Abstract
We have prospectively analyzed the DNA fingerprints of Mycobacterium tuberculosis strains from a random sample of patients with newly diagnosed tuberculosis in Windhoek, Namibia. Strains from 263 smear-positive patients in whom tuberculosis was diagnosed during 1 year were evaluated, and the results were correlated with selected epidemiological and clinical data. A total of 163 different IS6110 fingerprint patterns were observed among the 263 isolates. Isolates from a high percentage of patients (47%) were found in 29 separate clusters, with a cluster defined as isolates with 100% matching patterns. The largest cluster included isolates from 39 patients. One predominant strain of M. tuberculosis caused 15% of cases of smear-positive pulmonary tuberculosis in Windhoek. That strain was also prevalent in the north of the country, suggesting that in contrast to other African countries with isolates with high levels of diversity in their DNA fingerprint patterns, only a restricted number of different strains significantly contribute to the tuberculosis problem in Namibia.
Control of tuberculosis relies on well-defined tools such as case finding, contact tracing, completion of successful treatment, and vaccination with Mycobacterium bovis BCG (6, 20). The impact of control measures on the course of the tuberculosis epidemic in high-incidence countries is often difficult to assess.
An alternative approach to describe the transmission dynamics of tuberculosis has been based on identification of chains of infection by molecular typing of the organism. This method has been very successful in outbreak investigations (12, 14, 17, 22, 28), and recently, DNA fingerprinting of Mycobacterium tuberculosis has been applied to analyze the spread of tuberculosis within circumscribed populations (3, 7, 21, 26).
We applied this approach using DNA fingerprinting by mixed-linker PCR (9) during a prospective study of patients with smear-positive tuberculosis in Windhoek, Namibia. The incidence of tuberculosis in Namibia is about 400/100,000 population, with 38% smear-positive cases, corresponding to an estimated annual risk of infection of 4% (13). The aim of this study was to determine the number of new cases due to recent infection and to identify hot spots of transmission, thus analyzing the transmission dynamics of the current tuberculosis epidemic in Namibia.
MATERIALS AND METHODS
Study population.
The study was performed in Windhoek, the capital of Namibia. Namibia has an estimated population of 1.41 million. Of these, 82% live in rural areas and 12% live in Windhoek. All cases of tuberculosis in the Windhoek region are referred for treatment and registration to the Tuberculosis Unit of the Windhoek Central Hospital.
All patients with newly diagnosed smear-positive tuberculosis that were registered between 1 April 1995 and 31 March 1996 were included in a prospective study. To exclude chronic carriers and treatment failures, only patients with no prior history of tuberculosis treatment within the preceding 6 months were admitted to the study.
Patient data concerning sex, age, smear result, previous history of tuberculosis, human immunodeficiency virus (HIV) infection status, and hometown were collected at admission by using standard questionnaires.
Mycobacterium strains.
Culture isolation was carried out at the Windhoek Tuberculosis Reference laboratory. The sputum was confirmed to be smear positive by direct microscopy, decontaminated by the NaOH method, and cultured for 4 to 8 weeks on Löwenstein-Jensen slants (with and without glycerol) by standard protocols (11). Samples of the colonies were placed into tubes containing 500 μl of DNA-free, sterile water, heat killed for 45 min at 80°C in a water bath, and shipped to the Molecular Genetic Laboratory at Heidelberg University. Cross-contamination between samples was controlled for by inclusion of mock samples at the steps of decontamination and cell sampling.
