Abstract
Ribosome binding to eukaryotic mRNA is a multistep process which is mediated by the cap structure [m7G(5′)ppp(5′)N, where N is any nucleotide] present at the 5′ termini of all cellular (with the exception of organellar) mRNAs. The heterotrimeric complex, eukaryotic initiation factor 4F (eIF4F), interacts directly with the cap structure via the eIF4E subunit and functions to assemble a ribosomal initiation complex on the mRNA. In mammalian cells, eIF4E activity is regulated in part by three related translational repressors (4E-BPs), which bind to eIF4E directly and preclude the assembly of eIF4F. No structural counterpart to 4E-BPs exists in the budding yeast, Saccharomyces cerevisiae. However, a functional homolog (named p20) has been described which blocks cap-dependent translation by a mechanism analogous to that of 4E-BPs. We report here on the characterization of a novel yeast eIF4E-associated protein (Eap1p) which can also regulate translation through binding to eIF4E. Eap1p shares limited homology to p20 in a region which contains the canonical eIF4E-binding motif. Deletion of this domain or point mutation abolishes the interaction of Eap1p with eIF4E. Eap1p competes with eIF4G (the large subunit of the cap-binding complex, eIF4F) and p20 for binding to eIF4E in vivo and inhibits cap-dependent translation in vitro. Targeted disruption of the EAP1 gene results in a temperature-sensitive phenotype and also confers partial resistance to growth inhibition by rapamycin. These data indicate that Eap1p plays a role in cell growth and implicates this protein in the TOR signaling cascade of S. cerevisiae.
Translation initiation in eukaryotes requires several polypeptide initiation factors which serve to direct the sequential assembly and positioning of the ribosome at the AUG initiation codon on the mRNA (for a review, see reference 59). Most eukaryotic mRNAs are thought to be translated in a cap-dependent manner whereby the heterotrimeric complex, eukaryotic initiation factor 4F (eIF4F), interacts directly with the cap structure. eIF4F is composed of eIF4E (the cap-binding subunit), eIF4A (an RNA helicase), and eIF4G, which serves as a scaffold protein (59). It is thought that eIF4F acts along with free eIF4A and eIF4B to unwind local secondary structure at the 5′ terminus of the mRNA to facilitate ribosome binding (31, 45, 59, 67, 71).
eIF4E is critical for cap-dependent translation and is a key target for regulatory pathways which control protein synthesis rates (36, 71, 72). Mechanisms by which eIF4E activity is modulated in the mammalian cell include transcriptional regulation of the eIF4E gene, alteration in the phosphorylation status of eIF4E, and the interaction of eIF4E with a family of polypeptides known as 4E-BPs (eIF4E-binding proteins) (reviewed in references 36 and 72). To date, three members of the mammalian 4E-BP family have been described (56, 62, 63), all of which share a small amino acid motif with eIF4G (YXXXXLΦ, where Φ denotes a hydrophobic residue, usually L, M, or F) that interacts with eIF4E (58). Binding of 4E-BPs to eIF4E precludes eIF4E interaction with eIF4G, thereby blocking assembly of the eIF4F complex and repressing cap-dependent translation (39). This repression can be alleviated through phosphorylation of 4E-BP, which decreases its affinity for eIF4E (32, 56, 62).
The finding that 4E-BPs are phosphorylated in response to a variety of growth factors and hormones links the control of cap-dependent translation to extracellular signaling pathways involved in major biological processes such as cell growth and proliferation, differentiation, and development (reviewed in references 36, 55, and 71). In mammalian cells, phosphorylation of 4E-BP1 is modulated, in part, by the mammalian target of rapamycin/FK506-binding protein (FKBP)-rapamycin-associated protein (mTOR/FRAP) (14, 19, 20, 35, 42). The TOR signaling pathway in the budding yeast Saccharomyces cerevisiae shares common features with the mTOR/FRAP cascade of higher cells. Two TOR genes (TOR1 and TOR2) in S. cerevisiae encode structurally and functionally similar phosphatidylinositol kinase homologs (43, 44, 52, 84). The FKBP-rapamycin complex binds to the yeast TOR proteins and inhibits their shared function, inducing growth arrest in early G1 and a severe reduction in protein synthesis (43, 44, 52, 84). The loss of TOR function in yeast causes an early and dramatic inhibition of translation initiation (11), and several lines of evidence indicate that the G1 cell cycle arrest is a consequence of this translational defect (11, 27). These data suggest that the TOR signaling pathway controlling cell growth is similar in yeast and higher eukaryotes and involves the modulation of translation initiation downstream of the TOR proteins (reviewed in references 25 and 77).
The yeast eIF4F complex is composed of the cap-binding subunit, eIF4E (encoded by CDC33) (17), eIF4G (TIF4631 and TIF4632, encoding two similar proteins termed eIF4G1 and eIF4G2, respectively) (37, 76), and eIF4A, which is bound weakly to eIF4G (60; M. Altmann and H. Trachsel, personal communication) and therefore dissociates from the complex during purification. Both eIF4G1 and eIF4G2 contain the canonical 4E-binding motif (58). No structural homologs of the 4E-BPs exist in yeast. However, a small polypeptide, p20 (encoded by CAF20) (4, 54), has been identified which contains a consensus eIF4E-binding domain, competes directly with eIF4G1 (6) for binding to a partially shared site on eIF4E (66), and specifically inhibits cap-dependent translation in cell extracts (6). In addition, p20 is a phosphoprotein (82) and acts as a general negative regulator of translation in vivo (24), suggesting that it may constitute an ortholog of 4E-BPs in yeast. In the present work, we characterize a novel eIF4E-associated protein (termed Eap1p) which also blocks cap-dependent translation via competition with eIF4G. Disruption of the EAP1 gene results in a temperature-sensitive phenotype and confers partial resistance to the growth-inhibitory properties of rapamycin, implicating Eap1p in the TOR signaling pathway controlling cap-dependent translation in S. cerevisiae.
MATERIALS AND METHODS
Yeast strains, genetic methods, and plasmids.
