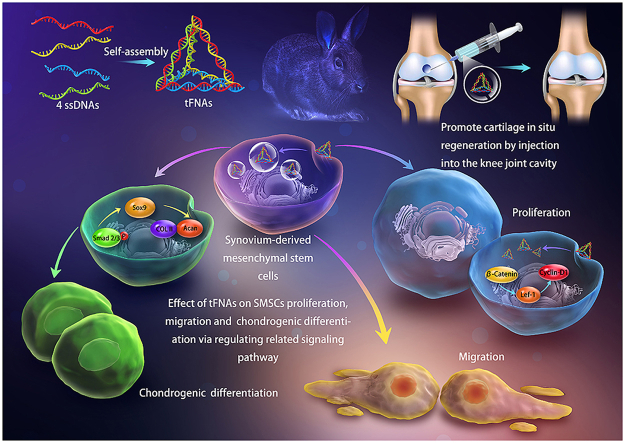
Abstract
Many recent studies have shown that joint-resident mesenchymal stem cells (MSCs) play a vital role in articular cartilage (AC) in situ regeneration. Specifically, synovium-derived MSCs (SMSCs), which have strong chondrogenic differentiation potential, may be the main driver of cartilage repair. However, both the insufficient number of MSCs and the lack of an ideal regenerative microenvironment in the defect area will seriously affect the regeneration of AC. Tetrahedral framework nucleic acids (tFNAs), notable novel nanomaterials, are considered prospective biological regulators in biomedical engineering. Here, we aimed to explore whether tFNAs have positive effects on AC in situ regeneration and to investigate the related mechanism. The results of in vitro experiments showed that the proliferation and migration of SMSCs were significantly enhanced by tFNAs. In addition, tFNAs, which were added to chondrogenic induction medium, were shown to promote the chondrogenic capacity of SMSCs by increasing the phosphorylation of Smad2/3. In animal models, the injection of tFNAs improved the therapeutic outcome of cartilage defects compared with that of the control treatments without tFNAs. In conclusion, this is the first report to demonstrate that tFNAs can promote the chondrogenic differentiation of SMSCs in vitro and enhance AC regeneration in vivo, indicating that tFNAs may become a promising therapeutic for AC regeneration.
Keywords: Tetrahedral framework nucleic acids, Articular cartilage regeneration, Mesenchymal stem cells, Chondrogenic differentiation
Graphical abstract
Highlights
-
•
Tetrahedral framework nucleic acids (tFNAs) can promote SMSCs proliferation by activating the Wnt/β-catenin pathway.
-
•
tFNAs can promote SMSCs migration in vitro and vivo.
-
•
tFNAs can promote SMSCs chondrogenic differentiation by regulating the TGF/Smad2/3 signaling pathway.
-
•
tFNAs show improved articular cartilage in situ regeneration activity in vivo.
1. Introduction
Due to the lack of vascular, nervous and lymphatic systems, the repair of damaged articular cartilage (AC) has always been a major challenge in clinical research and regenerative medicine [1,2]. Traditional treatment methods, such as bone marrow stimulation, autografts, or autologous chondrocyte implantation, have achieved some success, but these strategies are limited by various issues, such as fibrocartilage generation and insufficient graft resources, and the long-term effect is unsatisfactory [[3], [4], [5], [6]]. In recent years, mesenchymal stem cell (MSC)-based tissue engineering strategies have shown promising results in the regeneration of AC [[7], [8], [9]]. MSCs are easily isolated, fibroblast-like, multipotent cell populations with a self-renewing capacity and are considered to be a promising cell type in the field of tissue engineering [7,10]. To date, MSCs can be isolated from many adult tissues, such as bone marrow, synovium, adipose tissue, and skeletal muscle [9]. In particular, the synovium is a thin layer of tissue lining the surface of cartilage or tendons, which maintains a cavity filled with synovial fluid [11,12]. Since 2001, when De Bari et al. first successfully isolated synovium-derived mesenchymal stem cells (SMSCs) from human synovial tissue, SMSCs have been widely used for cartilage regeneration due to their greater chondrogenic differentiation potential in vitro than MSCs from other tissues [[13], [14], [15]]. However, the applications of exogenous MSCs are limited by abnormal cell phenotypes, reduced differentiation potential and poor self-renewal capacity in in vitro culture [[7], [8], [9]]. In addition, regenerated cartilage often exhibits hypertrophy and fibrosis after transplantation of exogenous MSCs into cartilage defects, which seriously affect the functions of newly regenerated AC [8,9,16].
To solve these problems, scientists have recently focused on the in situ regeneration of AC based on endogenous MSCs [17,18]. Some studies have shown that the migration of native joint-resident MSCs is essential for chondrogenesis during embryogenesis, and SMSCs may be the main driver of cartilage repair in adults [19]. However, both the insufficient number of MSCs and the lack of an ideal regenerative microenvironment in the defect area will seriously affect the regeneration of AC [20,21]. Thus, exploring and developing a strategy to induce more native joint-resident MSCs to migrate to defect areas and then alter the regenerative microenvironment to promote cell proliferation, differentiation and secretion of extracellular matrix could be a potential solution for cartilage repair [22,23].
Here, we used tetrahedral frame nucleic acids (tFNAs), novel DNA nanomaterials [24], to promote in situ regeneration of AC. tFNAs are stable tetrahedral three-dimensional DNA nanomaterials composed of four predesigned single strands of DNA (ssDNAs), which have shown strong potential in the field of biomedical science [[25], [26], [27]]. tFNAs can regulate cell biological functions through caveolin-mediated endocytosis to enter cells and influence different signaling pathways [28,29]. Previous studies have proven that tFNAs can promote the proliferation and osteogenic differentiation of adipose-derived MSCs (ADSCs) [30]. Furthermore, tFNAs can regulate the phenotype and proliferation of chondrocytes [31]. However, the direct effect of tFNAs on AC regeneration has not been reported.
To explore whether tFNAs have positive effects on AC in situ regeneration, in this study, we first successfully synthesized tFNAs and then demonstrated that tFNAs can be abundantly taken up by SMSCs. Next, the effects of tFNAs on the biological functions of SMSCs in vitro, including cell proliferation, cell migration, and cell chondrogenic differentiation, were investigated. Furthermore, through phospho-antibody array and Western blot analysis, the signaling pathways through which tFNAs may play a role in the chondrogenic differentiation of SMSCs were determined. Finally, we injected tFNAs into the articular cavity of rabbit cartilage defect models to investigate the tFNA-mediated enhancement of in situ AC regeneration in vivo. We believe that our findings will pave the way for the application of tFNAs in the field of AC regeneration.