Cell membranes were disintegrated by freezing-thawing the specimen three times in an 80°C water bath and a −60°C methanol-dry ice bath. Subsequently, 4 μl of the lysate was subjected to mixed-linker PCR fingerprinting as described previously (9). In brief, mycobacterial DNA was digested with the restriction enzyme HhaI (Gibco Life Technologies, Gibco-BRL, Eggenstein, Germany), and the resulting fragments were ligated (T4 ligase; Gibco Life Technologies, Gibco-BRL) to the mixed linker, which consists of a double-stranded 20-bp oligonucleotide, with one strain containing uracil instead of thymidine. For correct ligation the mixed linker has a GC overhang at the 3′ end complementary to that at the HhaI-cutting site. In the next step, uracil is removed by adding uracil-N-glycosylase (Gibco Life Technologies, Gibco BRL). Thus, in a first PCR only fragments containing the 3′ end of IS6110 are amplified with a primer compatible with a 20-bp fragment at the 3′ end of IS6110 and a primer complementary to the uracil-containing oligonucleotide strand. Finally, a seminested PCR with a second primer specific for IS6110 is performed to increase the specificity of the amplification. The mixed-linker PCR products were separated by gel electrophoresis on a 8% polyacrylamide gel together with three external size markers (100-bp DNA ladder; Gibco Life Technologies, Gibco-BRL). The resulting DNA fingerprints were visualized under UV light by staining with 0.5 mg of ethidium bromide per ml and were photographed with black-and-white Polaroid film (Polaroid Corp., St. Albans, United Kingdom).
rRNA gene 16S PCR and IS6110 PCR.
Identification of the species of the isolates that were negative by mixed-linker PCR was carried out as published elsewhere (8). The presence of the IS6110 insertion sequence was demonstrated by using a highly sensitive diagnostic PCR assay based on the amplification of a 123-bp target sequence within IS6110, as described previously (19).
Spoligotyping.
Spoligotyping was performed as previously described by Kamerbeek et al. (10). Briefly, 10 ng of mycobacterial DNA was amplified with 5′ biotinylated oligonucleotides designated DRa and DRb. PCR was performed with PCR buffer containing 50 mM KCl, 10 mM Tris-HCl, (pH 8.3), each deoxynucleoside triphosphate at a concentration of 200 mM, 20 pmol of each primer, and 0.5 μl of AmpliTaq DNA polymerase (Perkin-Elmer, Weiterstadt, Germany). A total of 20 μl of the amplification product was hybridized with 43 oligonucleotides covalently bound to a membrane (Isogen Bioscience B. V., Maarssen, The Netherlands). Each of these oligonucleotides represented a known spacer sequence. Detection of the hybridized PCR product was performed with a streptavidin-horseradish peroxidase-enhanced chemiluminescence system according to the recommendation of the manufacturer (Amersham Buchler GmbH & Co. KG, Braunschweig, Germany). Analysis of spoligotypes was done by eye.
Computer-assisted analysis of DNA fingerprints.
Polaroid pictures of the gels were digitized with an HPIIc scanner (Hewlett-Packard, Hopkins, Minn.). Conversion of the digitized gels, normalization, and cluster analysis were carried out with GelCompar software (Applied Maths BVBA, Kortrijk, Belgium). The similarities of the strains were calculated by the unweighted pair group method with arithmethic averages with the Dice coefficient and 0.7% position tolerance. Clusters were defined as isolates with 100% identical patterns. Cluster frequencies were calculated as [(number of isolates in clusters − number of index patients)/number of patients] (18).
RESULTS
Between April 1995 and March 1996, 378 of 577 patients diagnosed with smear-positive pulmonary tuberculosis fulfilled the inclusion criteria of the study. Of the patients included in the study, 99 were culture negative, and no DNA fingerprint pattern could be produced for isolates from 16 patients. Among these 16 isolates, sequencing of the 16S rRNA gene confirmed that 14 isolates were M. tuberculosis and that 1 isolate was Mycobacterium kansasii. One sample was negative by both PCR methods, probably due to a sampling error or inhibition of the amplification reaction. The presence of IS6110 was shown for the 14 M. tuberculosis isolates by using a diagnostic PCR assay based on the detection of this insertion sequence, excluding the possibility of an IS6110-negative cluster. Thus, the DNA fingerprinting results and data could be analyzed for isolates from 263 (70%) patients. Comparison of the available data for culture-negative and culture-positive patients in the study group showed no significant difference (data not shown). The distribution of female-to-male patients was 1:2.6, with a mean age at the time of diagnosis of 36.0 years. Of the analyzed patients, 82% lived in Windhoek; however, only 27% were born there. HIV infection status data were available for 212 (81%) patients, and the rate of 34% HIV-M. tuberculosis coinfection was in good agreement with the results of other studies (15a). A prior history of treatment for tuberculosis was reported for 103 (40%) patients.
