Abstract
Although laboratory diagnosis of respiratory viruses has been widely studied, there is a relative insufficiency of literature examining the impact of specimen type on the laboratory diagnosis of influenza A and B. In a clinical study comparing the FLU OIA test with 14-day cell culture, clinical specimens from nasopharyngeal swabs, throat swabs, nasal aspirates, and sputum were obtained from patients experiencing influenza-like symptoms. A total of 404 clinical specimens were collected from 184 patients. Patients were defined as influenza positive if the viral culture of a specimen from any sample site was positive. Patients were defined as influenza negative if the viral cultures of specimens from all sample sites were negative. By this gold standard, culture and FLU OIA test results for each sample type were compared. For each of the four specimen types, the viral culture and FLU OIA test demonstrated equal abilities to detect the presence of influenza A or B virus or viral antigen. Sputum and nasal aspirate samples were the most predictive of influenza virus infection. Throat swabs were the least predictive of influenza virus infection, with both tests failing to detect influenza virus in nearly 50% of the throat samples studied.
Influenza is an epidemic illness that occurs during the fall and winter months. Symptoms of influenza are nonspecific and may include the sudden onset of cough, fever, weakness, and myalgia. The duration of the illness is typically 5 to 7 days, although some symptoms, most notably cough, may persist for 2 to 3 weeks (9, 21).
Influenza is usually diagnosed on clinical grounds alone, but this method of diagnosis has been demonstrated to be both insensitive and nonspecific (3, 10, 23). The gold standard of laboratory diagnosis is 14-day cell culture with one of a variety of cell lines that can support the growth of influenza virus (2, 18). Unfortunately, this technique has limited clinical utility, as results are obtained too late in the clinical course for effective patient intervention.
Because more immediate laboratory diagnosis of influenza could prove useful in patient management, rapid diagnostic tests by shell vial culture, direct immunofluorescence, and enzyme immunoassay techniques have been developed and widely studied. Although each technique has demonstrated a high degree of specificity for the influenza viruses, their sensitivities have been poor when compared to that of 14-day cell culture (6, 14). Additionally, while there is a wealth of literature establishing nasal aspirates and washes as being superior to nasopharyngeal swabs for diagnosis of respiratory syncytial virus (1, 11, 17, 19, 24), there is a relative insufficiency of literature examining the impact of sample type on the laboratory diagnosis of influenza (4, 7, 13).
The FLU OIA test (BioStar, Inc., Boulder, Colo.) is an optical immunoassay (OIA) designed to detect the presence of influenza virus A or B antigen from a variety of clinical specimen types. In this study, we collected single or multiple sample types from patients exhibiting influenza-like illness and performed the FLU OIA test and viral culture on each. In an effort to assess the impact of sample type on virus or viral-antigen recovery, we defined patients as being positive for influenza virus if they were viral-culture positive with any specimen type. Patients were defined as negative for influenza virus if they were viral-culture negative with all specimen types. We then compared the FLU OIA test or culture result from any single sample site against this gold standard. In addition, we examined the sensitivity and specificity of the FLU OIA test as directly compared to those of 14-day culture for each specimen type.
(A preliminary report of this work was presented at the International Symposium on Influenza and Other Respiratory Viruses, 4 to 6 December 1998.)
MATERIALS AND METHODS
Patients.
Patients were enrolled in the study from January 1998 to April 1998. Patients came from three cities in the Midwest, Southwest, and Rocky Mountain regions of the United States. Patient enrollment was conducted at emergency rooms, physician offices, employee clinics, and urgent-care facilities. Patients were enrolled in the study based on the following clinical criteria: the onset of illness within the past 36 h, a temperature of ≥100°F, and at least two influenza-like symptoms, including, but not limited to, cough, eye or ear pain, headache, sore throat, myalgia, congestion, malaise, and chills. Patients who had previously received an influenza vaccine were not excluded from this study.
Clinical specimens.
Any combination of throat swab, nasopharyngeal swab, and nasal aspirate and/or sputum specimens were collected from each patient. Specimens were collected by the following techniques.
(i) Throat swabs.
Two sterile rayon swabs (Hardwood Products LP, Guilford, Maine) were vigorously rubbed on both tonsillar surfaces and the posterior pharynx. One swab was then inserted, tip down, into the original paper wrapper for the OIA test; the second swab was inserted into 1.5 ml of Multi-Microbe Medium (M4; MicroTest, Inc., Lilburn, Ga.) for culture.