The S. cerevisiae strains used are listed in Table 1. Standard procedures for yeast culture, mating, sporulation, and tetrad analysis were used (50). Yeast transformation was performed by the lithium acetate method (33). The compositions of rich medium (YPD) and synthetic glucose medium (SD) complemented with the appropriate nutrients for plasmid maintenance were as described elsewhere (38). Rapamycin (provided by Sandoz Pharma, Basel, Switzerland) was diluted into medium from a stock solution of 1 mg/ml in 90% ethanol–10% Tween 20.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Description | Source and/or reference |
|---|---|---|
| YCG323 | leu2-3,112 ura3-52 his4-917δ | Haploid progeny of YCG312 (37) |
| YCG324 | YCG323 tif4631::LEU2 | Haploid progeny of YCG312 (37) |
| YCG325 | YCG323 tif4632::URA3 | Haploid progeny of YCG312 (37) |
| YMA-4B | Wild type | 6 |
| YMA-2A | caf20::URA3 | 6 |
| JK9-3da/α | MATa/MATα leu2-3,112 ura3-52 trp1 his4 rme1 HMLa (isogenic) | |
| JK9-3da | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa | |
| JK9-3dα | MATα leu2-3,112 ura3-52 trp1 his4 rme1 HMLa | |
| JH6-1C | JK9-3da TRP1+ | 43 |
| JH11-1C | JK9-3da TOR1-1 | 44 |
| SH12-1A | JK9-3dα caf20::URA3 | This study |
| YGC034 | JK9-3da eap1::TRP1 | This study |
| YGC047 | JK9-3da eap1::TRP1 caf20::URA3 | This study |
| AS93-2A | JK9-3da tor1::LEU2 TRP1+ | This study |
| TS6-5A | JK9-3da tor1::LEU2 eap1::TRP1 | This study |
The bacterial expression vectors pAR(ΔRI)[59/60] and pGEX-HMK (16), carrying the heart muscle kinase (HMK) recognition motif fused to the Flag epitope and glutathione S-transferase (GST), respectively, were gifts of M. Blanar (University of California, San Diego). Vectors were linearized with EcoRI, and 5′-overhangs were filled in with the Klenow fragment of Escherichia coli DNA polymerase (New England Biolabs). The yeast eIF4E gene was PCR amplified from pVTrp-eIF4E (5) using primers which introduced EcoRV sites 3 nucleotides upstream and downstream of the eIF4E open reading frame (ORF). Following digestion with EcoRV, the amplified fragment was subcloned into EcoRV-cut plasmid Bluescript KS (Stratagene), and a clone was selected which placed the 5′ end of the ORF proximal to the ClaI site on the KS polylinker. eIF4E was reisolated from the KS plasmid with ClaI/PstI, overhangs were filled in, and the fragment was ligated to each of the vectors described above to yield pAR(ΔRI)[59/60]-eIF4E or pGEX/HMK-eIF4E.
Full-length EAP1 cDNA was recovered from a λgt11 clone identified by the eIF4E interaction screen. The cDNA spanned genome coordinates 53690 to 55754 from S. cerevisiae chromosome XI. The EAP1 gene was isolated from λgt11 arms using EcoRI and was subcloned into an EcoRI-digested KS vector to yield KS-EAP1. The in-frame N-terminal deletion mutants, mut.(108-632) and mut.(164-632), were generated by first introducing an NcoI site at the initiating ATG codon of KS-EAP1 to yield KS-[NcoI]EAP1. Truncated fragments of EAP1 were PCR amplified using primers which contained NcoI sites at nucleotide positions 321 and 490, respectively. The NcoI/StyI fragment from KS-[NcoI]EAP1 was then released and replaced with similarly digested truncated fragments to generate KS-[NcoI]EAP1mut.(108-632) and KS-[NcoI]EAP1mut.(164-632). The 5′-flanking region of the EAP1 gene was PCR amplified from cosmid clone pEKG086 (a gift of B. Dujon, Institut Pasteur, Paris, France), using primers spanning genome coordinates 53428 to 54033 and which placed an EcoRI site at the 5′ end of the amplified fragment. This fragment was subcloned into KS-EAP1 using EcoRI/NdeI to generate KS-5′+EAP1. For expression of Eap1p in yeast, KS-5′+EAP1 was digested with EcoRI and the EAP1 gene was ligated to similarly linearized YEp352 plasmid to yield YEp352-5′+EAP1. The triple hemagglutinin epitope (HA) tag was PCR amplified from the pACTAG-2 vector using primers which placed PflMI sites at both ends of the amplified fragment. YEp352-5′+EAP1 was partially digested with PflMI, and the three-HA fragment was ligated in-frame to generate YEp352-5′+3xHA/EAP1. Tyrosine-109 was mutated to alanine by PCR and was subcloned into KS-EAP1, using unique NdeI/StyI sites. The mutated fragment was reisolated with PstI/BsiWI and subcloned into similarly digested YEp352-5′+3xHA/EAP1 to yield YEp352-5′+HA/EAP1 [Y109A]. Plasmid pTS115, expressing EAP1 under control of its own promoter, was constructed by subcloning the 2.9-kb EcoRI/HindIII fragment of PCR-amplified EAP1 into YCplac33 (CEN URA3) (34). To construct pTS117, an internal 400-bp PstI/SphI fragment of pTS115 was replaced with the corresponding fragment encoding the Y109A mutation from YEp352-5′ +3xHA/EAP1[Y109A]. EAP1 was subcloned into the baculovirus transfer vector pVL1392flagHMK (40) from KS-EAP1 at cohesive EcoRI sites.
The targeting vector for disruption of the EAP1 gene was generated by deletion of the 1.7-kb PflMI fragment from KS-5′+EAP1 followed by blunt-end ligation with the 0.8-kb BglII/EcoRI fragment of pJH-W1 (a gift of H. Bussey, McGill University) containing full-length TRP1. The TRP1-disrupted EAP1 gene was then isolated as a 1.2-kb EcoRI DNA fragment for transformation into diploid strain JK9-3da/α. The targeting vector for the CAF20 disruption was made by digestion of a genomic fragment containing CAF20 with BclI at the CAF20 start codon and insertion of a BglII URA3 cassette. Strain JK93d a/α was transformed with a 3.3-kb EcoRI fragment containing caf20::URA3.
Far-Western analysis and cloning of EAP1.
E. coli BL21(DE3)pLysS was transformed with pAR(ΔRI)[59/60]-eIF4E, grown in liquid culture, and induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG; Boehringer Mannheim) as described elsewhere (74). Cells were recovered by centrifugation, and the pellet was taken up in 2 volumes of 50 mM Tris-HCl (pH 8.0)–1 mM EDTA–100 mM NaCl–1 mM phenylmethylsulfonyl fluoride–1 mM dithiothreitol–10% glycerol. Cells were then disrupted by sonication, and eIF4E was purified by using Flag immunoaffinity resin (IBI) according to the manufacturer's instructions. Radiolabeling of the HMK domain was carried out essentially as previously described (16) except that 5 μg of protein was used in the labeling reaction.
Yeast cells were grown to exponential phase in either rich or synthetic selection medium, and whole-cell extracts were prepared in disruption buffer (20 mM Tris-HCl [pH 7.5]–100 mM KCl–1 mM EDTA–5% glycerol–1 mM dithiothreitol–1 mM phenylmethylsulfonyl fluoride) using the glass bead lysis method (10). Soluble proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted to nitrocellulose membranes, and probed with radiolabeled eIF4E according to the published protocol (16). Far-Western screening of a commercial S. cerevisiae λgt11 cDNA expression library (Clontech no. YL1008b) was conducted according to published protocols (10, 16).
