Abstract
The ribosomal region spanning the two internal transcribed spacer (ITS) regions and the 5.8S ribosomal DNA region was sequenced for asexual, anthropophilic dermatophyte species with morphological similarity to Trichophyton rubrum, as well as for members of the three previously delineated, related major clades in the T. mentagrophytes complex. Representative isolates of T. raubitschekii, T. fischeri, and T. kanei were found to have ITS sequences identical to that of T. rubrum. The ITS sequences of T. soudanense and T. megninii differed from that of T. rubrum by only a small number of base pairs. Their continued status as species, however, appears to meet criteria outlined in the population genetics-based cohesion species concept of A. R. Templeton. The ITS sequence of T. tonsurans differed from that of the biologically distinct T. equinum by only 1 bp, while the ITS sequence of the recently described species T. krajdenii had a sequence identical to that of T. mentagrophytes isolates related to the teleomorph Arthroderma vanbreuseghemii.
Recently, the advent of molecular biological techniques has made possible a much more detailed examination of species structure and evolution in the dermatophytes and dermatophytoids. Studies to date have tended to support the existing concepts of the geophilic and zoophilic species delineated by mating studies (8, 12, 18–20, 26). Among the asexual anthropophilic dermatophytes a more complex picture has emerged, and some species concepts have been called into question. For example, a phylogenetic comparison of large-subunit ribosomal DNA (rDNA) sequences by Leclerc et al. (20) showed that the asexual anthropophile Microsporum audouinii could not be distinguished from the sexual zoophile Microsporum canis while another asexual anthropophile, Microsporum ferrugineum, deviated minimally. The same study did not distinguish the endemic African endothrix tinea capitis agent Trichophyton soudanense, an asexual anthropophile, from the morphologically distinct, cosmopolitan asexual anthropophile T. rubrum. A phylogeny derived from the study of mitochondrial DNA restriction types (26) failed to distinguish the distinctive aconidial, asexual agent of human favus, T. schoenleinii, from the heavily conidial T. mentagrophytes var. quinckeanum, a group of rodent-infecting isolates reported to be interfertile with the teleomorph Arthroderma benhamiae (9, 31). The profound epidemiological, morphological, and physiological differences among the species which were not distinguished in these molecular studies indicate that the exact techniques used, while appropriate for putatively anciently evolved geophiles and zoophiles, may lack sufficient resolving power to discern evolutionary trends in at least some of the more recently evolved anthropophilic dermatophytes (29). The foregoing statement presupposes that any specifically anthropophilic species will probably have arisen within the relatively recent evolution of the hominid lineage, while zoophilic species may approximately share the evolutionary age of the broad mammalian clades with which they primarily associate, e.g., rodents and lagomorphs for the T. mentagrophytes complex and canids and felids for M. canis.
In molecular studies of recently evolved species complexes, e.g., the study of Lake Victoria cichlid fish (24) and the study of fungal crop pathogens in the genus Sclerotinia (3), it has been found that only the most highly variable genetic regions are likely to be suitable for investigating the delimitation and phylogeny of species. The present study therefore investigated whether a highly variable region of fungal DNA, the ribosomal region consisting of internal transcribed spacer sequences (ITS) 1 and 2 and their intermediary 5.8S rDNA, exhibited adequate variability to resolve the diversity of anthropophilic species and to determine their phylogeny. Some reports in which sequences are given for the ITS1 region of certain dermatophytes have recently appeared, giving broad overviews of dermatophytes (22) and of the T. mentagrophytes complex (23). The present study focused in greater detail on species known or thought to be related to T. rubrum, including some zoophilic lineages that may be ancestral to this common anthropophile. Isolates representing the recently described segregate species T. fischeri, T. raubitschekii, and T. kanei, placed in the T. rubrum species complex by Kane (14), were included, including those used in preparing the original descriptions of T. fischeri and T. raubitschekii. In addition, in order to give a complete survey of T. rubrum-like dermatophytes and potentially aid in the development of diagnostic probes, some isolates showing strong phenotypic convergence with T. rubrum were included, such as a blood red-pigmented T. mentagrophytes isolate and representative isolates of the recently described species T. krajdenii, a T. mentagrophytes segregate that may strongly resemble yellow variants of T. rubrum. While this study was in review, a general overview of dermatophyte phylogeny based on a ribosomal region strongly overlapping that used in the present study was published (7); that study excluded many of the taxa and variants included here.
MATERIALS AND METHODS
Fungal cultures.
The origins of the Trichophyton isolates examined are given in Table 1. All isolates were grown at 22.5°C on Sabouraud's peptone-glucose agar supplemented with cycloheximide (100 μg/ml), chloramphenicol (100 μg/ml), and gentamicin (50 μg/ml).
TABLE 1.