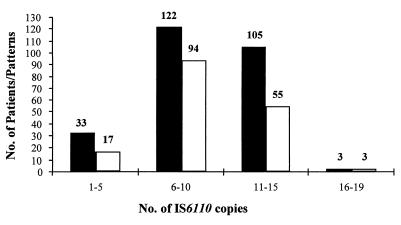
The DNA fingerprints of the 263 patient isolates were initially separated according to the number of IS6110 insertion elements. No isolates without IS6110 were identified, and a majority (87.5%) of the strains exhibited more than five IS6110 copies. The number of isolates per-pattern ratio was highest for isolates with 1 to 5 IS6110 copies and those with 11 to 15 IS6110 copies (1.94 and 1.90, respectively), corresponding to a low variability of patterns, especially among low- and high-copy-number strains (Fig. 1).
FIG. 1.
Distribution of isolates (black columns) and patterns (white columns) according to number of IS6110 insertion elements. Numbers over the columns indicate numbers of isolates. Number of isolates per pattern ratio = number of isolates/number of patterns.
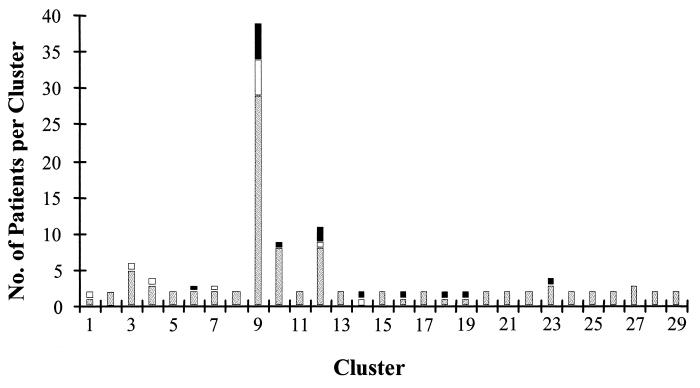
Pattern analysis revealed 123 isolates (47%) in 29 clusters with identical DNA fingerprints, corresponding to a cluster frequency of 36%. The sizes of clusters varied between 2 and 39 isolates, with nine clusters containing more than 2 isolates (Table 1). Comparison of isolates from patients with known HIV status demonstrated a significant predominance of isolates from HIV-positive patients in clusters with small (fewer than three) numbers of isolates (15 of 29 versus 18 of 67 isolates, respectively; P = 0.02). HIV infection status data were available for 78% of patients whose isolates were in clusters, and the number of isolates from patients whose HIV infection status was known did not differ significantly between clusters with 2 isolates and clusters with larger numbers of isolates. No difference between small and large clusters was found for sex, age, and prior treatment for tuberculosis.
TABLE 1.
Band frequency and size of 29 clusters found in the study population
| No. of clusters | No. of isolates per cluster | No. of IS6110 bands |
|---|---|---|
| 20 | 2 | 3–14 |
| 3 | 3 | 2–7 |
| 2 | 4 | 4–7 |
| 1 | 7 | 5 |
| 1 | 9 | 12 |
| 1 | 11 | 10 |
| 1 | 39 | 11 |
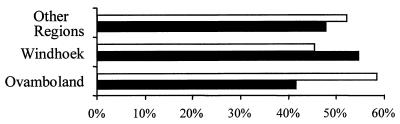
Of the patients living in Windhoek, 45% were infected with a strain in a cluster and 55% were infected with individual isolates (isolates not in a cluster). The proportion of clustered strains among isolates from patients from other areas did not differ significantly (Fig. 2).
FIG. 2.
Percentage of patients infected with strains in clusters (white columns) and patients infected with strains showing individual patterns (black columns) in three different regions of Namibia.
Isolates in 16 of the 29 clusters were most likely from local outbreaks, as the affected patients lived in the Windhoek area. These clusters included isolates from 33 patients, corresponding to 27% of patients whose isolates were in clusters. The remaining clustered strains were not restricted to local outbreaks. All clusters with isolates from more than three patients contained at least one isolate from a patient who lived outside of Windhoek (Fig. 3).
FIG. 3.
