Fig. 1), although there was also a trend for the LA to pTGC areas to be reduced in the Slc6a4 KO placentas relative to WT placenta (p = 0.06).
Table 1.
Morphological comparisons of WT vs. Slc6a4 (SERT) KO placenta.
| WT Placenta | Slc6a4 (SERT) KO Placenta | P value | |||
|---|---|---|---|---|---|
|
| |||||
| Variable | Mean (Pixels) | SEM | Mean (Pixels) | SEM | Significance |
|
| |||||
| LA | 3386269.25 | 764900.34 | 3452322.00 | 199735.67 | N.S. |
| SpongioTB | 3928196.75 | 679603.32 | 3116243.67 | 249725.72 | N.S. |
| pTGC | 453818.25 | 48274.79 | 1639780.00 | 237970.99 | 0.002 |
| LA:pTGC ratio | 7.5950000 | 1.8281328 | 2.2175667 | 0.3870017 | 0.06 |
| pTGC:SpongioTB ratio | 0.1244750 | 0.0215799 | 0.5323000 | 0.0871145 | 0.003 |
| LA:SpongioTB ratio | 0.8520250 | 0.0896600 | 1.1177667 | 0.0895804 | N.S. |
N.S.- not significant
Fig. 1.
Placental histology of SERT KO and WT mice. Panels A and B show the placental histology for WT and SERT KO conceptuses, respectively. C) Depicts the results of the spongioTB to pTGC in both placentas. As shown, this ratio is reduced in SERT KO vs. WT placenta, which is likely attributed to increase thickness of the pTGC in the former. LA= labyrinth; spongioTB= spongiotrophoblast; pTGC= parietal trophoblast giant cells. N = 3 SERT KO and 3 WT placenta.
3.2. RNAseq results
RNAseq was performed on placentas of Slc6a4 KO and WT conceptuses at E 12.5. The average number of paired-end reads was 253,694,156, the average percent alignment 93.8%, and the average number of mapped paired-end reads 224,893,694 (Supplementary Table 1). These numbers are considered more than sufficient for eukaryotic transcriptomes, and considerably greater than our past transcriptome studies with the mouse placenta [27, 44].
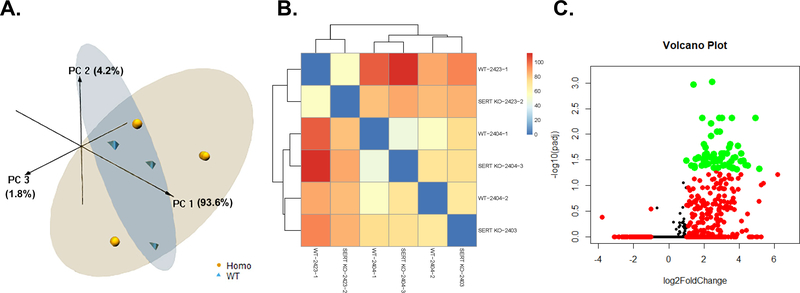
Principal Component Analyses (PCA) of Slc6a4 KO vs. WT placenta revealed that the PC1, PC2, and PC3 axes accounted for 93.6, 4.2, and 1.8%, respectively, of the variation (Fig. 2A). While no clear separation was noted in heatmap analysis (Fig. 2B), a volcano plot based on an adjusted P values (false discovery rate, FDR) revealed several genes that were considered differentially expressed, all of which were upregulated in the Slc6a4 KO vs. WT placenta (Fig. 2C).
Fig. 2.
Placental gene expression in SERT KO vs. WT conceptuses. A) 3D PCA plot of RNAseq results from placenta of SERT KO males vs. WT males. B) Heatmap based on all transcripts identified in placenta of SERT KO males vs. WT males. d. C) Volcano plot analyses of gene transcripts identified in placenta of SERT KO males vs. WT males. Transcripts represented by Orange: FDR < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: FDR < 0.05 AND log2FoldChange < −1 or > 1. N = 3 SERT KO and 3 WT placenta.
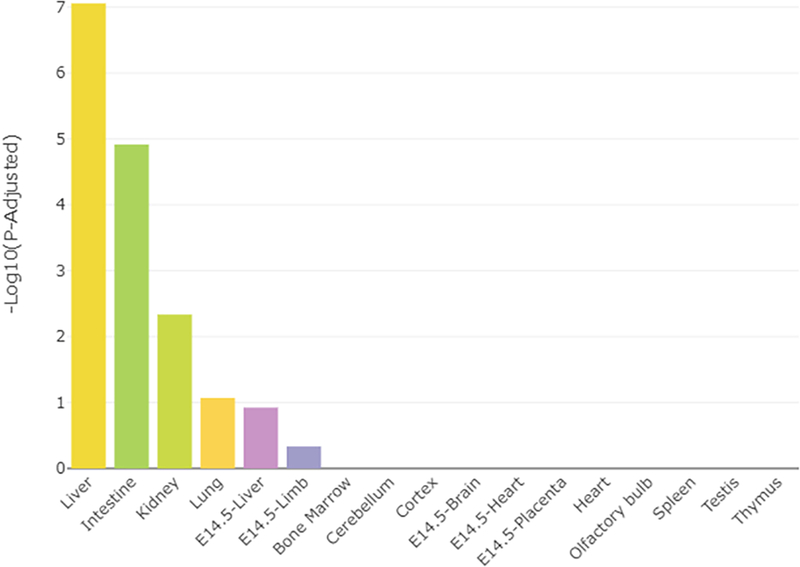
The full list of all the DEG identified in Slc6a4 KO vs. CTL placentas is provided in Supplementary Dataset 1. The top 10 DEG based on FDR are listed in Table 1. The TissueEnrich program [37] was then used to determine whether or not these DEG had placenta-specific gene expression patterns. They did not. As shown in Fig. 3, they were more closely associated with liver, lung, and kidney function and linked to nutrient absorption and metabolism functions.