2. Materials and methods
2.1. Synthesis of tFNAs
tFNAs were prepared on the basis of previous studies [24,32]. Four predesigned ssDNA sequences (Table 1) stored at −20 °C were centrifuged at 10,000 g for 10 min at 4 °C and dissolved in DNase-free water to a concentration of 100 μM. Next, 1 μl of each ssDNA at a concentration of 100 μM was added to 96 μl of TM buffer solution containing 10 mM Tris-HCl and 50 mM MgCl2 (pH = 8.0) in a 200 μl centrifuge tube. After the tube was mixed by gentle vortexing, the mixture was denatured at 95 °C for 10 min and then cooled rapidly at 4 °C for 30 min. Finally, the solution was kept in a thermal cycler for 20 min to maintain a tetrahedral structure.
Table 1.
Base sequence of single-stranded DNAs (ssDNAs) used to synthesize tFNAs.
| ssDNA | Direction | Base Sequence |
|---|---|---|
| S1 | 5′→3′ | ATTTATCACCCGCCATAGTAGACGTATCACCAGGCAGTTGAGACGAACATTCCTAAGTCTGAA |
| S2 | 5′→3′ | ATTTATCACCCGCCATAGTAGACGTATCACCAGGCAGTTGAGACGAACATTCCTAAGTCTGAA |
| S3 | 5′→3′ | ACTACTATGGCGGGTGATAAAACGTGTAGCAAGCTGTAATCGACGGGAAGAGCATGCCCATCC |
| S4 | 5′→3′ | ACGGTATTGGACCCTCGCATGACTCAACTGCCTGGTGATACGAGGATGGGCATGCTCTTCCCG |
2.2. Characterization of tFNAs
We used polyacrylamide gel electrophoresis (PAGE) and transmission electron microscopy (TEM) to confirm the successful synthesis of tFNAs. In addition, the particle size and electric potential were measured by dynamic light scattering and zeta potential assays.
2.3. Cell culture
The study was approved by the Ethical Committee of PLA. Isolation and culture of SMSCs were performed according to a previous method [33,34]. In general, articular synovial tissue was obtained from the knee joints of male New Zealand rabbits (1 month old, 500–800 g) on a sterile bench. The synovial tissue was rinsed with PBS containing 0.5% penicillin/streptomycin (Sigma, USA), shredded with ophthalmic scissors and digested in Dulbecco's modified Eagle's medium/F12 (Corning, USA) containing 0.3% collagenase I (Gibco, USA) for 30 min. The SMSCs were centrifuged and resuspended in growth medium [DMEM/F12 with 10% fetal bovine serum (FBS, Gibco)]. The SMSCs were then transferred to a 25 cm2 culture flask (Corning) and cultured. When the cells reached 90% confluence, they were subcultured at a rate of 1:2. Third-passage SMSCs were used for the subsequent experiments.
2.4. Uptake of tFNAs by SMSCs
tFNAs loaded with cyanine 5 (Cy5-tFNAs) and ssDNAs loaded with Cy5 (Cy5-ssDNAs) were added to SMSCs to confirm the effect of the interaction between tFNAs and SMSCs. SMSCs were seeded on cell slides in a 24-well plate at 1 × 104 cells per well and cultured in normal growth medium for 1 day. Then, the cells were cultured with Cy5-tFNAs (250 nM) or Cy5-ssDNAs (250 nM) in fresh DMEM/F12 with only 1% FBS, and each group had 3 replicates. After 12 h of treatment, the cell samples were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.5% Triton X-100 for 30 min. Next, FITC-labeled phalloidin (1:60; Beyotime, Shanghai, China) was used to stain the cell cytoskeleton for 40 min. DAPI (4′,6-diamidino-2-phenylindole) (1:200; Beyotime, Shanghai, China) was then used to stain the cell nucleus for 10 min. Finally, images of all samples were captured via fluorescence microscopy.
2.5. Cell proliferation assay
We used Cell Counting Kit-8 (CCK-8) assays, EdU imaging and cell cycle assay kits to assess the impact of tFNAs on SMSC proliferation. For the CCK-8 assay, SMSCs were seeded in a 96-well plate at a density of 8000 cells per well and cultured in conventional proliferation medium for 24 h. The normal medium was replaced with medium with tFNAs at different concentrations (0 nM, 62.5 nM, 125 nM, 250 nM and 375 nM), and then, fresh CCK-8 reagent was added to each well and incubated for 2 h before measurement. Each concentration had 4 replicates. For EdU staining, we used a Cell-Light EdU in vitro kit (RiboBio, Guangzhou, China). SMSCs were cultured on cell slides in a 24-well plate with approximately 1 × 104 cells per well for 24 h, followed by treatment with DMEM/F12 with 1% FBS and tFNAs at different concentrations for another 24 h. Then, the medium was replaced by a 50 mM EdU solution, and the samples were incubated. EdU and DNA staining was then performed using Apollo and Hoechst solutions as per standard procedures. Each group of EdU staining had 3 replicates. The cell cycle was assessed via flow cytometry. In brief, we used trypsin solution to digest and collect treated cells and then used 70% ethanol to fix the cells at 4 °C for 24 h. After PBS washes, 500 μL of propidium iodide (PI) staining solution was added to each sample at 37 °C for 30 min. Subsequently, the percentages of G1-, S-, and G2-phase cells were measured using a flow cytometer (BD FACSCelesta, USA) and analyzed by FlowJo software. Each group had 3 replicates.
2.6. In vitro and in vivo cell migration assays
2.6.1. In vitro cell migration assay
Parallel cell migration (scratch wound healing experiments) and vertical cell migration assays (Transwell chamber experiments) were used to assess cell migration. For scratch wound healing experiments, SMSCs were seeded in a 6-well plate at approximately 1 × 105 cells per well. When the confluence of the seeded SMSCs in a 6-well plate reached 80%–90%, two crossing linear scratches were generated by a pipette tip. The cells were then cultured in DMEM/F12 with tFNAs at concentrations of 0 nM, 125 nM and 250 nM and 1% FBS. Wound healing images were acquired after 0, 12 and 24 h, and the size of the scratches was recorded, measured and analyzed. Transwell chamber assays were performed using Transwell permeable plates (Corning, USA) consisting of polycarbonate Transwell inserts (8 μm pore diameter) and a 24-well plate. The cells (2 × 104) were seeded in the upper half of the insert membrane and cultured with tFNAs (0 nM, 62.5 nM, 125 nM and 250 nM) for 24 h. We used DMEM/F12 with 5% FBS as a chemoattractant in the lower compartment of the 24-well plate, and in the upper half of the insert membrane, SMSCs were cultured in DMEM/F12/tFNAs with 1% FBS. The cells were fixed with 4% paraformaldehyde for 30 min. After the cells were rinsed with PBS, DAPI (1:200; Beyotime, Shanghai, China) was used to stain them for 10 min. Images of migrating SMSCs in the lower half of each insert were observed by fluorescence microscopy, and the number of cells that migrated to the lower part of the insert was counted in three microscopic views per well. Subsequently, the difference in the mean cell number per well was analyzed. Each group had 3 replicates.