Protein interactions.
Eap1p deletion mutants were transcribed in vitro with T3 RNA polymerase (Promega), using linearized KS-[NcoI]EAP1 as a template. Resulting mRNAs were translated in rabbit reticulocyte lysate in the presence of [35S]methionine (ICN) as instructed by the manufacturer (Promega). Wild-type Eap1p, mut.(1-124), and mut.(1-106) were generated by linearization of KS-EAP1 DNA with BamHI, BsiWI, and NdeI, respectively. N-terminal deletion mutants mut.(108-632) and mut.(164-632) were generated by linearization of the corresponding deletion in KS-[NcoI]EAP1 with BamHI. GST-HMK and the GST-HMK-eIF4E fusion protein were synthesized in E. coli and purified using glutathione-Sepharose beads as instructed by the manufacturer (Pharmacia Biotech). In vitro coprecipitation analyses were carried out as previously described (22).
In vivo coimmunoprecipitation analyses were conducted on yeast derived from YGC034 transformed with YEp352, YEp352-5′+3xHA/EAP1, or YEp352-5′+3xHA/EAP1[Y109A]. Whole-cell extracts were prepared as described above, and 100 μg of protein was incubated with 5 μl of the anti-HA antibody HA.11 (BAbCo) for 60 min at 4°C. Protein G-Sepharose beads (Pharmacia Biotech) were added for an additional 30 min. Following extensive washings with coimmunoprecipitation buffer (50 mM Tris-HCl [pH 7.5]–150 mM NaCl–1 mM EDTA–0.1% Nonidet P-40), Laemmli buffer was added; samples were resolved by SDS-PAGE and electroblotted to nitrocellulose membranes. HA-tagged Eap1p was decorated with anti-HA antibody, and yeast eIF4E was decorated with anti-eIF4E monoclonal antibody 9B12. Proteins were revealed by enhanced chemiluminescence (Amersham Corp.).
Coprecipitation using m7GDP-coupled agarose resin (30) was conducted on samples equivalent to those described above for coimmunoprecipitation analysis. Following incubation with yeast extract for 120 min at 4°C, the m7GDP-resin was washed extensively with disruption buffer supplemented with 0.1 mM ATP and 0.1 mM GTP. Bound proteins were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and probed by Western blotting with anti-eIF4E antibody 9B12 or by far-Western blotting with 32P-labeled eIF4E as described above.
In vitro translation.
Translation-grade yeast extract was prepared and cell-free translation was performed as previously described (3, 6, 7). Vectors pJII-2 (CAT [chloramphenicol acetyltransferase]) and pJII-102 (Ω CAT) (70) were generous gifts of M. Altmann (University of Bern). Capped CAT and Ω CAT mRNAs were transcribed from the BglII-linearized vectors using SP6 RNA polymerase in the presence of 50 μM GTP and 500 μM m7GpppG. Flag-Eap1p fusion protein was expressed in Spodoptera frugiperda (Sf9) insect cells using recombinant baculovirus generated with the transfer vector pVL1392flagHMK-EAP1 as described previously (40) and was purified from insect cell lysate using Flag immunoaffinity resin (IBI) according to the manufacturer's instructions.
RESULTS
eIF4E-interacting proteins in S. cerevisiae.
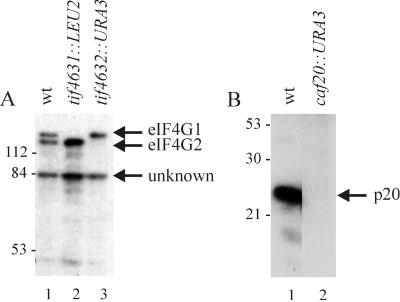
To identify novel proteins which interact with yeast eIF4E, a far-Western blotting assay was carried out using a crude S. cerevisiae extract. Total cellular proteins were resolved by PAGE and transferred to nitrocellulose membranes, where upon they were probed using 32P-labeled HMK-eIF4E. The eIF4E probe interacted efficiently with four yeast proteins in the wild-type extract (Fig. 1). The apparent molecular weights of three proteins correspond to the known eIF4E-interacting factors eIF4G1, eIF4G2, and p20, while a fourth polypeptide migrated at approximately 84 kDa. Denaturation and refolding of the immobilized proteins on the membrane using guanidine hydrochloride (79) did not alter the binding pattern (data not shown). To ascertain the identity of the eIF4E-binding proteins, additional far-Western analyses were carried out using extracts from haploid yeast strains with targeted gene disruptions for the eIF4G1, eIF4G2, and p20 genes. In each case, deletion of the gene resulted in the loss of the signal for the predicted band in the far-Western blot (Fig. 1). These data confirm that eIF4G1, eIF4G2, and p20 interact directly with yeast eIF4E and demonstrate the utility of the far-Western approach to identify the unknown 84-kDa eIF4E-associated protein.
FIG. 1.
Interaction of S. cerevisiae proteins with eIF4E. Yeast strains were grown to exponential phase and lysed by the glass bead method. Total protein (30 μg) from the clarified extracts was fractionated by SDS-PAGE and transferred to nitrocellulose membranes, which were then probed with 32P-labeled HMK-eIF4E. (A) SDS-PAGE (8% gel). Strains used: lane 1, YCG323 (wild type [wt]); 2, YCG324 (tif4631::LEU2); 3, YCG325 (tif4632::URA3). (B) SDS-PAGE (15% gel). Lane 1, YMA-4B (wild type); 2, YMA-2A (caf20::URA3). The deduced identity of each of the eIF4E-interacting proteins is indicated with an arrow on the right. Positions of molecular mass standards (in kilodaltons) are marked on the left.
Cloning of EAP1.
To identify the unknown 84-kDa eIF4E-interacting protein, a commercial S. cerevisiae λgt11 expression library (Clontech) was screened using 32P-labeled HMK-eIF4E as a probe. Screening of 2 × 106 plaques yielded 39 positive clones, 3 of which corresponded to eIF4G1 and 24 of which corresponded to eIF4G2. The remaining positive clones contained overlapping sequences corresponding to a chromosome XI hypothetical ORF YKL204w (GenBank accession number Z28204) (29; T. M. Pohl and F. M. Pohl, unpublished data), which we designated EAP1 (for eIF4E-associated protein 1). The predicted amino acid sequence of Eap1p (Fig. 2A) is 632 amino acids (aa) in length, with a calculated molecular mass of 69,762 Da. A search of the Eap1p sequence against the PROSITE database (Swiss Institute of Bioinformatics (SIB) [http://www.expasy.ch]) (9) revealed a putative bipartite nuclear targeting sequence (28) and a Walker A consensus motif (81) (Fig. 2A), indicating the potential for purine nucleotide binding. The Eap1p polypeptide also contains a proline-rich domain in its C terminus, comprised of three stretches of four or more consecutive proline residues (Fig. 2A).
