Abstract
Reprogramed cellular metabolism is one of the most significant hallmarks of cancer. All cancer cells exhibit increased demand for specific amino acids, and become dependent on either an exogenous supply or upregulated de novo synthesis. The resultant enhanced availability of amino acids supports the reprogramed metabolic pathways and fuels the malignant growth and metastasis of cancers by providing energy and critical metabolic intermediates, facilitating anabolism, and activating signaling networks related to cell proliferation and growth. Therefore, pharmacologic blockade of amino acid entry into cancer cells is likely to have a detrimental effect on cancer cell growth. Here we developed a nanoplatform (LJ@Trp-NPs) to therapeutically target two transporters, SLC6A14 (ATB0,+) and SLC7A5 (LAT1), that are known to be essential for the sustenance of amino acid metabolism in most cancers. The LJ@Trp-NPs uses tryptophan to guide SLC6A14-targeted delivery of JPH203, a high-affinity inhibitor of SLC7A5. In the process, SLC6A14 is also down-regulated. We tested the ability of this strategy to synergize with the anticancer efficacy of lapatinib, an inhibitor of EGFR/HER1/HER2-assocated kinase. These studies show that blockade of amino acid entry amplifies the anticancer effect of lapatinib via interference with mTOR signaling, promotion of apoptosis, and suppression of cell proliferation and metastasis. This represents the first study to evaluate the impact of amino acid starvation on the anticancer efficacy of widely used kinase inhibitor.
Keywords: Amino acid delivery; ATB0,+ targeting; Nanoparticles; Cancer therapy; mTOR
Graphical abstract
Highlights
-
•
Blockade of amino acid uptake synergizes Lapatinib for enhanced anticancer therapy.
-
•
Tryptophan-conjugated nanoparticles target SLC6A14 for precise cancer drug delivery.
-
•
SLC6A14 was downregulated in the uptake of SLC6A14-targeted nanoparticles.
-
•
JPH203 inhibits SLC7A5 to deactivate mTOR signaling.
-
•
Nanoparticle block amino acid delivery to starve cancer cells.
1. Introduction
Cancer is a major public health problem worldwide. There are many methods to treat cancers, including surgery, radiation therapy, chemotherapy, and immunotherapy. Surgery and radiotherapy are usually applied for primary or large metastases [1]. Chemotherapy is used for disseminated tumors or as an adjunctive treatment. However, many chemotherapeutics have experienced clinical failure due to the intrinsic/acquired resistance [2]. The tolerance and desensitization make cancer refractory. Combination chemotherapy is used to overcome resistance, but multidrug resistance still remains a hurdle in effective cancer therapy [3]. Lapatinib, an inhibitor of tyrosine kinase associated with EGFR/HER1/HER2, has produced a remarkable response in clinics for the treatment of breast cancer, but the development of drug resistance has become a challenge [4]. A combination of chemotherapeutics with non-identical molecular targets might avoid or delay the occurrence of resistance and synergistically enhance anticancer therapy.
Cancer cells reprogram the metabolic pathways to optimize and benefit their survival, growth, and proliferation [[5], [6], [7], [8], [9], [10]]. Apart from the well-known Warburg effect [11], cancer cells also have an increased metabolic demand for amino acids, which generates ATP by glutaminolysis, maintains the intracellular redox homeostasis by NADPH and glutathione synthesis, provides carbon and nitrogen in the synthesis of purines, pyrimidines, lipid and cholesterol, activates specific signaling pathways, and promotes protein synthesis [11]. This increased demand is met by upregulating specific transporters for enhanced entry of amino acids into cells [10]. Sodium- and chloride-dependent neutral and basic amino acid transporter B(0+) (ATB0,+, SLC6A14) is a unique amino acid transporter with capacity for the concentrative influx of 18 of the 20 proteinogenic amino acids [12]. It is upregulated in multiple cancers compared to corresponding normal tissues [13]. Inhibition of ATB0,+ in cancer cells could result in amino acid deprivation, interference with mTOR and HIF1α signaling, and promotion of autophagy followed by apoptosis, with net suppression of cell proliferation. L-type/large neutral amino acid transporter-1 (LAT1, SLC7A5) is an obligatory exchanger for neutral amino acids; it is also upregulated in many types of tumors [14]. Both ATB0,+ and LAT1 transport leucine, which stimulates the kinase activity of mTOR and initiates a signaling cascade, facilitating protein synthesis and cell growth/survival [15]. Targeting these transporters to block amino acid entry into cancer cells could lead to cell starvation, impaired redox homeostasis, disrupted signaling networks, and even cell death.
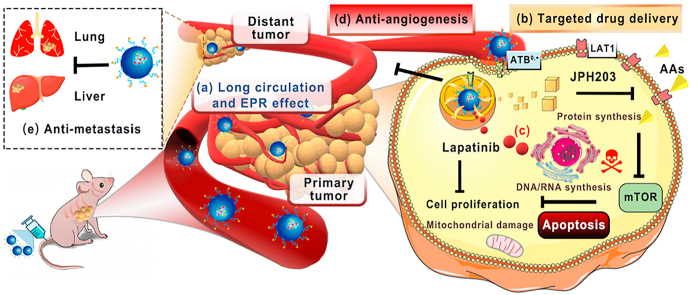
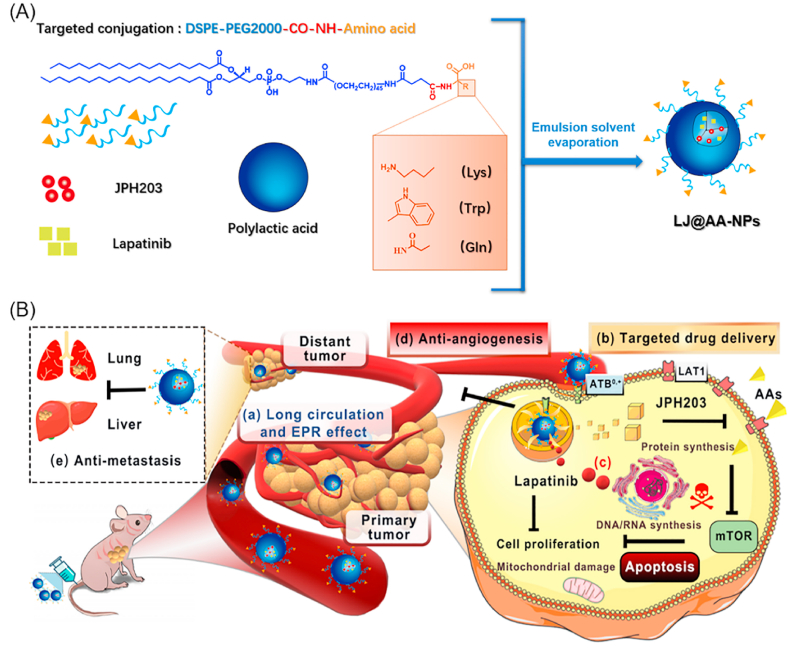
Besides as the pharmacological targets, the plasma membrane transporters with differential expression patterns in tumors compared to normal tissues could also provide novel routes for nano drug delivery systems [16,17]. We have demonstrated ATB0,+-targeted nanoparticles for tumor-specific drug delivery [18,19]. What's interesting is that the application of blank ATB0,+-targeted nanoparticles in cancer cells transiently down-regulates ATB0,+ [18,19]. In most cases, high-level expression of specific nutrient transporters in tumors is related to the poor prognosis [[20], [21], [22], [23]]. We hypothesized that, with ATB0,+-targeted nanoparticles, we could not only suppress the transport function of ATB0,+ but also block the function of any other transporter by delivering a high-affinity inhibitor for the latter as a cargo in the nano-delivery system. In the present study, we chose LAT1 as the target for the ATB0,+-targeted delivery of a selective inhibitor in a nano-formulation. We also used this approach to reset the cancerous amino acid metabolism to determine if this could synergize with a tyrosine kinase inhibitor in providing robust anticancer efficacy (Fig. 1).
Fig. 1.
Schematic illustration for the design of ATB0,+-targeted nanoparticles and its application for cancer therapy. (A) The design and construction of lapatinib/JPH203-coloaded amino acid-conjugated nanoparticles (LJ@AA-NPs). (B) The anticancer mechanism of LJ@AA-NPs. (a) Upon injection, LJ@AA-NPs exhibit prolonged half-life in circulation and accumulate in tumors. (b) The conjugated amino acid interacts with ATB0,+ on tumor-cell surface and mediates the enhanced uptake of NPs; ATB0,+ protein gets down-regulated in the process. (c) After endocytosed into cells, lapatinib and JPH203 are released inside the cells; JPH203 blocks LAT1-mediated amino acid delivery, which inactivates mTOR function. The decreased function of ATB0,+ due to its down-regulation also contributes to amino acid starvation in cancer cells. Lapatinib exerts anticancer effect by itself by blocking the growth-promoting signaling of EGFR/HER1 and HER2. LJ@AA-NPs also (d) prevent angiogenesis and (e) exert anti-metastatic effects.
2. Materials and methods
2.1. Materials
Lapatinib was obtained from Aladdin Biochemical Co., Ltd. (Shanghai, China). JPH203 was obtained from AZBIOCHEM Co., Ltd. (Shanghai, China). DSPE-PEG2000 and DSPE-PEG2000-NHS were purchased from AVT (Shanghai) Pharmaceutical Co., Ltd. Annexin V-fluorescein isothiocyanate (FITC), propidium iodide (PI), and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were purchased from Beyotime Biotechnology Co., Ltd. Primary antibodies to mTOR (7C10), p-mTOR (D9C2), Bax (2772S), Bcl-2 (3498S), and β-actin (4970S) were purchased from Cell Signaling Technology Inc. (Denver, CO, USA). Primary antibody to SLC6A14 (99102) was purchased from Abcam (Shanghai, China). Primary antibody to SLC7A5 (DF8065) was purchased from Affinity Biosciences (Pottstown, PA, USA). 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiR) and l-Glycine were purchased from Dalian Meilun Biotechnology Co., Ltd. (Dalian, China). l-Lysine (56-87-1), l-Glutamine (56-85-9) and 2-Aminobicyclo[2.2.1]heptane-2-carboxylic acid (BCH) were obtained from Aladdin Biochemical Co., Ltd. (Shanghai, China). L-Tryptophan (231123) was obtained from J&K Scientific Co., Ltd. (Beijing, China). All other chemicals and reagents were of analytical grade.
2.2. Cell lines and animals
MCF7, T47D, MB231, and human umbilical vein endothelial cells (HUVEC) were obtained from the cell bank of type culture collection of Chinese Academy of Sciences (Shanghai, China). MCF7, T47D, and MB231 cells were grown in RPMI medium; HUVEC cells were cultured in Dulbecco's modified Eagle's medium (DMEM). All media were supplemented with 10% FBS, 50 units/mL streptomycin, and 100 units/mL penicillin. Cells were all cultured at 37 °C in an incubator with 5% CO2.
