Abstract
The ELVIS HSV Id test kit (an enzyme-linked virus-inducible system) (Diagnostic Hybrids, Inc.) uses genetically engineered BHK cells to produce a detectable enzyme, beta-galactosidase, upon infection with either herpes simplex virus (HSV) type 1 (HSV-1) or HSV-2. Twenty six ELVIS-positive clinical specimens were selected for study by PCR and with monoclonal antibodies because they were originally low-titer HSV-positive specimens by ELVIS but HSV antibody nonreactive upon follow-up staining of the ELVIS monolayer. Twenty-one of 26 specimens were frozen, thawed, and retested with ELVIS without removing the cellular debris from the specimen; 18 were ELVIS positive and 3 were ELVIS negative on retesting. A typing result was provided upon retesting for 14 of 18 ELVIS-positive specimens (11 were HSV-1 and 3 were HSV-2) with HSV-specific monoclonal antibodies; no antibody signal was observed for 4 of 18 ELVIS-positive specimens. Sixteen of 26 specimens were subjected to blinded PCR analysis with two different primer sets, including all those that were repeat tested with ELVIS without success and those that had insufficient quantity for repeat testing. All 16 specimens analyzed were PCR positive with primer set 1; 15 of 16 were also positive with primer set 2, with the HSV type identified for all specimens (7 were HSV-1 and 8 were HSV-2). These results indicate that the original ELVIS result with these low-titer specimens was correct and further confirm the sensitivity and specificity of ELVIS HSV Id as a rapid, cell culture-based kit for the detection of HSV.
The dramatic increase in herpesvirus infections over the past decade emphasizes the continuing need for highly sensitive, specific, rapid, and cost-effective virus detection methods (1). The enzyme-linked virus-inducible system (ELVIS; Diagnostic Hybrids, Inc. [DHI], Athens, Ohio) is a new cell culture-based, viral diagnostic technology that uses cells genetically engineered with virus-inducible reporter genes to detect herpesviruses (3). Induction of the reporter gene occurs rapidly after viral infection and produces relatively large quantities of the enzyme beta-galactosidase that can provide an amplified detection signal. By use of a sensitive in situ histochemical staining method, ELVIS technology can detect a single infected cell in 12 to 24 h postinfection (7). ELVIS cells have been used for detection of herpes simplex virus (HSV) in clinical specimens (8), for antiviral susceptibility testing (9), and in in vitro neutralization assays (2).
The first commercial kit that has used this technology, ELVIS HSV Id, has been shown to be a sensitive, specific, and rapid alternative to traditional tube culture methods for the identification of HSV (4). An HSV-typing capability was recently added by modifying the cell fixative to an acetone-water-based formulation. This change enhances color formation in ELVIS-positive cells, preserves certain HSV-specific antigens, and enables the routine use of polystyrene multiwell plates with antibody-based assays. Specimens can now be routinely screened for the presence of blue-stained cells from ELVIS-induced beta-galactosidase expression and then subsequently stained directly on the ELVIS monolayer with type-specific, fluorescein-labeled monoclonal antibodies.
Using this combined ELVIS typing algorithm, however, laboratory technologists observed on occasion specimens that produced blue-stained cells by the ELVIS protocol but that could not be directly typed due to a lack of a fluorescent-antibody signal. These untypeable specimens were most often characterized by the presence of few blue ELVIS cells in the entire monolayer, suggesting that these specimens contained relatively low levels of HSV, generally less than five blue stained cells. Traditional culture was not viewed as the best method to study these low-titer specimens because reduced or no recovery of infectious virus after freezing and thawing of the specimens was likely. The current study used PCR analysis and ELVIS retesting with HSV-specific monoclonal antibodies from two commercial suppliers to reexamine 26 clinical specimens originally submitted for testing for HSV.
MATERIALS AND METHODS
Specimens.
Twenty-six nonconsecutive genital specimens (designated specimens 1 to 26) submitted for HSV typing were included in the study. These specimens had previously tested ELVIS positive but antigen nonreactive after staining of the ELVIS monolayer with Syva HSV Id/Typing reagents (Behring Diagnostics, Palo Alto, Calif.). All specimens were frozen at −70°C after primary testing until thawed for secondary analyses with ELVIS and/or by PCR.
ELVIS HSV Id retest categories.
Specimens were divided into the following categories depending upon the volume available for each. Category A specimens (>0.6 ml) were inoculated into three ELVIS wells and were typed with both Syva and DHI typing reagents. Category B specimens (>0.4 ml) were inoculated into two ELVIS wells and were typed with Syva reagents alone. Category C specimens (>0.2 ml) were inoculated into one ELVIS well and were typed with DHI reagents only. Category D specimens (<0.2 ml) had insufficient volume for testing with ELVIS HSV Id.