Fungal cultures used for sequence analysis in this studya
| Species | Strain no. | Sourceb | GenBank accession no. |
|---|---|---|---|
| Arthroderma spp. | |||
| A. vanbreuseghemii | |||
| − mating type | RV 27961 | R. Vanbreuseghem | AF 170453 |
| + mating type | RV 27960 | R. Vanbreuseghem | AF 170452 |
| A. simii | |||
| − mating type | UAMH 2944 | University of Alberta | AF 170455 |
| + mating type | UAMH 2943 | University of Alberta | AF 170454 |
| A. benhamiae | |||
| − mating type, European | RV 26680 | R. Vanbreuseghem | AF 170457 |
| + mating type, African | RV 30000 | R. Vanbreuseghem | AF 170456 |
| Trichophyton spp. | |||
| T. mentagrophytes | UAMH 7339 | University of Alberta | AF 170467 |
| UAMH 8543 | OMH | AF 170465 | |
| UAMH 8544 | OMH | AF 170466 | |
| T. krajdenii | UAMH 8546 | OMH | AF 170461 |
| UAMH 8539 | OMH | AF 170462 | |
| T. tonsurans | UAMH 8549 | OMH | AF 170476 |
| UAMH 8550 | OMH | AF 170477 | |
| UAMH 8551 | OMH | AF 170478 | |
| UAMH 8552 | OMH | AF 170479 | |
| T. equinum | NOMH 1051 | OMH | AF 170458 |
| T. yaoundei | UAMH 7388 | LSPQ/OMH | AF 170480 |
| T. simii | NOMH 898 | OMH | AF 170475 |
| T. raubitschekii | UAMH 8545 | OMH | AF 170468 |
| NOMH 789 | OMH | AF 170469 | |
| ATCC 42631 | OMH | AF 170470 | |
| T. rubrum | UAMH 8547 | OMH | AF 170471 |
| ATCC 28188 | ATCC | AF 170472 | |
| T. kanei | UAMH 8538 | OMH | AF 170460 |
| T. fischeri | ATCC 32871 | OMH | AF 170459 |
| T. megninii | UAMH 8542 | OMH | AF 170463 |
| ATCC 12106 | ATCC | AF 170464 | |
| T. soudanense | NOMH 831 | Ontario Ministry of Health | AF 170473 |
| UAMH 8548 | LSPQ/OMH | AF 170474 |
Strains and species determined to have identical sequences in this study are grouped without spacing in this table.
Most strains were isolated at the Mycology Laboratory, Laboratories Branch, OMH. Many were deposited in the University of Alberta Microfungus Collection and Herbarium (UAMH), the American Type Culture Collection (ATCC), or the OMH collection (NOMH), including some isolates originating at the Laboratoire de Santé Publique de Québec (LSPQ) and received as reference cultures at OMH. Other isolates are listed according to the collections which originally supplied them to OMH.
DNA isolation.
Each isolate was inoculated into 10 ml of Sabouraud's peptone-glucose broth and incubated at 22.5°C for 14 to 28 days. Mycelia were collected by centrifugation for 10 min at 13,000 × g and resuspended in 300 μl of a buffer consisting of 50 mM Tris-HCl (pH 8.2), 20 mM EDTA, and 1% mercaptoethanol. To the mycelial suspension was added 50 to 100 mg of glass beads. Each sample was homogenized for 5 min with a motorized pestle (Sigma Chemical Co., St. Louis, Mo.). All homogenized samples were incubated for 30 min at 60°C. Each homogenate was extracted with an equal volume of a cloroform-isoamyl alcohol (24:1) mixture followed by buffer-saturated phenol. The top (aqueous) phase was mixed with 2 volumes of 100% ethanol and stored at −20°C for 30 min. DNA was precipitated by centrifugation for 15 min at 13,000 × g and 4°C. The DNA pellet was washed with 70% ethanol, centrifuged, and then resuspended in 100 μl of Tris-EDTA buffer.
PCR.
At the Ontario Ministry of Health (OMH), PCR amplifications were carried out in 100-μl volumes. Each reaction mixture contained 1 μl of the prepared DNA from an isolate and 99 μl of the following PCR master mixture: 10 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 20 mM MgCl, and 0.01% [wt/vol] gelatin), 2 μl of a mixture of 10 mM deoxynucleoside triphosphates (equimolar concentrations of dATP, dCTP, dUTP, and dGTP), 1 μl of a 50 μM stock of the NS9 primer, 1 μl of a 50 μM stock of the ITS6 primer, 2 μl of a 25 mM MgCl2 solution, and 0.5 μl (2.5 U) of AmpliTaq polymerase (Perkin-Elmer Cetus Corp., Norwalk, Conn.), with sterile distilled water making up the remaining volume.
To prevent contamination and to ensure assay reproducibility, large batches of the PCR master mixture (excluding the AmpliTaq polymerase) were prepared in advance and stored at −20°C until they were needed, at which time sample DNA templates and the Taq DNA polymerase were added and the reaction mixture was amplified. Positive-displacement pipettes were used in preparing all PCR mixtures. The amplification was carried out in an isolated room, using a GeneAmp 9600 PCR system (Perkin-Elmer Cetus Corp., Foster City, Calif.), and consisted of 30 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min. A final extension was done at 72°C for 10 min.