Female BALB/c-null mice (4–5 weeks old) and Sprague-Dawley (SD) rats (200–220 g) were purchased from Experimental Animal Center of Wenzhou Medical University. All animals were fed under a specific pathogen-free condition and housed in a barrier facility. All animal studies were carried out in accordance with the Guidelines for Animal Experimentation of Wenzhou Medical University, and the Animal Ethics Committee of the institution approved the protocol.
2.3. Synthesis and characterization of amino acid-conjugated polymers
Tryptophan, lysine and glutamine were selected as the ligand candidates for targeting cancer cells that show robust expression of ATB0,+. Briefly, each ligand candidate and DSPE-PEG2000-NHS (10:1, molar ratio) were dissolved in dimethyl sulfoxide, and the solution was stirred overnight at room temperature under nitrogen. Trimethylamine, equimolar to DSPE-PEG2000-NHS, was also added to facilitate the reaction. The reaction mixture was then dialyzed against double-distilled water for purification, and the resultant solution was freeze-dried. The structure of the prepared amino acid-conjugated PEG2000-DSPE was confirmed by a Bruker 1H NMR (AVANCE III, Rheinstetten, Germany).
2.4. Preparation of dual-drug loaded ATB0,+-targeting nanoparticles
Both lapatinib and JPH203 are hydrophobic. Therefore, a modified emulsion evaporation method was used to prepare the amino acid-conjugated nanoparticles [24,25]. Briefly, poly(lactic acid) (PLA), amino acid conjugated-PEG2000-DSPE, lapatinib (L) and JPH203 (J) with the mass ratio of 10:1:0.6:0.6 were dispersed in a solution of formic acid/dichloromethane (1/100, v/v). Formic acid was added to improve the solubility of JPH203 in organic solvent. This solution was added into 1% (w/v) polyvinyl alcohol (PVA) solution at a volume ratio of 1:3, and then immediately sonicated on ice with an ultrasonic homogenizer (Xinzhi JY92, Ningbo, China). The resultant emulsion was stirred for 6 h at room temperature in a fume hood to evaporate the organic solvent. The nanoparticle solution was then washed thrice with double-distilled water and then freeze-dried. The achieved nanoparticles were weighted, and the yield was calculated. Tryptophan-, lysine-, and glutamine-conjugated nanoparticles were denoted as LJ@Trp-NPs, LJ@Lys-NPs, and LJ@Gln-NPs, respectively. The dual-drug loaded nanoparticles without conjugation (LJ@NPs) were prepared by replacing amino acid-conjugated-PEG2000-DSPE with DSPE-PEG2000. When coumarin 6 (C6) or DiR was used as a probe, the nanoparticles were prepared by replacing lapatinib and JPH203 with C6 or DiR.
2.5. Characterization of amino acid-conjugated NPs
Particle size, size distribution, and zeta potential were determined by a NanoZetasizer (Zetasizer Nano, Malvern, UK). The morphology of NPs was visualized by transmission electron microscope (TEM) (JEM 1200EX, JEOL, Japan). A drop of sample solution was placed onto the carbon-coated copper grid and taped with a filter paper. Phosphotungstic acid (0.01%, v/v) was used for negative staining. The ultraviolet (UV) absorption spectra of nanoparticles ranging from 250 nm to 600 nm was detected by a UV spectrophotometer (UV6100S, MAPADA, Shanghai, China). Lapatinib or JPH203 was dissolved in DMSO (20 μg/mL), and the same drug concentration in the form of J@NPs, L@NPs, LJ@NPs, LJ@Trp-NPs, LJ@Lys-NPs and LJ@Gln-NPs was used for detection.
The loaded amount of lapatinib and JPH203 in NPs was measured with a high-performance liquid chromatography (HPLC) system (Agilent 1290 Infinity, USA), equipped with a Zorbax Elipse XDB C18 column (5 μm, 4.6 × 150 mm, Agilent, USA) at a column temperature of 35 °C. The mobile phase was composed of acetonitrile, methanol and water (50/30/20, v/v/v), and the flow rate was set as 1 mL/min, with detect wavelength at 284 nm. The drug encapsulation efficiency (EE) and drug load (DL) was calculated according to the following equations:
The in vitro release behaviors of lapatinib and JPH203 were measured using a dialysis method [26]. In brief, the freeze-dried nanoparticles were firstly dispersed in double-distilled water to prepare NP solution (1 mg/mL of lapatinib). 2 mL NP solution was added into the dialysis bag and sealed. Then the dialysis bag was immersed in 10 mL solution medium and shaken at 100 rpm in a 37 °C oscillator (88A, Nuoji, Changzhou, China). At specified time intervals, 4 mL of sample was taken from the release medium. After filtering with a 0.22 μm filter membrane (Jinteng, Tianjin, China), the drug concentration in medium was determined by HPLC as described above. The in vitro release test was performed in PBS, pH 7.4 and pH 5.5. The stability of NPs in PBS (pH 7.4) or PBS containing 10% FBS (simulation in vivo) was investigated by monitoring the changes in particle size and PDI. The dilution stability was also studied.
Hemolysis assay was carried out to evaluate the biosafety of NPs when used in animals. Briefly, whole blood, freshly collected from rats, was washed thrice with saline by centrifuging at 1500 rpm for 5 min until the supernatant had no red color, and an appropriate amount of red blood cells was diluted to 2% (v/v) for use. Equal volumes of NP solutions were mixed with the red blood cell suspensions and incubated for 12 h at 37 °C in a water bath shaker. The mixture was then centrifuged at 1500 rpm for 10 min and photographed. 0.09% NaCl solution and 0.9% NaCl solution (saline) were used as positive and negative control, respectively. The degree of cell lysis was quantified by measuring the absorbance at 570 nm.
2.6. RNA extraction and polymerase chain reaction (PCR)
Total RNA was isolated from MCF7 and T47D cells with Trizol (Invitrogen, Carlsbad, CA, USA), and reverse transcription of total RNA was performed with Tli RNaseH Plus Reverse Transcription Kit (RR820A, TaKaRa, Japan). After cDNA was amplified in Thermal Cycler (T100, Bio-Rad, Hercules, CA, USA), the products were electrophoresed with Ultra GelRed Nucleic Acid Stain (GR501-01, vazyme, Nanjing), and then analyzed by a chemiluminescence imager (12003153, BIORAD). Quantitative PCR (qPCR) was performed with FastSYBR Green qPCR Master Mix (Life Technologies) and a fluorescence quantitative real-time PCR machine (Roche LightCyele480, USA). HPRT1 mRNA was used as a reference control. Primers were: hSLC6A14: 5′- CAAATCGTCTGGCAAGGTGG-3’ (sense) and 5′-GCAACTAAGCCACCCCAA -3’ (antisense); hSLC7A5: 5′- GACTACGCCTACATGCTGGA-3’ (sense) and 5′- GTGAGCAGCAGCACGCA-3’ (antisense); hHPRT1: 5′- GCGTCGTGATTAGCGATGAAC-3’ (sense) and 5′- CCTCCCATCTCCTTCATGACATCT-3’ (antisense).
2.7. MTT assay
The synergistic effect of lapatinib and JPH203 was evaluated in MCF7 and T47D cells to optimize the L/J drug ratio formulated in NPs. Briefly, MCF7 and T47D cells were seeded in a 96-well plate and allowed to grow for 12 h for adhesion. L, J, and L + J (L/J ratio ranging from 0.5 to 4) were incubated with the cells for 24 h. Afterward, 10 μL of MTT reagent (5 mg/mL) was added to each well and incubated for 4 h, and then 150 μL of DMSO was added to dissolve the resultant formazan. The absorbance at 490 nm was measured using a microplate reader (Infinite M200 pro, TECAN, Switzerland). The combination index (CI) of lapatinib and JPH203 was calculated as follows:
DL or DJ represents the combined concentration for lapatinib or JPH203 to achieve X% cytotoxicity, and DL.X or DJ.X indicates the concentration of single drug to achieve X% cytotoxicity. The in vitro anticancer effect of drug-loaded NPs was also assessed by MTT assay. Briefly, MCF7 or T47D cells were seeded into a 96-well plate. After 24-h culture, the cells were treated with NPs at different concentrations (0.02–20 μg/mL) in an incubator for 24 h. The rest of the procedure was as same as that described above. The IC50 value was calculated using GraphPad Prism 8.0.2.
2.8. In vitro cellular uptake assay
Coumarin 6 (C6) was selected as a probe to investigate the uptake profile of amino acid-conjugated NPs. In brief, MCF7 and T47D cells were seeded in 12-well plates containing coverslips with a density of 4 × 104 cells/well. After the cells grew to the appropriate density, the cells were washed and incubated with free coumarin 6 (C6), C6-loaded nanoparticles (C6@NPs) and C6-loaded amino acid-conjugated nanoparticles (C6@Trp-NPs, C6@Gln-NPs and C6@Lys-NPs) at the same C6 concentration (4 μg/mL) in culture medium. After 0.5 h, 1 h or 12 h treatment, the uptake was stopped by washing the cells thrice with cold PBS. The coverslips were placed sample-side down onto ProLong Diamond Antifade Mountant with DAPI (ThermoFisher, USA). The slides were kept in dark at room temperature for 6 h. The samples were visualized using Leica fluorescence microscope (DM2500, Lycra, Germany). The mean fluorescence intensity was quantitated using Image J software.
To investigate the interaction of nanoparticles with specific target, the colocalization assay was performed as previously reported [27]. Briefly, MCF7 cells were cultured on 12-mm cover glasses in 12-well plate with a density of 3 × 105 cells/well for 24 h. Subsequently, the medium was removed, and the cells were replenished with fresh DMEM containing C6@NPs, C6@Trp-NPs, C6@Gln-NPs or C6@Lys-NPs, respectively, at the same C6 concentration (4 μg/mL). After 1 h incubation at 37 °C, the cells were washed with PBS thrice, fixed with 4% paraformaldehyde for 20 min at 4 °C and incubated with 0.1% Triton X-100 for 10 min at room temperature. After wash another three times with PBS, the cells were incubated with primary antibody to LAT1 or ATB0,+ for 12 h at 4 °C, and then incubated with Alexa Fluor 594-labeled goat anti-rabbit IgG secondary antibody (ab150080) for half-hour at room temperature. The cover slides were then placed sample-side down onto ProLong Diamond Antifade Mountant with DAPI (ThermoFisher, USA) and kept in dark at room temperature for 6 h. The samples were then visualized using Nikon fluorescent microscope (Nikon, Japan).