2.6.2. In vivo endogenous cell migration study
Six male New Zealand rabbits (6 months old, 3.0–3.3 kg) were prepared and randomly divided into 2 groups: the negative control group and the tFNA group. Then, we established the same rabbit cartilage defect models as described in section 2.12 to investigate the ability of tFNAs to enhance endogenous SMSC migration in vivo. In brief, a cylindrical cartilage defect (3.5 mm in diameter and 1 mm in depth) was drilled at the trochlear groove of both limbs. After surgery, the animals in the control group were injected with 100 μL of 0.9% saline solution in the knee joint cavity daily during the first seven days. For the tFNA group, the knee joint cavity was injected with tFNAs (250 nM) prepared with 0.9% saline solution daily. After 7 days, the rabbits were euthanized, and the regenerated tissue in the damaged area was sampled. CD73 and CD105 were defined as MSC-specific markers, and the effects of tFNAs on SMSC migration in vivo were determined by immunofluorescence staining. Briefly, the samples were fixed with 4% paraformaldehyde for 30 min. Triton X-100 (0.5%) was used to permeabilize regenerated tissue for 30 min. After PBS washes, the samples were blocked with immune blocking solution (Beyotime, Shanghai). The samples were then incubated overnight with primary antibodies against CD73 (1:500, Novus Biologics) and CD105 (1:500, Novus Biologics) at 4 °C, followed by secondary antibodies conjugated with Alexa Fluor 488 and Fluor 594 (1:100, Abcam, Cambridge, UK) for 1 h and DAPI (1:200; Beyotime, Shanghai, China) for 10 min. The number of CD73 and CD105 double-positive cells was determined by TCS-SP8 (Leica, Germany) confocal microscopy.
2.7. Accelerating effect of tFNAs on chondrogenic differentiation of SMSCs
Chondrogenic differentiation of SMSCs was induced through pellet aggregate culture according to previous studies [35]. Briefly, 3 × 105 SMSCs were centrifuged at 250 g for 5 min in a 15-mL conical tube to form pellets. Then, the SMSC pellets were maintained in basal media at 37 °C with 5% CO2 for 24 h and cultured in stem cell chondrogenic complete differentiation medium (MSCgoTM, Biological Industries, Israel) for up to 14 and 21 days. For confirmation of the effect of tFNAs on SMSC chondrogenic differentiation, induction medium with a final concentration of 250 nM tFNAs was used. The same volume of TM buffer was added to the induction medium for the control groups. The medium was changed every 2 days. After 14 and 21 days of induction, the cell pellets were sectioned into 5 μm frozen slices and then stained with hematoxylin and eosin (H&E), safranin O, Alcian blue and immunohistochemistry (IHC) for collagen II. The samples were stained with H&E, safranin O, and Alcian blue following the manufacturer's standard protocols. The procedures for immunohistochemical staining of type II collagen were as follows: in brief, after the section was dewaxed and washed, endogenous peroxidase was removed with hydrogen peroxide. Then, 0.5% Triton X-100 was used for permeabilization. After PBS washes, the samples were blocked. The sections were incubated with anti-collagen II primary antibody (1:200, Novus, NY, USA) overnight at 4 °C. Eventually, an immunohistochemical secondary antibody was added, and a chromogenic agent was used. After dewaxing and sealing, the slides were observed and photographed under a microscope. In addition, total RNA and proteins were extracted for RT-qPCR and western blotting (WB), and detailed experimental procedures are available in sections 2.10, 2.9. Each group had three replicates at each time point.
2.8. Phospho-antibody array analysis
For comparison of the activation and expression of phosphoproteins involved in chondrogenic differentiation, SMSCs were seeded in a 6-well plate at 1 × 105 per well and then treated with 0 nM tFMAs (control group) and 250 nM tFNAs in stem cell chondrogenic differentiation medium for 24 h. Then, the samples were treated with cell lysis buffer (Full Moon BioSystems, USA), and at least 3 million cells were collected for each group. Next, phosphatase inhibitor and protease inhibitor were added to each sample at a volume ratio of 1:50, and each sample was treated with a tube of Full Moon's cell lysis beads. After the above steps, the proteins of the samples were extracted, and then, Phospho Explorer Antibody Arrays (Full Moon BioSystems, USA) were used to react with the protein according to the standard procedures provided by Full Moon. The array contained 1318 antibodies, and each of them had 2 replicates. In brief, the proteins of samples were labeled with biotin and then diluted with 6 mL of Coupling Solution (Full Moon BioSystems, USA). Next, the biotin-labeled proteins were incubated with the antibody microarray for 2 h. Finally, the related proteins were identified using a Cy3-chain avidin detection system. Wayen Biotechnologies (Shanghai) performed the experiment and analyzed the data. The extent of protein phosphorylation was calculated by the following equation: phosphorylation ratio = phosphorylated value/unphosphorylated value.
2.9. qPCR
For analysis of the expression of genes related to bulbous chondrogenic differentiation of pellets at 14 and 21 days, total RNA was extracted from the SMSC pellets after 14 and 21 days of culture using a Cell Total RNA Isolation Kit (Foregene, Chengdu, China) and then converted to complementary DNA using 5x RT Master Mix (Toyobo, Osaka, Japan). RT-PCR was performed on a StepOneTM Real-Time PCR system (Applied Biosystems, USA) using 2x RealStar Green Fast mixture (Genstar, Beijing, China) according to the manufacturer's protocols. The gene primer sequences for Col2al, Col1a1, Sox9, Aggrecan and GAPDH are listed in Table S1. To test the reliability of the primers, we established fusion curves for each reaction system, and there was no nonspecific amplification in the dissolution curves. Relative gene expression was normalized to that of a housekeeping gene (GAPDH) and calculated by the 2-ΔΔCT method.