Hometowns of 123 patients infected with clustered isolates. Hatched columns represent patients from Windhoek, black columns represent patients from Ovamboland, and white columns represent patients from other regions of Namibia.
No difference in age and sex distribution, history of prior treatment for tuberculosis, and HIV-M. tuberculosis coinfection was observed between clustered patients and patients infected with individual strains. These data and demographic parameters are summarized in Table 2.
TABLE 2.
Demographic and clinical data for patients infected with unique isolates and clustered strains with identical mixed-linker PCR DNA fingerprints
| Characteristic | All isolates (n = 263) | Isolates with unique patterns (n = 140) | Isolates in clusters (n = 123) |
|---|---|---|---|
| Sex (no. of men/no. of women [%]) | 189/74 (70/30) | 98/42 (74/26) | 91/32 (72/28) |
| Age (yr) | 35.9 | 36.39 | 35.45 |
| No. (%) of patients with previous treatment for tuberculosis | 103/262 (40) | 58/139 (42) | 45/123 (37) |
| No. (%) of HIV-seropositive patients | 73/212 (34) | 40/116 (34) | 33/96 (24) |
| Current hometown (no. [%] of patients) | |||
| Windhoek | 214 (82) | 117 (84.8) | 97 (78.9) |
| Ovamboland | 24 (9.2) | 10 (7.2) | 14 (11.4) |
| Other regions | 23 (8.8) | 11 (8.0) | 12 (9.7) |
| Place of birth (no. [%] of patients) | |||
| Windhoek | 50 (26.5) | 25 (26.3) | 25 (26.6) |
| Ovamboland | 76 (40.2) | 33 (34.7) | 43 (45.7) |
| Other regions | 63 (33.3) | 37 (39.0) | 26 (27.7) |
| No. (%) of patient isolates with the following no. of bands per pattern | |||
| 1–5 | 33 (12.5) | 9 (6.4) | 24 (19.5) |
| >5 | 230 (87.5) | 131 (93.6) | 99 (80.5) |
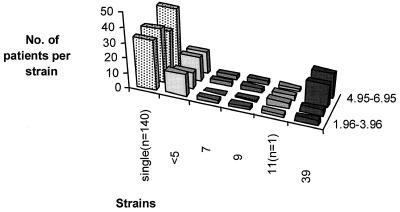
The strain that formed the largest cluster comprised isolates from 39 patients with 100% identical DNA fingerprint patterns by mixed-linker PCR. Inclusion of 26 strains with similar patterns that differed by one IS6110 fragment increased the size of this main cluster so that it comprised 24% of all isolates from study patients.
Secondary typing revealed a clonal spoligotype for 58 of these 65 isolates. Two isolates in the main cluster could not be analyzed by spoligotyping due to a lack of DNA. All but 3 of the remaining 37 isolates in the main cluster were clonal by both typing methods. Remarkably, these three isolates exhibited identical spoligotypes. A total of 92% (24 of 26) of the isolates with one band difference had the same spoligotype as that for the isolates in the main cluster.
Comparison of patients whose isolates were in the main cluster with patients who harbored individual strains did not reveal any significant differences with respect to age, sex, prior tuberculosis treatment, HIV infection status, or demographic data. Of the patients whose isolates were in the main cluster, 82% of patients received a diagnosis of tuberculosis within the first 6 months of the study period, whereas the intake of all patients in that time period corresponded to 53% of the study patients (P < 0.001) (Fig. 4). The appearance of this large cluster was not due to cross-contamination, as all but nine isolates were cultured on 28 different days. Together with five of these nine isolates, other samples with isolates with different DNA fingerprints were decontaminated on the same date. In addition, negative controls included with the samples showed no growth after 8 weeks. The equal distribution of positive cultures for all patients during the study period further excludes a bias due to changing sensitivity in the culture isolation methods applied in the study.
FIG. 4.
Time distribution of clustered strains versus single isolates. The number of isolates is depicted by quarter of study intake (indicated on the right as month.year-month.year); for simplification, the distribution of clustered strains isolated from less than five patients have been combined.