The competition assay was conducted to investigate the effect of ATB0,+ substrate or LAT1 blocker on the uptake of C6@Trp-NPs, C6@Gln-NPs and C6@Lys-NPs. Briefly, MB231, MCF7 or T47D cells were seeded in 24-well plates with a density of 2 × 105 cells/well. After 12 h for attachment, the cells were pretreated with glycine (1 mM) or BCH (1 mM) for half hour, and then incubated with C6@Trp-NPs, C6@Gln-NPs or C6@Lys-NPs for 1 h. After wash three times with PBS, the cells were lysed by lysis buffer, and the fluorescence of these samples was measured by a microplate reader (Excitation/Emission = 466/504) (Infinite M200 pro, TECAN, Switzerland). The uptake of nanoparticles in normal medium was taken as a control, and the relative uptake was calculated.
To investigate the intracellular traffic of NPs, MCF7 and T47D cells were seeded on coverslips in 12-well plate. Upon confluence, fresh medium containing C6-labeled NPs was added and incubated for 0.5 h for uptake; the plate was kept at 37 °C for 1 h or 3 h to allow the intracellular trafficking of NPs. After washing with cold PBS, the slides were stained with Lyso-Tracker Red (C1046, Beyotime, China) for 20 min and DAPI (blue) for 15 min for lysosomes and nuclei, respectively. The samples were then subjected to confocal laser scanning microscopy (CLSM) (SP8, Leica, Germany).
2.9. Colony formation assay
Colony formation assay was performed to study the inhibitory effect of NPs on cell proliferation. Briefly, MCF7 and T47D cells were seeded into 6-well plates (2000 cells/well). The cells were then treated with lapatinib (L) (0.1 μg/mL), L (0.1 μg/mL) + J (0.1 μg/mL), L@NPs, LJ@NPs, and LJ@Trp-NPs. The medium was changed every two days until the colonies were visible. Subsequently, the cells were washed with cold PBS and fixed with 4% paraformaldehyde for 20 min. The colonies were stained with 1X Giemsa Stain (G1015, Solarbio) (2 mL per well) for 40 min and then washed thrice with PBS, and then photographed. Finally, 1 mL lysis buffer was added to dissolve the stained colonies, and the buffer was collected to determine the absorbance at 630 nm using a microplate reader (Infinite M200 pro, TECAN, Switzerland).
2.10. Wound-healing assay
Wound healing experiment was conducted to explore the inhibitory effect of NPs on cancer cell migration. MCF7 and T47D cells were seeded into 6-well plates at a density of 4 × 105 cells/well and incubated for 24 h at 37 °C to grow a monolayer. Next, a linear wound was generated in the center of the well surface with a pipette tip. The cells were then incubated with different treatments in media containing 2.5% FBS for 36 h. The cells were washed with PBS, and the wound gap was photographed by inverted microscope (Ti, Nikon, Japan) at 0 h and 24 h.
2.11. Angiogenesis assay
The anti-angiogenesis effect of NPs was investigated using HUVEC cells. First, the cell migration assay was conducted. Briefly, MCF7 and T47D cells were seeded in the lower well of transwell plates (2 × 104 cells/well) and treated with different formulations for 12 h. After that, the medium was refreshed as an inductive agent. HUVECs were then seeded onto the upper well of the transwell plates (1 × 104 cells/well) and incubated with serum-free medium for another 24 h. HUVEC cells in the upper well which did not migrate were removed by wiping with a cotton swab, and the migrated cells were fixed with 4% paraformaldehyde and then stained with 1X Giemsa Stain for 30 min. The cells were washed twice with PBS and photographed under a microscope (Ti, Nikon, Japan).
Tube formation assay was also performed. HUVEC cells were seeded into a 96-well plate (2 × 104 cells/well), which was pre-coated with 60 μL of matrigel (3 mg/mL) (BD Biosciences), and then treated with different drug formulations in medium for 24 h. The number of tubes were observed under the microscope and photographed, and also counted by Image J software.
2.12. Apoptosis analysis by flow cytometry
Apoptosis in cells after various treatments was determined by flow cytometry. Briefly, MCF7 and T47D cells were seeded into 6-well plates (5 × 105 cells/well) and treated with L, LJ, L@NPs, LJ@NPs, and LJ@Trp-NPs, at the drug concentration of 10 μg/mL for 24 h. Subsequently, cells were washed, digested, collected and stained with Annexin V/FITC and PI (BD Biosciences, San Jose, CA, USA), and then analyzed by flow cytometry (Beckman coulter, CytoFLEX S, US).
2.13. Western blot
MCF7 cells were seeded in 6-well plate (6 × 105 cells/well). After adhesion, these cells were treated with L, LJ, L@NPs, LJ@NPs, and LJ@Trp-NPs for 24 h. The treated cells were washed thrice with cold PBS and lysed by RIPA buffer containing protease/phosphatase inhibitor cocktails. Total protein was determined by a BCA assay kit (Beyotime, China). The lysed samples (30 μg protein) were loaded on SDS-PAGE, and the separated proteins were transferred to the PVDF membrane (1704156, BIORAD). The membrane was blocked with TBST with 5% bovine serum albumin (BSA) (A8020, Solarbio) for 2 h, and then probed with appropriate primary antibodies overnight at 4 °C. The membrane was washed thrice with TBST and incubated the HRP-coupled secondary antibody for 2 h at room temperature. Immunoreactive bands were soaked with developer solution (BeyoECL Moon, P0018FS, Beyotime) and visualized using a gel imaging analysis system (12003153, BIORAD). β-Actin was used as a control.
To investigate the effect of endocytosis of LJ@Trp-NPs in ATB0,+ expression, the uptake was conducted in 4 °C because the low temperature could suppress the endocytosis process. Briefly, MCF7 cells were seeded in 6-well plate (6 × 105 cells/well). After adhesion, these cells were treated with LJ@Trp-NPs for 24 h at 4 °C and 37 °C, respectively. The following process was the same as described above. In addition, the expression of ATB0,+ in MCF7, T47D, and MB231 cells was also tested.
2.14. Metabolomics study
MCF7 cells were seeded in 10-cm dish at a density of 1 × 105 cells/dish and cultured 12 h. The cells were then treated with different formulations for 48 h (L, 1 μg/mL; J, 1 μg/mL), and the metabolites were extracted as described previously [28]. Briefly, extracellular medium was collected and centrifuged (1500 rpm, 5 min) to prepare the supernatant. 50 μL supernatant, 50 μL distilled water, and 900 μL ice-cold acetonitrile were mixed, vortexed, and allowed to settle down for 60 min. The mixture was then centrifuged at 12000 rpm for 20 min at 4 °C, and the supernatant was collected as the extracellular sample. In addition, the cells were collected into a 1.5 mL tube after centrifugation. The cell pellets were then resuspended in 200 μL ice-cold water and probe-sonicated on ice for 10 sec. 225 μL ice-cold acetonitrile was added to the cell suspension, and the mixture was vortexed and kept on ice for 60 min. After centrifugation (12000 rpm, 10 min), the supernatant was collected as intracellular samples. The samples were analyzed by an ultra-performance liquid chromatography-dual mass spectrometry (UPLC-MS/MS) (Agilent 6400, G6420A QQQ, Agilent, USA).
2.15. Biodistribution of NPs in mice
DiR was utilized as a probe to investigate the in vivo biodistribution of NPs in MCF7 tumor-bearing mice. Briefly, MCF7 cells (100 μL, 5 × 106 cells per mouse) were inoculated onto the left back side of female nude BALB/c mice. After 9 days, the MCF7 tumor-bearing mice were randomly divided into 3 groups. Each group received i.v. injection of free DiR, DiR@NPs or DiR@Trp-NPs, with a DiR dose of 2 mg/kg. The mice were imaged at predetermined time intervals by an in vivo imaging system (Flex, Maestro). After 12 h, the mice were killed, and the tumors and the specific organs were collected for further imaging by the imaging system. Quantitative analysis for each image was performed by the matched software.
2.16. Pharmacokinetic study
The pharmacokinetic profiles of lapatinib or JPH203 from NPs were investigated using female SD rats (200–220 g). Nine rats were randomly divided into three groups, and free LJ, LJ@NPs and LJ@Trp-NPs (2 mg/kg for lapatinib or JPH203) were intravenously injected. Blood samples were collected into the heparinized tubes from the orbital plexus at predetermined intervals and then centrifuged for 10 min at 4 °C (13000 rpm) to obtain plasma. The drug concentration in plasma was determined by UPLC-MS/MS (Agilent 6400, G6420A QQQ, Agilent, USA). The mobile phase was a mixture of 0.1% formic acid in water and 100% acetonitrile in a volume ratio of 80:20. The column temperature was 40 °C, and the flow rate was 0.4 mL/min. Protein precipitation extraction was applied for JPH203 and lapatinib. Compounds were monitored via electrospray ionization positive ion mode (ESI+) using the following conditions: JPH203, m/z 472.0 → 224.2, and lapatinib, m/z 581.2 → 365.1. the standard curves were linear (r2 ≥0.99) within the range of 1–5000 ng/mL.
2.17. In vivo therapy efficacy in animal models
The MCF7 tumor-bearing nude mice were generated as above. When the tumor volume reached ~200 mm3, the mice were randomly divided into six groups (n = 6): saline, L, L + J, L@NPs, LJ@NPs and LJ@Trp-NPs. Each formulation was intravenously injected with a dose of 2 mg/kg on day 0, 2, 4, and 6. The weight and tumor volume were recorded every two days throughout the procedure. The tumor volume was calculated with the formula: tumor volume (mm3) = 0.52 × length × width2. After 18 days, the mice were killed to collect whole blood, tumors and other major organs (heart, liver, spleen, lung, kidney) for further assessment. The blood samples were used to determine the levels of aspartate transaminase (AST, also known as glutamate-oxaloacetate transaminase), alanine transaminase (ALT, also known as glutamate-pyruvate transaminase), blood urea nitrogen (BUN), and creatinine by serum biochemical analysis. The metastatic lesions in lungs were counted and photographed. The tumors were photographed and weighed. The tumors and organs were fixed by 4% paraformaldehyde, embedded and sliced for H&E staining. The tumor sections were also used for immunohistochemical analysis and transferase dUTP nick end labeling (TUNEL) analysis.
2.18. Statistical analysis
Data were analyzed and plotted using GraphPad Prism 8.0.2. Statistical analysis was performed by Student's t-test or one-way ANOVA. Data are presented as means ± SD (n ≥ 3). Statistically, significant differences compared to the indicated group were defined as *, p < 0.05, **, p < 0.01, ***, p < 0.001.
3. Results and discussion
In our previous study, we developed ATB0,+-targeted nanoparticles (NPs) by conjugating glycine, aspartate, and lysine on the surface of NPs, and found lysine-conjugated NPs displayed the best targeting efficiency [18,19]. It has been well established that ATB0,+-mediated glutamine entry into cells fuels glutaminolysis in specific cancers [13]. α-Methyl-tryptophan, a derivative of tryptophan, is a specific blocker of ATB0,+ [28]. Therefore, to further optimize the targeting efficiency of NPs, we conjugated lysine, glutamine, and tryptophan to the hydrophilic end of DSPE-PEG2000 to modify NPs. DSPE-PEG2000 conjugated with these three amino acids were synthesized and confirmed by 1H NMR (Fig. S1).