2.10. Western blot
The protein expression of β-catenin, Lef-1, cyclin D, chondrogenic-related proteins (Sox9, collagen II and aggrecan), Smad 2/3, phospho-Smad2, and phospho-Smad3 was examined by western blotting. SMSCs seeded in 6-well plates were treated with and without exposure to 250 nM tFNAs for 24 h and used for the detection of proteins related to proliferation and chondrogenic differentiation. Cartilage pellets with and without exposure to tFNAs for 14 and 21 days were used to detect chondrogenic-related proteins. Total proteins of all samples were collected using cellular protein extraction reagent (Beyotime, Shanghai, China). A mixture of protein samples and a 1/4 volume of 5 × loading buffer was prepared and then boiled for 10 min. Next, the target proteins were separated by 10% SDS-PAGE gels. After the relevant proteins were transferred to polyvinylidene fluoride (PVDF) membranes, 5% skim milk was used to block PVDF membranes for 1 h. Relevant membranes were then incubated with primary antibodies consisting of anti-β-actin (1:5000, Immunoway, TX, USA), anti-β-catenin (1:2000, Abcam, Cambridge, England), anti-Lef-1 (1:1000, Novus, NY, USA), anti-cyclin D1 (1:1000, Novus, NY, USA), anti-collagen II (1:1000, Novus, NY, USA), anti-Sox9 (1:1000, Abcam, Cambridge, England), anti-Aggrecan (1:2000, Invitrogen, CA, USA), anti-phospho-Smad2 (1:1000, Cell Signaling Technology, MA, USA), anti-phospho-Smad3 (1:1000, Cell Signaling Technology, MA, USA), and anti-Smad 2/3 (1:1000, Cell Signaling Technology, MA, USA) overnight at 4 °C. After washing and incubation with the secondary antibody (Abcam, Cambridge, England) for 1 h, the protein bands were detected using an enhanced chemiluminescence (ECL) system. β-actin was used as the internal control.
2.11. Immunofluorescence
For immunofluorescence analysis, SMSCs were seeded onto 24-well plates at 1 × 104 cells per well and then treated with tFNAs (0, 250 nM). After 24 h of treatment, the cell samples were rinsed with PBS three times and fixed in cold 4% formaldehyde solution for 20 min. After 3 washes with PBS again, Triton X-100 (0.5%) was used to permeabilize the SMSCs for 30 min, and then, the samples were blocked with immune blocking solution (Beyotime, Shanghai, China) for 30 min. Fixed SMSCs that were rinsed with PBS were then incubated with anti-β-catenin (1:200, Abcam, Cambridge, England), anti-Lef-1 (1:200, Novus, NY, USA), anti-cyclin D1 (1:200, Novus, NY, USA), anti-phospho-Smad2 (1:2000, Cell Signaling Technology, MA, USA), and anti-phospho-Smad3 (1:200, Cell Signaling Technology, MA, USA) primary antibodies overnight at 4 °C. Fluorescence-conjugated secondary antibody was then combined with primary antibodies for 1 h at 37 °C. DAPI (1:200; Beyotime, Shanghai, China) was applied to stain the cell nucleus for 10 min, and phalloidin (1:60; Beyotime, Shanghai, China) was used to stain F-actin for 40 min. Finally, cell images were observed by fluorescence microscopy (Keyence, Osaka, Japan).
2.12. Animal models
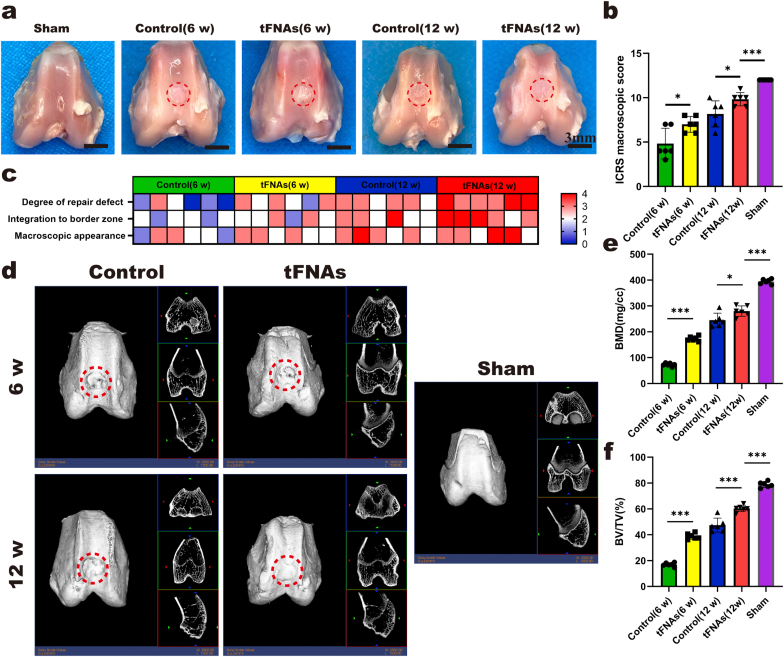
Animal experiments were approved by the Institutional Animal Care and Use Committee at PLA General Hospital. Twenty-four male New Zealand rabbits (6 months old, 3.0–3.3 kg) were prepared and randomly allocated to three groups, with 8 rabbits in each group: the negative control group, the tFNA group and the sham group. After anesthetization by intramuscular injections of ketamine (50 mg/kg) and xylazine (4 mg/kg), the knee was exposed by a medial parapatellar approach. Lateral dislocation of the patella was followed by flexion of the knee. For the control group and the tFNA group, a cylindrical cartilage defect (3.5 mm in diameter and 1 mm in depth) was drilled at the trochlear groove with a trephine, which was then removed and rinsed to remove debris. For the sham group, the patella was immediately reset after dislocation, and then, the joint capsule was sutured. After surgery, the animals in the control group and sham group were injected with 100 μL of 0.9% saline solution in the knee joint cavity daily during the first seven days. For the tFNA group, the knee joint cavity was injected with tFNAs (250 nM) prepared with 0.9% saline solution daily. After the first seven days, we changed to weekly injection of 500 μL of 0.9% saline solution or tFNAs (250 nM) to minimize the risk of intraarticular injection. After 6 and 12 weeks, four rabbits per group were euthanized at each time point, and the femurs of the rabbits were sampled. The cylindrical cartilage defect area (3.5 mm in diameter 1 mm in depth) in the femur samples was observed and photographed. For macroscopic evaluation, images were given to a researcher with extensive experience in cartilage regeneration and with no knowledge of the grouping to score according to the International Cartilage Repair Society (ICRS) macroscopic evaluation guidelines [36].
2.13. Microcomputed tomography (micro-CT) analysis
After the macro evaluation, the samples were placed and scanned in the General Electric (GE) Explorer Locus SP system (GE, Boston, MA, USA). After scanning, the appropriate sagittal and coronal cross-sections of cartilage regeneration were adjusted in the software, 3D reconstruction was performed on each femur sample, and a columnar region of interest (ROI) (diameter 3.5 mm, height 1 mm) was selected at the sample defect regeneration area. Then, the bone mineral density (BMD) and bone volume/tissue volume (BV/TV) of the ROI of each sample were analyzed.