(ii) Nasopharyngeal swabs.
Two Dacron nasopharyngeal swabs (Hardwood Products LP) were inserted beneath the inferior turbinate of either nare and vigorously rubbed and rolled against the mucosal surface. One swab was then inserted, tip down, into the original paper wrapper for the OIA test; the second swab was inserted into 1.5 ml of M4 transport medium for culture.
(iii) Nasal aspirates.
A depressed bulb syringe (Bard, Atlanta, Ga., or Owen & Minor, Denver, Colo.) was deeply inserted into either nare and suctioned while being withdrawn. The specimen was expelled into a sterile cup and thoroughly mixed with a rayon throat swab. The swab was subsequently used for testing by the FLU OIA test. M4 medium (1.5 ml) was then added to the remaining nasal aspirate specimen for culture.
(iv) Sputum specimens.
Sputum specimens were obtained after either a spontaneous deep cough or a deep cough by mechanical induction with a throat swab. The specimen was collected in a sterile cup and thoroughly mixed with a rayon throat swab. The swab was subsequently used for testing by the FLU OIA method. M4 medium (1.5 ml) was then added to the remaining sputum specimen for culture.
All specimens were labeled with the patient identification data, date, and exact time of specimen collection. Specimens were stored at 2 to 8°C for up to 24 h until testing by the FLU OIA method or culture could be performed.
FLU OIA test.
OIA technology enables the direct visual detection of a physical change in the optical thickness of molecular thin films (5, 12, 20, 22). This change is a result of macromolecular binding on an optical surface (silicon wafer). When extracted specimen is placed directly on the optical surface, the immobilized specific capture of the target analyte increases the thickness of the film. This change in thickness alters the reflected light path and is visually perceived as a color change. Slight changes in optical thickness produce a distinct visible color change. A positive result appears as a blue to purple spot on the predominant gold background. When analyte is not present in the specimen, no binding takes place; therefore, the optical thickness remains unchanged and the surface retains the original gold color, indicating a negative result. Internal procedural control dots are visible in a valid test result. If a procedural control dot is not visible, the test was not performed correctly and the result is considered invalid.
The FLU OIA test is a 15-min antibody-based assay for the detection of influenza virus types A and B nucleoprotein antigen from clinical specimens. The FLU OIA test was performed according to the manufacturer's instructions provided in the package insert. Reagents were removed from refrigerated storage and allowed to warm to room temperature (18 to 30°C). All extraction tubes and test devices were labeled with a patient identification number.
For the antigen extraction procedure, 3 drops of sample diluent and 2 drops of extraction reagent were added to an extraction tube. A throat swab (used for pharyngeal specimens or to absorb nasal aspirate and sputum specimens) or a nasopharyngeal swab was added to the extraction tube, thoroughly mixed in the solution, and incubated for 3 to 5 min. One drop of conjugate was then added to the extraction tube, and the solution was thoroughly mixed with the swab.
For the assay procedure, 1 drop of the solution containing extracted antigen and conjugate was added to the test surface and incubated for 6 to 7 min. After the test surface was then washed and blotted, 1 drop of substrate was added to the center of the test surface and incubated for an additional 6 to 7 min. The test surface was then again washed and blotted, and the results were interpreted. Results were interpreted as positive for influenza virus A or B antigen if a blue or purple reaction zone was visible in the center of the test surface. Results were interpreted as negative for influenza virus A or B antigen if no color change was visible. The upper blotters of the test device were removed to retain the permanent result of the test.
Cell culture.
Specimens were vigorously vortexed and centrifuged at 400 × g for 10 min. The supernatant was used for cell culture inoculation. Supernatant was filtered through a 0.45-μm-pore-size filter, and 0.2 ml of filtrate was inoculated onto each of two pRMK tubes (Viromed Laboratories, Minneapolis, Minn.) containing serum-free Eagle's minimal essential medium. All inoculated tubes were incubated at 33 to 35°C and examined for cytopathic effects (CPE) on alternating days for a period of up to 14 days. CPE-negative tubes were tested by hemadsorption on day 3 and day 7 with 0.25% guinea pig erythrocytes. Cells from tubes demonstrating CPE or positive for hemadsorption were harvested, and slides for influenza virus A and B direct fluorescent antibody staining (Imagen; Dako Diagnostics Ltd.) were prepared. Cells from tubes still negative on day 14 were stained to confirm the absence of influenza virus A or B antigen.