Our goal was to investigate the anticancer efficacy of lapatinib, an inhibitor of the EGFR (HER1)/HER2 receptor kinase, and JPH203, a high-affinity inhibitor of the amino acid transporter LAT1, if formulated as a cargo in NPs with selective targeting to cancer cells via ATB0,+. As shown in Fig. S2, the proportion of lapatinib/JPH203 was optimized by MTT assay in T47D and MCF7 cells, both being positive for robust expression of ATB0,+ [12]. Four different proportions ranging from 1:0.5 to 1:4 were evaluated. JPH203 by itself showed less toxicity towards both cells compared with lapatinib. But, JPH203 significantly increased the anticancer effect of lapatinib in both T47D and MCF7 cells (Figs. S2A and B), corroborated by the relative IC50 values (Figs. S2C and D). This shows that inhibition of LAT1-mediated amino acid supply to cancer cells synergizes with the anticancer effect of lapatinib. The synergistic effect was further confirmed by the analysis of the combination index (CI) for lapatinib and JPH203 (Figs. S2E and F). The CI values for all four different ratio formulations were smaller than 0.9, strongly indicating synergism. As there was very little difference in CI values among the four different ratios, we selected the 1:1 ratio for subsequent studies with NP formulations.
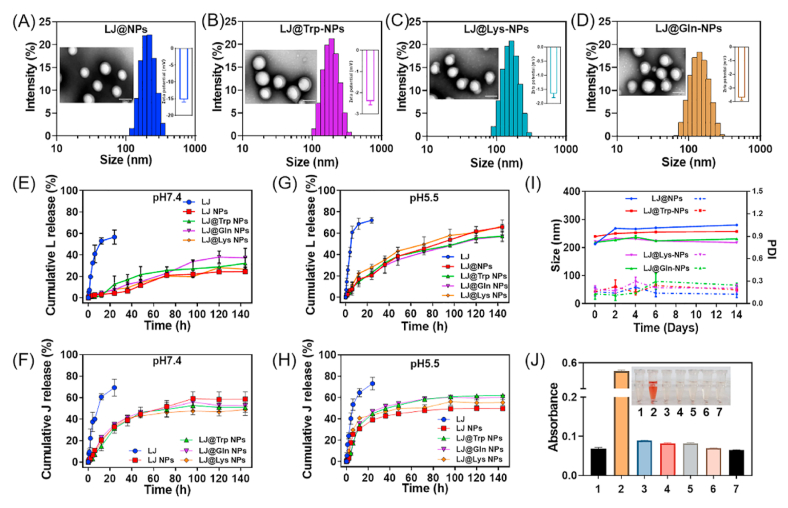
Lapatinib (L) and JPH203 (J)-coloaded glutamine (Gln)-, lysine (Lys)-, and tryptophan (Trp)-conjugated NPs (LJ@Gln-NPs, LJ@Lys-NPs, and LJ@Trp-NPs) were prepared by a modified emulsified method. DSPE-PEG2000 without conjugation with amino acids was used to prepare control NPs (LJ@NPs). These four types of NPs showed narrow size distribution and sphere shape (Fig. 2A–D). The hydrodynamic diameter was ~150 nm, and the PDI value was smaller than 0.1 (Table S1), indicating uniform distribution. The surface potential of LJ@NPs was −15.30 ± 0.71 mV (Fig. 2A and Table S1). Conjugation with amino acids decreased the potential close to zero (Fig. 2B–D and Table S1). The yield of four type of nanoparticles all exceeded 80% (Table S1). Both lapatinib and JPH203 showed excellent loading efficiency in NPs; the encapsulation efficiency (EE) for each drug was higher than 70%, and the drug load (DL) surpassed 4% (Table S1). The UV-VIS spectroscopy was used to confirm similar drug loading in different NP formulations (Fig. S3). The in vitro release profile of lapatinib and JPH203 from NPs was investigated in PBS at pH 7.4 and pH 5.5 (Fig. 2E–H). All NPs exhibited sustained drug release profiles, while JPH203 was released slightly faster than lapatinib from NPs. Additionally, the lapatinib and JPH203 released at a faster rate at pH 5.5 as compared to released at pH 7.4, indicating their pH-sensitive release characteristics. What should be mentioned is that JPH203 is more soluble in acid solution, which contributed to accelerated nanoparticle dissociation and drug release. Thereby, JPH203 and lapatinib were released faster in an acid medium as compared to a neutral medium. The stability assay showed that the NPs remained stable over a period of time under various experimental conditions, evidenced by the negligible changes of particle size and PDI (Fig. 2I and S4). We assessed the biocompatibility of these NPs by hemolysis assay. As shown in Fig. 2J, the NPs showed very little hemolysis compared with the positive control, indicating good biocompatibility and hence suitability for intravenous administration.
Fig. 2.
Characterization of lapatinib/JPH203-coloaded amino acid-targeted NPs. The size distribution, TEM image (Scale bar = 100 nm) and zeta potential of (A) LJ@-NPs, (B) LJ@Trp-NPs, (C) LJ@Lys-NPs, and (D) LJ@Gln-NPs. The cumulative release of (E) Lapatinib (L) or (F) JPH203 (J) from free LJ, LJ@NP, LJ@Trp-NP, LJ@Gln-NP, LJ@Lys-NP in pH 7.4 PBS at 37 °C. The cumulative release of (G) Lapatinib (L) or (H) JPH203 (J) from free LJ, LJ@NP, LJ@Trp-NP, LJ@Gln-NP, LJ@Lys-NP in pH 5.5 PBS at 37 °C. (I) The stability of LJ@NP, LJ@Trp-NP, LJ@Gln-NP, LJ@Lys-NP was indicated by the changes of particle size and polydispersity index (PDI) in pH 7.4 PBS at 37 °C. (J) The absorbance of the supernatant after incubation of blood cells with the following formulations: 1. saline, 2. 0.09% NaCl solution, 3. blank NP, 4. LJ@NP, 5. LJ@Lys-NP, 6. LJ@Trp-NP, and 7. LJ@Gln-NP. The results were presented as mean ± SD (n = 3).
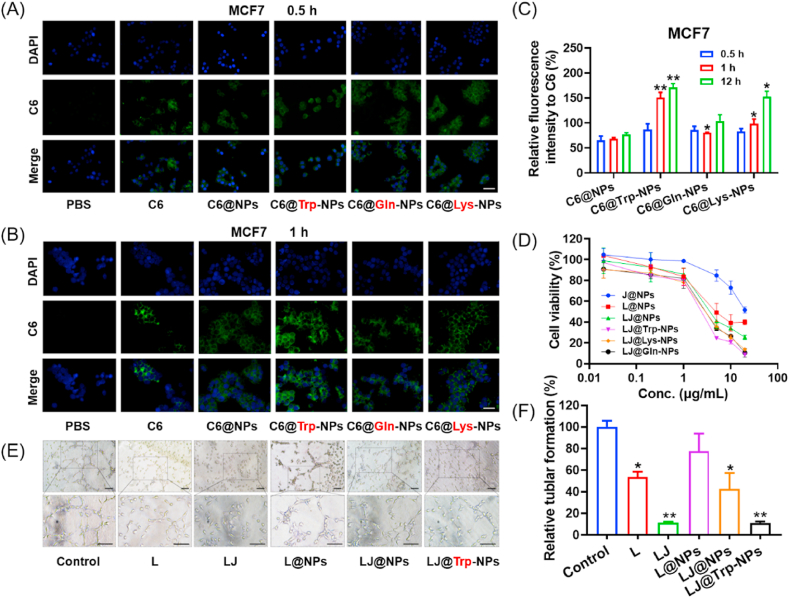
The uptake and anticancer effect of these four types of NPs were assessed in MCF7 and T47D cells. Both cell lines have robust expression of ATB0,+ [12]. We confirmed this again by RT-PCR and qPCR; in addition, we also evaluated the expression of LAT1 (Fig. S5). Both cell lines expressed high levels of ATB0,+ and LAT1. Therefore, these two cell lines were used for the following in vitro study. To investigate the uptake of NPs, coumarin 6 (C6) was selected as a fluorescence probe. As shown in Fig. 3A and B, weak fluorescence was observed for all four groups of NPs in MCF7 cells after 0.5 h incubation, and there was no significant difference among the different formulations. After 1 h incubation however, the fluorescence increased, and C6@Trp-NPs displayed the highest intensity compared to other groups. Lys- and Gln-conjugation also guided the increased uptake of NPs, but to a lesser degree than Trp conjugation. When the incubation time was increased to 12 h (Fig. S6), all three conjugations showed further enhanced fluorescence intensity, and C6@Trp-NPs still hold the highest signal. Quantitative analysis with normalization to the level of free C6 confirmed the superiority of Trp-NPs for uptake into these cells (Fig. 3C). The results were similar in T47D cells (Fig. S7). MTT assay was then performed for the anticancer efficacy of the NPs in vitro. As shown in Fig. 3D, dual drug loaded NPs showed enhanced anticancer effect than the single drug loaded NPs. In addition, all three NPs, LJ@Trp-NPs. LJ@Gln-NPs, and LJ@Lys-NPs, showed much lower IC50 values than bare LJ@NPs (Fig. S8C). LJ@Trp-NPs showed the lowest IC50 value, which was consistent with the uptake assay, indicating that Trp conjugation is superior to glutamine or lysine for optimal targeting efficiency and outcome. MTT results in T47D cells were consistent with those in MCF7 cells (Figs. S8A and B), further confirming the enhanced anticancer efficacy of LJ@Trp-NPs.
Fig. 3.
In vitro performance of amino acid-conjugated NPs in MCF7 cells and HUVEC cells. The uptake profiles of amino acid-conjugated NPs in MCF7 cells after incubation with different formulations for (A) 0.5 h and (B) 1 h. Scale bar = 50 μm. (C) Quantitative analysis of uptake normalized to free C6. C6@NPs group was selected as a control for comparison. (D) MTT assay in MCF7 cells. (E) HUVEC tube formation in Matrigel with various drug treatments (scale bar = 100 μm); (F) quantitative analysis; the untreated group was selected as a control for comparison. Data are presented as mean ± SD (n = 3). *, p < 0.05, **, p < 0.01, compared with the control group.
