Abstract
Intestinal nematode infections common during pregnancy have recently been shown to have impacts that extend to their uninfected offspring including altered brain gene expression. If maternal immune signals reach the neonatal brain, they might alter neuroimmune development. We explored expression of genes associated with four distinct types of T cells (Th1, Th2, Th17, Treg) and with leukocyte transendothelial migration and endocytosis transport across the blood–brain barrier (BBB) in the postnatal brain of offspring of nematode-infected mice, through secondary analysis of a whole brain gene expression database. Th1/Th17 expression was lowered by maternal infection as evidenced by down-regulated expression of IL1β, Th1 receptors and related proteins, and of IL22 and several Th17 genes associated with immunopathology. In contrast, Th2/Treg related pathways were upregulated as shown by higher expression of IL4 and TGF-β family genes. Maternal infection also upregulated expression of pathways and integrin genes involved in transport of leukocytes in between endothelial cells but downregulated endosome vesicle formation related genes that are necessary for endocytosis of immunoglobulins across the BBB. Taken together, pup brain gene expression indicates that maternal nematode infection enhanced movement of leukocytes across the neonatal BBB and promoted a Th2/Treg environment that presumably minimizes the proinflammatory Th1 response in the pup brain.
Subject terms: Developmental biology, Immunology, Neuroscience, Zoology
Introduction
Chronic gastrointestinal (GI) nematode infections are extremely important in low-income countries where human hookworm infections exacerbate anemia during pregnancy1 and in ruminants where GI nematodes lower birth weight2. On the other hand, GI nematode infections have been observed to enhance immune development in mouse3,4 and human5 studies and enhance immunity against non-infectious Th2-related conditions in human6,7 and mouse8 neonates, at least in part through transfer of cytokines and immunoglobulins through breast milk. Moreover, enhanced neonatal systemic immunity in response to maternal GI nematodes has been shown to promote long-lasting immunity against nematode infection in the offspring4. Thus, GI nematodes might have both harmful and beneficial consequences for the next generation, and benefits might be a consequence of the Th2/Treg responses typically induced by GI nematodes that dampen Th1/Th17 immunopathology9,10.
Infection of pregnant and lactating mice with the GI nematode Heligmosomoides bakeri (= Heligmosomoides polygyrus, Nematospiroides dubius) has been shown to alter brain gene expression in the late-term fetus11 and the 7-day old (P7) neonates12 relative to offspring of uninfected mothers. Particularly intriguing was the upregulation of long-term potentiation (LTP) and related pathways in the P7 pup brain. LTP promotes synaptogenesis, spatial learning and memory13–15 and is observed when the neonatal brain is exposed to Th2 conditions16 but impaired in IL4 knock-out mice17. Thus, the upregulated expression of LTP in response to maternal H. bakeri infection not only indicated a possible benefit for the pups of infected dams but also raised the possibility that the well-documented Th2/Treg response to H. bakeri in the infected host18 might be reflected in the brains of their uninfected pups.
Endocytosis19 and transendothelial leukocyte migration20 allow immune elements such as antigens, immunoglobulins, cytokines, and leukocytes obtained from breast milk21 or produced by the neonate to cross the blood–brain barrier (BBB). Cytokines22 and immunoglobulins23 bind to surface receptors on endothelial cells and the resulting complex triggers an endocytosis signalling cascade that induces vesicle formation, budding, and intracellular transport24. Although leukocytes might also enter the brain through endocytosis, activated leukocytes more commonly enter between endothelial cells through diapedesis20. Proteins such as integrins on the surface of leukocytes dock with cell adhesion molecules (CAMs) and other proteins on the surface of endothelial cells25. The resulting movement of the actin cytoskeleton26 inside endothelial cells loosens the tight junctions, allowing leukocytes to squeeze between endothelial cells27 and enter the brain. This process temporarily compromises junction integrity27 after which junction gene expression is upregulated to restore endothelial cell adherence and re-establish normal BBB integrity28.
Given that H. bakeri induces a Th2/Treg response in the infected host18, that maternal immune elements are transferred to the neonate, and that Th2 responses favour LTP, we hypothesized that neonatal brain gene expression in response to maternal infection might reflect a heightened Th2/Treg profile and dampened Th1/Th17 associated neuro-inflammation, especially if movement of either leukocytes or immunoglobulins across the BBB was compromised. The goal of this secondary analysis was to explore an existing brain gene expression database for evidence that maternal H. bakeri infection altered the profile of T helper cell responses in the P7 brain. The first specific objective was to determine whether or not expression of genes related to innate and/or adaptive immune responses was shifted toward a Th2/Treg profile. The second objective was to determine whether or not maternal infection altered expression of genes involved in transport of immune elements across the BBB, with a focus on endocytosis and associated vesicle transport of immunoglobulins, and on transendothelial migration and regulation of junctions involved in transport of leukocytes. Rather than relying on pathway analysis, we manually explored context relevant cascades within pathways by inspecting expression data for ligands and receptors to assess pathway activation and by inspecting products to assess function. This allowed us to identify differentially expressed sets of genes that would be expected to alter function.
Results
Impact of Maternal Infection on Immune-Related Genes in the Neonatal Brain
Differential expression of immune related pup brain genes was examined for evidence that maternal intestinal nematode infection might have altered components of the innate immune system and/or the adaptive immune system. Given that H. bakeri infection typically upregulates Th2/Treg responses and downregulates Th1/Th17 responses, genes related to these responses were of particular interest.
Limited negative impact on expression of innate immune genes
The innate immune response to pathogens involves the hematopoietic cell lineage, the complement and coagulation cascade and platelet activation pathways as well as several receptor-mediated (Toll, Imd, NOD and RIG-like) signalling pathways that recognize molecular signals of pathogens and activate an adaptive response. Based on the previous KEGG pathway analysis12, only the RIG-I like receptor signalling KEGG pathway (Table 1) that activates an innate immune response to viral pathogens was downregulated. We also observed downregulated expression of seven of the eight differentially expressed cell surface markers and all nine of the differentially expressed chemokine ligand genes involved in the myeloid cell lineage that generates innate immune cells (Table 2). Thus, the maternal infection had a modest negative impact on the neonatal brain innate immune system.
Table 1.