FIG. 2.
(A) Predicted amino acid sequence of Eap1p in single-letter code. The p20 homology region is boxed, a potential bipartite nuclear localization sequence is in bold, a Walker A motif is underlined, and proline stretches in the C-terminal region are indicated by bullets above the sequence. (B) Limited sequence alignment between Eap1p and p20. Identical (black box) and conserved (shaded box) amino acids are highlighted. The alignment of critical residues common to mammalian 4E-BPs is shown below (x, any amino acid; Φ, hydrophobic residue).
A BLAST search (8) failed to detect significant homologies to any protein in all available databases (National Center for Biotechnology Information, National Library of Medicine [http://www.ncbi.nlm.nih.gov]), Swiss-Prot (SIB [see above]), and (Saccharomyces Genome Database [SGD], Stanford University [http://genome-www.stanford.edu/Saccharomyces]). However, upon visual inspection we noted that a sequence of 13 aa could be aligned with a highly similar sequence at the N terminus of p20 (Fig. 2). This short alignment is remarkable because it contains the 4E-binding motif (Fig. 2B), which is phylogenetically conserved from yeast to humans (36, 58, 63).
EAP1 gene disruption.
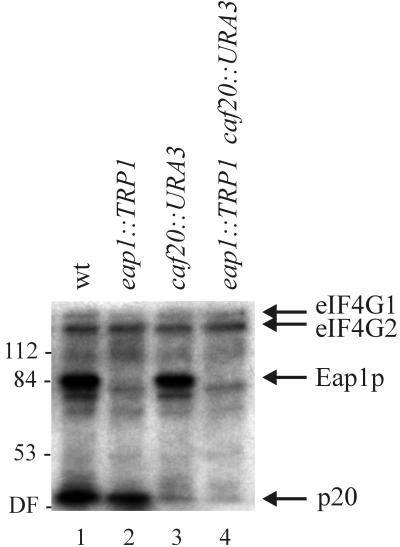
Disruption of one copy of EAP1 in the diploid strain JK9-3da/α was performed by substituting approximately 90% of the ORF (including the putative initiator codon) with TRP1. The appropriate integration of the targeting construct was confirmed by Southern analysis (data not shown). The targeted gene disruption resulted in four viable meiotic products upon sporulation (data not shown), demonstrating that EAP1 is not an essential gene. The deletion of Eap1p in extracts derived from the eap1::TRP1 haploid strain (YGC034) was verified by far-Western analysis (Fig. 3). As described above, the yeast eIF4E probe interacted with four proteins in wild-type yeast extract. In contrast, the signal corresponding to the 84-kDa protein was not observed in cells containing the EAP1 disruption (Fig. 3, compare lanes 1 and 2). These data confirm that EAP1 encodes a novel eIF4E-interacting protein.
FIG. 3.
EAP1 encodes a novel 84-kDa eIF4E-interacting protein. Yeast extracts were prepared and analyzed by far-Western blotting as described for Fig. 1 except that samples were resolved by SDS-PAGE on a 10% gel. Strains used: lane 1, JK9-3da (wild type [wt]); 2, YGC034 (eap1::TRP1); 3, SH12-1A (caf20::URA3); 4, YGC047 (eap1::TRP1 caf20::URA3). The eIF4E-interacting proteins are indicated by arrows on the right. Positions of molecular mass standards (in kilodaltons) are marked on the left. DF, dye front.
Disruption of EAP1 did not affect yeast growth at 30°C on rich or defined medium or on mating and subsequent meiosis (data not shown). An eap1 caf20 double deletion was generated by mating the eap1::TRP1 haploid strain with an isogenic strain carrying a caf20::URA3 disruption. Both proteins, Eap1p and p20, were absent in haploid progeny that were prototrophic for Trp and Ura (Fig. 3, lane 4). No synergistic effects on the growth of yeast containing the double gene disruption were observed in comparison to deletion of either Eap1p or p20 alone (data not shown).
Interaction with yeast eIF4E.
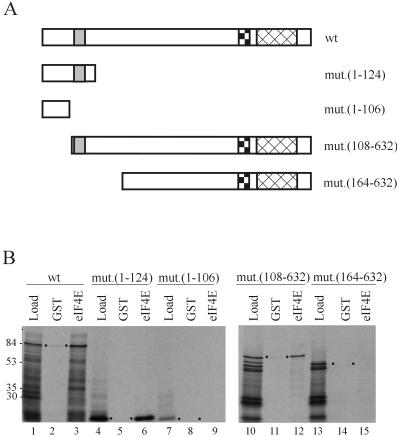
Based on the presence of the eIF4E-binding motif, we reasoned that the N-terminal region of Eap1p (containing aa 109 to 115 [Fig. 2]) would mediate the interaction with yeast eIF4E. To investigate this, the interaction between in vitro-translated Eap1p truncation mutants and GST-eIF4E was examined in a coprecipitation assay. Figure 4A shows the truncation mutants tested in this assay. As expected, full-length Eap1p coprecipitated with GST-eIF4E on glutathione-Sepharose resin, whereas GST bound only weakly to Eap1p (Fig. 4B, lanes 1 to 3). These data confirm the interaction detected for these proteins in the far-Western assay. Deletion of the C-terminal 508 aa of Eap1p (mut.1-124) had no effect on the interaction with GST-eIF4E, demonstrating that neither the Walker A motif nor the proline-rich C-terminal region was necessary for the interaction. However, elimination of an additional 18 aa from the C terminus (mut.1-106) resulted in the complete loss of eIF4E binding (Fig. 4B, lanes 4 to 6 and 7 to 9). Note that the amino acid residues deleted in the mut.1-106 encompass the p20 homologous sequence (Fig. 2). N-terminal deletions were also generated and tested in the coprecipitation assay. mut.(108-632) complexed efficiently with GST-eIF4E (Fig. 4B, lanes 10 to 12), whereas further deletion from the N terminus [mut.(164-632)] resulted in the complete loss of the interaction between the two proteins (Fig. 4B, lanes 13 to 15). These data demonstrate that the region of Eap1p encompassing the p20 homology domain and comprising the prototypic eIF4E-binding motif is necessary for the interaction with eIF4E.
FIG. 4.
Mapping of the eIF4E interaction domain of Eap1p in vitro. Full-length (wild-type [wt]) EAP1 and deletion mutants of EAP1 were translated in vitro and incubated with either purified GST or GST-eIF4E prior to the addition of glutathione-Sepharose beads. Following extensive washing, the bound material was eluted by boiling in Laemmli buffer and resolved by SDS-PAGE (10% gel). (A) Schematic diagram of deletion mutants used in the study. The p20 homology region (aa 109 to 121) is indicated as a shaded box, the Walker A motif is shown as a stippled box, and the proline-rich region is cross-hatched. (B) Coprecipitation analysis. Load, one-fifth of total radiolabeled Eap1p used in the coprecipitation; GST, Eap1p coprecipitated by GST alone; GST-eIF4E, Eap1p coprecipitated by GST-eIF4E fusion protein. Full-length translation products are indicated by dots. Sizes of standards are indicated in kilodaltons.