At the Environmental Protection Agency, PCRs were carried out under somewhat different conditions, briefly noted below. In a model 480 thermocycler (Perkin-Elmer), purified, extracted DNA, primers, and deoxynucleoside triphosphates overlaid with mineral oil were heated at 94°C for 5 min and held at 72°C during addition of premixed enzyme (High Fidelity enzyme mix; Boehringer Mannheim, Indianapolis, Ind.), 1.5 mM MgCl2, 7% glycerol, and buffer provided with the enzyme. The complete mixture was step cycled 30 times at 94°C for 1 min, 52°C for 15 min, and 68°C for 4 min, with a 7-min 68°C extension following the final cycle.
One-microliter subsamples of each PCR amplification mixture were electrophoresed through a 1.2% agarose gel in standard 1× Tris-borate-EDTA buffer. Gels were stained with a 0.5-μg/ml ethidium bromide solution for 30 min, destained in distilled water, and photographed with Polaroid type 57 film.
PCR product yields were estimated by comparison of the fluorescence signals of the products with those of a series of known-mass standards (Gibco/BRL, Grand Island, N.Y.), using a model SI fluorimager (Molecular Dynamics, Sunnyvale, Calif.).
DNA sequencing.
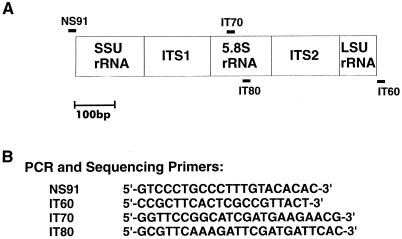
At OMH, the PCR fragments were cleaned and purified by using Gene Max DNA purification cartridges (GIBCO Life Technology, Mississauga, Ontario, Canada). At the Environmental Protection Agency, they were subjected to three cycles of dilution with distilled water and concentration by centrifugation in Centricon 100 concentrators (Millipore Corp., Bedford, Mass.) according to the vendor's instructions for removal of unincorporated primers. Sequencing reactions containing the purified, double-stranded PCR products as templates were performed with ABI PRISM DyeTerminator cycle sequencing kit reagents (Applied Biosystems Inc.) according to the vendor's instructions. The sequencing strategy and the primers used are shown in Fig. 1A and B, respectively. Purification of the extension products was performed by either a phenol-chloroform extraction procedure or direct ethanol precipitation in accordance with the protocols provided by the vendor of the sequencing kits. Electrophoresis and automated analyses of sequence ladders were performed with a model 373A DNA sequencer (Perkin-Elmer). Compilation and editing of multiple sequences generated from each template were performed with the SeqMan analysis program (Perkin-Elmer).
FIG. 1.
Map of the rDNA regions sequenced in this study and descriptions of PCR and sequencing primers. (A) Target sites of the primers on the rDNA map. Forward primers are shown above the map, and reverse primers are shown below it. (B) Sequences of the primers.
Sequence alignment and phylogenetic analysis.
Sequences were initially aligned and examined for redundancy by using the MegAlign program within the Lasergene Biocomputing Software for Windows package (DNASTAR Inc., Madison, Wis.). Unique sequences were realigned with MultAlin 4.0 (F. Corpet, Centre de Recherches, INRA, Toulouse, France), using default parameters. Manual editing of alignment gaps and exclusion of positions having ambiguous alignments were performed with MacClade version 3.0 (21). Phylogenetic trees were constructed from the aligned sequence data by parsimony analysis using the branch and bound search option in PAUP version 3.0 (30). The branch and bound search option was also used for bootstrap analysis (5) of 1,000 replicate samplings of the data in PAUP 3.0. Nucleotide substitutions were equally weighted and unordered, and alignment gaps were treated as missing information in the phylogenetic analysis.