List of immune related KEGG pathways considered in this study.
| Classification | Pathway name | Differential expression1 | Reference number |
|---|---|---|---|
| General immunity | Hematopoietic cell lineage | N/A | 04640 |
| Cytosolic DNA sensing | N/A | 04623 | |
| Intestinal immune network for IgA production | Downregulated | 04672 | |
| Innate immunity | Fc gamma R mediated phagocytosis | Upregulated | 04666 |
| Antigen processing and presentation | N/A | 04612 | |
| Complement and coagulation cascade | N/A | 04610 | |
| Platelet activation | N/A | 04611 | |
| Toll-like receptor signaling | N/A | 04620 | |
| Toll and Imd signaling | N/A | 04624 | |
| NOD-like receptor signaling | N/A | 04621 | |
| RIG-I-like receptor signaling | Downregulated | 04622 | |
| Adaptive immunity | Chemokine signaling | Upregulated | 04062 |
| Cytokine-cytokine receptor interaction | Downregulated | 04060 | |
| Fc epsilon RI signaling | Upregulated | 04664 | |
| C-type lectin receptor signaling – polarize T cell responses | N/A | 04625 | |
| Natural killer cell mediated cytotoxicity / adaptive induces apoptosis | N/A | 04650 | |
| T cell receptor signaling | Upregulated | 04660 | |
| B cell receptor signaling | Upregulated | 04662 | |
| Th1 and Th2 cell differentiation | N/A | 04658 | |
| Th17 cell differentiation | N/A | 04659 | |
| IL-17 signaling | N/A | 04657 | |
| BBB related | Leukocyte transendothelial migration | Upregulated | 04670 |
| Tight junction | Upregulated | 04530 | |
| Adheren junction | Upregulated | 04520 | |
| Endocytosis | Upregulated | 04144 | |
| Regulation of actin cytoskeleton2 | Upregulated | 04810 |
1Differential regulation as reported by Haque et al.12.
2Subpathway of the Leukocyte transendothelial migration pathway.
Table 2.
List of innate immune system related genes differentially expressed in the pup brain, in response to maternal H. bakeri infection.
| Classification | Gene name | Gene symbol | p value | Log 2 fold change |
|---|---|---|---|---|
| CD cell surface markers | CD302 antigen | CD302 | 7.17E−12 | − 1.8988 |
| CD40 antigen | CD40 | 4.33E−10 | − 1.7484 | |
| CD83 antigen | CD83 | 3.03E−08 | − 1.2487 | |
| CD300A antigen | CD300A | 4.65E−08 | − 1.4979 | |
| CD209f antigen | CD209F | 9.01E−07 | − 1.4813 | |
| CD93 antigen | CD93 | 1.05E−05 | 1.3519 | |
| CD200 receptor 1 | CD200R1 | 1.99E−05 | − 1.5711 | |
| CD209g antigen | CD209G | 3.77E−05 | − 1.9848 | |
| Chemokines | chemokine (C–C motif) ligand 9 | CCL9 | 8.11E−13 | − 2.2198 |
| chemokine (C–C motif) ligand 6 | CCL6 | 1.49E−10 | − 1.9152 | |
| chemokine (C–C motif) receptor 1 | CCR1 | 6.32E−08 | − 1.5864 | |
| chemokine (C–X–C motif) ligand 1 | CXCL1 | 5.24E−08 | − 1.9013 | |
| chemokine (C–C motif) ligand 25 | CCL25 | 1.74E−08 | − 1.6614 | |
| chemokine (C–C motif) ligand 12 | CCL12 | 7.17E−07 | − 1.9695 | |
| chemokine-like factor | CKLF | 3.37E−06 | − 1.2909 | |
| chemokine (C–C motif) ligand 24 | CCL24 | 9.86E−05 | − 1.2683 | |
| chemokine (C–C motif) ligand 7 | CCL7 | 3.00E−05 | − 1.6164 |
Differential regulation as reported by Haque et al.12.
Differential expression of adaptive immune genes
Consistent with upregulation of KEGG pathway maps for T cell receptor signaling and B cell receptor signaling (Table 1;12), many pup brain genes of the adaptive immune system were differentially expressed. Among those associated with leukocyte, lymphocyte, and immunoglobulin superfamilies (Table 3), expression of four nuclear factors of activated T cells (NFAT5, NFATC1, NFATC2, NFATC3), a T cell transcription factor (TCF7L1), and early B cell factor 3 (EBF3) was upregulated (Table 3) and expression of leukocyte transcript (LST1), lymphocyte antigens (LY86, LY6G6D), cytotoxic T-lymphocyte associated proteins (CTLA2A, CTLA2B), T cell proliferation (MTCP1), T cell linkers of activation (LAT, LAT2) and a B cell receptor associated protein (BCAP29) was downregulated. Within the immunoglobulin superfamily, IGSF3 was upregulated and GM4926 expression was downregulated. All differentially expressed CD cell surface markers and 5 of 6 chemokines were downregulated (Table 3). Taken together, these results highlight the responsiveness of adaptive immune genes in the pup brain to maternal nematode infection.
Table 3.
List of adaptive immune system related genes differentially expressed in the pup brain, in response to maternal H. bakeri infection.
| Classification | Gene name | Gene symbol | p value | Log 2 Fold change |
|---|---|---|---|---|
| Leukocytes | Leukocyte specific transcript 1 | LST1 | 3.30E−08 | − 1.4504 |
| Lymphocytes | Lymphocyte antigen 86 | LY86 | 1.55E−08 | − 1.6468 |
| Lymphocyte antigen 6 complex, locus G6D | LY6G6D | 7.51E−05 | − 2.0459 | |
| Lymphocyte protein tyrosine kinase | LCK | 3.31E−10 | − 1.2848 | |
| Cytotoxic T lymphocyte-associated protein 2 alpha | CTLA2Α | 1.71E−11 | − 1.9618 | |
| Cytotoxic T lymphocyte-associated protein 2 beta | CTLA2Β | 3.60E−09 | − 2.041 | |
| Transcription factor 7 like 1 (T cell specific, HMG box) | TCF7L1 | 3.83E−07 | 1.5258 | |
| Mature T cell proliferation 1 | MTCP1 | 7.32E−07 | − 1.3497 | |
| Linker for activation of T cells family, member 2 | LAT2 | 3.64E−06 | − 1.9848 | |
| Linker for activation of T cells | LAT | 2.73E−05 | − 1.9806 | |
| Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 | NFATC1 | 9.49E−07 | 1.3363 | |
| Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 | NFATC2 | 1.11E−05 | 1.2136 | |
| Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 | NFATC3 | 6.84E−09 | 1.2199 | |
| Nuclear factor of activated T cells 5 | NFAT5 | 1.68E−14 | 2.378 | |
| X-linked lymphocyte-regulated complex | XLR | 1.26E−06 | − 1.7171 | |
| B cells | Early B cell factor 3 | EBF3 | 3.53E−15 | 1.6944 |
| B cell receptor associated protein 29 | BCAP29 | 3.70E−09 | − 1.4942 | |
| Immunoglobulin superfamily | Immunoglobulin superfamily, member 3 | IGSF3 | 2.69E−08 | 1.7284 |
| T-cell immunoglobulin and mucin domain containing 2 pseudogene | GM4926 | 1.69E−06 | − 1.5855 | |
| CD cell surface markers | CD1d1 antigen | CD1D1 | 1.60E−05 | − 1.2071 |
| CD48 antigen | CD48 | 5.13E−07 | − 1.8838 | |
| CD52 antigen | CD52 | 1.50E−05 | − 1.7423 | |
| CD53 antigen | CD53 | 1.94E−10 | − 1.5542 | |
| CD59a antigen | CD59A | 4.72E−08 | − 1.7165 | |
| CD63 antigen | CD63 | 9.54E−08 | − 1.4596 | |
| CD84 antigen | CD84 | 6.02E−07 | − 1.4359 | |
| CD86 antigen | CD86 | 2.09E−07 | − 1.4856 | |
| CD320 antigen | CD320 | 3.60E−11 | − 1.3617 | |
| Chemokine | Chemokine (C–X–C motif) ligand 11 | CXCL11 | 4.45E−05 | − 1.7877 |
| Chemokine (C–C motif) ligand 24 | CCL24 | 9.86E−05 | − 1.2683 | |
| Chemokine (C–C motif) ligand 25 | CCL25 | 1.74E−08 | − 1.6614 | |
| Chemokine (C–C motif) ligand 27A | CCL27A | 4.27E−09 | − 1.8182 | |
| Chemokine (C–X–C motif) receptor 5 | CXCR5 | 6.26E−05 | 1.7003 | |
| Chemokine-like factor 1 | CKLF | 3.37E−06 | − 1.2909 |
Differential regulation as reported by Haque et al.12.