Direct fluorescent antibody staining.
Cells harvested from culture tubes were spotted onto two glass microscope slides. Specimens were air dried and acetone fixed for 10 min. Staining was performed according to the manufacturer's directions. A 25-μl drop of reagent containing fluorescein-conjugated influenza virus A and B monoclonal antibodies was used to cover cell spots. Slides were incubated for 15 min at 37°C, washed for 5 minutes in phosphate-buffered saline, air dried, covered with a coverslip, and read with a fluorescence microscope.
PCR.
If excess specimen was available after culture and the OIA test were performed, the sample was frozen for reverse transcriptase PCR-enzyme hybridization assay (RT-PCR-EHA) detection of influenza virus A and B nucleic acids in OIA test-positive, culture-negative samples. All samples tested by the RT-PCR-EHA were sent off-site (Prodesse, Inc., Waukesha, Wisc.) and tested by Hexaplex (Prodesse, Inc.) technology as previously described (8).
Statistical methods.
The percentage of specimens positive by the FLU OIA test or by viral culture was calculated by using one of the following formulae: (number of positive samples detected by the FLU OIA test for a specific specimen type/total number of samples for that specimen type, positive or negative, from influenza virus-positive patients) × 100 and (number of positive samples detected by viral culture for a specific specimen type/total number of samples for that specimen type, positive or negative, from influenza virus-positive patients) × 100. Fisher's exact test was used to compare the results of the two tests for each specimen type.
RESULTS
Patients and clinical specimens.
One hundred eighty-four patients, ranging in age from 2 months to 76 years, fulfilled the clinical criteria for inclusion in this study. Fifty-eight patients were 16 years of age or younger, 117 patients were between the ages of 17 and 54 years, and 8 patients were 55 years of age or older. Age was not recorded for one patient. Each patient donated 1 to 4 specimens, with the mean number of specimens donated per patient being 2.2.
The frequency with which a particular type of specimen was obtained varied by patient age. Nasal aspirates were more commonly obtained from pediatric patients (age, <17 years) than from older patients (P < 0.001). Sputum specimens were more commonly obtained from the elderly (age, ≥55 years) than from younger patients (P < 0.01) (Table 1).
TABLE 1.
Number and distribution of each specimen type donated and collected from each age group
| Age (years) | No.a of specimens of indicated type donated
|
Total no. (%) of specimens | |||
|---|---|---|---|---|---|
| Nasal aspirate | Nasopharyngeal swab | Throat swab | Sputum | ||
| 0–16 | 45 (40.9) | 36 (32.7) | 18 (16.4) | 11 (10) | 110 (27.3) |
| 17–54 | 33 (11.9) | 102 (36.8) | 90 (32.5) | 52 (18.8) | 277 (68.7) |
| ≥55 | 1 (6.3) | 4 (25.0) | 4 (25.0) | 7 (43.8) | 16 (4.0) |
| Total | 79 (19.6) | 142 (35.2) | 112 (27.8) | 70 (17.4) | 403 (100)b |
The percentages of specimens by age group and sample type are in parentheses.
A total of 404 specimens were donated. One patient report did not have an age provided; therefore, this specimen was not included in the table.
Ninety-two (50%) of the patients were positive for influenza virus by culture of at least one specimen type. When we examined only the patients that were culture positive for influenza virus A or B from testing of at least one sample site, a total of 54 nasal aspirate, 65 nasopharyngeal swab, 56 throat swab, and 41 sputum specimens were included in our analysis.
Culture and FLU OIA test results.