RESULTS
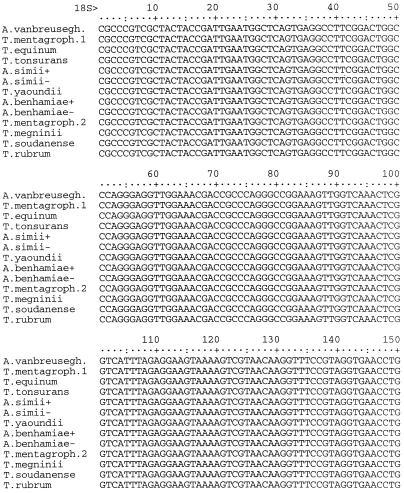
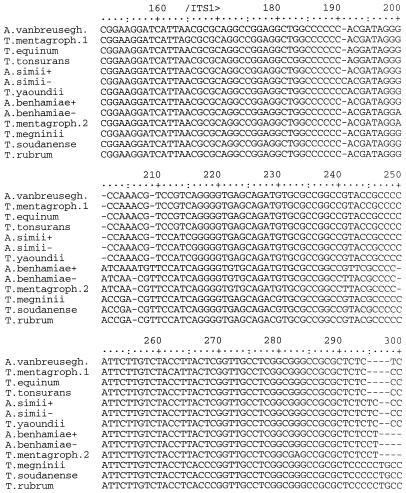
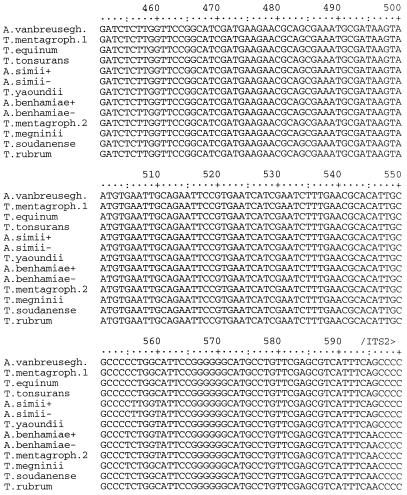
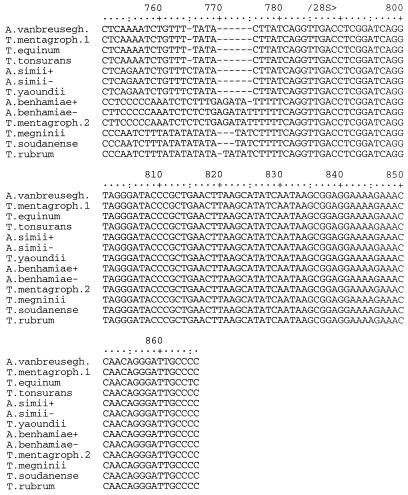
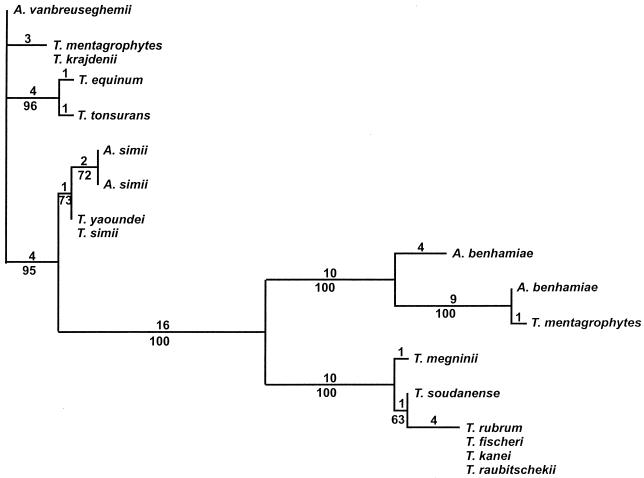
The segment of rDNA sequenced in this investigation included 164 bases at the 3′ end of the small-subunit RNA gene, the entire ITS1 and ITS2 regions and the 5.8S gene, and 81 bases at the 5′ end of the large-subunit RNA gene (Fig. 1). Among the 30 strains and 15 species examined, a total of 13 different sequences were identified (Table 1). The final alignment of these sequences contained a total of 866 nucleotide positions (Fig. 2). Of these, 27 were manually excluded from parsimony analysis due to uncertain alignment and another 787 positions were either constant or uninformative with regard to parsimony. Of the 52 parsimony-informative positions, 3 occurred in the 5.8S gene, 33 occurred in the ITS1 region, and 16 occurred in the ITS2 region. The two most parsimonious trees of 72 steps were generated from a phylogenetic analysis of the alignment data, using the branch and bound search method in PAUP version 3.0 (Fig. 3). The results showed that the ITS sequence of T. rubrum is identical to that of its recent segregates T. raubitschekii, T. kanei, and T. fischeri. It is also highly similar to that of T. soudanense and T. megninii. These species form a well-supported clade with an apparent relationship to the A. benhamiae lineage of T. mentagrophytes isolates. One anthropophilic endothrix tinea capitis species, T. yaoundei, was shown to be closely allied with the simian-parasitizing species Arthroderma simii, while another, T. tonsurans, was shown to be very closely related to the morphologically and physiologically dissimilar equine dermatophyte T. equinum. T. equinum and T. tonsurans, in turn, are on a well-supported branch that contains another major lineage with anamorphs traditionally included in T. mentagrophytes, namely, the Arthroderma vanbreuseghemii lineage. Very closely related to A. vanbreuseghemii and associated anthropophilic T. mentagrophytes anamorphs is the recently described species (16) T. krajdenii.
FIG. 2.
Aligned rDNA sequences of Trichophyton and Arthroderma species. Abbreviations: A. vanbreusegh., A. vanbreuseghemii; T. mentagroph., T. mentagrophytes. The symbols + and − designate mating types, as shown in Table 1. T. mentagrophytes has two sequence types, here designated as T. mentagroph.1 and T. mentagroph.2, exemplified by strains UAMH 8543 and 7339, respectively. Positions 295 to 296, 325 to 329, 339, 386, 389, 639, 641 to 643, 754 to 761, 763, 765, 767, 769, and 771 of this alignment were determined to contain ambiguously aligned nucleotides and were not included in phylogenetic analyses.
FIG. 3.