Downregulated Th1/Th17 gene expression
The original KEGG pathway analysis12 revealed that the Th1 and Th2 cell differentiation pathway was not differentially regulated (Table 1) but, as this pathway generates both Th1 and Th2 responses, our in-depth exploration of gene expression revealed several intriguing results (Table 4). We observed upregulated expression of the intermediate complex MAML1 (Supplementary Table 1) and receptor IL12RB2 both of which activate Th1 cell differentiation. Although this hints at a heightened Th1 response, expression of four Th1 interleukins (IL1B, IL15, IL15RA, IL18) was downregulated. In addition, one TNF superfamily alpha inducible gene (TNFAIP8L2), one TNF superfamily receptor (TNFRSF12A), three INF inducible or induced proteins (IFI27L2A, IFI47, IFI35) and one INF stimulated protein (ISG20) were also downregulated (Table 4). Furthermore, in depth analysis of the Th17 signalling KEGG pathway revealed downregulation of its product (IL22) and four genes associated with immunopathology (CCL7, S100A8, S100A9, MMP13) (Table 4) but upregulation of IL17RD, a negative regulator of inflammation29. Together, these observations indicate that maternal H. bakeri infection might have limited Th1/Th17 inflammation and immunopathology.
Table 4.
List of differentially expressed cytokine related genes classified by immune response in the pup brain, in response to maternal H. bakeri infection.
| Immune response | Classification | Gene name | Gene symbol | p value | Log 2 fold change |
|---|---|---|---|---|---|
| Th1 | Interferon | Interferon, alpha-inducible protein 27 like 2A | IFI27L2A | 2.51E−09 | − 2.5516 |
| Interferon gamma inducible protein 47 | IFI47 | 6.30E−07 | − 1.6666 | ||
| Interferon-induced protein 35 | IFI35 | 2.15E−06 | − 1.3033 | ||
| Interferon-stimulated protein | ISG20 | 7.81E−07 | − 1.442 | ||
| Tumor necrosis factor | Tumor necrosis factor, alpha-induced protein 8-like 2 | TNFAIP8L2 | 5.60E−08 | − 1.6267 | |
| Tumor necrosis factor receptor superfamily, member 11a | TNFRSF11A | 2.39E−08 | 1.2215 | ||
| Tumor necrosis factor receptor superfamily, member 12a | TNFRSF12A | 6.00E−06 | − 1.2305 | ||
| Interleukin | Interleukin 1 beta | IL1B | 1.33E−07 | − 3.1953 | |
| Interleukin-1 receptor-associated kinase 1 binding protein 1 | IRAK1BP1 | 3.38E−10 | − 1.5769 | ||
| Interleukin 15 | IL15 | 6.30E−06 | − 1.662 | ||
| Interleukin 15 receptor, alpha chain | IL15RA | 4.79E−05 | − 1.3734 | ||
| Interleukin 18 | IL18 | 4.49E−09 | − 1.4295 | ||
| Interleukin 12 receptor, beta 2 | IL12RB2 | 1.32E−06 | 2.0879 | ||
| Th2 | Interleukin | Interleukin 4 | IL4 | 1.81E−07 | 1.2171 |
| Interleukin 13 receptor, alpha 2 | IL13RA2 | 1.62E−09 | − 1.9619 | ||
| Interleukin enhancer binding factor 2 | ILF2 | 6.79E−08 | − 1.2226 | ||
| Treg | Transforming growth factor | Transforming growth factor, beta 2 | TGFB2 | 2.55E−07 | 1.02481 |
| Transforming growth factor, beta receptor III | TGFBR3 | 4.69E−08 | 1.2645 | ||
| Transforming growth factor, beta receptor associated protein 1 | TGFBRAP1 | 8.92E−07 | 1.3063 | ||
| Latent transforming growth factor beta binding protein 3 | LTBP3 | 1.35E−07 | 1.3637 | ||
| Latent transforming growth factor beta binding protein 4 | LTBP4 | 3.43E−06 | 1.6984 | ||
| Transforming growth factor alpha | TGFA | 1.30E−06 | 1.2669 | ||
| Interleukin | Interleukin 10-related T cell-derived inducible factor beta | ILTIFB | 3.20E−07 | − 2.3961 | |
| Th17 | Interleukin | Interleukin 17 receptor D | IL17RD | 4.41E−10 | 1.9374 |
| Interleukin 22 | IL22 | 9.41E−06 | − 2.162 | ||
| Chemokine | Chemokine (C–C motif) ligand 7 | CCL7 | 3.00E−05 | − 1.6164 | |
| Related proteins | S100 calcium binding protein A8 (calgranulin A) | S100A8 | 3.07E−08 | − 2.4356 | |
| S100 calcium binding protein A9 (calgranulin B) | S100A9 | 2.39E−07 | − 2.0865 | ||
| Matrix metallopeptidase 13 | MMP13 | 4.26E−05 | − 1.7194 |
Differential regulation as reported by Haque et al.12.
1Borderline significant value.
Upregulated Th2/Treg gene expression
As expected given the cross-regulation between Th1/Th17 and Th2/Treg responses, maternal H. bakeri infection upregulated Th2/Treg gene expression in the pup brain, as evidenced by the relatively consistent pattern of upregulation from receptor (Notch1/2) to its intermediate complex MAML1 (see Supplementary Table 1) to IL4 (Table 4), the hallmark Th2 product of the Th1and Th2 cell differentiation pathway. Among Treg-related genes (Table 4), expression of TGF-β receptor 3 (TGFBR3), TGF-β receptor-associated protein (TGFBRAP1), latent TGF-β binding proteins (LTBP3, LTBP4) and TGFA were upregulated, and TGFB2 expression was upregulated in response to maternal infection although it did not meet our log 2 fold change cut-off. These findings are consistent with a dominant Th2/Treg bias in response to maternal H. bakeri infection, a response that might play an important role in modulating inflammation and auto-immune responses in the brains of the uninfected neonates.
Impact of Maternal Infection on Genes Involved in Transport of Immune Signals across the BBB
Based on our previous KEGG pathway analysis12, there was evidence that maternal H. bakeri infection altered the expression of several pathways involved in transport of immune signals across the BBB. Five pathways (transendothelial migration, regulation of actin cytoskeleton, adheren junction, tight junction, endocytosis) were upregulated whereas two pathways (cytokine-cytokine receptor interaction, soluble N- ethylmaleimide-sensitive factor attachment protein receptor (SNARE) interactions for vesicular transport) were downregulated (Table 1)12. To gain better clarity, gene expression data were probed to more precisely define functions by which a maternal nematode infection might have altered transendothelial migration of leukocytes and receptor-mediated endocytosis of cytokines and immunoglobulins.