Fig. 3.
TissueEnrich results for SERT KO vs. WT placentas. Bar plot distribution of the tissue enrichment of all DE genes.
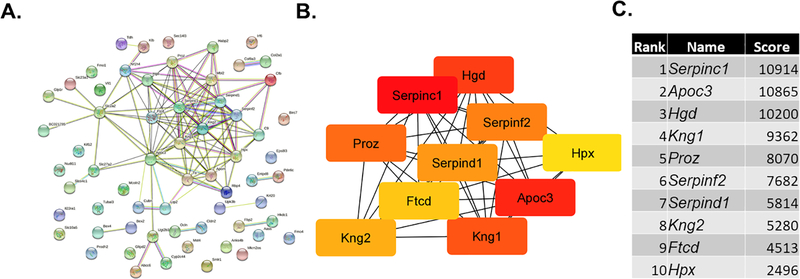
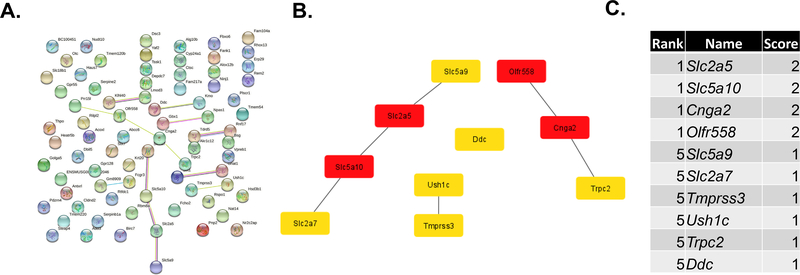
The STRING program was then used to examine potential protein-protein interaction pathways among the protein products encoded by the DEG, and this information imported into the cytohubba app [39] in Cytoscape [40] to identify the top hub genes. Both the STRING and cytohubba analyses revealed one main cluster that included 10 hub genes Serpinc1, Apoc3, Hgd, Kng1, Proz, Serpinf2, Serpind1, Kng2, Ftcd, and Hpx (Fig. 4).
Fig. 4.
STRING and hub gene analyses for SERT KO males vs. WT males. The protein-protein-interactions (PPI) were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [39] in Cytoscape [40] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [39].
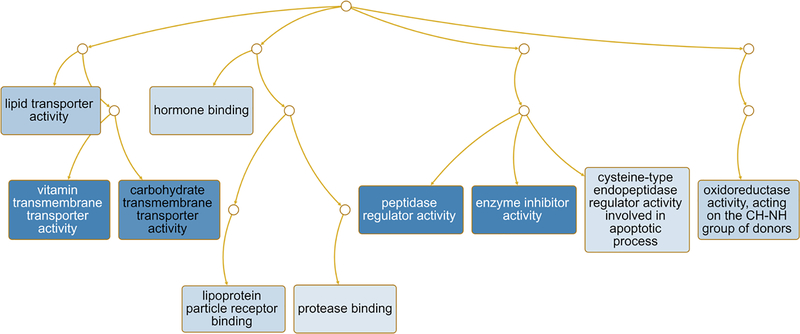
The WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) [41] and GO molecular biological terms was also performed on the DEG. This analysis revealed that the main pathways likely to be altered in the placenta of Slc6a4 KO conceptuses were lipid transporter activity, vitamin transmembrane transporter activity, carbohydrate transmembrane transporter activity, hormone binding, lipoprotein particle receptor binding, protease binding, peptidase regulator activity, enzyme inhibitor activity, cysteine-type endopeptidase regular activity involved in apoptotic process, and oxidoreductase activity acting on the CH-NH group of donors (Fig. 5).
Fig. 5.
Pathways predicted to be affected in SERT KO vs. WT placenta. Functional enrichment analyses with the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) [41] and GO molecular function terms was performed based on DE genes to determine which pathways might be affected in SERT KO relative to WT placenta. As shown in this Fig., many of the pathways involve nutrient, enzyme, hormone binding activity, including for vitamins, lipids, carbohydrates, and proteins. Those categories surrounded by a dark blue box are significant based on a false discovery rate (FDR) ≤ 0.05, whereas categories surrounded by a light blue box are significant based on a p value ≤ 0.05. Connecting lines indicate inter-relationships between categories.
3.3. qPCR results
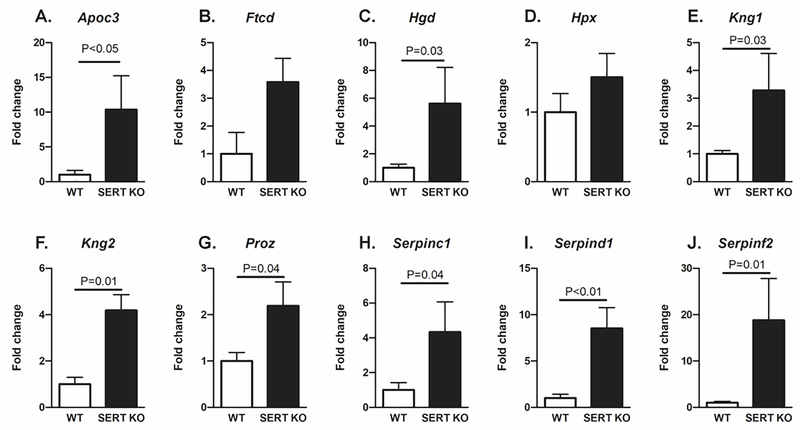
For the qPCR analyses, we selected genes that were determined to be hub genes based on the above analyses. In general, the qPCR results confirmed the RNAseq data. Apoc3, Hgd, Kng1, Kng2, Proz, Serpinc1, Serpind1, and SerpinF2 were significantly upregulated in placentas from Slc6a4 KO mice relative to WT (Fig. 6). Ftcd and Hpx transcripts also tended to be elevated in Slc6a4 KO but the considerable variance in values between individual mice precluded assigning significance.