ELVIS HSV Id culture.
Specimens were inoculated into ELVIS HSV Id cells in 24-well plates (Falcon) according to the manufacturer's instructions (DHI), with a preincubation time of 18 h and a postinoculation time of 20 h. The specimens were rapidly thawed in a 35°C water bath. Shipping medium was gently aspirated from the cell wells and was replaced with 1 ml of ELVIS Replacement Medium. The specimen was briefly vortexed, and 0.2 ml of specimen was added per well. The multiwell plates were centrifuged at 700 × g for 60 min at 30°C.
ELVIS HSV Id and Syva typing staining protocol.
Culture medium was removed by aspiration, and 0.25 ml of ELVIS Solution 1 (Cell Fixative; acetone-based) was added for 1 to 2 min. Solution 1 was removed and 0.25 ml of ELVIS Solution 2 (Staining Buffer; buffered 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]) was added. The plates were incubated for 60 min at 37°C. Cell monolayers were viewed for blue-stained cells with an inverted light microscope. If blue cells were observed, Solution 2 was removed and 0.25 ml of either HSV type 1 (HSV-1) or HSV-2 typing solution (Syva), each of which was used at 1:4 (vol/vol) in phosphate-buffered saline (PBS), was added to an appropriate well. The plates were incubated for 30 min at 37°C and rinsed twice with PBS, mounting fluid was added, and the monolayers were viewed under a Nikon epifluorescent microscope for fluorescent cells.
ELVIS HSV Id/Typing (DHI) staining protocol.
Culture medium was removed by aspiration, and 0.25 ml of ELVIS Solution 1 (Cell Fixative; acetone-based) was added for 1 to 2 min. Solution 1 was removed, and 0.25 ml of ELVIS Solution 2T (Staining Buffer; buffered X-Gal plus two fluorescein isothiocyanate [FITC]-conjugated HSV-2-specific monoclonal antibodies and two unlabeled HSV-1-specific monoclonal antibodies) was added. The plates were incubated for 60 min at 37°C. The cell monolayers were viewed for blue-stained cells with an inverted light microscope. If blue cells were observed, Solution 2T was removed, the monolayers were rinsed once with distilled water, ELVIS mounting fluid was added, and the wells were viewed for fluorescent cells. If fluorescent cells were observed, the HSV isolate was reported as HSV-2. If no fluorescing cells were observed, the mounting fluid was removed by rinsing twice with distilled water, and 0.25 ml of ELVIS Solution 3 was added (FITC-conjugated goat anti-mouse antibodies). The plate was incubated for 15 min at 37°C and rinsed twice with distilled water, mounting fluid was added, and the monolayer was reviewed for fluorescing cells. If fluorescence was observed after Solution 3 was added, the isolate was reported as HSV-1. If no fluorescing cells were observed after staining with Solution 3, then the isolate was reported as HSV positive, no type available.
PCR analyses.
Selected specimens from categories A to D were tested by PCR analysis under blind code at the Molecular Microbiology Laboratory, Johns Hopkins Medical Institutions, Baltimore, Md. These specimens included eight specimens that were, upon retesting, ELVIS positive and fluorescence positive, three specimens that were negative, and five specimens for which insufficient volume was available for retesting. DNA was extracted from all samples with the QIAamp Blood Kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Two primer sets were used: HSV primer set 1 is a non-type-specific set (5), and HSV primer set 2 is a type-specific set (6).
(i) Primer set 1 is a pair of 21-base oligonucleotides with homology to regions of the DNA polymerase genes of HSV-1 and HSV-2. Samples (0.01 ml of extracted specimen in a PCR mixture volume of 0.1 ml) were initially heated at 95°C for 5 min and were then exposed to 40 cycles of amplification (94°C for 45 s, 62°C for 45 s, and 72°C for 45 s), with a final 3-min extension at 72°C. The PCR products were analyzed by electrophoresis through a 4% (wt/vol) NuSieve agarose gel, and a 179-bp band was detected by staining with ethidium bromide. Southern blot hybridization with a 32P-labeled probe was performed for confirmation as well to enhance sensitivity and specificity.
(ii) Primer set 2 is a pair of 18-base oligonucleotides with homology to regions of the DNA polymerase genes of HSV-1 and HSV-2. Samples (0.01 ml of extracted specimen in a PCR volume of 0.1 ml) were initially heated at 72°C for 3 min and were then exposed to 35 cycles of amplification (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s). The 211-bp PCR products were then analyzed as described above for primer set 1. The identity of the amplified reaction product was confirmed by Southern blot hybridization with a 29-base, 32P-labeled probe. PCR products were also treated with the restriction enzyme BamHI to determine the HSV type. The amplified segment of HSV-2, but not that of HSV-1, contains a BamHI site that yields products of 121 and 90 bp.