DISCUSSION
In a prospective study of patients with smear-positive tuberculosis in Windhoek, demographic data were available and isolates could be obtained by culture for 279 (74%) of 378 patients. The finding of smear-positive samples without growth on culture media was probably due to the early start of antituberculosis treatment before obtaining samples for microbiologic analysis. More than 94% of the culture-positive isolates produced a clear DNA fingerprint pattern by mixed-linker PCR. This loss of typeable isolates is comparable to the results obtained by standard restriction fragment length polymorphism analysis with IS6110 (3, 18). The demographic data obtained for our study population including age and sex distribution and HIV-M. tuberculosis coinfection were compatible with data available from the Ministry of Health and Social Services and were in accordance with data from published studies conducted in other countries in the South African region (13, 25).
DNA fingerprint analysis of the cultured isolates showed a cluster frequency of 36%. In comparison, cluster frequencies obtained for isolates from other developing countries ranged between 20 and 30% (24, 27). In contrast to other studies, the cluster analysis in this study was based on a 100% identity of patterns. Thus, the specificity of the analysis was increased, while the sensitivity of our results was sacrificed. As the number of isolates analyzed corresponded to about 46% of all isolates from patients with smear-positive tuberculosis in the study period (including patients with reinfections and chronic infections) the cluster frequency determined in the present study reflects the minimal degree of ongoing spread of tuberculosis.
Typing of the isolates showed less than six IS6110 fragments in 10% of strains or 13% of patient isolates, respectively. In a study by Warren et al. (24) from South Africa, 5% of strains were low-copy-number strains. Studies from other geographic regions revealed that up to 40% of strains had one to five copies of IS6110 (16, 27). The number of isolates-per-pattern ratio showed that the variability of patterns was lowest in this group. However, the small number of isolates in this group did not significantly influence the overall cluster frequency.
A total of 29 clusters with 100% identical DNA fingerprint patterns were identified, with cluster sizes ranging between 2 and 39 isolates. Among these clusters, 9 (31%) included more than 2 isolates (Table 1). Remarkably, the percentage of isolates from HIV-positive patients in small clusters with two isolates was significantly higher than the percentage in clusters with larger numbers of isolates (P = 0.02). However, for patients, infection with a clustered strain was independent of HIV infection status (P = 1.0). In contrast, in industrialized countries HIV-positive patients have predominantly been found to be involved in large outbreaks (1, 4, 5, 18). This difference might be explained by incomplete sampling or the poor prognosis for patients with HIV-M. tuberculosis coinfection in developing countries. In addition, patients were not differentiated according to the stage of HIV disease in this study.
All clusters with more than three isolates included isolates from patients living in different areas of Namibia, even though 82% of all patients reported Windhoek as their hometown (P < 0.001). This observation together with small regional variations in cluster frequencies suggests a lack of hot spots of tuberculosis transmission. This might be due to the high prevalence of tuberculosis. Also, the important function of Windhoek as an economic and administrative center leads to the periodic presence of large numbers of Namibian citizens and might be contributing to the spread of tuberculosis.
A striking result was the detection of an unsuspected outbreak involving 15% of the 263 patients in the study. The isolates in this cluster exhibited 100% identical patterns consisting of 11 IS6110 fragments, thus providing a characteristic and specific DNA fingerprint (23). Patients affected by this outbreak did not differ significantly in their demographic characteristics from patients whose isolates were in other clusters or patients who harbored individual strains. The finding of 25 strains with highly related DNA fingerprint patterns raises the possibility that this outbreak resulted from a strain of tuberculosis endemic to Namibia, as has been described in the context of a low-prevalence country (2). This is further supported by the finding of isolates with identical DNA fingerprints in a rural health district in the north of the country (data not shown). The results of secondary typing by spoligotyping analysis further prove that we have identified the ongoing transmission of a strain endemic to Namibia. This hypothesis of the spread of an endemic strain could be tested by the development of a rapid test for the specific detection of this strain, similar to the approach taken for the W strain in New York City (15). Rapid recognition of patients infected with this highly transmissible strain might represent an important step in breaking the chain of transmission of smear-positive tuberculosis.