To further investigate the targeting efficiency of amino acid-conjugated nanoparticles, colocalization experiments were conducted to test the interaction of nanoparticles with SLC6A14. As shown in Fig. S9A, C6@Lys-NPs and C6@Trp-NPs showed significant overlap of green and red (yellow), indicating the interaction of nanoparticles with SLC6A14. Specifically, Trp-NPs showed the strongest yellow signal, indicating the best targeting efficiency. These results were consistent with the cellular uptake assay. In addition, amino acid transporters usually have overlapped substrates, which might result in one ligand targeting two transporters. Therefore, we further tested the interaction of these nanoparticles with other transporters, and SLC7A5 was selected (Fig. S9B). There is also a slight overlap of green and red, but comparable distinct, indicating the interaction between nanoparticles and SLC7A5 was also exist but weak. These results suggested that Trp-NPs possessed strong interaction activity with SLC6A14, which contributed to SLC6A14 (ATB0,+) targeted delivery. Additionally, Trp-NPs could also interact with SLC7A5 in a relatively weak way as the reviewer guessing. Following that, the uptake of these nanoparticles in the presence of glycine (a substrate of ATB0,+) or BCH (a blocker of LAT1) was investigated. In both MCF7 (Fig. S10C) and T47D (Fig. S10D) cells, glycine could decrease the uptake of Trp-NPs and Lys-NPs, but not significant for Gln-NPs; BCH could also slightly decrease the uptake of Trp-NPs to some extent, but this difference was not statistically significant. These data suggested that the enhanced uptake of Trp-NPs was mainly attributed to the ATB0,+-targeting property, which was consistent with the colocalization study. In MB231 cells, an ATB0,+-negative cell line (Fig. S10A), the uptake profiles of all three amino acid-conjugated nanoparticles were similar to that of bare nanoparticles (Fig. S10B), further confirming the dominant role of ATB0,+ in the uptake of Trp-NPs. Based on these data, we selected LJ@Trp-NPs as the ATB0,+-targeted nanoparticles for further evaluation.
ATB0,+-targeted NPs enter cells via a endocytosis. Therefore, it was critical to determine whether the NPs release the cargo into cytoplasm instead of limiting the release to the endosomes. For this, we tracked the intracellular trafficking of the NPs by CLSM. C6 (green) was used to track Trp-NPs, Lysotrack (red) was used to label lysosomes, and DAPI (blue) was used to label nuclei. As shown in Fig. S11, most of C6@Trp-NPs was found attached to the plasma membrane in the first 0.5 h, and only a small portion entered cells via endocytosis and was found located in endosomes (evidenced by the very slight yellow signal). The C6@Trp-NPs were then removed, and the attached nanoparticles were allowed for their subsequent intracellular trafficking. At 1 h, most C6@Trp-NPs were co-localized with lysotrack (yellow signal), indicating that C6@Trp-NPs were endocytosed. After 2 h, the yellow signal almost disappeared, indicating the exit of the NPs from the endosomes into the cytoplasm.
The anti-proliferation and anti-metastasis properties of LJ@Trp-NPs were then evaluated by colony formation assay and wound-healing assay, respectively. As shown in Fig. S12A, free LJ displayed significant anti-proliferation effect in both MCF7 and T47D cells. LJ@Trp-NPs showed prominent anti-proliferation effect compared to other groups in both cell lines. The quantitative analysis further confirmed these results (Fig. S12B). The enhanced anti-proliferation efficacy of LJ@Trp-NPs compared to bare LJ@NPs could be attributed to the ATB0,+-mediated uptake. The drug-loaded NPs showed a slightly lesser effect than the corresponding free drug, which might be due to the slow release of the drugs from NPs. Wound healing assay in Fig. S13 showed that LJ@Trp-NPs significantly suppressed the closure rate in both cell lines compared to control and LJ@NPs, suggesting potentially enhanced anti-metastasis properties. We believe that the synergism between lapatinib and JPH203 and the ATB0,+-targeting of NPs contributed to the significantly enhanced inhibitory effect on cancer cell migration. Taking collectively, LJ@Trp-NPs exhibit robust anticancer effect by synergistic inhibition of cell proliferation and cell migration.
HUVEC cells form capillary-like structures in culture. We used these cells to perform endothelial cell tube formation assay to study the anti-angiogenesis effect of LJ@Trp-NPs. As shown in Fig. 3E, the control group showed capillary-like structures, and the combination of lapatinib and JPH203 suppressed this process significantly. LJ@Trp-NPs also caused significantly greater inhibition of tube formation compared to the control group and LJ@NPs. The inhibition was ~90%, close to the inhibitory effect of free lapatinib + JPH203. Tumor microenvironment also affects the vascular properties. We investigated the effect of LJ@Trp-NPs on tumor cell-elicited vascular differentiation, elongation, and migration. As shown in Fig. S14, LJ@Trp-NPs significantly suppressed the HUVEC migration. These results suggest that LJ@Trp-NPs could also inhibit angiogenesis in the tumor microenvironment, which would potentiate the amino acid deprivation effect and consequently amplify the anticancer effect of LJ@Trp-NPs.
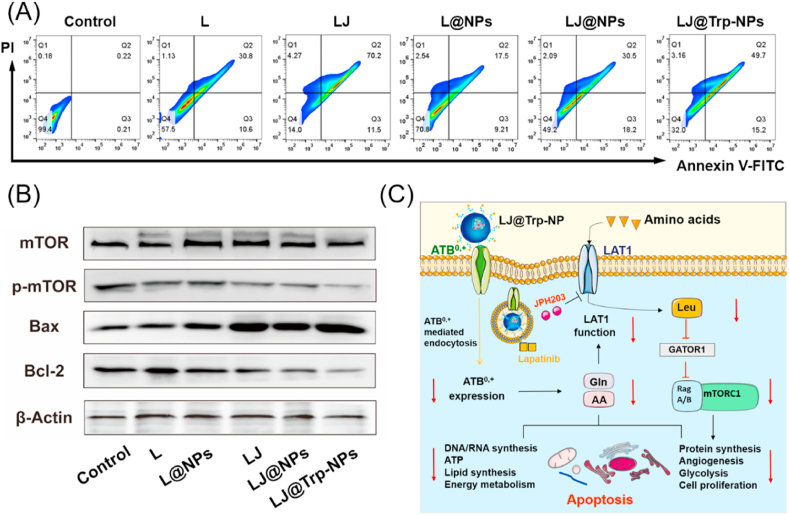
Cell apoptosis was investigated by Annexin V-FITC/PI staining to evaluate its potential involvement as an anticancer mechanism of LJ@Trp-NPs. As shown in Fig. 4A and S15C, L@NPs induced 17.5% late apoptosis and 9.2% early apoptosis in MCF7 cells; when both lapatinib and JPH203 were encapsulated, the early and late apoptosis rate increased up to 18.2% and 30.5%, respectively. ATB0,+-targeted LJ@Trp-NPs could further increase the late apoptosis to 49.7%. A similar trend was observed in T47D cells (Fig. S15A&B). The relatively milder effect observed with NPs compared to the corresponding free drugs might be due to the slow release of drugs from the NP formulation. We further assessed the expression of apoptosis-related proteins under different treatment conditions. As shown in Fig. 4B, LJ@Trp-NPs significantly upregulated the expression of Bax, a proapoptotic protein, but downregulated the expression of Bcl-2, an antiapoptotic protein. The ratio of Bax/Bcl-2 was considered as a parameter in the regulation of apoptosis. The increased Bax/Bcl-2 ratio with LJ@Trp-NPs indicates the ability of this NP formulation to promote apoptosis in cancer cells.
Fig. 4.
Anticancer mechanisms of LJ@Trp-NPs in MCF7 cells. (A) Apoptosis analysis using flow cytometry. (B) Expression pattern of key apoptotic proteins and mTOR phosphorylation under various treatment conditions. (C) A schematic diagram showing the actions of LJ@Trp-NPs on amino acid entry into tumor cells and consequent changes in cellular signaling and metabolism.
LJ@Trp-NPs were designed to interfere with amino acid entry into cancer cells with consequent detrimental changes in amino acid metabolism and associated cellular signaling and biological processes. Firstly, the ATB0,+ downregulation in conjunction with ATB0,+-targeted endocytosis of NPs would interfere with the entry of amino acids via this transporter. This was verified in this study. The expression of ATB0,+ in MCF7 cells was monitored under various treatment conditions (Fig. S16). LJ@Trp-NPs significantly decreased the ATB0,+ expression at protein level; other formulations had little or no effect on ATB0,+ protein levels. To further investigate the relationship of this phenomenon with endocytosis, MCF7 cells were treated by LJ@Trp-NPs under 4 °C (Fig. S17). When the endocytosis was suppressed by the low temperature [29], the expression of ATB0,+ in MCF7 cells was not affected. These results suggested that ATB0,+-mediated endocytosis might result in the downregulation of ATB0,+, which was consistent with our previous study [19]. ATB0,+ is a broad spectrum amino acid transporter with ability to transport 18 of the 20 proteinogenic amino acids; therefore, the observed downregulation of ATB0,+ in MCF7 cells when exposed to LJ@Trp-NPs would certainly decrease the amino acid supply. In addition, the cargo contained JPH203, an effective inhibitor of LAT1, another broad-spectrum amino acid transporter, thus enhancing amino acid starvation in these cells even further. Both ATB0,+ and LAT1 transport leucine, an activator of mTOR signaling [30]. Therefore, we monitored the activation of mTOR after LJ@Trp-NPs treatment. As shown in Fig. 4B, exposure of the cells to LJ@Trp-NPs did not affect the expression of total mTOR but decreased the content of phospho-mTOR. This mode of anticancer effect of LJ@Trp-NPs by limiting the supply of amino acids to cancer cells is likely to amplify the anticancer efficacy of lapatinib and possibly deter development of resistance to lapatinib.