RESULTS
Retesting with ELVIS.
Twenty-six specimens were collected for retesting and were placed into one of four categories depending upon the specimen volume available for analyses. Due to insufficient volume for retesting, specimens in category D were not retested with ELVIS.
Among the eight specimens in category A, five were ELVIS positive in the wells used for either the Syva typing reagents or the DHI typing reagents. Three of five of the ELVIS-positive specimens from category A showed one or more fluorescing cells. Two specimens were HSV-1 and one specimen was HSV-2, as determined with both typing reagent sets. Two ELVIS-positive specimens did not show evidence of fluorescing cells with either typing reagent set. Three of eight specimens were ELVIS negative after reculture.
All seven specimens in category B were ELVIS positive in the wells used for the Syva typing reagents. Six of the seven specimens showed one or more fluorescing cells; five specimens were HSV-1 and one specimen was HSV-2. One ELVIS-positive specimen did not show evidence of fluorescing cells.
All six specimens in category C were ELVIS positive in the single-well format used for the DHI typing test. Five of the six specimens showed one or more fluorescing cells; four specimens were HSV-1 and one specimen was HSV-2. One ELVIS-positive specimen did not show evidence of fluorescing cells.
PCR analyses.
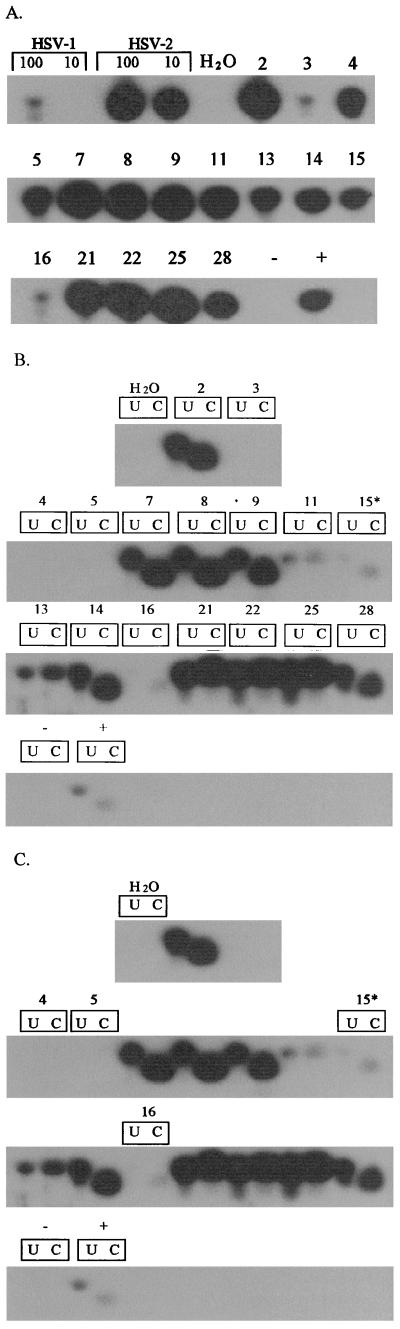
Sixteen specimens were evaluated by PCR analyses. Figure 1A is an autoradiograph that shows an HSV-specific PCR product that was produced for all 16 specimens tested with PCR primer set 1. The identical specimens were also tested with PCR primer set 2, a HSV typing primer set. Figure 1B shows that 12 of 16 specimens were PCR positive with these primers and an exposure time of 4 h. Extension of the autoradiograph exposure time to 17 h resulted in the identification of three additional positive specimens (Fig. 1C, specimens 4, 5, and 16). Specimen 3 was the only specimen that did not produce an HSV-specific PCR product with this primer set. When the PCR products from the 15 positive specimens were subjected to restriction endonuclease digestion with BamHI, 7 specimens showed a banding profile on the autoradiograph characteristic of HSV-1 and 8 showed a banding profile characteristic of HSV-2.
FIG. 1.
PCR analysis of 16 specimens from categories A, B, C, and D with nontyping (primer set 1) and typing (primer set 2) HSV primer sets. (A) PCR with HSV primer set 1 and an exposure time of 4 h; 100 and 10, numbers of copies of HSV DNA; H2O, reagent control; and − and +, viral transport medium controls without and with 1 PFU of HSV-2, respectively. (B) PCR with HSV primer set 2 and an exposure time of 4 h; U, PCR product only; C, PCR product treated with BamHI. (C) Extended exposure time of the autoradiograph (17 h) in panel B to enhance visualization of specimens 4, 5, 15, and 16.