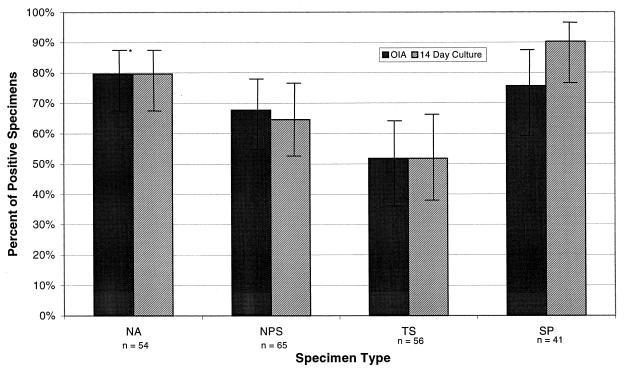
The percentages of positive specimens detected by viral culture and the FLU OIA test were calculated for each specimen type. In culture, sputum specimens most accurately predicted influenza virus infection, with 90.2% of infected patients being identified. Nasal aspirates were the second best sample, with 79.6% of those infected being identified. Nasopharyngeal swabs were the third best sample, with 64.6% of those infected being identified. Throat swab specimens (51.8%) were the least predictive of influenza virus infection and were significantly less predictive than sputum (P < 0.001) and nasal aspirate (P < 0.01) but not nasopharyngeal swab (P = 0.15) specimens (Fig. 1).
FIG. 1.
Percentages of specimens positive by the FLU OIA test or 14-day culture taken from patients who were culture positive for influenza virus with any specimen type. NA, nasal aspirate; NPS, nasopharyngeal swab; TS, throat swab; SP, sputum; ∗, 95% confidence intervals.
In the FLU OIA test, nasal aspirates were the most likely to accurately predict influenza virus infection, with 79.6% of infected patients being identified, followed by sputum specimens (75.6%), nasopharyngeal swab specimens (67.7%), and throat swab specimens (51.8%). As with viral culture, throat swab specimens were significantly worse than sputum (P < 0.02) and nasal aspirates (P < 0.01) but not nasopharyngeal swab specimens (P = 0.07) in detection of infection (Fig. 1).
For each of the four specimen types, the FLU OIA test demonstrated statistical equivalence to 14-day viral culture in its ability to detect influenza virus or viral antigen from patients considered positive for influenza A or B. This equivalence was most evident with nasal aspirates (P = 1.0), throat swab specimens (P = 1.0), and nasopharyngeal swab specimens (P = 0.71) but was less evident for sputum specimens (P = 0.08).
In Table 2, we compared the sensitivity and specificity of the FLU OIA test to those of viral culture for each specimen type. The sensitivity of the FLU OIA test ranged from 88.4% for nasal aspirates to 62.1% for throat swab specimens. The specificity ranged from 79.5% for throat swab specimens to 51.5% for sputum specimens. With a sample of specimens of sufficient volume that were OIA test positive and culture negative, the RT-PCR-EHA was performed on 32 OIA test-positive and culture-negative specimens. Twenty-one (66%) of these specimens were positive by RT-PCR-EHA, indicating that a high percentage of those specimens classified as false positives when their EHA results were compared to those of 14-day viral culture alone actually contained influenza virus RNA and are therefore more properly classified as culture false negatives.
TABLE 2.
Comparison of the FLU OIA test to 14-day cell culture
| Specimen type | Total no. of specimens | No. of specimens that were:
|
Sensitivitya | Specificitya | |||
|---|---|---|---|---|---|---|---|
| OIA test+ and culture+ | OIA test+ and culture− | OIA test− and culture+ | OIA test− and culture− | ||||
| Nasal aspirate | 79 | 38 | 11 | 5 | 25 | 88.4 (74.9–96.1) | 69.4 (51.9–83.7) |
| Nasopharyngeal swab | 143 | 35 | 24 | 7 | 77 | 83.3 (68.6–93.0) | 76.2 (66.7–84.1) |
| Throat swab | 112 | 18 | 17 | 11 | 66 | 62.1 (42.3–79.3) | 79.5 (69.2–87.6) |
| Sputum | 70 | 30 | 16 | 7 | 17 | 81.1 (64.8–92.0) | 51.5 (33.5–69.2) |
| Total | 404 | 121 | 68b | 30 | 185 | 80.1 (72.9–86.2) | 73.1 (67.2–78.5) |
Sensitivities and specificities are shown as percentages; 95% confidence intervals (by the exact binomial method) are shown in parentheses.
Thirty-two of these specimens were available for the RT-PCR-EHA; 21 of 32 were positive for influenza virus nucleic acid.
DISCUSSION
In this study, we compared with four specimen types the abilities of the FLU OIA test and 14-day viral culture to detect influenza virus A or B in patients who were found to be influenza virus A or B positive from any sample type. By so doing, we sought to determine which combinations of diagnostic test and sample type could yield the highest rates of influenza detection among patients experiencing influenza-like illness.