Fig. 6.
qPCR results for genes identified to be hub genes. The qPCR results largely confirmed the RNAseq data. Apoc3, Hgd, Kng1, Kng2, Proz, Serpinc1, Serpind1, and SerpinF2 were significantly upregulated in SERT KO mice relative to WT. Ftcd and Hpx also showed a trend to be elevated in SERT KO vs. WT mice. N = 3 SERT KO and 3 WT placenta.
3.4. WGCNA results
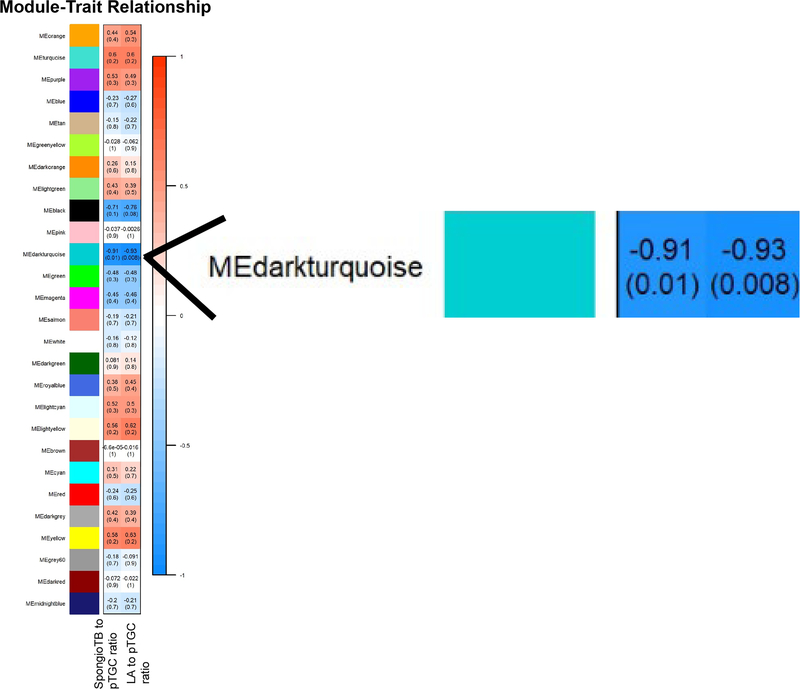
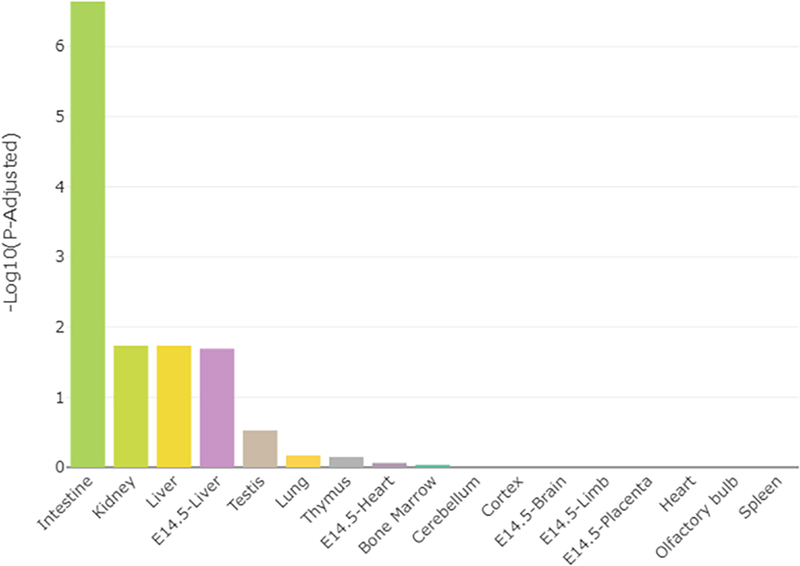
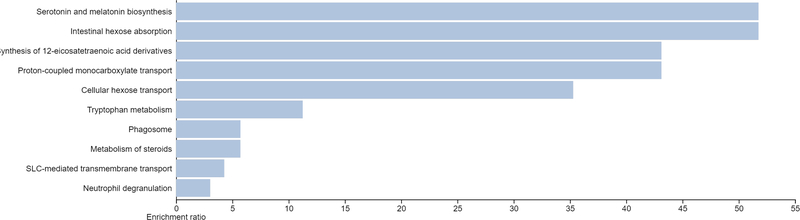
WGCNA [43] was used to link genes identified as differentially expressed between Slc6a4 KO and WT placentas and placental morphometric analyses (LA to pTGC ratio and spongioTB to pTGC ratio). For these analyses, 23,558 genes for Slc6a4 KO vs. WT placenta were used for gene module and placenta morphometrics correlation analyses. Individual modules were indicated by different colors, and module eigengenes (ME) were then correlated with the histological indices above. Different color modules are defined as clusters of densely inter-connected genes as determined by the program [43]. The grey color module, however, includes those genes that do not fit into the other modules. Supplementary Fig. 1 shows the dendrogram based on the different color modules based on the genes identified in the Slc6a4 KO vs WT placenta. As shown in the heatmap for ME and trait relationships, only the ME dark turquoise was significantly and inversely correlated with these histological assessments (Fig. 7). Thus, we further examined the genes within this ME as listed in Supplementary Dataset 2. TissueEnrich analyses of genes in the ME dark turquoise module revealed that they most closely align with gene expression profiles of the intestine, kidney, and liver (Fig. 8). STRING analyses revealed relatively loose associations between genes within this module, but cytohubba analyses revealed some commonalities within the top 10 hub genes: Slc2a5, Slc5a10, Cnga2, Olf558, Slc5a9, Slc2a7, Tmprss3, Ush1, Trpc2, and Ddc (Fig. 9) with most of these genes regulating nutrient and ion transfer. One notable exception is Ddc, dopa decarboxylase, which catalyzes the decarboxylation of L-3,4-dihydroxyphenylalanine (DOPA) to dopamine and L-5-hydroxytryptophan to 5-HT. While Ddc was not significant based on an FDR, it was significantly upregulated in Slc6a4 KO based on p value alone (p = 0.002, Supplementary Dataset 1). The WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) [41] and KEGG and REACTOME analyses of genes in the ME dark turquoise module revealed that the top pathways likely to be affected were serotonin and melatonin biosynthesis, intestinal hexose absorption, synthesis of 12-eicosatetraenoic acid derivatives, proton-coupled monocarboxylate transport, cellular hexose transport, and tryptophan metabolism (Fig. 10).