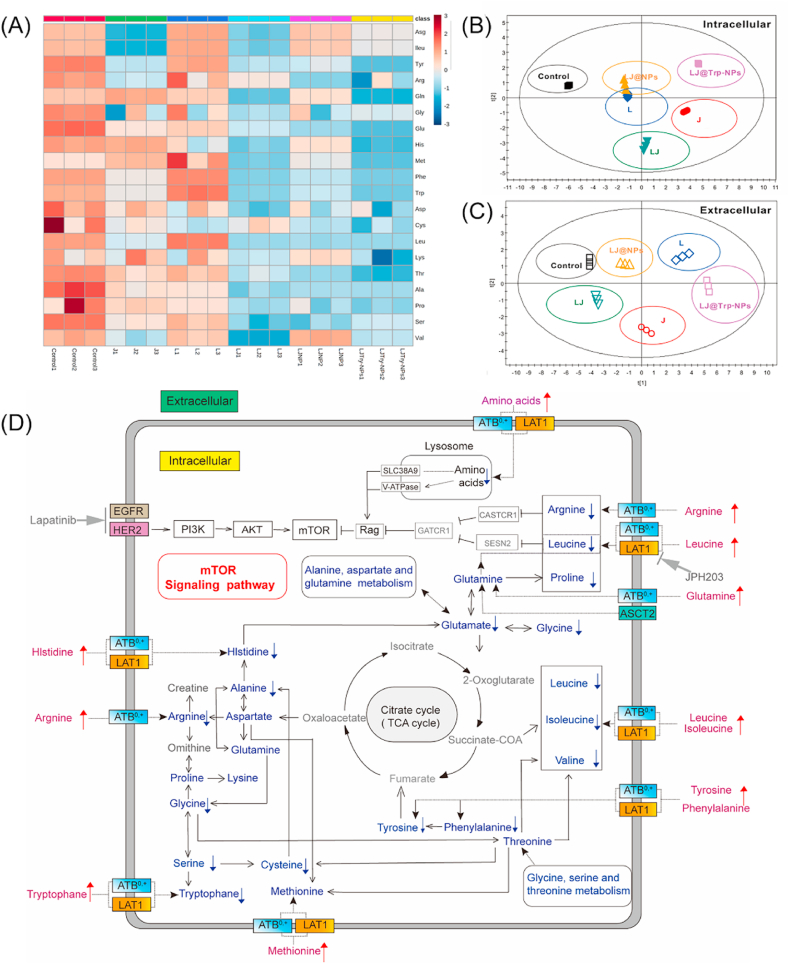
To understand effects of LJ@Trp-NPs on amino acid metabolism in tumor cells in a greater detail, we conducted a targeted study of metabolomics in MCF7 cells. The 20 proteinogenic amino acids were identified in the intracellular and extracellular media, and quantitative analysis was performed to show the metabolomic changes in MCF7 cells. The results are presented in Fig. S18 (intracellular medium) and Fig. S19 (extracellular medium) and also as heatmaps (Fig. 5A for intracellular metabolites, Fig. S20 for extracellular metabolites). It was clear that LJ@Trp-NPs significantly restricted the uptake of most of the amino acids in comparison with other formulations; this resulted in a decrease in intracellular amino acids and an increase in extracellular amino acids. These metabolomic data were then subjected to partial least-squares discriminate analysis (PLS-DA) (Fig. 5B and C). Distinct metabolomic profiles in response to different treatments were observed. The resulting PLS-DA score plots of both the intracellular metabolites (Fig. 5B) and the extracellular metabolites (Fig. 5C) clearly showed that the metabolomic profiles for the control, JPH203, lapatinib, lapatinib + JPH203, LJ@NPs, and LJ@Trp-NPs groups were distinct and separable. LJ@Trp-NPs showed the strongest modulation effect. These studies provide evidence for LJ@Trp-NPs causing marked interference with the amino acid metabolism in MCF7 cells, which is expected from the observed changes in the intracellular and extracellular levels of amino acids (Fig. 5D). LJ@Trp-NPs treatment significantly decreased the uptake of a broad spectrum of amino acids and suppressed many of the related metabolic signaling pathways. For example, mTOR pathway, which is responsible for cell proliferation, angiogenesis, bioenergetics through nutrient availability and revving up anabolic pathways, and cell survival through DNA repair and decreasing apoptosis [31,32], was significantly suppressed. It has been reported that the activation of mTOR signaling pathway contributes to lapatinib resistance [33]. Therefore, mTOR inhibition would avoid the development of resistance to lapatinib and enhance the treatment outcome if administered in the form of LJ@Trp-NPs. Taken collectively, LJ@Trp-NPs elicit their anticancer effect by blocking the entry of amino acids into cancer cells and consequently interfering with cellular metabolism and signaling pathways. As lapatinib is included in this formulation as a cargo, the amino acid starvation caused by the blockade of the two critical amino acid transporters synergizes with lapatinib to maximize the anticancer effect.
Fig. 5.
Targeted metabolomic study focusing on amino acids in MCF7 cells with exposure to various NP formulations. (A) Heatmap of intracellular amino acid content in MCF7 cells treated with different formulations. PLS-DA score plot was used to identify the variations of (B) the intracellular metabolomic profile and (C) the extracellular metabolomic profile among the six groups. (D) The pathways were deduced based on the KEGG database and the small molecule pathway database (SMPDB). The arrows denote significant differences (increase or decrease) in amino acid metabolite levels in MCF7 cells exposed to the LJ@Trp-NP treatment compared with control.
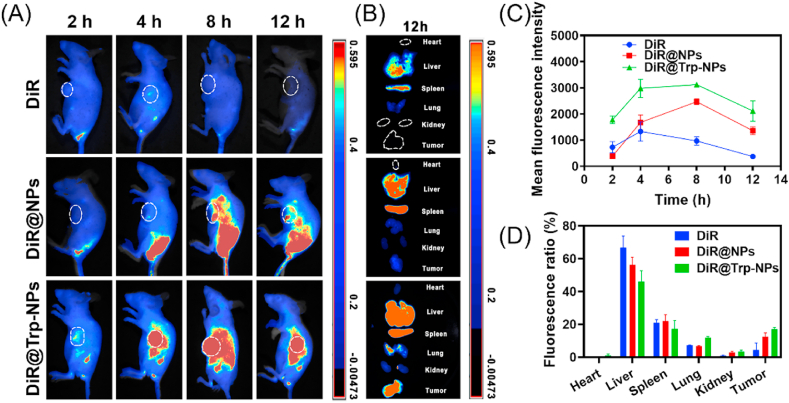
The in vivo behavior of LJ@Trp-NPs was investigated by a pharmacokinetic study in female rats and a biodistribution assay in tumor-bearing mice. The concentration curves of lapatinib and JPH203 from LJ@Trp-NPs are displayed in Fig. S21 with LJ@NPs and free LJ as controls, and the calculated pharmacokinetic parameters are listed in Tables S2 and S3. The AUC0-t values of lapatinib from LJ@NPs and LJ@Trp-NPs were 518.1 and 512.9 μg min·mL−1, respectively, which were 4.22 and 4.17 folds higher than that from free LJ, indicating the prolonged circulation. In addition, the half-life of lapatinib increased from 93.6 min to 351.5 min and 380.1 min after encapsulated into LJ@NPs and LJ@Trp-NPs, further confirming that NP formulation resulted in enhancement of the drug half-life in circulation compared to free drugs. A similar profile was seen for JPH203 from LJ@NPs and LJ@Trp-NPs, displaying increased AUC and decreased t1/2 compared to the free from. These results show that both LJ@Trp-NPs and LJ@NPs increase the half-life of the drugs in circulation, a feature beneficial for increased tumor accumulation. The biodistribution of LJ@Trp-NPs was investigated by an in vivo imaging system using DiR as a probe. As shown in Fig. 6A, very little free DiR accumulated in tumors, but NP formulation resulted in much more accumulation in tumors than free DiR. DiR@Trp-NPs displayed the strongest and the most persistent signals in the tumors, which could be explained by the ATB0,+-mediated targeting to tumor cells. The quantitative analysis shown in Fig. 6C further confirmed the enhanced and persistent distribution of DiR@Trp-NPs compared to free DiR or DiR@NPs. After 12 h, the mice were killed, and the major organs were collected and imaged (Fig. 6B). The ex vivo results further confirmed that DiR@Trp-NPs exhibited the most tumor accumulation. Moreover, the quantitative analysis suggested that the DiR signal in tumors for DiR@Trp-NPs was close to 20% of the total signal, demonstrating the ideal distribution for loaded drugs (Fig. 6D). However, significant accumulation of DiR@Trp-NPs in liver and spleen was also observed. Liver has a robust capacity to extract amino acids from the circulation using a multitude of amino acid transporters; therefore, LJ@Trp-NPs may be recognized by one or more of these transporters, thus facilitating the liver uptake of the NP formulation. In addition, it was acknowledged that nanoparticles could be easily sequestered by the liver and spleen after i.v. injection since the liver is the largest vascularized organ and the spleen is an important component of the immune system [[34], [35], [36]]. The nanoparticles were exogenous and could be captured by the spleen, even under the protection of PEG. This phenomenon was also observed in many other studies [[37], [38], [39]]. Further studies are needed to assess the consequences of the accumulation of the drug cargo in the liver in terms of toxicity. Nonetheless, it should be noted that DiR@Trp-NPs displayed significant superiority over other formulations to cause accumulation of the drug cargo in tumors, suggesting potential superior outcome in terms of anticancer effect.
Fig. 6.
Biodistribution of DiR@Trp-NPs in MCF7 tumor-bearing mice. (A) In vivo fluorescence images of MCF7 tumor-bearing mice at 2, 4, 8, and 12 h after i.v. injection of free DiR, DiR@NPs and DiR@Trp-NPs. (B) Fluorescence images of vital organs, collected at 12 h. (C) Quantitative analysis of the fluorescence signals in the tumor site at different time points. (D) Fluorescence ratio of each organ as percent of total fluorescence (all organs and tumor). Data are means ± SD (n = 3).
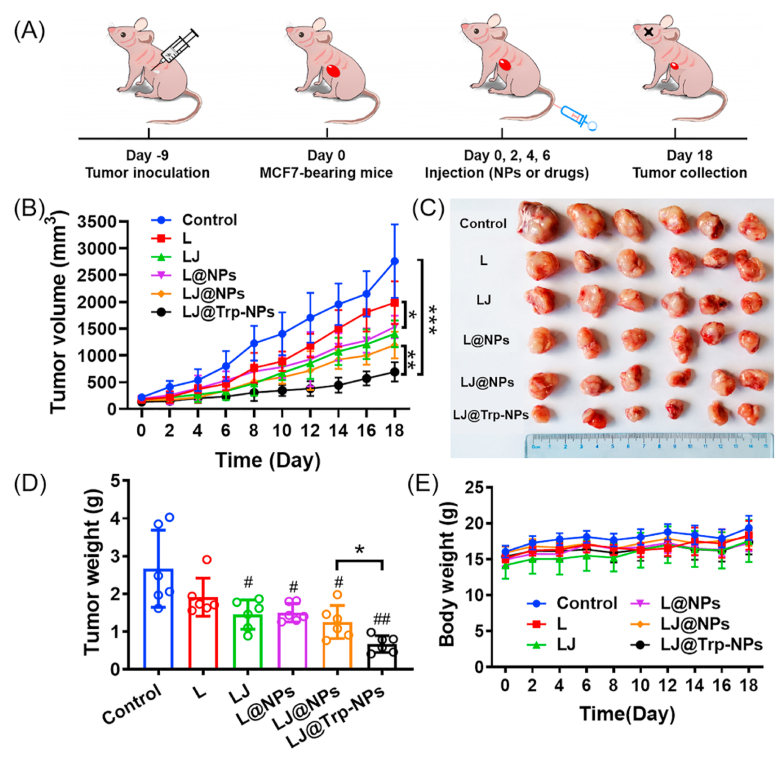
The in vivo anticancer efficacy of LJ@Trp-NPs was studied in MCF7 tumor-bearing mice. To assess the anticancer effect of LJ@Trp-NPs, the tumors were allowed to grow for 9-days following inoculation (Fig. 7A). The tumor-bearing mice were then randomly divided into 6 groups to receive i.v. administration of saline, free lapatinib, free lapatinib and JPH203 (L + J), L@NPs, LJ@NPs, and LJ@Trp-NPs on day 0, 2, 4, and 6, respectively. The length and width of the subcutaneous tumors were recorded to calculate the tumor volume, and the mouse weight was also monitored. As shown in Fig. 7B, the growth profiles of the tumors demonstrated that LJ@Trp-NPs significantly suppressed the tumor growth compared to other groups. Specifically, the combination of lapatinib and JPH203 displayed an enhanced anticancer effect compared to lapatinib alone, suggesting that LAT1 blockade amplified the therapeutic outcome of lapatinib, which might be associated with the suppression of mTOR signaling (Fig. 4B). LJ@Trp-NPs significantly suppressed the tumor growth compared to L@NPs, demonstrating the potentiating effect of amino acid starvation and the resultant changes in amino acid metabolism on the anticancer efficacy of lapatinib, consistent with the in vitro data (Fig. 5). The amino acid starvation is caused not only by the blockade of LAT1 (Fig. 4C) but also by the down-regulation of ATB0,+ resulting from the transporter-targeted endocytosis (Fig. S16). LJ@Trp-NPs also showed higher anticancer efficacy than LJ@NPs, suggesting that Trp-mediated ATB0,+ targeting benefited the treatment efficacy, which could be explained by the enhanced distribution (Fig. 6) and ATB0,+ downregulation (Fig. S16). As such, the enhanced targeting feature and the ability to block amino acid entry into tumor cells enabled LJ@Trp-NPs to elicit the strongest anticancer effect in vivo. After 18 days, the mice were killed, and the tumors were collected, photographed (Fig. 7C), and weighed (Fig. 7D). These results were consistent with that shown in Fig. 7A. The body weight of the mice during treatment did not decrease with any of the formulations (Fig. 7E).