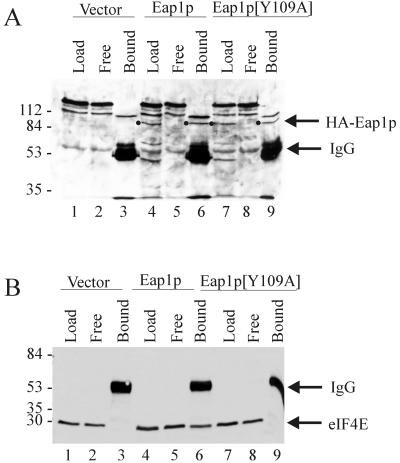
To demonstrate an interaction between Eap1p and yeast eIF4E in vivo, coimmunoprecipitations were performed on extracts from the eap1::TRP1 strain (YGC034) transformed with a yeast expression vector carrying HA-tagged EAP1. Expression of the tagged protein was confirmed by Western blotting with anti-HA monoclonal antibody (Fig. 5A, compare lanes 4 and 1). Immunoprecipitations were carried out using an anti-HA antibody followed by Western blotting using anti-HA or anti-eIF4E monoclonal antibody to reveal the bound proteins. HA-Eap1p was quantitatively immunoprecipitated from yeast lysate by the anti-HA antibody (Fig. 5A, lane 6). Approximately 45% of endogenous eIF4E was coimmunoprecipitated with Eap1p, as measured by densitometric scanning of the chemiluminescent signal on the Western blot (Fig. 5B, lane 6). In contrast, eIF4E was not coimmunoprecipitated from cells transformed with the expression vector alone (Fig. 5B, lane 3). These results show that a significant amount of eIF4E is associated with Eap1p in the cell.
FIG. 5.
In vivo interaction of Eap1p with eIF4E is dependent on the 4E-binding motif. Yeast strain YGC034 (eap1::TRP1) was transformed with vector YEp352 alone or with the same vector expressing either HA-Eap1p or the HA-Eap1p[Y109A] mutant. Yeast were grown to exponential phase and lysed by the glass bead method. Protein (100 μg) was incubated with anti-HA monoclonal antibody before the addition of protein G-Sepharose beads. Following extensive washings, the bound proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted against anti-HA antibody (to detect HA-Eap1p; indicated by a dot) (A) or anti-yeast eIF4E monoclonal antibody 9B12 (B). Load, an equal amount of extract used for the coimmunoprecipitation loaded directly onto the gel; Free, proteins remaining in supernatant following immunoprecipitation; Bound, proteins adsorbed to the resin. IgG, immunoglobulin G heavy chain. Sizes of standards are indicated in kilodaltons.
To show that the interaction between Eap1p and eIF4E occurs through the canonical eIF4E-binding motif, Tyr-109 was substituted by alanine and the mutant protein was examined in the coimmunoprecipitation assay. The corresponding mutation in other eIF4E-binding proteins abolishes the interaction with eIF4E (58, 63). The Y109A mutant was expressed in eap1::TRP1 cells (Fig. 5A, lane 7) and was efficiently immunoprecipitated by the anti-HA antibody (Fig. 5A, lane 9). The Y109A point mutation abolished eIF4E coimmunoprecipitation (Fig. 5B, compare lane 9 to lane 6). Consistent with these results, 32P-labeled HMK-eIF4E also failed to bind HA-Eap1p[Y109A] in whole-cell lysates as determined by far-Western analysis (data not shown). Taken together, these data demonstrate that the interaction of Eap1p and yeast eIF4E occurs both in vitro and in vivo and is dependent on the integrity of the 4E-binding consensus sequence located between aa 109 and 115.
Eap1p competes with eIF4G and p20 for binding to eIF4E in vivo.
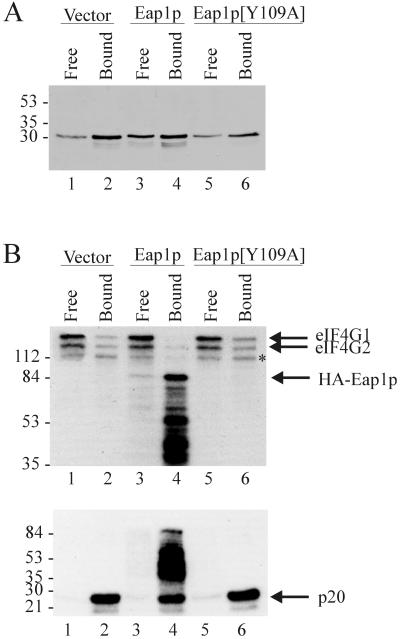
As Eap1p shares an eIF4E-binding motif with eIF4G and p20, we reasoned that Eap1p and eIF4G should compete for interaction with eIF4E. To investigate this, endogenous eIF4E was precipitated from extracts of the eap1::TRP1 strain transformed with either wild-type or Y109A mutant of HA-tagged EAP1, using an m7GDP-agarose resin. Bound proteins were revealed by Western blotting using anti-eIF4E antibody or by far-Western analysis using 32P-labeled HMK-eIF4E as a probe. The amounts of eIF4E precipitated by the m7GDP-resin were similar for all of the yeast strains tested, indicating that Eap1p does not affect eIF4E interaction with the cap structure (Fig. 6A). In the presence of wild-type Eap1p, the amount of eIF4G1, eIF4G2, and p20 that coprecipitated with eIF4E was reduced by 67, 57, and 38%, respectively, compared to cells transformed with vector alone (Fig. 6B, compare lanes 4 and 2). The competition was contingent on the interaction of Eap1p with eIF4E, as the amounts of eIF4G and p20 that coprecipitated in the presence of the Eap1p Y109A mutant were equal to those observed using extracts derived from vector control cells (Fig. 6B and C, compare lane 6 to 2).
FIG. 6.
Eap1p competes with eIF4G and p20 for binding to eIF4E. Yeast extract was prepared as described for Fig. 5. Protein (100 μg) was incubated with m7GDP-agarose, and bound proteins were resolved by SDS-PAGE. (A) Western blotting was performed using anti-eIF4E monoclonal antibody 9B12. (B) Far-Western blotting was conducted using 32P-labeled HMK-eIF4E as a probe. The eIF4E-interacting proteins are identified with arrows on the right. The asterisk indicates a degradation product of eIF4G2 which was observed sporadically in crude yeast extracts. Note that Eap1p[Y109A] mutant is not revealed by this analysis (see text). Free, proteins remaining in supernatant following immunoprecipitation; Bound, proteins adsorbed to the resin. Sizes of standards are indicated in kilodaltons.