Table 1 summarizes the comparative study results by retesting with ELVIS and PCR. All specimens (26 of 26) were found to be HSV positive by at least one of the three methods used for retesting.
TABLE 1.
Comparative study results by retesting with ELVIS and PCR
| PCR result | No. of specimens with the following result by retesting with ELVIS:
|
||
|---|---|---|---|
| Positive | Negative | Not tested | |
| Positive | 8 | 3 | 5 |
| Negative | 0 | 0 | 0 |
| Not Tested | 10 | 0 | 0 |
DISCUSSION
The ELVIS HSV test was previously shown to be an effective alternative to long-term culture for the rapid diagnosis of HSV infection (4). The specimens reevaluated in this study represent a small collection of specimens that produced a very low number of blue-stained ELVIS cells upon initial testing but that failed to show fluorescent ELVIS cells after staining with Syva's fluorescent antibody reagents. The correlation of the primary ELVIS finding, the secondary ELVIS reculture results, and the presence of HSV DNA in all specimens analyzed by PCR strongly indicate that the original finding of blue-stained cells was very sensitive (in some cases, detecting a single infected cell) and specific for HSV.
This study also used two different typing reagent approaches depending upon the specimen volume available. The new ELVIS HSV Id/Typing reagent uses a pool of HSV-1 and HSV-2 antibodies in the ELVIS Staining Buffer, Solution 2T. These antibodies are capable of binding simultaneously to HSV-infected cells during the 60-min color development period. The HSV-2-specific antibodies are conjugated directly with FITC, while the HSV-1-specific antibodies are unlabeled. If fluorescence is observed directly after finding blue-stained cells, the isolate is reported as HSV-2. If no fluorescence is directly observed, FITC-conjugated goat anti-mouse polyclonal antibody is added to bind to the unlabeled HSV-1-specific monoclonal antibodies. A fluorescent signal by this indirect method indicates that the isolate is HSV-1. The primary benefits of using the DHI typing reagents are that HSV can be detected by inoculating a single well or vial, and in cases of a negative result (no blue observed) or a result indicating that the isolates are HSV-2 (blue-stained cells plus direct fluorescence), the test is completed simultaneously with two specific detection technologies.
Why a type result was not obtained with 14 specimens upon initial testing is uncertain. The fluorescent signal can be quenched for ELVIS-positive cells that are heavily stained blue by the X-Gal chromogen and number less than five blue-stained cells (unpublished observation). The specimens included in this study generally had low virus titers, i.e., only a few individually stained cells with little evidence of virus amplification. Thus, it is possible that the fluorescent signal was present for only a few cells and that those cells were heavily stained blue.
There are two possible explanations for why the specimens were more amenable to typing when they were retested. First, a routine procedure used in many clinical laboratories is to clarify the specimen by centrifugation to remove specimen toxicity. This may result in the potential removal of cell-associated HSV from the inoculum. When specimens were retested, each sample was briefly vortexed and inoculated directly. While a lack of removal of debris could lead to increased toxicity to the cell monolayer, cell toxicity was noted in only 1 of 21 specimens retested with ELVIS. This one specimen (specimen 7) was unique in that a relatively large number of blue-stained cells were observed, but no typing signal was observed with monoclonal antibodies (identified as HSV-2 by PCR). Second, some specimens may be compromised by freezing and thawing, e.g., specimens 2, 3, and 8. However, it is also possible that freezing and thawing may promote cell-associated virions and aggregates of virus to dissociate, creating more virus particles in some specimens and thus an increased number of antigen-expressing cells, resulting in improved antibody detection.