2.14. Histomorphological evaluation
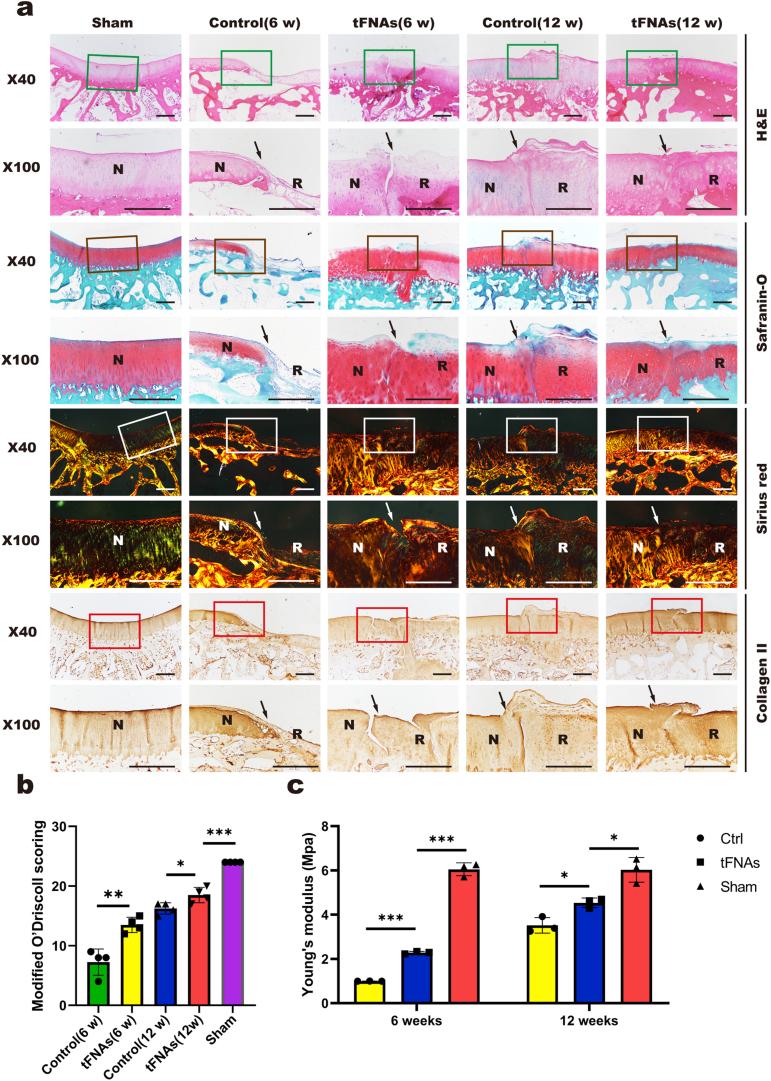
After micro-CT analysis, the femoral samples from the three groups were fixed in paraformaldehyde for 3 days, decalcified for 1 month and continuously trimmed during the process. After decalcification and dehydration, the samples were embedded in paraffin, sectioned into 5 μm slices and stained with H&E, safranin O, and Sirius red following standard protocols. The procedures for immunohistochemical staining of type II collagen were as follows: in brief, after the section was dewaxed and washed, endogenous peroxidase was removed with hydrogen peroxide. Then, 0.5% Triton X-100 was used for permeabilization. After PBS washes, the samples were blocked. The sections were incubated with anti-collagen II primary antibody (1:200, Novus, NY, USA) overnight at 4 °C. Eventually, an immunohistochemical secondary antibody was added, and a chromogenic agent was used. After dewaxing and sealing, the slides were observed and photographed under a microscope. All images were given to a researcher with extensive experience in histopathology of cartilage and with no knowledge of the grouping to score according to the elevated O'Driscoll score system evaluation guidelines [21].
2.15. Biomechanical testing
The mechanical properties of the samples at 6 and 12 weeks were tested by a biomechanical testing machine (Bose, 5100) after surgery. Young's modulus, reflecting mechanical strength, was calculated using the following formula: E=(FL)/(SΔL), where F is the applied pressure and ΔL is the deformation variable.
2.16. Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 statistical software with one-way ANOVA and Student's t-test. *p < 0.05, **p < 0.01, and ***p < 0.001 represent thresholds for statistical significance. All quantitative data are shown as the mean ± standard deviation (SD).
3. Results
3.1. Characterization of tFNAs and cellular uptake
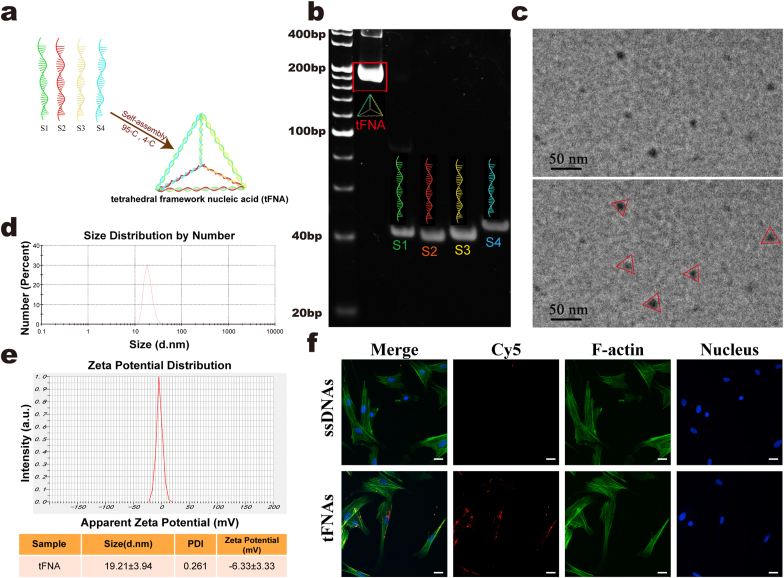
tFNAs were successfully assembled from four predesigned ssDNAs, as shown in Table 1, via specific base pairing. A schematic diagram of tFNA synthesis is shown in Fig. 1a. Each ssDNA self-assembled into a triangular facet and then hybridized with the adjacent sides of the other three ssDNA-forming facets. Finally, a tetrahedral framework was formed. The PAGE results revealed that the four ssDNA molecules formed tFNAs (Fig. 1b). The morphology of the tFNAs was observed using TEM, and the triangle shape of the tFNAs is shown in Fig. 1c. The results of PAGE and TEM confirmed that we successfully synthesized tFNAs. In addition, the average particle size of the tFNAs was 19.21 ± 3.94 nm, as assessed by dynamic light scattering (Fig. 1d). The zeta potential assay revealed that the surface potential of the tFNAs was −6.33 ± 3.33 mV (Fig. 1e), which indicated that the tFNAs were stable in TM buffer solution.
Fig. 1.