Heightened leukocyte transendothelial cell migration
Leukocyte migration involves integrins that allow docking and diapedesis and the dynamically responsive actin cytoskeleton that allows endothelial cells to expand and contract as leukocytes pass in between them. Expression of three integrin alpha genes (ITGA3,4,11) and integrin beta (ITGB4) was upregulated, and expression of integrin beta 1 binding protein 1 (ITGB1BP1) was downregulated by maternal infection (Table 5). Among the matrix metallopeptidases (MMPs) that are regulated by actin cytoskeleton remodeling and involved in formation of transcellular channels, maternal H. bakeri infection upregulated MMMP15 and downregulated MMP13 (Table 5).
Table 5.
List of cell adhesion molecules and related genes involved in leukocyte transendothelial cell migration that are differentially expressed in the pup brain, in response to maternal H. bakeri infection.
| Classification | Gene name | Gene symbol | p value | Log 2 fold change |
|---|---|---|---|---|
| Leukocyte transendothelial cell migration | Integrin alpha 3 | ITGΑ3 | 1.16E−06 | 1.2366 |
| Integrin alpha 4 | ITGΑ4 | 3.81E−08 | 1.2737 | |
| Integrin alpha 11 | ITGΑ11 | 2.20E−08 | 1.7196 | |
| Integrin alpha E, epithelial-associated | ITGΑE | 3.47E−06 | − 2.3048 | |
| Integrin beta 4 | ITGΒ4 | 1.71E−09 | 1.8016 | |
| Integrin beta 1 binding protein 1 | ITGΒ1BP1 | 6.09E−11 | − 1.9182 | |
| Calcium and integrin binding 1 (calmyrin) | CIB1 | 1.04E−09 | − 1.4908 | |
| Calcium and integrin binding family member 2 | CIB2 | 6.26E−09 | − 1.3909 | |
| Matrix metallopeptidase 13 | MMP13 | 4.26E−05 | − 1.7194 | |
| Matrix metallopeptidase 15 | MMP15 | 4.67E−05 | 1.295 | |
| Adheren junctions | Cadherin 3 | CDH3 | 1.01E−08 | 1.6134 |
| Cadherin 4 | CDH4 | 4.83E−06 | 1.3775 | |
| Cadherin 5 | CDH5 | 2.41E−05 | 1.2137 | |
| Cadherin 6 | CDH6 | 5.87E−07 | 1.6177 | |
| Cadherin 23 (otocadherin) | CDH23 | 8.92E−09 | 1.9526 | |
| Cadherin, EGF LAG seven-pass G-type receptor 1 | CELSR1 | 2.34E−11 | 2.5308 | |
| Cadherin, EGF LAG seven-pass G-type receptor 2 | CELSR2 | 2.81E−08 | 2.8609 | |
| Cadherin, EGF LAG seven-pass G-type receptor 3 | CELSR3 | 5.52E−10 | 2.3788 | |
| Catenin (cadherin associated protein), delta 2 | CTNND2 | 7.83E−07 | 1.5838 | |
| Catenin (cadherin associated protein), delta 1 | CTNND1 | 2.55E−06 | 1.2385 | |
| Desmoglein 2 | DSG2 | 2.09E−05 | 1.2256 | |
| Tight junctions | Vinculin | VCL | 2.16E−07 | 1.7986 |
| Tight junction protein 1 | TJP1 | 9.28E−09 | 1.234 | |
| Cingulin | CGN | 1.11E−05 | 1.3057 | |
| Cingulin-like 1 | CGNL1 | 8.38E−11 | 1.9121 | |
| Occludin/ELL domain containing 1 | OCEL1 | 1.48E−09 | − 1.2976 | |
| Claudin 10 | CLDN10 | 3.11E−09 | − 1.5952 |
Transport of leukocytes between endothelial cells is regulated by adheren junctions, tight junctions and gap junctions. Among the genes involved in adheren junctions, maternal infection upregulated expression of eight cadherins (CDH3,4,5,6,23; CELSR1,2), two catenins (CTNND1, 2) and desmogliein (DSG2) (Table 5) providing a strong indication that the function of these junctions was heightened in response to a maternal nematode infection. With respect to tight junctions, expression of vinculin (VCL), tight junction protein 1 (TJP1) and two cingulins (CGN, CGNL1) involved in actin binding was upregulated whereas expression of genes associated with sealing tight junctions (the occludin, OCEL1 and the claudin, CLDN10) was downregulated (Table 5). Of note, we found no evidence of differential expression of genes associated with gap junctions.
Taken together, these gene expression data provide evidence of more dynamic interactions between junctions and the actin cytoskeleton at the BBB of neonates of infected mothers which would be consistent with heightened migration of leukocytes between endothelial cells.
Endocytosis limited by impaired intracellular trafficking
Receptor-mediated endocytosis involves initiation and signalling as well as vesicle migration and endosome formation. We observed that maternal infection upregulated expression of eight genes involved in initiation and signalling including three involved in TGF-β transport (TGFβ, TGFβR3, SMAD3) as well as dynamin genes (DNM3, DNMBP) that are critical for vesicle budding, but downregulated expression of calveolin 2 (CAV2), a clathrin (CLTA) and epidermal growth factor receptor (EGFR) (Table 6). Importantly, however, we found evidence that intracellular trafficking was impaired based on downregulation of four sorting nexin family genes (SNX1,2,5,7), one vacuolar protein sorting gene (VSP29), one coiled-coil domain gene (CCDC53), and one charged multivesicular body protein gene (CHMP2A) that are all involved in vesicular migration and endosome formation. Among differentially expressed genes involved in intracellular trafficking, only the early endosome antigen 1 gene (EEA1) was upregulated by maternal H. bakeri infection (Table 6). Notably, the programmed cell death 6 (PDCD6) gene was downregulated as was the apoptosis pathway which suggests that maternal nematode infection might have downregulated apoptosis in the neonatal brain to further protect neural development.
Table 6.