Four specimens were ELVIS positive but remained untypeable (no type available) upon retesting in a rapid format. The isolates in these specimens may be untypeable because of low titers or slowed and/or inhibited viral replication in the ELVIS cells. Reduced antigen production in fewer cells is likely to reduce the amount of bound antibody and, subsequently, a reduced detectable fluorescent signal. Two specimens (specimens 5 and 11) contained only one blue cell per well and one specimen (specimen 15) contained only three blue cells, suggesting that these specimens indeed contained very low virus titers. The inability to type the HSV isolate in specimen 7 by ELVIS reculture cannot be explained by the presence of too few HSV-infected cells. This specimen had retested ELVIS positive in duplicate wells with a relatively large number of blue-stained cells (17 and 34, respectively). Some toxicity was noted when viewing this specimen for blue-stained cells. Neither a cytopathic effect nor viral foci were evident. However, the color in the stained cells was not noticeably reduced. The possibility that the specimen contained a virus which nonspecifically induced the expression of the ELVIS cell reporter gene is most unlikely since PCR analyses with two different primer sets identified the specimen as HSV positive. It is possible that with an extended incubation time, i.e., greater than 20 h postinoculation, virus spread would have been enhanced and the typing reactivity would be improved. Alternatively, the presence of an antiviral agent such as acyclovir in a specimen at a sufficient concentration could lead to such a finding. The induction of the infected cell protein 6 promoter in HSV-infected ELVIS HSV Id cells occurs prior to DNA synthesis and virus yield. Thus, the presence of acyclovir during rapid ELVIS HSV Id culture could result in blue-stained cells but minimal to no HSV glycoprotein antigen production. We have shown that when the virus is amplified for longer periods in tube culture and supernatant is inoculated onto ELVIS HSV Id cells, typing with either Syva or ELVIS HSV Id/Typing results in a strong, easy-to-read fluorescent signal. The degree, relative frequency, and potential effect of antiviral agents on culture detection of HSV are the subject of further study.
In summary, this retrospective study of 26 low-titer specimens has shown that ELVIS HSV Id is a very sensitive and specific test system for the rapid detection of HSV infections. ELVIS HSV-positive, antigen-nonreactive specimens containing low virus titers were confirmed to be true HSV-positive specimens by retesting with ELVIS or by PCR. The HSV types for all except one of the specimens were resolved by combining retesting with ELVIS with two different antibody reagent kits and PCR. Additionally, the development of the ELVIS HSV Id/Typing kit offers the distinct advantage of being able to use a single ELVIS HSV Id monolayer with specimens submitted for HSV typing. The single-well procedure provides a cost-effective, labor-saving alternative to the use of double shell vials or the two-well ELVIS typing method with Syva reagents.
ACKNOWLEDGMENTS
We thank Steven Lobel for helpful editorial discussions and are grateful to SmithKline Beecham Clinical Laboratories for purchase of the ELVIS HSV Id kits from DHI for use in this study.
REFERENCES
- 1.Arvin A M, Prober C G. Herpes simplex virus type 2—a persistent problem. N Engl J Med. 1997;337:1158–1159. doi: 10.1056/NEJM199710163371609. [DOI] [PubMed] [Google Scholar]
- 2.Ashley R L, Dalessio J, Sekulovich R E. A novel method to assay herpes simplex virus neutralizing antibodies using BHKICP6LacZ-5 (ELVIS®) cells. Viral Immunol. 1997;10:213–220. doi: 10.1089/vim.1997.10.213. [DOI] [PubMed] [Google Scholar]
- 3.Lakeman F D, Whitley R J the National Institute of Allergy and Infectious Diseases Collaborative Antiviral Study Group. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlation with disease. J Infect Dis. 1995;171:857–863. doi: 10.1093/infdis/171.4.857. [DOI] [PubMed] [Google Scholar]
- 4.Olivo P D. Transgenic cell lines for detection of animal viruses. Clin Microbiol Rev. 1996;9:321–334. doi: 10.1128/cmr.9.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proffitt M R, Schindler S A. Rapid detection of HSV with an enzyme-linked virus inducible system (ELVIS®) employing a genetically modified cell line. Clin Diagn Virol. 1995;4:175–182. doi: 10.1016/0928-0197(95)00011-v. [DOI] [PubMed] [Google Scholar]
- 6.Schlesinger Y, Buller R S, Brunstrom J E, Moran C J, Storch G A. Expanded spectrum of herpes simplex encephalitis in childhood. J Pediatr. 1995;126:234–241. doi: 10.1016/s0022-3476(95)70550-3. [DOI] [PubMed] [Google Scholar]
- 7.Stabell E C, Olivo P D. Isolation of a cell line for rapid and sensitive histochemical assay for the detection of herpes simplex virus. J Virol Methods. 1992;38:195–204. doi: 10.1016/0166-0934(92)90110-y. [DOI] [PubMed] [Google Scholar]
- 8.Stabell E C, O'Rourke S R, Storch G A, Olivo P D. Evaluation of a genetically engineered cell line and a histochemical beta-galactosidase assay to detect herpes simplex virus in clinical specimens. J Clin Microbiol. 1993;31:2796–2798. doi: 10.1128/jcm.31.10.2796-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tebas P, Scholl D, Jollick J, McHarg K, Arens M, Olivo P D. A rapid assay to screen for drug-resistant herpes simplex virus. J Infect Dis. 1998;177:217–220. doi: 10.1086/517357. [DOI] [PubMed] [Google Scholar]