Eap1p inhibits cap-dependent translation.
The effect of Eap1p on cap-dependent translation was investigated using two different capped CAT mRNAs in a cell-free yeast system (6). The two reporter mRNAs differ only by insertion of the 67-nucleotide Ω sequence from tobacco mosaic virus mRNA in the 5′ untranslated region (70). The Ω sequence decreases the requirement for eIF4E in translation (2). Baculovirus-generated recombinant Flag-tagged Eap1p interacted with eIF4E in vitro (Fig. 7A). Translation extracts prepared from yeast null for Eap1p (YGC034) were programmed with either capped CAT or capped Ω CAT mRNA in the presence of [35S]methionine and Eap1p. Addition of increasing amounts of recombinant Eap1p resulted in a graded inhibition of translation (up to 10-fold inhibition in the presence of 2.5 μg of Eap1p; Fig. 7B and C). In contrast, equivalent levels of Eap1p had a much smaller effect on Ω CAT mRNA translation (approximately twofold inhibition [Fig. 7B and C]). These data are similar to those obtained in another study showing that p20 preferentially inhibits cap-dependent versus cap-independent translation in yeast (6).
FIG. 7.
In vitro translation in yeast cell extract. (A) Recombinant Flag-tagged Eap1p was immunopurified from insect cells. The purified protein was resolved by SDS-PAGE and revealed by Coomassie staining or by far-Western analysis using 32P-labeled eIF4E. Sizes of molecular weight (MW) markers are indicated in kilodaltons. (B) Translation reactions in an extract generated from YGC034 (eap1::TRP1) were conducted as described in Materials and Methods. Increasing amounts of recombinant Flag-Eap1p were added to the reaction mixtures, and translation was initiated with 100 ng of capped CAT mRNA containing or lacking the Ω sequence. Samples were fractionated by SDS-PAGE, and CAT protein was revealed by autoradiography. (C) Data shown in panel B were quantitated by densitometry and normalized to the amount of CAT synthesis in the absence of added Eap1p. The results are a representative of two independent experiments which did not vary significantly.
Disruption of EAP1 confers partial resistance to rapamycin and temperature-sensitive growth.
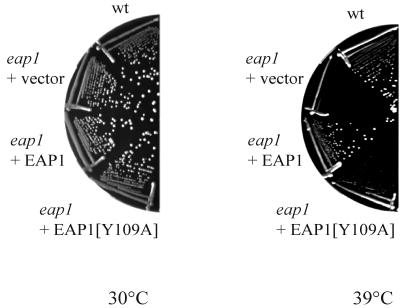
As noted above, disruption of the EAP1 gene had no effect on yeast growth under standard conditions (i.e., incubation at 30°C). However, at elevated temperatures (39°C), growth of the eap1 strain was substantially impaired (Fig. 8). The temperature-sensitive phenotype could be reverted by introduction of the wild-type EAP1 gene on a low-copy-number vector but only weakly by the mutant Y109A (Fig. 8). These data suggest that the temperature-sensitive phenotype is engendered by the deficiency in Eap1p-eIF4E complex formation.
FIG. 8.
Disruption of EAP1 confers a temperature-sensitive phenotype. Yeast strain YGC034 (eap1) was transformed with either YCplac33 (vector), pTS115 (EAP1), or pTS117 (EAP1[Y109A]). Resulting transformants and wild-type strain JH6-1C (wt) were streaked on YPD media and incubated at either 30 or 39°C for 2 and 3 days, respectively.
Because the macrolide antibiotic rapamycin blocks the TOR signaling pathway and inhibits cap-dependent translation in yeast and mammals, it was of interest to investigate whether loss of EAP1 could maintain growth of cells treated with this compound (11, 14, 75). The eap1 strain grew better than the isogenic wild-type strain on medium containing low concentrations (20 ng/ml) of rapamycin (Fig. 9A). However, the differential growth effect was not observed at higher drug concentrations (50 ng/ml [data not shown]). In comparison, TOR1-1 cells, which carry a dominant mutation in TOR1, rendered yeast resistant to drug concentrations as high as 200 ng/ml (44). Also, TOR1-1 cells grew more efficiently than eap1 cells in the presence of 20 ng of rapamycin per ml (Fig. 9A). The rapamycin resistance of eap1 cells indicates that the absence of Eap1p partially relieves the inhibition of protein synthesis caused by rapamycin, thus allowing cell growth on medium containing the drug.
FIG. 9.
Disruption of EAP1 confers partial resistance to rapamycin. (A) Yeast strains JH11-1C (TOR1-1), JH6-1C (wild type [wt]), and YGC034 (eap1) were streaked on YPD alone and on YPD containing rapamycin (20 ng/ml) and incubated at 30°C. (B) Indicated yeast strains were transformed with YCplac33 (vector), pTS115 (EAP1), or pTS117 (EAP1[Y109A]). Resulting transformants were streaked on SD-Ura medium and SD-Ura medium containing rapamycin (1 ng/ml) and then incubated at 30°C. tor1, strain AS93-2A; tor1 eap1, strain TS6-5A.
We also tested the effect of EAP1 deletion in a yeast strain which is hypersensitive to the growth-inhibitory effects of rapamycin due to reduced TOR function as a result of the targeted disruption of the TOR1 gene (57). The tor1 eap1 strain grew better than the isogenic tor1 strain on medium containing low concentrations (1 ng/ml) of rapamycin (Fig. 9B). Furthermore, transformation of the tor1 eap1 strain with a plasmid expressing wild-type EAP1 under control of its own promoter completely abolished the rapamycin-resistant phenotype, demonstrating that this effect is due to loss of Eap1p. In contrast, the rapamycin-resistant phenotype was only weakly reverted by expression of the Y109A mutant of EAP1 (Fig. 9B), indicating that it is dependent on the efficient interaction of eIF4E and Eap1p in vivo.
It is noteworthy that strains deleted for CAF20 showed no resistance to rapamycin (data not shown), demonstrating that the partial rapamycin resistance of eap1 cells is a TOR-signaling-specific effect and not simply a general manifestation of increased cap-dependent translation. These results are consistent with a role for Eap1p as a regulator of cap-dependent translation in response to the TOR signaling pathway in yeast.
DISCUSSION
We have identified an ORF (YKL204w) in S. cerevisiae that encodes a novel eIF4E-interacting protein, which we termed EAP1. Eap1p competes with the eIF4Gs for binding to eIF4E and can inhibit cap-dependent translation in vitro, consistent with a role for this protein in translational control. However, Eap1p also contains potential functional domains which were not explored in the present work (i.e., a putative nuclear localization sequence and proline-rich C terminus). In particular, one interesting possibility is that Eap1p also provides a function related to the partial localization of yeast eIF4E in the nucleus (53, 65).