Fig. 7.
Relationship of WGCNA results and placental morphometric assessments for SERT KO vs. WT conceptuses. Modules identified in Supplementary Fig. 1 were correlated with placental morphometric assessments (Fig. 1). Each row corresponds to a Module Eigengene (ME), and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figure.
Fig. 8.
TissueEnrich results for genes in the ME dark turquoise module that was shown to be significantly correlated with the placental histology results.
Fig. 9.
STRING and hub gene analyses for genes in the ME dark turquoise module. The protein-protein-interactions (PPI) were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [39] in Cytoscape [40] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [39].
Fig. 10.
Pathways predicted to be affected based on gene in the ME dark turquoise module. Functional enrichment analyses with the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) [41] and screened with KEGG and REACTOME pathways. The top pathways likely to be affected based on gene-sets in this module are serotonin and melatonin biosynthesis, intestinal hexose absorption, synthesis of 12-eicosatetraenoic acid derivatives, proton-coupled monocarboxylate transport, cellular hexose transport, and tryptophan metabolism. All categories listed are significant based on a p-value of ≤ 0.05.
4. Discussion
The goals of the current experiments were, first, to examine how loss of Slc6a4 affected the morphology of the mouse placenta at E 12.5, when all of the layers of the placenta have developed and are presumably functional. Second, we sought to determine how absence of this gene affected global gene expression patterns in the placenta, given that 5-HT has been inferred to play a role in placental function in mice and humans [1, 10, 13–19]. Finally, integrative correlation analyses were performed to link phenotypic changes in the Slc6a4 KO placenta to gene expression profiles.
Histological analyses at E 12.5 revealed a reduction in the spongioTB to pTGC layer in Slc6a4 KO relative to WT placentas. This shift appeared to be due primarily to an increased thickness of the pTGC layer in Slc64a4 KO, although there was also a trend towards decreased area of the spongioTB. In this regard our study is consistent with a previous report that the placentas of Slc64a4/SERT KO mice at E18 were reduced in weight compared to controls and had increased apoptosis, areas of necrosis, and hemorrhage and fibrosis in the spongioTB region [12], although no changes in pTGC numbers or area were reported at that late stage of pregnancy. The placenta is a dynamic organ. Thus, it is plausible that initially the absence of SLC6A4 causes hypertrophy of the pTGC layer as a potential compensatory mechanism for its inability to acquire 5-HT from maternal sources [17]. The upregulation of Ddc, which encodes an enzyme essential for 5-HT synthesis, in the placentas of the Slc6a4 KO mice is consistent with this hypothesis. A reliance of spongioTB on the pTGC to supply 5-HT and other factors may also explain the reduced placental weight and necrosis within the spongioTB layer observed in the KO mice at E18.5 [12].
In previous work we showed that developmental exposure to the endocrine disruptors, bisphenol A and bisphenol S cause an apparent shrinkage in the number of pTGC, evident as an increase in the spongioTB to pTGC area ratio. There was also a concomitant drop in the placental content of 5-HT and a reduction of 5-HT immunoreactivity in the pTGC [12]. Since BPA-exposed conceptuses also demonstrate a reduction in placental size, as well as pathological lesions in the spongioTB [12], these data together are consistent with a role for 5-HT in governing placental homeostasis and strongly suggest that pTGC are the prime targets for endocrine disrupting chemicals in the mouse placenta.
RNAseq analyses of transcripts from Slc6a4-KO and WT revealed the surprising fact that every gene that had been differentially regulated based on FDR had, in fact, been upregulated, possibly as an acute compensatory response to loss of 5-HT. Moreover, based on a TissueEnrich analysis, these genes were largely known to be associated with liver, lung, and kidney functions. Thus, loss of the 5-HT transporter likely impacts nutrient absorption and related metabolic functions. Among those genes prominently upregulated in the Slc6a4 KO placenta were several hub genes (Serpinc1, Serpinf2, Serpind1, Kng1, Kng2, Apoc3, Hgd, Proz, Ftcd, and Hpx). Of these Serpinc1, Serpind1, Serpinf2, Kng1, and Kng2 are implicated in various arms of either the blood coagulation or de-coagulation pathways. SERPINC1, known as anti-thrombin III, inhibits thrombin, a protease responsible for the formation of fibrin clots. Similarly, SERPIND1 (heparin cofactor 2) also inhibits thrombin formation, while, by contrast, SERPINF2 is a plasmin inhibitor and prevents clot dissolution [45]. KNG1 and KNG2 are thiol protease inhibitors and have complex roles in controlling blood coagulation, as does PROZ, a vitamin K dependent plasma glycoprotein. These observations may explain why one of the pathological changes noted in E18 placenta of Slc6a4 KO is hemorrhage within the spongioTB layer [12]. Even at E12.5 (Fig. 1) our micrographs show some indications of capillary blood leakage in this tissue. We suggest, therefore, that serotonin has some role in controlling the balance between hemorrhage and clotting as placental blood flow to the maturing placenta increases.