The number of patients involved in the outbreak peaked within the first 6 months without increasing the number of smear-positive patients diagnosed with tuberculosis. Thus, the outbreak was missed by classical epidemiological tools such as case detection rate. Similar results have been reported for studies of tuberculosis transmission in low-prevalence countries, even including thorough epidemiological follow-up and contact tracing (18, 21). However, the dynamic of the outbreak with diagnosis of tuberculosis in more than 80% of patients within the first half of the study period is remarkable.
In conclusion, this molecular study of smear-positive tuberculosis in Windhoek, Namibia, confirmed the significant contribution of new infections to the burden of tuberculosis in a high-prevalence country, despite an estimated BCG vaccine coverage rate of more than 80%. In addition to the detection of large, unsuspected outbreaks, the results disproved the hot-spot hypothesis of transmission for Namibia that had been derived from the situation in low-prevalence countries. We suggest that determination of the cluster frequency might serve as a useful addition to other more sophisticated ways of estimating the risk of infection, thus providing an independent tool for the identification of changes in the efficacy of tuberculosis control. The equal distribution of cluster frequencies for different areas analyzed in this study will allow the design of follow-up studies at the community level, including classical epidemiological investigations, to test this hypothesis.
ACKNOWLEDGMENTS
This study was supported by research grant 01 KA 9301 from the Bundesministerium für Bildung, Wissenschaft und Technologie and grant HA 1921/3-2,3 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Beck-Sagué C, Dooley S W, Hutton M D, Otten J, Breeden A, Crawford J T, Pitchenik A E, Woodley C, Cauthen G, Jarvis W R. Hospital outbreak of multidrug-resistant Mycobacterium tuberculosis infections. JAMA. 1992;268:1280–1286. doi: 10.1001/jama.1992.03490100078031. [DOI] [PubMed] [Google Scholar]
- 2.Braden C R, Templeton G L, Cave M D, Valway S, Onorato I M, Castro K G, Moers D, Yang Z, Stead W W, Bates J H. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis. 1997;175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 3.Bradford W Z, Koehler J, El-Hajj H, Hopewell P C, Reingold A L, Agasino C B, Cave M D, Rane S, Yang Z, Crane C M, Small P M. Dissemination of Mycobacterium tuberculosis across the San Francisco Bay Area. J Infect Dis. 1998;177:1104–1107. doi: 10.1086/517405. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Nosocomial transmission of multidrug-resistant tuberculosis among HIV-infected persons—Florida and New York, 1988–1991. Morbid Mortal Weekly Rep. 1991;40:585–591. [PubMed] [Google Scholar]
- 5.Daley C L, Small P M, Schecter G F, Schoolnik G K, McAdam R A, Jacobs W R, Jr, Hopewell P C. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N Engl J Med. 1992;326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 6.Enarson D A. The International Union Against Tuberculosis and Lung Disease model National Tuberculosis Programmes. Tubercle Lung Dis. 1995;76:95–99. doi: 10.1016/0962-8479(95)90548-0. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez M C, Vincent V, Aubert D, Bizet J, Gaillot O, Lebrun L, Le-Pendeven C, Le-Pennec M P, Mathieu D, Offredo C, Pangon B, Pierre A C. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J Clin Microbiol. 1998;36:486–492. doi: 10.1128/jcm.36.2.486-492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas W H, Butler W R, Kirschner P, Plikaytis B B, Coyle M B, Amthor B, Steigerwalt A G, Brenner D J, Salfinger M, Crawford J T, Böttger E C, Bremer H J. A new agent of mycobacterial lymphadenitis in children: Mycobacterium heidelbergense sp. nov. J Clin Microbiol. 1997;35:3203–3209. doi: 10.1128/jcm.35.12.3203-3209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas W H, Butler W R, Woodley C L, Crawford J T. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J Clin Microbiol. 1993;31:1293–1298. doi: 10.1128/jcm.31.5.1293-1298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent P T, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. 1985. Atlanta, Ga: Public Health Service, U.S. Department of Health and Human Services; 1985. [Google Scholar]
- 12.Kline S E, Hedemark L L, Davies S F. Outbreak of tuberculosis among regular patrons of a neighborhood bar. N Engl J Med. 1995;333:222–227. doi: 10.1056/NEJM199507273330404. [DOI] [PubMed] [Google Scholar]
- 13.Ministry of Health and Social Services. National Tuberculosis Control Programme (NTCP): guidelines for tuberculosis control. Ministry of Health; 1995. and Social Services, Republic of Namibia. [Google Scholar]
- 14.Pearson M L, Jereb J A, Frieden T R, Crawford J T, Davis B J, Dooley S W, Jarvis W R. Nosocomial transmission of multidrug-resistant Mycobacterium tuberculosis. Ann Intern Med. 1992;117:191–196. doi: 10.7326/0003-4819-117-3-191. [DOI] [PubMed] [Google Scholar]
- 15.Plikaytis B B, Marden J L, Crawford J T, Woodley C L, Butler W R, Shinnick T M. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:1542–1546. doi: 10.1128/jcm.32.6.1542-1546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Rabbow, M. Personal communication.