Disruption of EAP1 confers partial resistance to the immunosuppressant macrolide, rapamycin. Rapamycin binds to the yeast immunophilin protein, FKBP, which then inhibits TOR1 and TOR2 activity, resulting in a block of translation initiation and arrest in early G1 phase of the cell cycle (11, 27, 43, 44, 52, 84). Several lines of evidence suggest that the cell cycle arrest is a consequence of reduced translation initiation: (i) reduction of cellular translation rates is an early effect detected upon inhibition of TOR by rapamycin (11); (ii) a specific block in translation through the mutation of initiation factors, including eIF4E, causes yeast cells to arrest in early G1 (12, 17, 41); and (iii) expression of the G1 cyclin gene, CLN3, under control of the 5′ untranslated region from polyubiquitin (UBI4), which confers reduced eIF4E dependence on translation (17), suppresses the G1 arrest induced either by rapamycin or by mutation of eIF4E (11, 23). This indicates that the block in translation initiation caused by the loss of TOR function is mediated through down-regulation of eIF4E function. Our present observation that deletion of Eap1p maintains growth in cells lacking TOR function (through treatment with rapamycin) extends the argument that the TOR pathway regulates translation initiation in yeast and that Eap1p may partially act as a functional homolog of mammalian 4E-BPs.
Rapamycin resistance conferred by a dominant TOR1-1 mutation is more pronounced than that observed with the eap1::TRP1 strain. This suggests that Eap1p contributes only partially to the TOR effects on cell growth and that additional TOR-dependent pathways exist. In mammalian cells, the mTOR signaling cascade bifurcates into two parallel pathways which control the phosphorylation of 4E-BP1 and ribosomal protein S6 (46, 80). Phosphorylation of S6 at multiple sites leads to activation of translation initiation (reviewed in reference 47). Thus, mTOR has the capacity to regulate translation via multiple mechanisms. However, phosphorylation of the yeast homolog of S6 (S10) has little effect on protein synthesis or cell growth (49, 51), suggesting that yeast TOR does not modulate translation initiation through ribosomal protein phosphorylation. Moreover, yeast mRNAs for ribosomal proteins do not contain a 5′-polypyrimidine tract which mediates the effect of S6 phosphorylation on translation (47). More recently, however, the TOR signal transduction pathway has been implicated in a broader range of metabolic activities which could modulate protein synthesis and cellular growth. These include control of amino acid transport (69), stability of eIF4G (15), cellular autophagy (61), RNA polymerase I and III transcription (83), ribosomal biogenesis (64), and transcriptional control of nutrient-regulated catabolic pathways (13). These mechanisms may function simultaneously with TOR-dependent regulation of Eap1p to modulate protein synthesis and cellular proliferation.
How might TOR signal to Eap1p? EAP1 (ORF YKL204w) is constitutively expressed at low levels (100 to 1,000 times less than actin mRNA [68]) and is not differentially regulated during batch growth, throughout the cell cycle, or during sporulation, based on yeast gene expression databases (21, 26, 68, 73, 78; SGD [see above]; P. O. Brown laboratory, Stanford University [http://cmgm.stanford.edu/pbrown/explore]). This suggests that regulation of Eap1p function occurs posttranslationally, possibly through reversible phosphorylation in analogy to 4E-BPs. Consistent with this idea, mass spectroscopy analysis shows that Eap1p is multiply phosphorylated in vivo (U. Schneider and P. Jenö [Department of Biochemistry, Biozentrum, University of Basel] unpublished data). Mammalian mTOR/FRAP phosphorylates 4E-BP1 in an in vitro immune kinase assay (18–20, 35). Yeast TOR1 is also capable of phosphorylating 4E-BP in vitro (1). In vivo, yeast TOR phosphorylates the essential protein, Tap42, thereby stimulating association of Tap42 with the catalytic subunit of type 2A protein phosphatases (PPH21 and PPH22) and a type 2A-related protein phosphatase (SIT4) (27, 48). It is conceivable that TOR-dependent regulation of the Tap42 complex could ultimately modify the phosphorylation state of Eap1p as has been proposed for the protein kinase NPR1 (69) and the transcription factor GLN3 (13). Future studies will be required to define the potential regulation and role of Eap1p phosphorylation in the control of translation initiation.
Eap1p and p20 share homology only in the 4E-binding domain, with no overall similarity between the proteins. Nevertheless, both could function as translational repressors in yeast by using a molecular mimicry mechanism in common with mammalian 4E-BPs. The differences between Eap1p and p20 may reflect their differential regulation by upstream effectors (evidence of which is provided by our finding that deletion of p20 had no effect on rapamycin sensitivity) and/or additional functions unrelated to translational control. Neither Eap1p nor p20 is essential for cell growth under standard laboratory conditions. Rather, these proteins may serve to modulate growth in response to adverse conditions in the natural environment (consistent with the temperature-sensitive phenotype observed for the eap1 strain). Further analysis of the pathways which modulate Eap1p and p20 should yield insights into the regulation of cellular growth through the function of eIF4E-associated proteins.
ACKNOWLEDGMENTS
We gratefully thank M. Altmann, M. Blanar, H. Bussey, B. Dujon, A. Schmidt, and H. Trachsel for providing reagents used in this work and C. Lister for providing excellent technical assistance.
This work was supported by a grant from the Medical Research Council of Canada and the Howard Hughes Medical Institute to N.S. and by grants from the Swiss National Science Foundation and the Canton of Basel to M.N.H. N.S. is a Distinguished Scientist of the Medical Research Council of Canada and Howard Hughes Medical Institute International Scholar. G.P.C. was supported by Bio-Méga Research Division, Boehringer Ingelheim (Canada) Ltd. T.S. was supported by the Boehringer Ingelheim Fonds.