Pathway enrichment analysis suggest that Slc6a4 KO placentas demonstrate a marked upregulation of pathways regulating nutrient uptake and metabolism (Fig. 5), in addition to blood coagulation. For example, among the upregulated genes in the Slc6a4 KO placenta that are correlated between 5-HT, nutrient absorption, and placental morphology (Fig. 7) are ones encoding sugar transporters Slc2a5, Slc2a7, Slc5a9, and Slc5a10, lipid metabolism (Apoe), and yet others implicated in nutrient sensing (Cnga2, Tmprss3, and Trpc2). Similar to other olfactory receptor genes, Olfr558 acts as a nutrient sensor. Thus, while 5-HT has been classically considered a neurotransmitter, emerging data, including our own, points to a possibly equally important role as an endocrine factor whose production is altered by nutrient status and in turn regulates nutrient transport and metabolism [46]. Intriguingly, Slc6a4 KO mice have increased enterocyte mass and enhanced capacity for intestinal absorption of glucose and peptides [47]. Also, in wild type mice, the SSRI, fluoxetine suppresses intestinal absorption of amino acids, especially leucine [48]. If, as it would appear, 5-HT is metabolic regulator common to both the intestine and placenta, we suggest that placental pTGC are functional analogs of the large, 5-HT-storing enterochromaffin cells of the gut, which, directly or indirectly, respond to nutrients and other metabolites by releasing 5-HT [49]. These cells also provide a local source of 5-HT for platelets and their blood clotting activities [50]. To carry this analogy further, we predict that the 5-HT released from pTGC has a role in differentiation, gene expression, and functional activities of spongiotrophoblast and labyrinth regions of the placenta and limits clot formation in local capillaries.
One other upregulated (p = 0.01) hub gene present in the complement of genes linked to the increased thickness of the pTGC layer (dark turquoise module, Fig. 7) is Ddc, which encodes the enzyme (dopa decarboxylase) that regulates the decarboxylation of L-3,4-dihydroxyphenylalanine (DOPA) to dopamine and L-5-hydroxytryptophan to 5-HT. Pathway enrichment analyses of genes within this module also provides robust support for another major metabolic pathway likely affected in Slc6a4 KO placenta in addition to nutrient acquisition and blood clotting, namely serotonin and melatonin biosynthesis. Like 5-HT, melatonin has been proposed to regulate aspects of placental function in an autocrine/paracrine manner in both the human [51, 52] and possibly the mouse placenta [53]. Whether the human placenta possesses cells with analogous function to rodent pTGC and gut enterochromaffin 5-HT-positive cells is unknown, but the possibility of their presence and a role in placental function is intriguing.
Both the mouse and human placenta express TPH1 and monoamine oxidase A and B (MAO-A and -B) that are involved in the synthesis and catabolism of 5-HT, respectively [8, 13, 15, 54–60]. However, the expression of the genes encoding these enzymes were not altered in SERT KO relative to WT placenta. Additionally, although genes that encode key enzymatic components of the 5-HT biosynthetic and catabolic pathways are expressed in the human and mouse placentas [8, 13, 15, 54–60] it still remains unclear whether or not the main source of fetal 5-HT is placental or maternal. Kliman et al. [17], for example, discount any significant synthesis of 5-HT by placental trophoblast and suggest that all that required by the placental-fetal unit is via uptake from the mother and involves the SLC6A4/SERT transporter. Moreover, they consider the source of the 5-HT to be chromaffin cells of the gut with platelets acting as intermediaries between the gut and the placenta. Although a requirement for accumulated placental 5-HT by the developing fetal brain has not been established, 5-HT does have modulating effects on the development and metabolic activities of trophoblast cells in vitro [22, 61, 62] and presumably during human pregnancy.
At the outset, we sought to examine both male and female placenta from Slc6a4 KO and WT mice. However, for reasons that are unclear we were only able to obtain sufficient number of male KO placentas. We recently found out that another whole body Slc6a4 KO mouse model has been generated by Dr. Jiying Sze at Albert Einstein College of Medicine [63]. Additionally, her laboratory generated Slc6a4fl/fl mice that offer the possibility of ablating this gene selectively in the placenta. Both strains are fully fertile, and we are in the process of obtaining both of them. Previous work has suggested that genetic background can influence behavioral phenotypes of Slc6a4 transgenic mice [64], among which might be maternal behaviors and general ability to care for offspring. These lines from the Sze laboratory should readily yield sufficient male and female placentas with specific deletion of SLC6A4/SERT in trophoblast to determine sexually dimorphic difference and how absence of SLC6A4/SERT in placenta affects brain and behavioral functions.
Another limitation of the current study is that it solely based on a transcriptomics approach. Additional experiments that utilize a variety of other omics procedures, particularly proteomics, will be essential for supporting these gene expression results and predictions on pathways that might be affected by lack of SLC6A4. The data reported herein are also based on a single timepoint in gestation, E 12.5, which was selected as it represents mid-trimester when the placental is fully developed and function. However, it is essential that other timepoints in gestation, spanning the initial stage of chorioallantoic placental formation through parturition are considered, as the role and source of 5-HT in the placenta might alter as pregnancy proceeds. While past work suggests that the high affinity SERT/SLC6A4 transporter may dominate transport of 5-HT [17], other transporters could be involved, as well. For instance, OCT3/SLC22A3 may be active transport of 5-HT within the fetus, and movement of 5-HT by this mechanism may be dependent of fetal sex [60, 65]. Intriguingly, SSRI anti-depressants also appear to block SLC22A3. Finally, it is conceivable that one compensatory mechanism for loss of SLC6A4 in placentas of Slc6a4−/− mice is upregulation of Slc22a3, although this was not evident from our transcriptomic data. In short, we cannot exclude the possibility that some 5-HT continues to be transported both within and into the placenta by means of lower affinity carriers, such as SLC22A3.