List of genes involved in endocytosis pathway that are differentially expressed in the pup brain, in response to maternal H. bakeri infection.
| Classification | Gene name | Gene symbol | p value | Log 2 fold change |
|---|---|---|---|---|
| Initiation and signalling | Transforming growth factor, beta 2 | TGFB2 | 2.55E−07 | 1.0248 |
| Transforming growth factor, beta receptor III | TGFBR3 | 4.69E−08 | 1.2645 | |
| MAD homolog 3 (Drosophila) | SMAD3 | 6.35E−08 | 1.6784 | |
| Dynamin 3 | DNM3 | 4.95E−09 | 1.6835 | |
| Dynamin binding protein | DNMBP | 2.17E−05 | 1.5473 | |
| Caveolin 2 | CAV2 | 6.89E−07 | − 1.2166 | |
| Clathrin, light polypeptide (Lca) | CLTA | 3.71E−08 | − 1.2065 | |
| EGF-like domain 7 | EGFL7 | 5.75E−10 | − 1.5489 | |
| Epidermal growth factor receptor | EGFR | 1.53E−11 | 1.4725 | |
| Adaptor protein complex AP-2, alpha 1 subunit | AP2A1 | 6.49E−05 | 1.3822 | |
| Protein kinase C, alpha | PRKCA | 3.03E−07 | 1.5849 | |
| Rous sarcoma oncogene | SRC | 5.11E−06 | 1.462 | |
| Vesicle migration and endosome formation | Sorting nexin 1 | SNX1 | 9.45E−08 | − 1.1603 |
| Sorting nexin 2 | SNX2 | 2.29E−07 | − 1.1769 | |
| Sorting nexin 5 | SNX5 | 3.31E−07 | − 1.234 | |
| Sorting nexin 7 | SNX7 | 1.98E−08 | − 1.1221 | |
| Vacuolar protein sorting 29 (S. pombe) | VPS29 | 2.19E−11 | − 1.8892 | |
| Coiled-coil domain containing 53 | CCDC53 | 2.87E−09 | − 1.4254 | |
| Charged multivesicular body protein 2A | CHMP2A | 5.63E−10 | − 1.6867 | |
| Early endosome antigen 1 | EEA1 | 1.71E−07 | 1.2971 | |
| RAB7, member RAS oncogene family-like 1 | RAB7L1 | 1.71E−09 | − 1.4527 | |
| CDC42 binding protein kinase beta | CDC42BPB | 7.17E−06 | 1.8055 | |
| Programmed cell death 6 | PDCD6 | 2.68E−10 | − 1.7176 | |
| Kinesin family member 5A | KIF5A | 2.32E−07 | 1.6266 | |
| ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) | ARFGEF2 | 1.21E−09 | 1.9556 | |
| WAS protein family, member 2 | WASF2 | 1.04E−07 | 1.5409 |
Differential regulation as reported by Haque et al.12.
Therefore, despite upregulation of the endocytosis KEGG pathway in the original study12, the observed downregulation of vesicle formation genes suggests that maternal infection might have impaired transport of immunoglobulins and cytokines across the BBB in the offspring of nematode infected dams.
Confirmation of sequencing data by qPCR
Using qPCR, we validated the gene expression data reported in our RNA Hi-seq sequencing (Supplementary Table 2). Interestingly two of the upregulated genes reported by RNA seq data showed a higher fold-change in qPCR analysis than RNA seq (TGFB2: 3.1 vs 1.02 and ITGA11: 2.3 vs 1.71) in brains of pups of infected compared with uninfected dams. In addition, VPS29 which was reportedly downregulated in RNA seq data showed a tendency to be lower in response to maternal nematode infection (p = 0.15).
Discussion
Our comprehensive interrogation of KEGG pathway-associated genes in our list of pup brain genes that were differentially expressed genes in response to maternal H. bakeri infection revealed three key findings. Unlike many maternal stressors that are associated with neonatal neuro-inflammation30–33, we showed that maternal nematode infection downregulated expression of only a few cell surface markers and chemokine ligand genes indicating a very limited impact on innate immune genes. However, this maternal nematode infection restricted to the maternal intestine led to widespread differential expression of genes of the adaptive immune response in the neonatal brain. Most notable was the upregulation of genes related to Th2 and Treg responses and downregulation of genes related to Th1 and Th17 responses. This is consistent with the Th2/Treg response typical in the host infected with H. bakeri18. We also found a gene expression signature of heightened leukocyte migration between endothelial cells of the BBB in response to maternal nematode infection. The upregulated expression of genes involved in the leukocyte transendothelial migration pathway and in expression of integrins and other junction genes indicated enhanced migration of leukocytes which likely included Th2 and Treg cells into the neonatal brain. In contrast, lowered expression of genes needed for vesicular transport indicated impaired endocytosis of immune elements including cytokines and immunoglobulins. Taken together, these findings indicate a Th2/Treg biased response in the pup brain perhaps driven more by T cell entry in between endothelial cells of the BBB than by immunoglobulin or cytokine endocytosis.
Innate and adaptive immune responses play important homeostatic roles in the developing brain that promote neurodevelopment, limit neuro-inflammation and neurological diseases, and ensure that any pathogens that cross the BBB are efficiently recognized and controlled34. With respect to innate responses, in addition to our previous report12 of downregulated expression of the RIG-I-like receptor signaling KEGG pathway involved in recognition of viral pathogens35, we found that maternal infection downregulated expression of several chemokines and CD cell surface markers, suggesting a limited negative impact on innate immunity. However, in exploring genes associated with vesicle mediated transport, we also observed differential expression of several genes in a direction that suggested reduced programmed cell death. Though not a focus of this study, this latter observation raises the intriguing possibility that maternal nematode infection might limit apoptosis in the uninfected neonatal brain.
In contrast to the innate immune system, our analysis provided considerable evidence that maternal infection not only altered expression of the adaptive immune response but also led to Th2/Treg bias in the pup brain. Upregulated expression of B cell and T cell receptor signaling KEGG pathways was previously reported12 and our current study showed differential expression of numerous genes needed for Th1, Th2, Treg and Th17 responses including genes involved in T and B cell differentiation, maturation, migration, activation as well as receptors, ligands, and signalling molecules. These findings strongly indicate that maternal H. bakeri infection affected adaptive immunity in the uninfected pup brain. Furthermore, the upregulated expression of the hallmark Th2 cytokine IL436,37 together with genes in its signaling cascade indicated a Th2 bias. As IL4 is an activator and recruiter of Th2 cells, downstream consequences might not yet be evident at P7, explaining why we did not detect differential expression of IL13, another hallmark Th2 cytokine37. In addition, the B cell receptor signaling pathway and the B cell development gene EBF3 were upregulated. They are important in initiating a heightened Th2 cell response38. Consistent with an upregulated Th2 response, upregulated expression of TGF family genes including receptors, binding proteins, and receptor associated proteins all point to an upregulated Treg response. Together, these results clearly highlight that this maternal nematode infection shifted gene expression toward a Th2/Treg response in the brain of the uninfected neonate.
Further evidence of a Th2/Treg bias was seen in the dampened expression of genes involved in the Th1/Th17 responses, an observation that is consistent with the cross-regulation of these two arms of the adaptive immune system10. As IL4 and TGF-β both have a negative effect on Th1 cytokine production9, it was not surprising to see the downregulated expression of Th1 interleukins as well as TNF and INF related proteins in the neonatal brain. In addition, the autoimmune Th17 response was downregulated as indicated by the downregulation of its hallmark IL22 coding gene as well as most autoimmune pathology genes in the IL17 signaling pathway.
The observed bias toward Th2/Treg adaptive immunity in response to H. bakeri is well documented in lymphoid tissues and blood of the infected mouse27,40 and there is evidence that a protective Th2 response against GI nematodes is transferred to the neonate through T cells and immunoglobulins in milk4,39. Transfer of H. bakeri specific IgG1 to the neonate has been shown to protect the pups from this infection39 and transfer of Th2 competent CD4 + T cells from mice infected with a related nematode (Nippostrongylus brasiliensis) has been shown to induce lasting protection against direct infection of the pup4. The maternal Th2/Treg bias extends beyond the intestine as seen in the lungs and spleen of neonates of H. bakeri infected mice4. Our results extend these systemic impacts of maternal nematode infection on immunity in the neonate to expression of the adaptive immune response in the pup brain.