- 16.Sahadevan R, Narayanan S, Paramasivan C N, Prabhakar R, Narayanan P R. Restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, India, by use of direct-repeat probe. J Clin Microbiol. 1995;33:3037–3039. doi: 10.1128/jcm.33.11.3037-3039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sepkowitz K A, Friedman C R, Hafner A, Kwok D, Manoach S, Floris M, Martinez D, Sathianathan K, Brown E, Berger J J, et al. Tuberculosis among urban health care workers: a study using restriction fragment length polymorphism typing. Clin Infect Dis. 1995;21:1098–1101. doi: 10.1093/clinids/21.5.1098. [DOI] [PubMed] [Google Scholar]
- 18.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 19.Smith K C, Starke J R, Eisenach K D, Ong L T, Denby M. Detection of Mycobacterium tuberculosis in clinical specimens from children using a polymerase chain reaction. Pediatrics. 1996;97:155–160. [PubMed] [Google Scholar]
- 20.Styblo K. Impact of present control measures on the overall tuberculosis situation. In: Broekmans J F, editor. Epidemiology of tuberculosis. The Hague, The Netherlands: Royal Netherlands Tuberculosis Association; 1991. pp. 81–89. [Google Scholar]
- 21.Torrea G, Offredo C, Simonet M, Gicquel B, Berche P, Pierre A C. Evaluation of tuberculosis transmission in a community by 1 year of systematic typing of Mycobacterium tuberculosis clinical isolates. J Clin Microbiol. 1996;34:1043–1049. doi: 10.1128/jcm.34.5.1043-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valway S E, Sanchez M P, Shinnick T F, Orme I, Agerton T, Hoy D, Jones J S, Westmoreland H, Onorato I M. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- 23.van Soolingen D, de Haas P E, Hermans P W, Groenen P M, van Embden J D. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren R, Hauman J, Beyers N, Richardson M, Schaaf H S, Donald P, van Helden P. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S Afr Med J. 1996;86:45–49. [PubMed] [Google Scholar]
- 25.Weyer K, Groenewald P, Zwarenstein M, Lombard C J. Tuberculosis drug resistance in the Western Cape. S Afr Med J. 1995;85:499–504. [PubMed] [Google Scholar]
- 26.Yang Z, Barnes P F, Chaves F, Eisenach K D, Weis S E, Bates J H, Cave M D. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z H, Mtoni I, Chonde M, Mwasekaga M, Fuursted K, Askgard D S, Bennedsen J, de Haas P E W, van Soolingen D, van Embden J D A, Andersen Å B. DNA fingerprinting and phenotyping of Mycobacterium tuberculosis isolates from human immunodeficiency virus (HIV)-seropositive and HIV-seronegative patients in Tanzania. J Clin Microbiol. 1995;33:1064–1069. doi: 10.1128/jcm.33.5.1064-1069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaza S, Blumberg H M, Beck-Sagué C, Haas W H, Woodley C L, Pineda M, Parrish C, Crawford J T, McGowan J E, Jarvis W R., Jr Nosocomial transmission of Mycobacterium tuberculosis: role of health care workers in outbreak propagation. J Infect Dis. 1995;172:1542–1549. doi: 10.1093/infdis/172.6.1542. [DOI] [PubMed] [Google Scholar]