Phylogenetic relationships of Arthroderma and Trichophyton species inferred from nucleotide sequences of the rDNA ITS1 and ITS2 regions and 5.8S gene. This unrooted phylogram is the consensus of the two most parsimonious trees (72 steps; consistency index = 0.944, retention index 0.971) found by a search conducted in PAUP 3.0 using the branch and bound method. Values above the branches are the total nucleotide changes assigned by the analysis, and values below the branches are the percentages of 1,000 bootstrap analysis replicates in which the branches occurred.
Sequence analysis also clearly distinguished the partially interfertile American-European and African races of A. benhamiae. This distinction has previously been shown in an ITS1 analysis by Makimura et al. (22). A very atypical T. mentagrophytes isolate with red colony reverse coloration and a growth requirement for exogenous inositol, UAMH 7339, clustered very closely to the American-European A. benhamiae but was not identical to A. benhamiae mating tester RV 26680.
DISCUSSION
Unlike some previous analyses based on less-variable nucleic acid characters such as the 18S rDNA sequence and mitochondrial restriction fragment length polymorphism markers (20, 26), ribosomal ITS analysis distinguished species within closely interrelated clades, such as T. rubrum-T. soudanense and T. tonsurans-A. vanbreuseghemii. The former pair of species is a particularly good example of lineages which, despite close phylogenetic affinity, have become highly distinct in all categories of phenotypic and habitat characters, including morphology, physiology, ecology, and pathogenicity. To summarize, T. soudanense is an endemic sub-Saharan Africa species that generally causes endothrix tinea capitis in children and tinea corporis in adults, producing few conidia in culture but with characteristic reflexive branching. It usually, but not always, has requirements for exogenous growth substances (17, 32). T. rubrum is a growth-factor-independent, cosmopolitan fungus, frequently, but not always, at least moderately sporulating, without reflexive branches; it is an agent of mainly tinea pedis and tinea unguium but also tinea corporis, tinea cruris, and tinea manuum. An unequivocal indicator of its differentiation from T. soudanense as a pathogen is that it never causes endothrix tinea capitis, in the typical childhood form of the disease or in any other, although in adults it rarely causes an atypical scalp infection in which infectious elements do not colonize the hair shaft either in the endothrix or the ectothrix pattern (1).
A recent study (23) has shown that the North African-Middle Eastern endemic endothrix tinea capitis species T. violaceum, which is also morphologically and physiologically distinct from T. rubrum, is likewise closely related to it as evidenced by ITS1 homology. In that report the authors state that they interpret their results as signalling that “these ‘species’ of dermatophytes have been overclassified.” We have argued in detail elsewhere, however (29), that the most comprehensible way to apply species concepts to the purely asexual anthropophilic dermatophytes is to adhere to the asexual population genetics concept of the cohesion species outlined by evolutionary phylogenist A. R. Templeton (31). In this concept, asexual isolates are held to be conspecific if they consist of a monophyletic lineage which, through sharing the same fundamental ecological niche, remains capable of generating mutation-bearing variants that can in theory replace (by random chance) or displace (by differential selection) previous and alternative variants throughout the entire population. Habitat disjunction, a measurable ecological character extrinsic to the organism itself (11) (and therefore, for example, not observable in individual cultures), is thus introduced as a criterion for determining whether phenotypic and genotypic differences between related asexual lineages have become significant at the species level. Within ecologically uniform species such as T. soudanense, it is clear that a new mutant, e.g., a hypothetical pathogenically fit white (unpigmented) variant, could, over evolutionary time and with sufficient normal human population interchange, predominate over and then replace the yellow and red T. soudanense variants known today. T. rubrum, however, never causes endothrix tinea capitis; it is thus highly unlikely to give rise to a genetic variant that could replace T. soudanense within its fundamental niche. Nor could a T. soudanense variant realistically be expected to replace T. rubrum throughout its niche. The ecological unity of clades such as T. soudanense and T. violaceum, encompassing populations subject to the asexual gene flow mechanisms of chance replacement or selective displacement (two concepts distinguished in detail by Templeton [31]), validates them as species biologically distinct from at least T. rubrum if not from each other.
This type of analysis, at least for species like anthropophilic dermatophytes, for which ecological distributions are exquisitely well documented, gives us an objective means to determine where observed differences in sequence analyses of variable genetic regions begin to reflect biologically functional species-level divergences in asexual clades. It avoids attributing species status to asexual clades on the basis of minor, evolutionarily recent divergences affecting areas of the genome with little or no relevance to the phenotypic functions which determine the structure of populations. At the same time, it also avoids the taxonomic fusing of completely ecologically and phenotypically diverged populations, likely distinguished by significant differences in DNA regions coding for externally interactive phenotypic characters, on the basis of similarities in more or less arbitrarily structured DNA regions. The latter DNA regions are those which are noncoding or which code for characters other than those giving rise to the strongly selected, ecologically interactive elements of phenotype. With highly related, recently adaptively radiated, sexually reproducing groups such as East African cichlids, the elucidation of mating barriers has shown that species divergence can take place during evolutionary time periods which allow for unexpectedly low levels of divergence in the most-hypervariable regions of DNA in genes, such as ribosomal genes, commonly used in phylogenetic analysis (24). Elucidation of niche separations yielding population-based “demographic exchangeability” (31) barriers (that is, barriers to new alleles pervading the entire population through displacement and/or replacement, as outlined above) may provide similar corrective information when comparable DNA sequences in asexual organisms must be analyzed.