Fig. 7.
The in vivo anticancer efficacy and safety of LJ@Trp-NPs in MCF7 tumor-bearing mice. (A) Experimental design. (B) Tumor growth curves following treatment with saline, lapatinib (L), lapatinib + JPH203 (LJ), L@NP, LJ@NP and LJ@Trp-NP. (C) Photograph of tumors collected at day 18. (D) Tumor weight of MCF7 tumor-bearing mice in different groups at day 18. (E) Body weight of MCF7 tumor-bearing mice in different groups following treatments. Data are presented as means ± SD (n = 6). *, p < 0.05, **, p < 0.01, ***, p < 0.001, indicate statistical difference between groups. #, p < 0.05, ##, p < 0.01, indicate statistical difference compared to the control group.
H&E staining and immunohistochemical analysis were further used to evaluate the in vivo anticancer efficacy of LJ@Trp-NPs (Fig. S22). H&E staining of tumor sections showed that LJ@Trp-NPs treatment caused extensive damage in tumor tissue, and comparatively less damage was observed in the groups of LJ, L@NPs, and LJ@NPs. There was little or no effect with the other groups. As a biomarker for cell proliferation, Ki-67 was monitored using immunohistochemical staining. LJ@Trp-NPs showed the lowest expression of Ki-67 in all six groups, while other treatments decreased positive staining to a much smaller extent. Quantitative analysis confirmed this observation (Fig. S22B). The expression of p-mTOR was measured using immunofluorescence analysis to evaluate the status of the mTOR signaling pathway. The control group showed the highest expression of p-mTOR, and the combination of lapatinib and JPH203 significantly decreased the p-mTOR expression (Figs. S22A and C). LJ@Trp-NPs treatment resulted in the lowest signals of p-mTOR, indicating the effective blocking effect on the cellular entry of mTOR activators such as leucine. TUNEL assay was used to monitor apoptosis induced by LJ@Trp-NPs (Figs. S22A and D). LJ@Trp-NPs group showed the strongest apoptosis. These results are consistent with the in vitro data. Taken collectively, LJ@Trp-NPs displayed the most prominent anticancer effect, which could be attributed to the multiple mechanisms, including suppressed cell proliferation, downregulated mTOR signaling, and enhanced cell death.
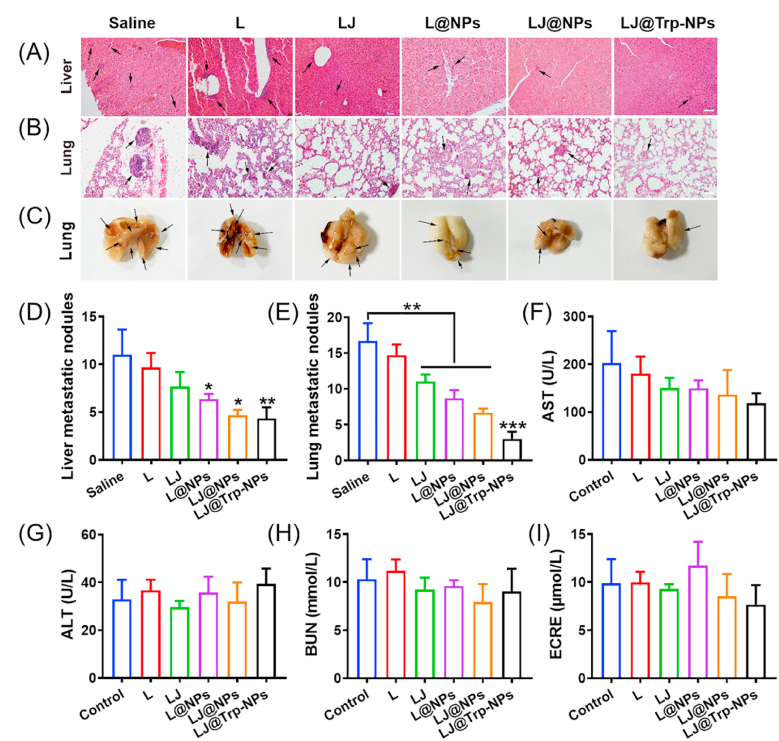
LJ@Trp-NPs showed a robust inhibitory effect on cell migration in vitro. Therefore, we evaluated the anti-metastasis effect of LJ@Trp-NPs in vivo. We detected the nodule formation and metastasis of MCF7 tumor cells in liver and lung tissues (Fig. 8A–E). The H&E staining showed distinct and large nodules in liver and lung tissues in the saline group (Fig. 8A&B); LJ@Trp-NPs treatment dramatically reduced the number of nodules compared to the other groups (Fig. 8D&E). The visible metastatic nodules on the lung surface of MCF7 tumor-bearing mice further confirmed the notion that LJ@Trp-NPs decreased the extent of lung metastasis. A more reasonable therapeutic regimen for early treatment might reach a better treatment effect and avoid metastasis occurrence. Due to the considerable liver accumulation of LJ@Trp-NPs observed in the biodistribution assay, the liver function of mice after various treatments was examined. The serum biochemical indicators, AST and ALT, were measured to assess the liver function (Fig. 8F and G). These two indicators did not change after LJ@Trp-NPs treatment, suggesting no significant injury to the liver. In addition, the BUN and creatinine levels also did not change after treatments, indicating intact renal function (Fig. 8H and I). We also assessed the safety of LJ@Trp-NPs by examining the histology of various organs (heart, spleen, and kidney) using H&E staining. As shown in Fig. S23, no evidence of injury was found in heart, spleen, and kidney of mice receiving LJ@Trp-NPs.
Fig. 8.
The anti-metastasis effect of LJ@Trp-NPs in MCF7 tumor-bearing mice. Representative H&E staining of (A) liver sections and (B) lung sections from each group. Scale bar = 50 μm, (n = 3). (C) Representative images of lung metastasis in each group. The arrow represents pulmonary metastatic nodule. The number of (D) liver and (E) lung metastatic nodules after various treatments. Serum biochemical analysis of (F) AST, (G) ALT, (H) BUN, (I) creatinine after various treatments (n = 6). *, p < 0.05, **, p < 0.01, ***, p < 0.001, statistical difference between groups or compared to the control group.
4. Conclusion
In this study, we developed, optimized, and characterized lapatinib- and JPH203-coloaded, ATB0,+-targeted nanoparticles (LJ@Trp-NPs) to block amino acid entry into tumor cells with resultant interference with amino acid metabolism and associated signaling pathways. The goal was to determine the impact of this change in amino acid metabolism on the anticancer efficacy of lapatinib, a chemotherapeutic that elicits its anticancer effect on a molecular target distinct from the amino acid entry pathways. LJ@Trp-NPs carry a drug cargo that contains not only lapatinib but also JPH203, a high-affinity inhibitor of the amino acid transporter LAT1. The NPs have tryptophan conjugated on the surface to target the NPs to another amino acid transporter, ATB0,+. Both LAT1 and ATB0,+ play an obligatory role in supplying most of the amino acids to tumor cells [13]. Endocytosis of LJ@Trp-NPs via ATB0,+ targeting results not only in the entry of NPs into tumor cells but also in the downregulation of the transporter on the cell surface, with resultant subsequent decrease in the entry of amino acids into tumor cells. The cargo delivers JPH203 into the cells to function as an inhibitor of LAT1. Thus, the function of two important amino acid transporters gets compromised by LJ@Trp-NPs treatment. The resultant amino acid starvation and accompanying changes in amino acid metabolism and intracellular signaling pathways synergizes with the anticancer effect of lapatinib. Thus, LJ@Trp-NPs offer a therapeutic strategy to potentiate the anticancer efficacy of lapatinib, which could also prevent the development of lapatinib resistance.
Author contributions
Q.Y. and R.C. conceived and designed the experiments. L.K. and X.J. performed most of the experiments and wrote the draft. Y.T, X.X. and Y.L. participated in cell culture and animal studies. A.C. helped with the metabolomic study. H.Z. contributed to flow cytometry analysis. H.Z. contributed to the data analysis and discussion. V.G., Q.Y., and R.C. edited the manuscript and prepared the final version. Q.Y. and R.C. supervised the whole project. All the listed authors have read and approved the manuscript.
CRediT authorship contribution statement
Longfa Kou: Conceptualization, Data curation, Formal analysis, Funding acquisition, Writing – original draft. Xinyu Jiang: Data curation, Formal analysis, Investigation, Methodology, Project administration. Yingying Tang: Methodology, Resources, Software. Xing Xia: Investigation, Methodology, Software. Yingtao Li: Resources, Software, Visualization. Aimin Cai: Methodology, Resources, Software. Hailun Zheng: Methodology, Resources. Hailin Zhang: Funding acquisition, Resources, Validation. Vadivel Ganapathy: Conceptualization, Writing – review & editing. Qing Yao: Conceptualization, Funding acquisition, Writing – review & editing. Ruijie Chen: Funding acquisition, Supervision, Validation.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
Financial support: This research was supported by the National Natural Science Foundation of China (81803443, and 81903551), Zhejiang Province Natural Science Foundation (LQ19H300001), Wenzhou Municipal Science and Technology Bureau (Y20180180), Xinmiao Talent Program of Zhejiang Province (Grant No. 2021R413073), and Excellent Young Scientist Training Program fund from Wenzhou Medical University.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.07.009.