Synthesis, characterization and SMSC uptake of tFNAs. a. A diagram for the synthesis of tFNAs. b. The results of PAGE showing the successful manufacture of tFNAs. c. TEM images indicating the average size and morphology of tFNAs. d. Size distribution of tFNAs. e. Zeta potential distribution of tFNAs. f. Cy5-ssDNAs and Cy5-tFNAs taken up by SMSCs. (nucleus: blue, F-actin: green, Cy5: red). Scale bars are 25 μm. Data are presented as mean ± SD (n = 3).
To investigate the ability of SMSCs to take up ssDNAs and tFNAs, we incubated the cells with Cy5-tFNAs or Cy5-ssDNAs at a concentration of 250 nM. After 12 h of treatment, we observed that the fluorescence signal of Cy5 in the tFNA-treated SMSCs was visibly stronger than that in the ssDNA-treated SMSCs (Fig. 1f). The results suggested that tFNAs can be easily taken up by SMSCs in abundance, while ssDNAs were not absorbed.
3.2. tFNAs promoted the proliferation of SMSCs by regulating the cell cycle
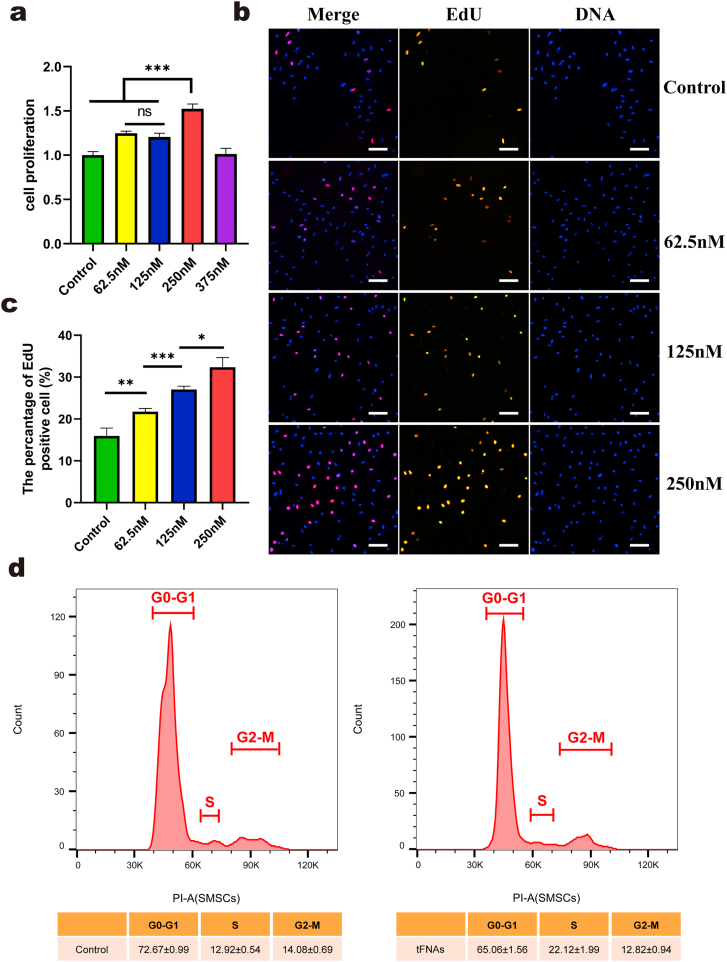
To determine whether tFNAs can affect the proliferation of SMSCs, we performed three assays. First, we used CCK-8 assays to initially investigate the effect on proliferation and screened for the optimal concentration. The results of CCK-8 assays (Fig. 2a) clearly showed that when the concentration of tFNAs ranged from 62.5 nM to 250 nM, SMSCs showed stronger proliferation than the control cells. In addition, tFNAs at a concentration of 250 nM had the optimal effect on SMSC proliferation. For the second assay, we used EdU staining to further confirm the effects of tFNAs on SMSC proliferation. The results of the EdU analysis (Fig. 2b and c) also indicated that after treatment with tFNAs, the number of EdU-positive SMSCs was greater than that of the control group. Furthermore, compared with those of both the 62.5 nM and 125 nM groups, the number of cells with EdU fluorescence in the 250 nM group was greater. Altogether, tFNAs could promote the proliferation of SMSCs, and 250 nM was the most suitable concentration.
Fig. 2.
The proliferation of SMSCs were promoted by exposure to tFNAs. a. CCK-8 results of SMSCs proliferation under the treatment of tFNAs with different concentrations. Data are presented as mean ± SD (n = 4). b. Immunofluorescence micrographs of EdU staining (EdU: orange, nucleus: blue). Scale bars are 100 μm. c. Semi-quantitative analysis of EdU positive cell ratio based on images. Data are presented as mean ± SD (n = 3). d. Flow-cytometry results of the cell cycle of SMSCs treated with or without 250 nM tFNAs for 24 h. Data are presented as the mean ± SD (n = 3). Statistical analysis: *p < 0.05, **p < 0.01, ***p < 0.001, n.s. represents no significant difference.
Therefore, we then chose 250 nM tFNAs to treat SMSCs and assessed the effect on the cell cycle after 24 h of treatment via flow cytometry. Based on the cell cycle results (Fig. 2d), we concluded that the proportion of SMSCs in S phase, which was vital for DNA replication after 250 nM tFNA treatment, was significantly greater than that of the control cells, while the proportion in G1 phase was decreased. Consequently, we demonstrated that tFNAs could promote the proliferation of SMSCs by regulating the cell cycle.