A second example of the applicability of cohesion species concepts to dermatophytes is provided within the present study by the close similarity of ITS1 and ITS2 sequences of T. tonsurans and T. equinum. The former species is a cosmopolitan, strictly anthropophilic endothrix tinea capitis agent. Cultures of T. tonsurans are distinguished by their thick microconidial pedicels, frequently inflated microconidia (“balloon forms”), and thiamine requirement. The latter dermatophyte is restricted to horses except for limited crossover to cause tinea corporis in humans (or rarely other dermatophytoses, but not childhood endothrix tinea capitis), usually after direct interspecies contact. It usually has a nicotinic acid requirement but is on rare occasions autotrophic, and it has uninflated, scarcely pedicellate (or, more often, nonpedicellate) microconidia. Clearly, however closely lineally coderived these species may be, with only a single-base-pair difference manifest in the variable regions assessed in this study, they not only have fixed a number of phenotypic apomorphies (that is, derived [i.e., novel] nonancestral characteristics [10]) within their populations but also have a sharply defined niche separation which prevents T. equinum variants from replacing or displacing T. tonsurans variants and vice versa. Therefore, they remain unequivocally separate species despite their highly similar ITS sequences. A similar argument should be made to maintain the separation between the monkey dermatophyte T. simii and the morphologically entirely distinct, endemic African anthropophilic endothrix tinea capitis agent T. yaoundei.
A recent ITS sequence study by Gräser et al. (7) has placed T. equinum near Epidermophyton stockdaleae in a lineage of nonpathogenic, geophilic dermatophytoids. This placement was based on a misidentified isolate deposited in the Centraalbureau voor Schimmelcultures culture collection (3a).
Besides encompassing some apparently well-defined cohesion species, the present study also encompasses some described species which do not appear to be supported by this species definition. For example, the ITS sequence of T. krajdenii is identical to that of some isolates of the A. vanbreuseghemii clade of T. mentagrophytes (a clade whose anamorphs are called T. interdigitale by some authors [25]). This recently described (16) species was traditionally known as the nodular variant of T. mentagrophytes subsequent to its informal description by Georg and Maechling in 1949 (6). (It should be noted that T. mentagrophytes var. nodulare, an ad hoc varietal name often used for T. krajdenii, is invalid under the International Code of Botanical Nomenclature, lacking both a type specimen and an explicit description with a Latin diagnosis.) Although it displays phenotypically distinct characters such as intense yellow pigmentation and a high proportion of nodular bodies, it is primarily an agent of human lower-body dermatophytoses (especially tinea pedis, tinea unguium, and tinea corporis), just like its relatives classified as anthropophilic strains of T. mentagrophytes sensu lato. In this case, clearly such variants, though distinct, could be ecologically replaced by other variants within their own monophyletic line (except in the highly unlikely event that in their growth on the human lower body they occupy niche space inaccessible to the other variants). T. krajdenii, therefore, despite its distinct morphological anomalies, is not currently supportable as a cohesion species. Further studies will be needed to determine its taxonomic status. If more-detailed genomic population analyses reveal that it is polyphyletic, it will represent only a variant phenotype that cannot be given a taxonomic rank. If, however, genomic population analyses reveal it to be monophyletic, and if it does not include isolates which intergrade with typical asexual, anthropophilic anamorphs of A. vanbreuseghemii (29), it will clearly be set apart as a distinct clade at some level. One proposed species definition, the phylogenetic species concept (PSC) of Nixon and Wheeler (27), would result in such a clade being classified as a species: “We define species as the smallest aggregation of … lineages (asexual) diagnosable by a unique combination of character states in comparable individuals. . . . . Under the PSC, when unique character combinations occur in asexual or clonal forms, these forms should be recognized as distinct species.” Such a definition, applied literally, might result in a large number of new species among the clonally diverse asexual dermatophytes. An alternative, if cohesion species concepts were adopted as suggested above, would be to regard such clades as Linnaean varieties.