Although very few articles have indicated sputum to be a recommended specimen type for the diagnosis of influenza (13, 15, 16), we found that viral culture of the sputum yielded the highest rate of influenza detection. Nasal aspirates were considered the best specimen for influenza detection by the FLU OIA test in this study. Influenza detection with sputum by the FLU OIA test was slightly less accurate. This study also demonstrated that sputum and nasal aspirates are better specimen types than throat swab specimens for detecting either influenza virus or viral antigen.
When an influenza virus-positive patient was used as the gold standard, the FLU OIA test demonstrated rates of influenza detection equivalent to those of 14-day viral culture with each of the four specimen types. Although 14-day viral culture remains the gold standard of diagnosis, the sensitivity of this technique must be considered, as was evident from the RT-PCR results in this study. Because the RT-PCR method detected influenza virus A or B nucleic acid from 66% of the culture-negative and FLU OIA test-positive patients, it appears that the false-negative rate of 14-day culture may be higher than previously appreciated.
Our study also demonstrated variability in the types of specimens collected depending on the age of the patient. It is unclear why this may be the case. Health care professionals may be more apt to collect a certain specimen type based on previous experience, training, or patient-care protocols. Patient willingness to undergo different collection procedures may vary with age. Reasons for the variation seen in this study and its generalizability merit further study.
Because physicians are unable to reliably make the diagnosis of influenza virus infection on clinical grounds alone and influenza virus culture is expensive, requires highly trained technical personnel, and gives results too late for effective pharmacologic intervention, there is a need for sensitive, rapid diagnostic tests for influenza virus A and B. The FLU OIA test is a 15-min point-of-care test that is relatively inexpensive, requires minimal training, and may be performed by nontechnical staff. When combined with clinical presentation, use of the FLU OIA test can lead to a more efficient, accurate diagnosis of influenza A and B. With the recent Food and Drug Administration approval of a new neuraminidase inhibitor for the treatment of influenza A or B, a rapid test for both types of influenza is required for the most jucicious use of these drugs.
ACKNOWLEDGMENTS
This work was supported by Biota Holdings, Ltd., Melbourne, Victoria, Australia.
Special thanks go to Sandra Butler at Viral Diagnostics Inc. (Dallas, Tex.) for editing the culture methods section of the manuscript.
REFERENCES
- 1.Ahlawalia G, Embree J, McNicol P, Law B, Hammond G W. Comparison of nasopharyngeal aspirate and nasopharyngeal swab specimens for respiratory syncytial virus diagnosis by cell culture, indirect immunofluorescence assay, and enzyme-linked immunosorbent assay. J Clin Microbiol. 1987;25:763–767. doi: 10.1128/jcm.25.5.763-767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts R F. Influenza virus. In: Mandell G L, Douglas R G Jr, Bennett J E, editors. Principle and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone, Inc.; 1995. pp. 1546–1567. [Google Scholar]
- 3.Carrat F, Tachet A, Housset B, Valleron A J, Rouzioux C. Influenza and influenza-like illness in general practice: drawing lessons for surveillance from a pilot study in Paris, France. Br J Gen Precut. 1997;47:217–220. [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz J R, Quinonez E, De Fernandez A, Peralta F. Isolation of viruses from nasopharyngeal secretions: comparison of aspiration and swabbing as means of sample collection. J Infect Dis. 1987;156:415–416. doi: 10.1093/infdis/156.2.415. [DOI] [PubMed] [Google Scholar]