Contributor Information
Qing Yao, Email: yqpharm@163.com.
Ruijie Chen, Email: crjpharm@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mun E.J., Babiker H.M., Weinberg U., Kirson E.D., Von Hoff D.D. Tumor-treating fields: a fourth modality in cancer treatment. Clin. Canc. Res. : Off. J. Am. Assoc. Canc. Res. 2018;24:266–275. doi: 10.1158/1078-0432.CCR-17-1117. [DOI] [PubMed] [Google Scholar]
- 2.Pritchard J.R., Lauffenburger D.A., Hemann M.T. Understanding resistance to combination chemotherapy. Drug Resist. Updates : Rev. Comment. Antimicrob. Anticanc. Chemother. 2012;15:249–257. doi: 10.1016/j.drup.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ke X., Shen L. Molecular targeted therapy of cancer: the progress and future prospect. Front. Lab. Med. 2017;1:69–75. [Google Scholar]
- 4.Gottfried E.K., Mark D.P., Natarajan V., Richard F., Guorong Y., Martina R., Michael U., David W.R., Glenn S., Robert J.M., Barry R.K., Tona M.G., Mark B., Karl C.P., Dennis J.S. Cancer research; 2006. Activity of the Dual Kinase Inhibitor Lapatinib (GW572016) against HER-2-Overexpressing and Trastuzumab-Treated Breast Cancer Cells; p. 66. [DOI] [PubMed] [Google Scholar]
- 5.Frezza C. Metabolism and cancer: the future is now. Br. J. Canc. 2020;122:133–135. doi: 10.1038/s41416-019-0667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park J.H., Pyun W.Y., Park H.W. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020:9. doi: 10.3390/cells9102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkoyluoglu-Cotul E., Arca A., Madak-Erdogan Z. Crosstalk between estrogen signaling and breast cancer metabolism. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2019;30:25–38. doi: 10.1016/j.tem.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Chong L.D. Exploiting cancer metabolism. Science. 2017;355:1036. doi: 10.1126/science.355.6329.1036-b. [DOI] [PubMed] [Google Scholar]
- 9.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alison N.M., Aimee L.E. Nutrient transporters: the Achilles' heel of anabolism. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2013;24 doi: 10.1016/j.tem.2013.1001.1002-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganapathy V., Thangaraju M., Prasad P.D. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol. Therapeut. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Karunakaran S., Ramachandran S., Coothankandaswamy V., Elangovan S., Babu E., Periyasamy-Thandavan S., Gurav A., Gnanaprakasam J.P., Singh N., Schoenlein P.V., Prasad P.D., Thangaraju M., Ganapathy V. SLC6A14 (ATB0,+) protein, a highly concentrative and broad specific amino acid transporter, is a novel and effective drug target for treatment of estrogen receptor-positive breast cancer. J. Biol. Chem. 2011;286:31830–31838. doi: 10.1074/jbc.M111.229518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhutia Y.D., Babu E., Prasad P.D., Ganapathy V. The amino acid transporter SLC6A14 in cancer and its potential use in chemotherapy. Asian J. Pharm. Sci. 2014;9:293–303. [Google Scholar]
- 14.Scalise M., Galluccio M., Console L., Pochini L., Indiveri C. The human SLC7A5 (LAT1): the intriguing histidine/large neutral amino acid transporter and its relevance to human health. Front. Chem. 2018;6:243. doi: 10.3389/fchem.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moldogazieva N.T., Mokhosoev I.M., Terentiev A.A. Metabolic heterogeneity of cancer cells: an interplay between HIF-1, GLUTs, and AMPK. Cancers. 2020:12. doi: 10.3390/cancers12040862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kou L., Bhutia Y.D., Yao Q., He Z., Sun J., Ganapathy V. Transporter-guided delivery of nanoparticles to improve drug permeation across cellular barriers and drug exposure to selective cell types. Front. Pharmacol. 2018;9:27. doi: 10.3389/fphar.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kou L., Yao Q., Zhang H., Chu M., Bhutia Y.D., Chen R., Ganapathy V. Transporter-targeted nano-sized vehicles for enhanced and site-specific drug delivery. Cancers. 2020;12:2837. doi: 10.3390/cancers12102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Q., Gong P., Sun M., Kou L., Ganapathy V., Jing Y., He Z., Sun J. Transporter occluded-state conformation-induced endocytosis: amino acid transporter ATB(0,+)-mediated tumor targeting of liposomes for docetaxel delivery for hepatocarcinoma therapy. J. Contr. Release : Off. J. Contr. Rel. Soc. 2016;243:370–380. doi: 10.1016/j.jconrel.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 19.Kou L., Huang H., Lin X., Jiang X., Wang Y., Luo Q., Sun J., Yao Q., Ganapathy V., Chen R. Endocytosis of ATB(0,+)(SLC6A14)-targeted liposomes for drug delivery and its therapeutic application for pancreatic cancer. Expet Opin. Drug Deliv. 2020;17:395–405. doi: 10.1080/17425247.2020.1723544. [DOI] [PubMed] [Google Scholar]
- 20.Mori Y., Tsukinoki K., Yasuda M., Miyazawa M., Kaneko A., Watanabe Y. Glucose transporter type 1 expression are associated with poor prognosis in patients with salivary gland tumors. Oral Oncol. 2007;43:563–569. doi: 10.1016/j.oraloncology.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Hafliger P., Charles R.P. The L-type Amino acid transporter LAT1-an emerging target in cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20102428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H., Xu Y., Wang F., Shen Z., Tuo X., Qian H., Wang H., Wang K. Clinical associations between ASCT2 and p-mTOR in the pathogenesis and prognosis of epithelial ovarian cancer. Oncol. Rep. 2018;40:3725–3733. doi: 10.3892/or.2018.6729. [DOI] [PubMed] [Google Scholar]
- 23.Bernhardt S., Bayerlová M., Vetter M., Wachter A., Mitra D., Hanf V., Lantzsch T., Uleer C., Peschel S., John J., Buchmann J., Weigert E., Bürrig K.F., Thomssen C., Korf U., Beissbarth T., Wiemann S., Kantelhardt E.J. Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors, Breast cancer research. BCR. 2017;19:112. doi: 10.1186/s13058-017-0905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kou L., Yao Q., Sun M., Wu C., Wang J., Luo Q., Wang G., Du Y., Fu Q., Wang J., He Z., Ganapathy V., Sun J. Cotransporting ion is a trigger for cellular endocytosis of transporter-targeting nanoparticles: a case study of high-efficiency SLC22A5 (OCTN2)-Mediated carnitine-conjugated nanoparticles for oral delivery of therapeutic drugs. Adv. Healthcare Mater. 2017;6 doi: 10.1002/adhm.201700165. [DOI] [PubMed] [Google Scholar]
- 25.Kou L., Sun R., Xiao S., Cui X., Sun J., Ganapathy V., Yao Q., Chen R. OCTN2-targeted nanoparticles for oral delivery of paclitaxel: differential impact of the polyethylene glycol linker size on drug delivery in vitro, in situ, and in vivo. Drug Deliv. 2020;27:170–179. doi: 10.1080/10717544.2019.1710623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qing Y., Zhi D., Jong Hoon C., Dongin K., Lin Z. Building stable MMP2-responsive multifunctional polymeric micelles by an all-in-one polymer-lipid conjugate for tumor-targeted intracellular drug delivery. ACS Appl. Mater. Interfaces. 2017;9 doi: 10.1021/acsami.1027b09511-32533. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Chi D., Wu X., Wang Y., Lin X., Xu Z., Liu H., Sun J., He Z., Wang Y. Tyrosine modified irinotecan-loaded liposomes capable of simultaneously targeting LAT1 and ATB(0,+) for efficient tumor therapy. J. Contr. Release : Off. J. Contr. Rel. Soc. 2019;316:22–33. doi: 10.1016/j.jconrel.2019.10.037. [DOI] [PubMed] [Google Scholar]
- 28.Cai A., Zheng H., Chen Z., Lin X., Li C., Yao Q., Bhutia Y.D., Ganapathy V., Chen R., Kou L. Synergism between SLC6A14 blockade and gemcitabine in pancreatic cancer: a 1H-NMR-based metabolomic study in pancreatic cancer cells. Biochem. J. 2020;477:1923–1937. doi: 10.1042/BCJ20200275. [DOI] [PubMed] [Google Scholar]
- 29.Kou L., Sun J., Zhai Y., He Z. The endocytosis and intracellular fate of nanomedicines: implication for rational design. Asian J. Pharm. Sci. 2013;8:1–10. [Google Scholar]
- 30.Nicklin P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., Myer V.E., MacKeigan J.P., Porter J.A., Wang Y.K., Cantley L.C., Finan P.M., Murphy L.O. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua H., Kong Q., Zhang H., Wang J., Luo T., Jiang Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mossmann D., Park S., Hall M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Canc. 2018;18:744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 33.D'Amato V., Raimondo L., Formisano L., Giuliano M., De Placido S., Rosa R., Bianco R. Mechanisms of lapatinib resistance in HER2-driven breast cancer. Canc. Treat Rev. 2015;41:877–883. doi: 10.1016/j.ctrv.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y.-N., Poon W., Tavares A.J., McGilvray I.D., Chan W.C.W. Nanoparticle–liver interactions: cellular uptake and hepatobiliary elimination. J. Contr. Release. 2016;240:332–348. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y., Sultan D., Liu Y. In: Theranostic Bionanomaterials. Cui W., Zhao X., editors. Elsevier; 2019. 2 - biodistribution, excretion, and toxicity of nanoparticles; pp. 27–53. [Google Scholar]
- 36.Sykes E.A., Dai Q., Tsoi K.M., Hwang D.M., Chan W.C. Nanoparticle exposure in animals can be visualized in the skin and analysed via skin biopsy. Nat. Commun. 2014;5:3796. doi: 10.1038/ncomms4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong Y., Su T., Shi Q., Feng Y., Tao Z., Huang Q., Li L., Hu L., Li S., Tan H., Liu S., Yang H. Co-administration of iRGD enhances tumor-targeted delivery and anti-tumor effects of paclitaxel-loaded PLGA nanoparticles for colorectal cancer treatment. Int. J. Nanomed. 2019;14:8543–8560. doi: 10.2147/IJN.S219820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson J., Rui Y., Kozielski K.L., Placone A.L., Choi O., Tzeng S.Y., Kim J., Keyes J.J., Bogorad M.I., Gabrielson K., Guerrero-Cazares H., Quinones-Hinojosa A., Searson P.C., Green J.J. Engineered nanoparticles for systemic siRNA delivery to malignant brain tumours. Nanoscale. 2019;11:20045–20057. doi: 10.1039/c9nr04795f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.