The species most closely related to T. rubrum form a complex picture. T. raubitschekii, the most commonly isolated of the T. rubrum segregates, is partially ecologically distinct from typical T. rubrum isolates. As shown prior to its elevation to species status by English (4) and subsequently by Kane et al. (15), it mainly causes tinea corporis and tinea cruris in tropical populations for which wearing of shoes has historically been uncommon. T. rubrum causes a statistically highly significantly greater proportion of foot dermatophytoses than other types of infections, and it mostly affects temperate-region populations that consistently use footwear (15). Typical T. rubrum isolates overlap with T. raubitschekii in frequently causing tinea corporis and tinea cruris, while a small but consistent proportion of T. raubitschekii infections are of the feet and nails. The latter fungus also has some distinct morphological and physiological characters, viz., heavy macroconidiation, the presence of profusely produced and often rounded microconidia, and production of a urease enzyme. In phylogenetic analysis, however, these are symplesiomorphies (shared ancestral-type characteristics, in this case consistent with putative ancestral lineages in the T. mentagrophytes complex) and do not support distinct species status the way a suite of synapomorphies (shared derived characteristics) would (10). The disjunction between populations and body sites supporting T. rubrum and T. raubitschekii, despite its high level of statistical support, is not so profound that population-wide genetic replacement or displacement over time cannot be envisioned. T. raubitschekii, unlike T. soudanense, does not have a significant portion of niche into which T. rubrum is physiologically unequipped to penetrate. Therefore, even though T. rubrum and T. raubitschekii have relatively high levels of morphological, physiological, and ecological distinctiveness, the retention of phylogenetic species status requires further evaluation. Analyses of other variable regions of DNA may aid in elucidating the exact nature of the genetic relationship between T. rubrum and T. raubitschekii.
T. kanei diverges from T. raubitschekii only by a single discrete apomorphy, namely the complete absence of the ability to differentiate microconidia, and by a quantitative character, an attenuation in the degree of urease activity. The sites of its isolation are consistent with sites of T. raubitschekii isolation. Its ultimate taxonomic assignment will clearly depend on that accorded to the latter organism.
T. fischeri was described as having a habitat different from that of T. rubrum. It has not been isolated as an agent of dermatophytosis, but only as a contaminant on laboratory media and uninfected skin and nails (13) and in sputum (28). Kane (13, 14) proposed that it is saprophyte distinct from T. rubrum. Its possession of an ITS sequence identical to that of T. rubrum appears to contradict this assessment. A detailed discussion of the ecological data on this taxon is beyond the scope of the present paper, but it should be noted that the consistently heavily conidial nature of T. fischeri strains is consistent with their regular isolation from uninfected body sites or environmental sources. They may represent a genetic type, within the T. rubrum lineage, possessing an elevated ability to produce aerially disseminable conidia during growth on environmental materials such as shed skin scales.
The ITS sequence data on dermatophytes confirm that ecologically and phenotypically strongly separated species may have only small numbers of differences even in this normally highly variable genetic region. It is clear that understanding population structure and making the best informed decision about species status in certain complex groups, such as the A. vanbreuseghemii anamorphs and the related T. krajdenii, as well as the taxa most closely related to T. rubrum, will require the study of additional loci. Multilocus genotyping studies, as have been done with other medically important fungi (2), must be recommended for further analysis of these important human pathogens.
ACKNOWLEDGMENTS
Maria Witkowska is thanked for technical support. Lynne Sigler (University of Alberta Microfungus Collection), Irene Weitzman, and Guy St.-Germain are thanked for supplying cultures used in this study. The collaboration made possible by A. Dufour is greatly appreciated.
REFERENCES
- 1.Bargman H, Kane J, Baxter M-L, Summerbell R C. Tinea capitis due to Trichophyton rubrum in adult women. Mycoses. 1995;38:231–234. doi: 10.1111/j.1439-0507.1995.tb00057.x. [DOI] [PubMed] [Google Scholar]
- 2.Burt A, Dechairo B M, Koenig G L, Carter D A, White T J, Taylor J W. Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Mol Ecol. 1997;6:781–786. doi: 10.1046/j.1365-294x.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 3.Carbone I, Kohn L M. Ribosomal DNA sequence divergence within internal transcribed spacer 1 of the Sclerotiniaceae. Mycologia. 1993;85:415–427. [Google Scholar]
- 3a.de Hoog, G. S. Personal communication.