In summary, our studies suggest a new paradigm in considering how 5-HT acts in the placenta, namely not so much as a classic neurotransmitter but as an endocrine factor regulating metabolic functions, blood clotting, and other processes through autocrine actions. We further suggest that the pTGC are functional analogs of the enterochromaffin 5-HT-positive cells of the gut, which have a role in controlling similar activities of intestinal epithelium.
Supplementary Material
Supplementary Fig. 1. Dendrogram and heatmap of WGCNA results for SERT KO vs. WT placental comparison. As detailed in [43], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image.
Highlights.
Parietal trophoblast giant cells (pTGC) accrue and/or produce serotonin (5-HT).
Ablation of Slc6a4, the 5-HT transporter, causes expansion of the pTGC layer.
Slc6a4 KO placentas display elevation of genes linked to metabolite transport.
Placental 5-HT may have a broad regulatory role unrelated to neurotransmitter function.
Acknowledgements
The work was supported by 1R01ES025547 (to CSR) and 1R01HD094937 (to RMR). We appreciate the assistance of several undergraduate students in taking care of the mice.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
Transcriptome data files have been deposited and will be available on the NCBI GEO Database.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Koren Z, Pfeifer Y, Sulman FG, Serotonin content of human placenta and fetus during pregnancy, Am J Obstet Gynecol 93 (1965) 411–5. [DOI] [PubMed] [Google Scholar]
- [2].Sato K, Placenta-derived hypo-serotonin situations in the developing forebrain cause autism, Med. Hypotheses 80(4) (2013) 368–72. [DOI] [PubMed] [Google Scholar]
- [3].Yang CJ, Tan HP, Du YJ, The developmental disruptions of serotonin signaling may involved in autism during early brain development, Neuroscience 267 (2014) 1–10. [DOI] [PubMed] [Google Scholar]
- [4].Ranzil S, Ellery S, Walker DW, Vaillancourt C, Alfaidy N, Bonnin A, Borg A, Wallace EM, Ebeling PR, Erwich JJ, Murthi P, Disrupted placental serotonin synthetic pathway and increased placental serotonin: Potential implications in the pathogenesis of human fetal growth restriction, Placenta (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ranzil S, Walker DW, Borg AJ, Wallace EM, Ebeling PR, Murthi P, The relationship between the placental serotonin pathway and fetal growth restriction, Biochimie 161 (2019) 80–87. [DOI] [PubMed] [Google Scholar]
- [6].Rosenfeld CS, The placenta-brain-axis, J Neurosci Res (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rosenfeld CS, Placental serotonin signaling, pregnancy outcomes, and regulation of fetal brain developmentdagger, Biol Reprod 102(3) (2020) 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Laurent L, Deroy K, St-Pierre J, Cote F, Sanderson JT, Vaillancourt C, Human placenta expresses both peripheral and neuronal isoform of tryptophan hydroxylase, Biochimie 140 (2017) 159–165. [DOI] [PubMed] [Google Scholar]
- [9].Vaillancourt C, Petit A, Gallo-Payet N, Bellabarba D, Lehoux JG, Belisle S, Labelling of D2-dopaminergic and 5-HT2-serotonergic binding sites in human trophoblastic cells using [3H]-spiperone, J Recept Res 14(1) (1994) 11–22. [DOI] [PubMed] [Google Scholar]
- [10].Huang WQ, Zhang CL, Di XY, Zhang RQ, Studies on the localization of 5-hydroxytryptamine and its receptors in human placenta, Placenta 19(8) (1998) 655–61. [DOI] [PubMed] [Google Scholar]
- [11].Viau M, Lafond J, Vaillancourt C, Expression of placental serotonin transporter and 5-HT 2A receptor in normal and gestational diabetes mellitus pregnancies, Reprod Biomed Online 19(2) (2009) 207–15. [DOI] [PubMed] [Google Scholar]
- [12].Hadden C, Fahmi T, Cooper A, Savenka AV, Lupashin VV, Roberts DJ, Maroteaux L, Hauguel-de Mouzon S, Kilic F, Serotonin transporter protects the placental cells against apoptosis in caspase 3-independent pathway, J Cell Physiol 232(12) (2017) 3520–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bonnin A, Levitt P, Fetal, maternal, and placental sources of serotonin and new implications for developmental programming of the brain, Neuroscience 197 (2011) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yavarone MS, Shuey DL, Sadler TW, Lauder JM, Serotonin uptake in the ectoplacental cone and placenta of the mouse, Placenta 14(2) (1993) 149–161. [DOI] [PubMed] [Google Scholar]
- [15].Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, Blakely RD, Deneris ES, Levitt P, A transient placental source of serotonin for the fetal forebrain, Nature 472(7343) (2011) 347–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cote F, Fligny C, Bayard E, Launay JM, Gershon MD, Mallet J, Vodjdani G, Maternal serotonin is crucial for murine embryonic development, Proc Natl Acad Sci USA 104(1) (2007) 329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kliman HJ, Quaratella SB, Setaro AC, Siegman EC, Subha ZT, Tal R, Milano KM, Steck TL, Pathway of maternal serotonin to the human embryo and fetus, Endocrinology 159(4) (2018) 1609–1629. [DOI] [PubMed] [Google Scholar]