- 5.Della-Latta P, Whittier S, Hosmer M, Agre F. Rapid detection of group A streptococcal pharyngitis in a pediatric population with optical immunoassay. Pediatr Infect Dis J. 1994;13:742–743. doi: 10.1097/00006454-199408000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Doing K M, Jerkofsky M A, Dow E G, Jellison J A. Use of fluorescent-antibody staining of cytocentrifuge-prepared smears in combination with cell culture for direct detection of respiratory viruses. J Clin Microbiol. 1998;36:2112–2114. doi: 10.1128/jcm.36.7.2112-2114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaldson A, Lewis F A, Kennett M I L, White J, Gust I D. The 1976 influenza epidemic in Melbourne. Med J Aust. 1978;2:45–49. doi: 10.5694/j.1326-5377.1978.tb131337.x. [DOI] [PubMed] [Google Scholar]
- 8.Fan J F, Henrickson K J, Savatski L L. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3, by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex) Clin Infect Dis. 1998;26:1397–1402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 9.Gill P W, Murphy A M. The diagnosis of influenza. Aust Fam Physician. 1991;20:1664–1665. [PubMed] [Google Scholar]
- 10.Govaert T H M E, Dinant G J, Knottnerus A K. The predictive value of influenza symptomatology in elderly people. Fam Pract. 1998;15:16–22. doi: 10.1093/fampra/15.1.16. [DOI] [PubMed] [Google Scholar]
- 11.Hall C B, Douglas R G. Clinically useful method for the isolation of respiratory syncytial virus. J Infect Dis. 1975;131:1–5. doi: 10.1093/infdis/131.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Hesterberg L K, Crosby M C. An overview of rapid immunoassays. Lab Med. 1996;27:41–46. [Google Scholar]
- 13.Horn M C, Reed S E, Taylor P. Role of viruses and bacteria in acute wheezy bronchitis in childhood: a study of sputum. Arch Dis Child. 1979;54:587–592. doi: 10.1136/adc.54.8.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston S L G, Seigel C S. A comparison of direct immunofluorescence, shell vial culture, and conventional cell culture for the rapid detection of influenza A and B. Diagn Microbiol Infect Dis. 1991;14:131–134. doi: 10.1016/0732-8893(91)90047-j. [DOI] [PubMed] [Google Scholar]
- 15.Kimball A M, Foy H M, Cooney M K, Allan I D, Matlock M, Plorde J J. Isolation of respiratory syncytial and influenza viruses from the sputum of patients hospitalized with pneumonia. J Infect Dis. 1983;147:181–184. doi: 10.1093/infdis/147.2.181. [DOI] [PubMed] [Google Scholar]
- 16.Kok T, Higgins G. Prevalence of respiratory viruses and mycoplasma pneumoniae in sputum samples from unselected adult patients. Pathology. 1997;29:300–302. doi: 10.1080/00313029700169135. [DOI] [PubMed] [Google Scholar]
- 17.Masters H B, Weber K O, Groothuis J R, Wren C G, Lauer B A. Comparison of nasopharyngeal washings and swab specimens for diagnosis of respiratory syncytial virus by EIA, FAT, and cell culture. Diagn Microbiol Infect Dis. 1987;8:101–105. doi: 10.1016/0732-8893(87)90156-8. [DOI] [PubMed] [Google Scholar]
- 18.McIntosh K. Diagnostic virology. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 401–430. [Google Scholar]
- 19.McIntosh K, Hendry R M, Fahnestock M L, Peirik L T. Enzyme-linked immunosorbent assay for detection of respiratory syncytial virus infection: application to clinical samples. J Clin Microbiol. 1982;16:329–333. doi: 10.1128/jcm.16.2.329-333.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostroff R, Maul D, Baouchi W. Thin film biosensors. Vacuum Thinfilm. 1999;1999(5):30–33. [Google Scholar]
- 21.Pachucki C T. The diagnosis of influenza. Semin Respir Infect. 1992;7:46–53. [PubMed] [Google Scholar]
- 22.Park C H, Ruprai D, Vandel N M, Hixon D L, Mecklenberg F E. Rapid detection of group B streptococcal antigen from vaginal specimens using a new optical immunoassay technique. Diagn Microbiol Infect Dis. 1996;24:125–128. doi: 10.1016/0732-8893(96)00013-2. [DOI] [PubMed] [Google Scholar]
- 23.Tannock G A, Reid A L A, Gillett S M, Herd R, Gillett R S, Hensley M J, Barry R D, Lawrance I P A, Nichols J, Adams M, Henry R L, Sauders N A. A study of respiratory infections in a healthy adult population during the 1987 Australian winter. Fam Pract. 1993;10:378–386. doi: 10.1093/fampra/10.4.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treuhaft M W, Soukup J M, Sullivan B J. Practical recommendations for the detection of pediatric respiratory syncytial virus infections. J Clin Microbiol. 1985;22:270–273. doi: 10.1128/jcm.22.2.270-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]