We had hypothesized that the neonatal brain may have received signals of maternal infection through movement of leukocytes in between endothelial cells of the BBB. Based on our analysis of leukocyte transendothelial migration, our data strongly suggest that paracellular movement of leukocytes into the brain was enhanced by maternal H. bakeri infection. In addition to the upregulated leukocyte transendothelial migration pathway, we observed upregulated expression of a variety of integrins that dock leukocytes to endothelial cells. Diapedesis also involves dynamic reshaping of endothelial cells by the actin cytoskeleton40 and transient loosening then tightening of tight junctions41,42. Gene expression data are consistent with both, as the actin cytoskeleton pathway was upregulated12 providing flexibility to the endothelial cells and as genes involved specifically in adheren junctions were upregulated. At first glance, the observed upregulation of junction unit pathways could be interpreted to reflect tightening of the BBB. However, we suggest that, as a response to the junction loosening caused by leukocyte infiltration, upregulated junction expression might restore the selective permeability that is critical for restoring and maintaining BBB integrity.
We had also hypothesized that maternal infection might have influenced transport of immunoglobulin and cytokine signals across the BBB. This transport typically occurs through vesicle mediated endocytosis of receptor-cytokine complexes through the endothelial cells via endosomes22–24. The endocytosis pathway was upregulated12 as was expression of ligands, receptors, signaling molecules, and endosome formation scissor genes. However, our data indicated that endosome formation was impaired given the downregulation of SNARE interactions for vesicular transport pathway and of several genes related to vesicle formation. This suggests that receptor mediated endocytosis of H. bakeri immune markers may not be functional despite it being a common pathway for immune signaling.
A variety of studies over the past decade have begun to detect ways in which GI nematode infections may provide benefits to the infected host7 and also to their uninfected offspring4,8. It was previously reported that maternal H. bakeri infection upregulated expression of LTP and synaptogenesis-related pathways in 7-day old pup brains12, pathways that are known to enhance learning and memory13–15. Our secondary analysis of gene expression data from these pups also revealed an upregulated Th2 response and upregulated expression of IL4, both of which are known to be necessary for memory and learning43. Furthermore, as Treg responses promote neural development by mediating axon specification and TGF-β receptor signaling guides neuronal axon initiation in the brain44, the observed upregulation of Treg responses would also have potentially positive impacts on learning and memory for the neonate of infected dams. An upregulated Treg response also plays an important role in dampening Th1 inflammation45 which would limit neuro-inflammation that in turn compromises the integrity of the BBB46. Thus, our results indicate that maternal infection might benefit the neonate by limiting neuro-inflammation and promoting a Th2/Treg environment that might stimulate learning and memory. Similar intergenerational findings may also be found for other maternal infections that induce a Th2/Treg response in the infected host.
A strength of this study was the identification of patterns of differential gene expression within KEGG pathway maps that are not evident by the previously published KEGG pathway analysis alone12. As the KEGG pathway analysis relies more on numeric evaluation of gene expression than on a logical analysis based on gene functionality, the results of KEGG pathway analysis can provide contradictory information whereby a pathway can be both upregulated and downregulated simultaneously. Pathway maps typically include three regions: pathway activation mediated by ligands and receptors, propagation of a signal, and formation of products that perform the function of the pathway. As signaling molecules and intermediates are highly redundant and shared among many pathways, our context dependent approach focused on ligands and receptors to assess pathway activation and on products to assess function. To minimize design bias, our search strategy included all possible immune-related genes taken from KEGG pathway maps and from a list of immune-related categories and processes identified from the literature. To lower false discovery rate, more stringent cut-offs for P-value and log 2 fold change were used than in the original gene expression database12. To minimize confirmation bias, we used every opportunity to receive critiques on the logic of our arguments. To ensure compatibility and consistency, comparisons were made with the few analogous studies. As a result, our approach overcame the limitation of relying only on KEGG pathway analysis and provided internally coherent observations that were consistent with the literature. Nevertheless, we acknowledge that we may have excluded important genes or included genes whose differential expression was of little functional importance. We also acknowledge the limitations associated with reliance on gene expression data with qPCR confirmation of only a few genes and without assaying protein concentrations or conducting functional assays to examine phenotypic effects.
In conclusion, our context relevant interrogation of gene expression in the neonatal brain indicated that a maternal H. bakeri infection might promote transendothelial migration of Th2/Treg cells across the BBB of the uninfected neonate and might induce a Th2/Treg response in the neonatal brain. As a Th2/Treg response could have potential benefits in reducing neuro-inflammation and promoting learning and memory, follow-up experimental studies to confirm the gene expression data and to explore neuro-immune development and behavioural responses in the pups of infected dams would be important.
Methodology
Source of data
This study was a secondary analysis of immune and BBB related genes that were differentially expressed in the neonatal brain in response to maternal H. bakeri infection (https://www.nature.com/articles/s41598-019-40729-w#Sec2)12. The original experiment used timed pregnant CD1 outbred mice that had been given a repeated (trickle) infection of 100 ± 3 L3 larvae of H. bakeri or a sham infection of distilled water through oral gavage on embryonic days E7, E12, E17, and postpartum day 3 (P3). The trickle infection protocol mimics natural transmission47 and allows both larvae and adults to be present simultaneously. Thus the immunoregulation induced by adult worms in the lumen is countered by the immunogenicity of the L4 larvae in the serosal musculature18. Pup brains were genotyped on P7 when most neurons have established synaptic connections and during the critical period of synaptogenesis48,49. Total brain RNA from one randomly selected male pup per litter (n = 5 per group) was sequenced in an Illumina HiSeq2000 sequencer. The sequence files were analysed using HT-seq50 and NetworkAnalyst51 to identify genes in the pup brain that were differentially expressed in response to maternal nematode infection with adjusted p value < 0.05 and log 2 fold change > 1. The original exploration of the KEGG pathway database in NetworkAnalyst12 provided a list of the differentially expressed pathways with biological significance.
Procedures for secondary analysis of differentially expressed genes in KEGG pathway maps
For our secondary analysis, we applied more stringent p value (< E−5) and log 2 fold change (> 1.2) cut-offs for differential gene expression than had been used in the original analysis12 to lower the false recovery rate52.
We matched this more stringent gene expression database against genes in KEGG pathway maps (Fig. 1). We focused on the ligand and receptor coding genes that activate the pathway and genes that code for final products rather than intermediates and signaling molecules that have high biological redundancy among pathways and less internal consistency in cascade expression. This allowed us to infer the implications of changes in gene expression for context relevant functions that occur within pathways and that may have been independent of overall differential expression of the KEGG pathway.
Figure 1.