- [18].Tuteja G, Chung T, Bejerano G, Changes in the enhancer landscape during early placental development uncover a trophoblast invasion gene-enhancer network, Placenta 37 (2016) 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Muller CL, Anacker AM, Rogers TD, Goeden N, Keller EH, Forsberg CG, Kerr TM, Wender C, Anderson GM, Stanwood GD, Blakely RD, Bonnin A, Veenstra-VanderWeele J, Impact of maternal serotonin transporter genotype on placental serotonin, fetal forebrain serotonin, and neurodevelopment, Neuropsychopharmacol 42(2) (2017) 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huybrechts KF, Palmsten K, Mogun H, Kowal M, Avorn J, Setoguchi-Iwata S, Hernandez-Diaz S, National trends in antidepressant medication treatment among publicly insured pregnant women, Gen Hosp Psychiatry 35(3) (2013) 265–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mitchell AA, Gilboa SM, Werler MM, Kelley KE, Louik C, Hernandez-Diaz S, Medication use during pregnancy, with particular focus on prescription drugs: 1976–2008, Am J Obstet Gynecol 205(1) (2011) 51.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Clabault H, Flipo D, Guibourdenche J, Fournier T, Sanderson JT, Vaillancourt C, Effects of selective serotonin-reuptake inhibitors (SSRIs) on human villous trophoblasts syncytialization, Toxicol. Appl. Pharmacol. 349 (2018) 8–20. [DOI] [PubMed] [Google Scholar]
- [23].Hudon Thibeault AA, Laurent L, Vo Duy S, Sauve S, Caron P, Guillemette C, Sanderson JT, Vaillancourt C, Fluoxetine and its active metabolite norfluoxetine disrupt estrogen synthesis in a co-culture model of the feto-placental unit, Mol. Cell. Endocrinol. 442 (2017) 32–39. [DOI] [PubMed] [Google Scholar]
- [24].Clabault H, Cohen M, Vaillancourt C, Sanderson JT, Effects of selective serotonin-reuptake inhibitors (SSRIs) in JEG-3 and HIPEC cell models of the extravillous trophoblast, Placenta 72–73 (2018) 62–73. [DOI] [PubMed] [Google Scholar]
- [25].Laurent L, Huang C, Ernest SR, Berard A, Vaillancourt C, Hales BF, In utero exposure to venlafaxine, a serotonin-norepinephrine reuptake inhibitor, increases cardiac anomalies and alters placental and heart serotonin signaling in the rat, Birth Defects Res A Clin Mol Teratol 106(12) (2016) 1044–1055. [DOI] [PubMed] [Google Scholar]
- [26].Levy M, Kovo M, Miremberg H, Anchel N, Herman HG, Bar J, Schreiber L, Weiner E, Maternal use of selective serotonin reuptake inhibitors (SSRI) during pregnancy-neonatal outcomes in correlation with placental histopathology, J Perinatol (2020). [DOI] [PubMed] [Google Scholar]
- [27].Mao J, Jain A, Denslow ND, Nouri MZ, Chen S, Wang T, Zhu N, Koh J, Sarma SJ, Sumner BW, Lei Z, Sumner LW, Bivens NJ, Roberts RM, Tuteja G, Rosenfeld CS, Bisphenol A and bisphenol S disruptions of the mouse placenta and potential effects on the placenta-brain axis, Proc Natl Acad Sci U S A 117(9) (2020) 4642–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Murphy DL, Lesch KP, Targeting the murine serotonin transporter: insights into human neurobiology, Nature Rev Neurosci 9(2) (2008) 85–96. [DOI] [PubMed] [Google Scholar]
- [29].Bengel D, Murphy DL, Andrews AM, Wichems CH, Feltner D, Heils A, Mössner R, Westphal H, Lesch KP, Altered brain serotonin homeostasis and locomotor insensitivity to 3, 4-methylenedioxymethamphetamine (“Ecstasy”) in serotonin transporter-deficient mice, Mol Pharmacol 53(4) (1998) 649–55. [DOI] [PubMed] [Google Scholar]
- [30].Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS, Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta, Proc Natl Acad Sci USA107(12) (2010) 5557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R, Male development of chromosomally female mice transgenic for Sry, Nature 351(6322) (1991) 117–21. [DOI] [PubMed] [Google Scholar]
- [32].Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal; Vol 17, No 1: Next Generation Sequencing Data Analysis DO - 10.14806/ej.17.1.200 (2011). [DOI] [Google Scholar]
- [33].Kim D, Langmead B, Salzberg SL, HISAT: a fast spliced aligner with low memory requirements, Nat Methods 12(4) (2015) 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics 30(7) (2014) 923–30. [DOI] [PubMed] [Google Scholar]
- [35].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol 15(12) (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jain A, Tuteja G, TissueEnrich: Tissue-specific gene enrichment analysis, Bioinformatics 35(11) (2019) 1966–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B, A map of the cis-regulatory sequences in the mouse genome, Nature 488(7409) (2012) 116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C, STRING v10: protein-protein interaction networks, integrated over the tree of life, Nucleic Acids Res 43(Database issue) (2015) D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y, Cytohubba: Identifying hub objects and sub-networks from complex interactome, BMC Syst Biol 8(4) (2014) S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res 13(11) (2003) 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang J, Duncan D, Shi Z, Zhang B, WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013, Nucleic Acids Res 41(Web Server issue) (2013) W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Solano ME, Thiele K, Kowal MK, Arck PC, Identification of suitable reference genes in the mouse placenta, Placenta 39 (2016) 7–15. [DOI] [PubMed] [Google Scholar]