REFERENCES
- 1.Alarcon C M, Heitman J, Cardenas M E. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol Biol Cell. 1999;10:2531–2546. doi: 10.1091/mbc.10.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmann M, Blum S, Wilson T M, Trachsel H. The 5′-leader sequence of tobacco mosaic virus RNA mediates initiation-factor-4E-independent, but still initiation-factor-4A-dependent translation in yeast extracts. Gene. 1990;91:127–129. doi: 10.1016/0378-1119(90)90173-o. [DOI] [PubMed] [Google Scholar]
- 3.Altmann M, Edery I, Sonenberg N, Trachsel H. Purification and characterization of protein synthesis initiation factor eIF-4E from the yeast Saccharomyces cerevisiae. Biochemistry. 1985;24:6085–6089. doi: 10.1021/bi00343a009. [DOI] [PubMed] [Google Scholar]
- 4.Altmann M, Krieger M, Trachsel H. Nucleotide sequence of the gene encoding a 20 kDa protein associated with the cap binding protein eIF-4E from Saccharomyces cerevisiae. Nucleic Acids Res. 1989;17:7520. doi: 10.1093/nar/17.18.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altmann M, Muller P P, Pelletier J, Sonenberg N, Trachsel H. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J Biol Chem. 1989;264:12145–12147. [PubMed] [Google Scholar]
- 6.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altmann M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Appel R D, Bairoch A, Hochstrasser D F. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 10.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 11.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes C A, Mackenzie M M, Johnston G C, Singer R A. Efficient translation of an ssa1-derived heat-shock mRNA in yeast cells limited for cap-binding protein and eIF-4F. Mol Gen Genet. 1995;246:619–627. doi: 10.1007/BF00298969. [DOI] [PubMed] [Google Scholar]
- 13.Beck T, Hall M N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 14.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 15.Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 17.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunn G J, Fadden P, Haystead T J, Lawrence J C J. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J Biol Chem. 1997;272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- 19.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 20.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 22.Craig A W, Cosentino G P, Donze O, Sonenberg N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem. 1996;271:24526–24533. doi: 10.1074/jbc.271.40.24526. [DOI] [PubMed] [Google Scholar]
- 23.Danaie P, Altmann M, Hall M N, Trachsel H, Helliwell S B. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem J. 1999;340:135–141. [PMC free article] [PubMed] [Google Scholar]
- 24.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis P B, Fumagalli S, Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 26.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 27.Di Como C, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 28.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 29.Dujon B, Alexandraki D, Andre B, Ansorge W, Baladron V, Ballesta J P, Banrevi A, Bolle P A, Bolotin-Fukuhara M, Bossier P, et al. Complete DNA sequence of yeast chromosome XI. Nature. 1994;369:371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- 30.Edery I, Altmann M, Sonenberg N. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene. 1988;74:517–525. doi: 10.1016/0378-1119(88)90184-9. [DOI] [PubMed] [Google Scholar]
- 31.Edery I, Lee K A, Sonenberg N. Functional characterization of eukaryotic mRNA cap binding protein complex: effects on translation of capped and naturally uncapped RNAs. Biochemistry. 1984;23:2456–2462. doi: 10.1021/bi00306a021. [DOI] [PubMed] [Google Scholar]
- 32.Fadden P, Haystead T A, Lawrence J C. Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J Biol Chem. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- 33.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 35.Gingras A C, Gygi S P, Raught B, Polakiewicz R D, Abraham R T, Hoekstra M F, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gingras A C, Raught B, Sonenberg N. eIF4F initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 37.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L J, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guthrie C, Fink G R, editors. Methods in enzymology. Vol. 194. 1991. Guide to yeast genetics and molecular biology, Academic Press, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 39.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanic-Joyce P J, Johnston G C, Singer R A. Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp Cell Res. 1987;172:134–145. doi: 10.1016/0014-4827(87)90100-5. [DOI] [PubMed] [Google Scholar]
- 42.Hara K, Yonezawa K, Kozlowski M T, Sugimoto T, Andrabi K, Weng Q P, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 43.Heitman J, Movva N R, Hall M N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 44.Helliwell S B, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall M N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaramillo M, Dever T E, Merrick W C, Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991;11:5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jefferies H B, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jefferies H B J, Thomas G. Ribosomal protein S6 phosphorylation and signal transduction. In: Hershey J W, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 389–409. [Google Scholar]
- 48.Jiang Y, Broach J R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson S P, Warner J R. Phosphorylation of the Saccharomyces cerevisiae equivalent of ribosomal protein S6 has no detectable effect on growth. Mol Cell Biol. 1987;7:1338–1345. doi: 10.1128/mcb.7.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 51.Kruse C, Johnson S P, Warner J R. Phosphorylation of the yeast equivalent of ribosomal protein S6 is not essential for growth. Proc Natl Acad Sci USA. 1985;82:7515–7519. doi: 10.1073/pnas.82.22.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 53.Lang V, Zanchin N I, Lunsdorf H, Tuite M, McCarthy J E. Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J Biol Chem. 1994;269:6117–6123. [PubMed] [Google Scholar]
- 54.Lanker S, Muller P P, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 55.Lawrence J C J, Fadden P, Haystead T A, Lin T A. PHAS proteins as mediators of the actions of insulin, growth factors and cAMP on protein synthesis and cell proliferation. Adv Enzyme Regul. 1997;37:239–267. doi: 10.1016/s0065-2571(96)00016-7. [DOI] [PubMed] [Google Scholar]
- 56.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C J. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 57.Lorenz M C, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 58.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merrick W C, Hershey J W. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–70. [Google Scholar]
- 60.Neff C L, Sachs A B. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol Cell Biol. 1999;19:5557–5564. doi: 10.1128/mcb.19.8.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 62.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 63.Poulin F, Gingras A C, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 64.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ptushkina M, Vasilescu S, Fierro-Monti I, Rohde M, McCarthy J E. Intracellular targeting and mRNA interactions of the eukaryotic translation initiation factor eIF4E in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1996;1308:142–150. doi: 10.1016/0167-4781(96)00096-6. [DOI] [PubMed] [Google Scholar]
- 66.Ptushkina M, von der Haar T, Vasilescu S, Frank R, Birkenhager R, McCarthy J E. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J. 1998;17:4798–4808. doi: 10.1093/emboj/17.16.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ray B K, Lawson T G, Kramer J C, Cladaras M H, Grifo J A, Abramson R D, Merrick W C, Thach R E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 68.Richard G F, Fairhead C, Dujon B. Complete transcriptional map of yeast chromosome XI in different life conditions. J Mol Biol. 1997;268:303–321. doi: 10.1006/jmbi.1997.0973. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sleat D E, Gallie D R, Jefferson R A, Bevan M W, Turner P C, Wilson T M. Characterisation of the 5′-leader sequence of tobacco mosaic virus RNA as a general enhancer of translation in vitro. Gene. 1987;60:217–225. doi: 10.1016/0378-1119(87)90230-7. [DOI] [PubMed] [Google Scholar]
- 71.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 72.Sonenberg N, Gingras A C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 73.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 75.Svitkin Y V, Hahn H, Gingras A C, Palmenberg A C, Sonenberg N. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J Virol. 1998;72:5811–5819. doi: 10.1128/jvi.72.7.5811-5819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarun S Z J, Sachs A B. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas G, Hall M N. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 78.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E J, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 79.Vinson C R, LaMarco K L, Johnson P F, Landschulz W H, McKnight S L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 80.von Manteuffel S, Dennis P B, Pullen N, Gingras A C, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zanchin N I, McCarthy J E. Characterization of the in vivo phosphorylation sites of the mRNA cap-binding complex proteins eukaryotic initiation factor-4E and p20 in Saccharomyces cerevisiae. J Biol Chem. 1995;270:26505–26510. doi: 10.1074/jbc.270.44.26505. [DOI] [PubMed] [Google Scholar]
- 83.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng X F, Florentino D, Chen J, Crabtree G R, Schreiber S L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]