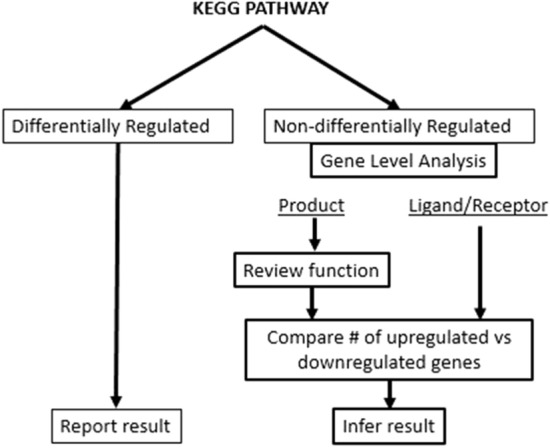
Schematic showing the approach for exploring KEGG pathway maps using the database of differentially expressed genes12.
Selection of immune system genes and related KEGG pathways
A list of differentially expressed immune system related genes was created using the more stringent cut-offs (Supplementary Table 1) in order to explore evidence that maternal infection altered expression of the different molecules and cells of the immune system based both on general categories of immune cells and molecules and on genes in immune KEGG pathways.
First, the original database was mined for genes using the categories of cells and molecules involved in any immune response. Cells explored included myeloid cell lineage for innate immune cells (monocyte, macrophage, microglia, dendritic cell, granulocyte, neutrophil, basophil, eosinophil, mast cell) and lymphoid cell lineage for adaptive immune cells (NK cell, lymphoid cell, lymphocyte, T cell, B cell, plasma cell, and leukocyte). Molecules explored included monocyte chemoattractant protein (MCP), colony stimulating factor (CSF), interferon (INF), interleukin (IL), chemokine, immunoglobulin (Ig), tumor necrosis factor (TNF), transforming growth factor (TGF), lymphotoxin, toll-like receptors, CD antigens, major histocompatibility complex (MHC), and selectin. Differential expression of genes in these categories provided the first insight into possible alterations to the immune system in the pup brain in response to maternal infection.
Second, we prepared a list of differentially expressed genes related to the immune system from each of the 21 immune related KEGG pathways (Table 2) (https://www.kegg.jp/kegg/pathway.html) regardless of whether or not they were reported as differentially regulated12. The list included inducible proteins, linkers for activation, subunits, inhibitors, activators, receptors, domains, binding proteins, related proteins, “like” genes, other members of the family, and all other intermediate molecules in the pathway.
Selection of blood brain barrier genes and related KEGG pathways
To determine whether mechanisms known to transport immune cells, cytokines, and immunoglobulins across the BBB were influenced by maternal infection, we made a list of all relevant genes from the KEGG pathway maps for endocytosis and leukocyte transendothelial cell migration. This list included cell adhesion molecules, junction proteins, ligands and receptors, and vesicle formation genes.
Validation of brain gene expression data
To validate the brain gene expression data obtained from the Illumina Hi-seq sequencing we performed real-time qPCR analysis of three representative genes (TGFB2, ITGA11 and VPS29) following the MIQE guidelines53. Already validated primer sequences were obtained from PrimerBank54 and purchased from Integrated DNA technologies (Supplementary Table 3). We used frozen whole brain RNA samples from the original experiment12 and taking 5ug of total RNA from five pups from control and infection group respectively, we synthesized cDNA using the iScript cDNA Synthesis kit (Bio-Rad, Canada) following instructions from the manufacturer. cDNAs were diluted (1:50) and used for qPCR in a CFX384 (BioRad) machine with the following protocol: initial denaturation at 95 °C for 3 min followed by 39 cycles at 95 °C for 15 s and 60 °C for 45 s for annealing, and finally 95 °C for 10 s. The data were normalized to the geometric mean expression levels of four reference genes (GADPH, L19, B2M, and SDHA).
Supplementary Information
Author contributions
N.E.A., M.H., K.K. and M.S. conceived and designed the study. N.E.A. was responsible for the methodology, data analysis, interpretation, and manuscript writing. M.H. performed the primary study from which we obtained the data. E.M. designed, conducted and analyzed the qPCR validation and incorporated the findings into the revised manuscript. M.S., K.K. and M.H. provided suggestions on the hypothesis and data interpretation as well as critical suggestions that were incorporated into the manuscript. M.S. obtained funding for the research.
Funding
Funding was provided by Natural Sciences and Engineering Research Council of Canada (Grant No. RGPIN/04563-2017).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-01510-0.
References
- 1.Awasthi S, Bundy D. Intestinal nematode infection and anaemia in developing countries. Br. Med. J. 2007;334:1065–1066. doi: 10.1136/bmj.39211.572905.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odiere MR, Koski KG, Weiler HA, Scott ME. Concurrent nematode infection and pregnancy induce physiological responses that impair linear growth in the murine foetus. Parasitology. 2010;137:991–1002. doi: 10.1017/S0031182009991764. [DOI] [PubMed] [Google Scholar]
- 3.Odiere MR, Scott ME, Leroux L-P, Dzierszinski FS, Koski KG. Maternal protein deficiency during a gastrointestinal nematode infection alters developmental profile of lymphocyte populations and selected cytokines in neonatal mice. J. Nutr. 2012;143:100–107. doi: 10.3945/jn.112.160457. [DOI] [PubMed] [Google Scholar]
- 4.Darby MG, et al. Pre-conception maternal helminth infection transfers via nursing long-lasting cellular immunity against helminths to offspring. Sci. Adv. 2019 doi: 10.1126/sciadv.aav3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masters S, Barrett-Connor E. Parasites and asthma–predictive or protective? Epidemiol. Rev. 1985;7:49–58. doi: 10.1093/oxfordjournals.epirev.a036285. [DOI] [PubMed] [Google Scholar]
- 7.Ponte EV, et al. Reduced asthma morbidity in endemic areas for helminth infections: A longitudinal ecological study in Brazil. J. Asthma. 2014;51:1022–1027. doi: 10.3109/02770903.2014.936454. [DOI] [PubMed] [Google Scholar]
- 8.Straubinger K, et al. Maternal immune response to helminth infection during pregnancy determines offspring susceptibility to allergic airway inflammation. J. Allergy Clin. Immunol. 2014;134:1271–1279. doi: 10.1016/j.jaci.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 9.Lazarski CA, Ford J, Katzman SD, Rosenberg AF, Fowell DJ. IL-4 attenuates Th1-associated chemokine expression and Th1 trafficking to inflamed tissues and limits pathogen clearance. PLoS ONE. 2013 doi: 10.1371/journal.pone.0071949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee GR. The balance of Th17 versus Treg cells in autoimmunity. Int. J. Mol. Sci. 2018;19:730. doi: 10.3390/ijms19030730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque M, Starr LM, Koski KG, Scott ME. Differential expression of genes in fetal brain as a consequence of maternal protein deficiency and nematode infection. Int. J. Parasitol. 2018;48:51–58. doi: 10.1016/j.ijpara.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Haque M, Koski KG, Scott ME. Maternal gastrointestinal nematode infection up-regulates expression of genes associated with long-term potentiation in perinatal brains of uninfected developing pups. Sci. Rep. 2019;9:4165–4165. doi: 10.1038/s41598-019-40729-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham WC, Jones OD, Glanzman DL. Is plasticity of synapses the mechanism of long-term memory storage? NPJ Sci. Learn. 2019;4:9. doi: 10.1038/s41539-019-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuchlik A. Dynamic learning and memory, synaptic plasticity and neurogenesis: an update. Front. Behav. Neurosci. 2014 doi: 10.3389/fnbeh.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez JL, Jr, Derrick BE. Long-term potentiation and learning. Annu. Rev. Psychol. 1996;47:173–203. doi: 10.1146/annurev.psych.47.1.173. [DOI] [PubMed] [Google Scholar]