- [43].Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis, BMC Bioinformatics 9 (2008) 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Green MT, Martin RE, Kinkade JA, Schmidt RR, Bivens NJ, Tuteja G, Mao J, Rosenfeld CS, Maternal oxycodone treatment causes pathophysiological changes in the mouse placenta, Placenta 100 (2020) 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shao X, Wang Y, Liu Y, Guo X, Li D, Huo R, Jia W, Cao G, Li YX, Liu M, Sha J, Zhao Y, Wang YL, Association of imbalanced sex hormone production with excessive procoagulation factor SerpinF2 in preeclampsia, J Hypertens 37(1) (2019) 197–205. [DOI] [PubMed] [Google Scholar]
- [46].Yabut JM, Crane JD, Green AE, Keating DJ, Khan WI, Steinberg GR, Emerging Roles for Serotonin in Regulating Metabolism: New Implications for an Ancient Molecule, Endocr Rev 40(4) (2019) 1092–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Greig CJ, Zhang L, Cowles RA, Potentiated serotonin signaling in serotonin re-uptake transporter knockout mice increases enterocyte mass and small intestinal absorptive function, Physiol Rep 7(21) (2019) e14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Urdaneta E, Idoate I, Larralde J, Drug-nutrient interactions: inhibition of amino acid intestinal absorption by fluoxetine, Br J Nutrition 79(5) (1998) 439–46. [DOI] [PubMed] [Google Scholar]
- [49].Lund ML, Egerod KL, Engelstoft MS, Dmytriyeva O, Theodorsson E, Patel BA, Schwartz TW, Enterochromaffin 5-HT cells - A major target for GLP-1 and gut microbial metabolites, Mol Metab 11 (2018) 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mawe GM, Hoffman JM, Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets, Nat Rev Gastroenterol Hepatol 10(8) (2013) 473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Iwasaki S, Nakazawa K, Sakai J, Kometani K, Iwashita M, Yoshimura Y, Maruyama T, Melatonin as a local regulator of human placental function, J Pineal Res 39(3) (2005) 261–5. [DOI] [PubMed] [Google Scholar]
- [52].Lanoix D, Beghdadi H, Lafond J, Vaillancourt C, Human placental trophoblasts synthesize melatonin and express its receptors, J Pineal Res 45(1) (2008) 50–60. [DOI] [PubMed] [Google Scholar]
- [53].Chuffa LGA, Lupi LA, Cucielo MS, Silveira HS, Reiter RJ, Seiva FRF, Melatonin promotes uterine and placental health: potential molecular mechanisms, Intl J Mol Sci 21(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Karahoda R, Abad C, Horackova H, Kastner P, Zaugg J, Cerveny L, Kucera R, Albrecht C, Staud F, Dynamics of tryptophan metabolic pathways in human placenta and placental-derived cells: Effect of gestation age and trophoblast differentiation, Front Cell Dev Biol 8 (2020) 574034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T, Derivation of human trophoblast stem cells, Cell Stem Cell 22(1) (2018) 50–63 e6. [DOI] [PubMed] [Google Scholar]
- [56].Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM, Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas, Proc Natl Acad Sci U S A 113(19) (2016) E2598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Laurent L, Deroy K, St-Pierre J, Côté F, Sanderson JT, Vaillancourt C, Human placenta expresses both peripheral and neuronal isoform of tryptophan hydroxylase, Biochimie 140 (2017) 159–165. [DOI] [PubMed] [Google Scholar]
- [58].Wu HH, Choi S, Levitt P, Differential patterning of genes involved in serotonin metabolism and transport in extra-embryonic tissues of the mouse, Placenta 42 (2016) 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Auda GR, Kirk SH, Billett MA, Billett EE, Localization of monoamine oxidase mRNA in human placenta, J Histochem Cytochem 46(12) (1998) 1393–400. [DOI] [PubMed] [Google Scholar]
- [60].Karahoda R, Horackova H, Kastner P, Matthios A, Cerveny L, Kucera R, Kacerovsky M, Duintjer Tebbens J, Bonnin A, Abad C, Staud F, Serotonin homeostasis in the materno-foetal interface at term: Role of transporters (SERT/SLC6A4 and OCT3/SLC22A3) and monoamine oxidase A (MAO-A) in uptake and degradation of serotonin by human and rat term placenta, Acta Physiologica 229(4) (2020) e13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Klempan T, Hudon-Thibeault AA, Oufkir T, Vaillancourt C, Sanderson JT, Stimulation of serotonergic 5-HT2A receptor signaling increases placental aromatase (CYP19) activity and expression in BeWo and JEG-3 human choriocarcinoma cells, Placenta 32(9) (2011) 651–656. [DOI] [PubMed] [Google Scholar]
- [62].Sonier B, Lavigne C, Arseneault M, Ouellette R, Vaillancourt C, Expression of the 5-HT2A serotoninergic receptor in human placenta and choriocarcinoma cells: mitogenic implications of serotonin, Placenta 26(6) (2005) 484–90. [DOI] [PubMed] [Google Scholar]
- [63].Chen X, Ye R, Gargus JJ, Randy D, Blakely K. Dobrenis, Sze Ji Y., Disruption of transient serotonin accumulation by non-serotonin-producing neurons impairs cortical map development, Cell Reports 10(3) (2015) 346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Holmes A, Lit Q, Murphy DL, Gold E, Crawley JN, Abnormal anxiety-related behavior in serotonin transporter null mutant mice: the influence of genetic background, Genes Brain Behav 2(6) (2003) 365–80. [DOI] [PubMed] [Google Scholar]
- [65].Baganz NL, Horton RE, Calderon AS, Owens WA, Munn JL, Watts LT, Koldzic-Zivanovic N, Jeske NA, Koek W, Toney GM, Daws LC, Organic cation transporter 3: Keeping the brake on extracellular serotonin in serotonin-transporter-deficient mice, Proc Natl Acad Sci USA 105(48) (2008) 18976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Dendrogram and heatmap of WGCNA results for SERT KO vs. WT placental comparison. As detailed in [43], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image.