- 4.English M P. Ecological aspects of dermatophytes regarded essentially as anthropophilic. In: Preusser H-J, editor. Medical mycology. Stuttgart, Germany: Gustav Fischer Verlag; 1980. pp. 53–59. [Google Scholar]
- 5.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Georg L K, Maechling E H. Trichophyton mentagrophytes (variety nodular) J Invest Dermatol. 1949;13:339–350. [PubMed] [Google Scholar]
- 7.Gräser Y, El Fari M, Vigalys R, Kuijpers A F A, de Hoog G S, Presber W, Tietz H-J. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med Mycol. 1999;37:105–114. [PubMed] [Google Scholar]
- 8.Harmsen D, Schwinn A, Weig M, Brocker E-B, Heesemann J. Phylogeny and dating of some pathogenic keratinophilic fungi using small subunit ribosomal RNA. J Med Vet Mycol. 1995;33:299–303. doi: 10.1080/02681219580000611. [DOI] [PubMed] [Google Scholar]
- 9.Hejtmanek M, Hejtmankova N. Hybridization and sexual stimulation in Trichophyton mentagrophytes. Folia Microbiol. 1989;34:77–79. doi: 10.1007/BF02821327. [DOI] [PubMed] [Google Scholar]
- 10.Hennig W. Phylogenetic systematics. Urbana: University of Illinois Press; 1979. [Google Scholar]
- 11.Horvath C D. Some questions about identifying individuals: failed intuitions about organisms and species. Philos Sci. 1997;64:654–658. [Google Scholar]
- 12.Ishizaki H. Fungal taxonomy based on mitochondria DNA analysis. Jpn J Med Mycol. 1993;34:243–251. [Google Scholar]
- 13.Kane J. Trichophyton fischeri sp. nov.: a saprophyte resembling Trichophyton rubrum. Sabouraudia. 1977;15:231–241. [PubMed] [Google Scholar]
- 14.Kane J. The biological aspects of the Kane/Fischer system for identification of dermatophytes. In: Kane J, Summerbell R C, Sigler L, Krajden S, Land G, editors. Laboratory handbook of dermatophytes. Belmont, Calif: Star Publishing; 1997. pp. 81–129. [Google Scholar]
- 15.Kane J, Krajden S, Summerbell R C, Sibbald G. Infections caused by Trichophyton raubitschekii: clinical and epidemiological features. Mycoses. 1990;33:499–506. doi: 10.1111/myc.1990.33.9-10.499. [DOI] [PubMed] [Google Scholar]
- 16.Kane J, Scott J A, Summerbell R C, Diena B. Trichophyton krajdenii, sp. nov.: an anthropophilic dermatophyte. Mycotaxon. 1992;45:307–316. [Google Scholar]
- 17.Kane J, Summerbell R C, Sigler L, Krajden S, Land G, editors. Laboratory handbook of dermatophytes. Belmont, Calif: Star Publishing; 1997. [Google Scholar]
- 18.Kawasaki M, Aoki M, Ishizaki H, Nishio K, Mochizuki T, Watanabe S. Phylogenetic relationships of the genera Arthroderma and Nannizzia inferred from mitochondrial DNA analysis. Mycopathologia. 1992;118:95–102. doi: 10.1007/BF00442537. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki M, Aoki M, Ishizaki H, Nishimura K, Miyaji M. Phylogeny of Epidermophyton floccosum and other dermatophytes. Mycopathologia. 1996;134:121–128. doi: 10.1007/BF00436718. [DOI] [PubMed] [Google Scholar]
- 20.Leclerc M C, Philippe H, Guého E. Phylogeny of dermatophytes and dimorphic fungi based on large subunit ribosomal RNA sequence comparisons. J Med Vet Mycol. 1994;32:331–341. doi: 10.1080/02681219480000451. [DOI] [PubMed] [Google Scholar]
- 21.Madison W P, Madison D R. MacClade: analysis of phylogeny and character evolution, version 3.0. Sunderland, Mass: Sinauer Associates; 1992. [Google Scholar]
- 22.Makimura K, Mochizuki T, Hasegawa A, Uchida K, Saito H, Yamaguchi H. Phylogenetic classification of Trichophyton mentagrophytes complex strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol. 1998;36:2629–2633. doi: 10.1128/jcm.36.9.2629-2633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makimura K, Tamura Y, Mochizuki T, Hasegawa A, Tajiri Y, Hanazawa R, Uchida K, Saito H, Yamaguchi H. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol. 1999;37:920–924. doi: 10.1128/jcm.37.4.920-924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer A, Kocher T D, Basasibwaki P, Wilson A C. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature. 1990;347:550–553. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki T K, Takada K, Watanabe S, Kawasaki M, Ishizaki H. Taxonomy of Trichophyton interdigitale (Trichophyton mentagrophytes var. interdigitale) by restriction enzyme analysis of mitochondrial DNA. J Med Vet Mycol. 1990;28:191–196. doi: 10.1080/02681219080000251. [DOI] [PubMed] [Google Scholar]
- 26.Nishio K, Kawasaki M, Ishizaki H. Phylogeny of the genus Trichophyton using mitochondrial DNA analysis. Mycopathologia. 1992;117:127–132. doi: 10.1007/BF00442772. [DOI] [PubMed] [Google Scholar]
- 27.Nixon K C, Wheeler Q D. An amplification of the phylogenetic species concept. Cladistics. 1990;6:211–223. [Google Scholar]
- 28.Rosenthal S A, Scott J S, Summerbell R C, Kane J. First isolation of Trichophyton fischeri in the United States. J Clin Microbiol. 1998;36:3389–3391. doi: 10.1128/jcm.36.11.3389-3391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summerbell R C, Li A, Haugland R. What constitutes a functional species in the asexual dermatophytes? Microbiol Cult Collect. 1997;13:29–37. [Google Scholar]
- 30.Swofford D L. PAUP: Phylogenetic Analysis Using Parsimony, version 3.0. Sunderland, Mass: Sinauer Associates; 1991. [Google Scholar]
- 31.Templeton A R. The meaning of species and speciation: a genetic perspective. In: Otte D, Endler J A, editors. Speciation and its consequences. Sunderland, Mass: Sinauer Associates Inc.; 1989. pp. 3–27. [Google Scholar]
- 32.Weitzman I, Salkin I F, Rosenthal S A. Evaluation of Trichophyton agars for identification of Trichophyton soudanense. J Clin Microbiol. 1983;18:203–205. doi: 10.1128/jcm.18.1.203-205.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]