- 16.Gadani SP, Cronk JC, Norris GT, Kipnis J. IL-4 in the brain: A cytokine to remember. J. Immunol. 2012;189:4213–4219. doi: 10.4049/jimmunol.1202246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derecki NC, et al. Regulation of learning and memory by meningeal immunity: A key role for IL-4. J. Exp. Med. 2010;207:1067–1080. doi: 10.1084/jem.20091419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maizels RM, et al. Immune modulation and modulators in Heligmosomoides polygyrus infection. Exp. Parasitol. 2012;132:76–89. doi: 10.1016/j.exppara.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith MW, Gumbleton M. Endocytosis at the blood-brain barrier: From basic understanding to drug delivery strategies. J. Drug Target. 2006;14:191–214. doi: 10.1080/10611860600650086. [DOI] [PubMed] [Google Scholar]
- 20.Carman CV. Mechanisms for transcellular diapedesis: Probing and pathfinding by ‘invadosome-like protrusions’. J. Cell Sci. 2009;122:3025–3035. doi: 10.1242/jcs.047522. [DOI] [PubMed] [Google Scholar]
- 21.Field CJ. The immunological components of human milk and their effect on immune development in infants. J. Nutr. 2005;135:1–4. doi: 10.1093/jn/135.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Pan W, et al. Cytokine signaling modulates blood–brain barrier function. Curr. Pharm. Des. 2011;17:3729–3740. doi: 10.2174/138161211798220918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippi M-D. Mechanism of diapedesis: Importance of the transcellular route. Adv. Immunol. 2016;129:25–53. doi: 10.1016/bs.ai.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao H, Shi W, Freund LB. Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA. 2005;102:9469–9474. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishara, N. The use of biomarkers for detection of early- and late-onset neonatal sepsis. In Hematology, Immunology and Infectious Disease: Neonatology Questions and Controversies (Second Edition) (eds. Ohls, R. K. & Maheshwari, A.) 303–315 (W.B. Saunders, 2012).
- 26.Schnoor M. Endothelial actin-binding proteins and actin dynamics in leukocyte transendothelial migration. J. Immunol. 2015;194:3535–3541. doi: 10.4049/jimmunol.1403250. [DOI] [PubMed] [Google Scholar]
- 27.Muller WA. Mechanisms of leukocyte transendothelial migration. Annu. Rev. Pathol. 2011;6:323–344. doi: 10.1146/annurev-pathol-011110-130224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen L. Tight junctions on the move: Molecular mechanisms for epithelial barrier regulation. Ann. NY Acad. Sci. 2012;1258:9–18. doi: 10.1111/j.1749-6632.2012.06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellett M, et al. Orphan receptor IL-17RD regulates Toll-like receptor signalling via SEFIR/TIR interactions. Nat. Commun. 2015;6:6669. doi: 10.1038/ncomms7669. [DOI] [PubMed] [Google Scholar]
- 30.Shanks N, et al. Early-life exposure to endotoxin alters hypothalamic-pituitary-adrenal function and predisposition to inflammation. Proc. Natl. Acad. Sci. USA. 2000;97:5645–5650. doi: 10.1073/pnas.090571897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes TM, Coe CL. Prenatal manipulations reduce the proinflammatory response to a cytokine challenge in juvenile monkeys. Brain Res. 1997;769:29–35. doi: 10.1016/s0006-8993(97)00687-2. [DOI] [PubMed] [Google Scholar]
- 32.Shanks N, Larocque S, Meaney MJ. Neonatal endotoxin exposure alters the development of the hypothalamic-pituitary-adrenal axis: Early illness and later responsivity to stress. J. Neurosci. 1995;15:376–384. doi: 10.1523/JNEUROSCI.15-01-00376.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanks N, Lightman SL. The maternal-neonatal neuro-immune interface: Are there long-term implications for inflammatory or stress-related disease? J. Clin. Invest. 2001;108:1567–1573. doi: 10.1172/JCI14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aarli JA. The immune system and the nervous system. J. Neurol. 1983;229:137–154. doi: 10.1007/BF00313738. [DOI] [PubMed] [Google Scholar]
- 35.Loo Y-M, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama T, et al. Th2 cells in health and disease. Annu. Rev. Immunol. 2017;35:53–84. doi: 10.1146/annurev-immunol-051116-052350. [DOI] [PubMed] [Google Scholar]
- 37.Bao K, Reinhardt RL. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine. 2015;75:25–37. doi: 10.1016/j.cyto.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddur MS, Bayry J. B cells drive Th2 responses by instructing human dendritic cell maturation. Oncoimmunology. 2015 doi: 10.1080/2162402X.2015.1005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris NL, et al. Mechanisms of neonatal mucosal antibody protection. J. Immunol. 2006;177:6256–6262. doi: 10.4049/jimmunol.177.9.6256. [DOI] [PubMed] [Google Scholar]
- 40.Prasain N, Stevens T. The actin cytoskeleton in endothelial cell phenotypes. Microvasc. Res. 2009;77:53–63. doi: 10.1016/j.mvr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavey M, Lecuit T. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harb. Perspect. Biol. 2009 doi: 10.1101/cshperspect.a002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber CR. Dynamic properties of the tight junction barrier. Ann. NY Acad. Sci. 2012;1257:77–84. doi: 10.1111/j.1749-6632.2012.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brynskikh A, Warren T, Zhu J, Kipnis J. Adaptive immunity affects learning behavior in mice. Brain Behav. Immun. 2008;22:861–869. doi: 10.1016/j.bbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-β signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Littringer K, et al. Common features of regulatory T cell specialization during Th1 responses. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SY, Buckwalter M, Soreq H, Vezzani A, Kaufer D. Blood-brain barrier dysfunction-induced inflammatory signaling in brain pathology and epileptogenesis. Epilepsia. 2012;53(Suppl 6):37–44. doi: 10.1111/j.1528-1167.2012.03701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brailsford TJ, Behnke JM. The dynamics of trickle infections with Heligmosomoides polygyrus in syngeneic strains of mice. Int. J. Parasitol. 1992;22:351–359. doi: 10.1016/s0020-7519(05)80013-x. [DOI] [PubMed] [Google Scholar]
- 48.Han X, et al. Transcriptome of embryonic and neonatal mouse cortex by high-throughput RNA sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:12741–12746. doi: 10.1073/pnas.0902417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semple BD, et al. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013;106–107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia J, Gill EE, Hancock REW. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 52.McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bustin SA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 54.Wang, X., Spandidos, A., Wang, H., & Seed, B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40(Database issue), D1144–9. 10.1093/nar/gkr1013 (2012). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