3.3. tFNAs affected the proliferation of SMSCs by activating the Wnt/β-catenin pathway
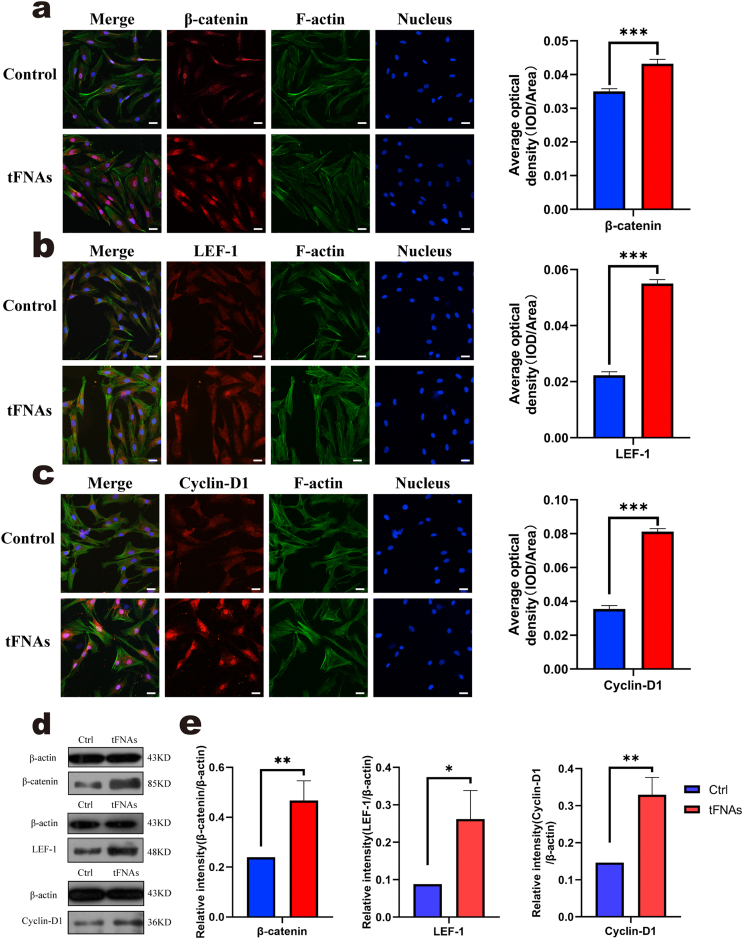
According to a previous study, tFNAs can promote the proliferation of rat chondrocytes by activating the classic Wnt/β-catenin pathway [31]. Therefore, in our study, we evaluated the expression of Wnt/β-catenin pathway-related proteins, including β-catenin, Lef-1 and cyclin D1, by immunofluorescence and Western blot analysis. For immunofluorescence, as shown in Fig. 3a, b, and c, the fluorescence intensity of these three proteins in the tFNA treatment group was significantly higher than that in the control group. Subsequently, WB (Fig. 3d) further confirmed that the expression of β-catenin, Lef-1 and cyclin D1 after 24 h of treatment with tFNAs was higher than that of the control group. In detail, the statistical analysis of the western blots showed that the upregulated expression of these proteins after exposure to 250 nM tFNAs was significant (β-catenin increased up to 1.95-fold, Lef-1 was 2.97-fold, cyclin D1 was 2.25-fold, Fig. 3e). Therefore, we concluded that tFNAs can promote the proliferation of SMSCs by activating the Wnt/β-catenin pathway.
Fig. 3.
tFNAs affected the proliferation of SMSCs by activating the Wnt/β-catenin pathway. a-c. Immunofluorescence detection of Wnt pathway-related proteins. (F-actin: green, nucleus: blue, and protein: red). Scale bars are 25 μm. Quantitative analysis of the average optical density of fluorescence images. Data are presented as the mean ± SD (n = 4). Statistical analysis: ***p < 0.001. d. WB analysis of the expression levels of Wnt pathway-related proteins (the internal control was β-actin). e. Quantitative analysis of the expression levels of Wnt pathway-related proteins. Data are presented as the mean ± SD (n = 3). Statistical analysis: *p < 0.05, **p < 0.01.
3.4. tFNAs promoted the migration of SMSCs both in vitro and in vivo
3.4.1. tFNAs increased SMSC migration in vitro
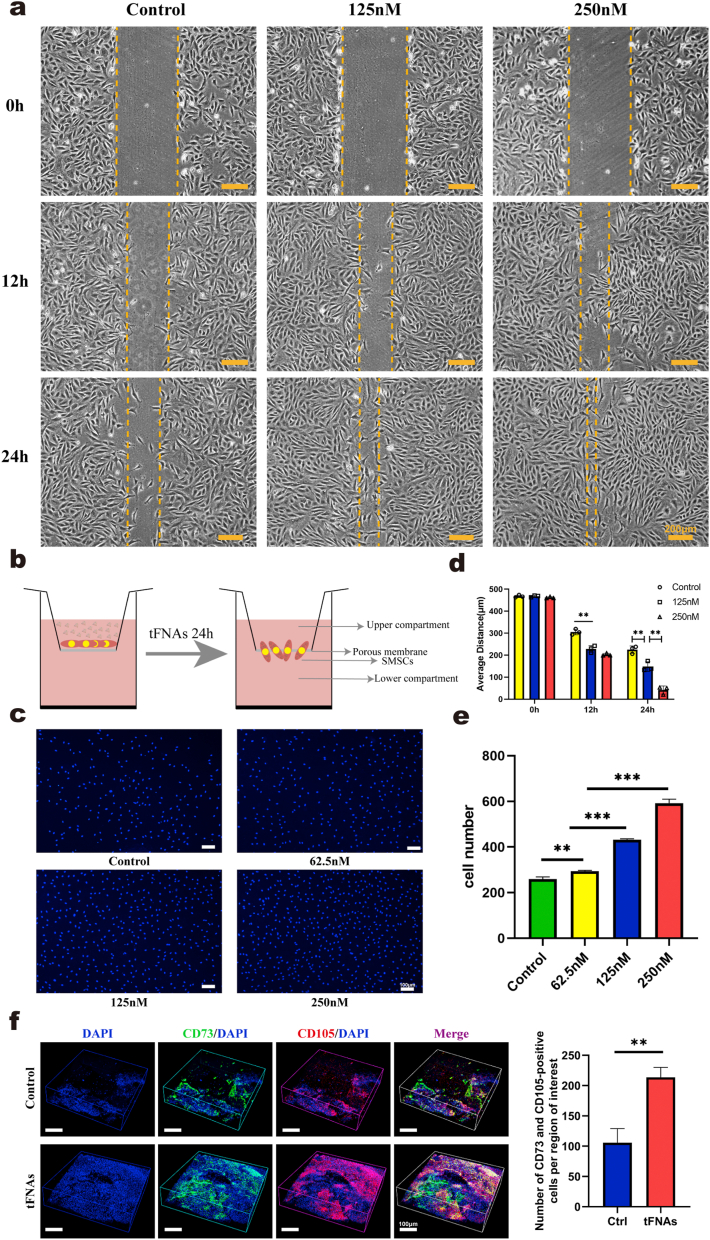
Scratch wound healing experiments and Transwell chamber assays were used to confirm the effect of tFNAs on the migration of SMSCs. As shown in Fig. 4a, images of SMSCs migrating to the scratches demonstrated that over time, the cells treated with tFNAs covered the scratch wound area earlier than the control cells, and the effect was most significant at a concentration of 250 nM, indicating that tFNAs could significantly enhance cell migration. Statistical analysis of the distance between scratches also confirmed that the scratches of the 250 nM tFNA group showed the best healing effect (Fig. 4d). Next, we used a Transwell chamber assay to analyze the effect of tFNAs on the vertical migration of SMSCs (Fig. 4b). Images of DAPI fluorescence staining revealed that after the cells in the upper compartment were exposed to tFNAs for 24 h, more migrated cells were observed on the lower surface of the inserts (Fig. 4c). Statistical analysis of the cell number showed that the migratory effect was gradually enhanced as the concentration increased, and the effect was strongest when the concentration of tFNAs was 250 nM (Fig. 4e). According to the results of two experiments, we concluded that tFNAs can significantly enhance cell migration, and further, 250 nM was the optimal working concentration based on these findings combined with the results of the previous proliferation experiments. Therefore, 250 nM was used as the experimental concentration in the subsequent experiments.
Fig. 4.
Effect of tFNAs on the migration of SMSCs. a. Scratch wound healing experiments verified the effect of tFNAs with different concentrations on promoting the migration of SMSCs at 0, 12, and 24 h. Scale bars are 200 μm. b. Structure diagram of Transwell chambers. c. Representative fluorescent images of transwell assay showed that tFNAs could effectively promote the migration of SMSCs (nucleus: blue). d. Quantitative analysis of the distance of the scratch wound healing experiment. Data are presented as the mean ± SD (n = 3). Statistical analysis: **p < 0.01. e. The number of cells migrated by SMSCs is shown in a histogram. Data are presented as the mean ± SD (n = 3). Statistical analysis: **p < 0.01, ***p < 0.001. f. Confocal 3D image of CD73+/CD105+ MSCs in two groups by immunofluorescence laser 7 days post-surgery and quantification of MSCs migrated to the injured sites. Data are presented as the mean ± SD (n = 3). Statistical analysis: **p < 0.01.
3.4.2. tFNAs increased SMSC migration in vivo
Then, we confirmed that tFNAs can also increase the migration of SMSCs in vivo. After 7 days of repair in animal models, immunofluorescence staining was performed on the regenerated AC tissue samples of the two groups. Immunofluorescence results showed that the number of CD73 and CD105 double-positive cells in the regenerated tissue of the tFNA group was significantly higher than that in the control group (Fig. 4f), indicating that more endogenous MSCs migrated to the defect area in the tFNA treatment group than in the control group. These results proved that tFNAs could significantly increase the migration of endogenous SMSCs to defect areas and enhance AC regeneration.
3.5. Effect of tFNAs on the chondrogenic differentiation of SMSCs
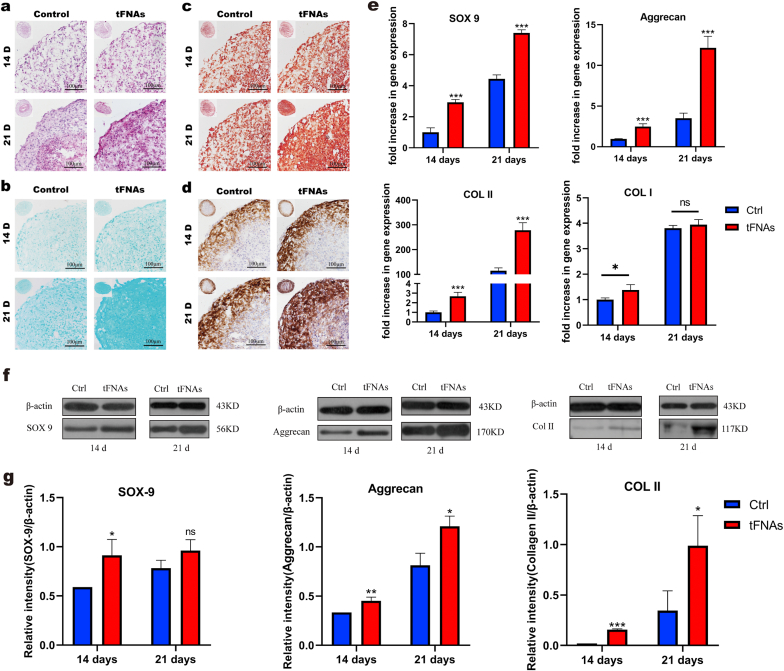
In our study, we used a pellet culture system to induce SMSC chondrogenic differentiation in vitro according to previous studies. After SMSCs were induced in chondrogenic differentiation medium with or without tFNAs (250 nM) for 14 and 21 days, we carried out histological and immunohistochemical staining to qualitatively evaluate chondrogenesis in the cell pellets. As shown in Fig. 5a, H&E staining revealed that SMSCs successfully differentiated into chondrocytes in all groups, and the tFNA group secreted more extracellular matrix than the control group. In addition, safranin O and Alcian blue staining showed that treatment with tFNAs increased proteoglycan production (Fig. 5b and c). Furthermore, IHC demonstrated that both the staining area and extent of type II collagen staining in the 250 nM tFNA treatment group were stronger than those in the control group, indicating that 250 nM tFNAs could significantly enhance the deposition of type II collagen in the pellets (Fig. 5d).
Fig. 5.
Effect of tFNAs on chondrogenic differentiation of SMSCs in vitro. a-d. H&E (a), Alcian blue (b), and safranin-O (c) staining and immunohistochemistry of collagen II (d) in SMSCs pellets that were cocultured with or without tFNAs. e. Gene expression of Col I, Col II, Sox9 and Aggrecan in SMSCs pellets at 14 and 21 days. Data are presented as the mean ± SD (n = 4). Statistical analysis: *p < 0.1, ***p < 0.001, n.s. indicates no significant difference. f. Protein expression of Aggrecan, SOX-9 and COL II in the SMSC pellets cocultured with or without tFNAs at 14 and 21 days. g. Quantitative analysis of the proteins expression levels of Aggrecan, SOX-9 and COL II. Data are presented as the mean ± SD (n = 3). Statistical analysis: *p < 0.05, **p < 0.01, ***p < 0.001, n.s. indicates no significant difference.
To further clarify the tFNA-mediated promotion of chondrogenic differentiation of SMSCs, we quantitatively detected typical chondrogenic-related differentiation markers, including collagen II, Sox9 and aggrecan, at the gene and protein levels. First, we used qPCR to determine the expression of the above markers at the gene level after SMSCs were treated with 250 nM tFNAs for 14 and 21 days. tFNAs significantly upregulated the expression of chondrogenic-specific genes (Fig. 5e). In addition, we detected the gene expression of collagen I and found that there was no significant difference between the two groups after 21 days of differentiation. Moreover, the expression of the abovementioned proteins was tested through Western blot analysis. As shown in Fig. 5f, the expression of these proteins after treatment with 250 nM tFNAs was significantly enhanced. Statistical analysis showed that collagen II expression was upregulated by 17.74-fold at day 14 and 50.73-fold at day 21, SOX9 expression was upregulated by 1.33-fold at day 14 day and 1.63-fold at day 21 and aggrecan expression was upregulated by 2.43-fold at day 14 and 3.62-fold at day 21 in the tFNA groups (Fig. 5g). Altogether, these results revealed that tFNAs strongly promoted chondrogenic differentiation of SMSCs.
3.6. Mechanism through which tFNAs regulate the chondrogenic differentiation of SMSCs
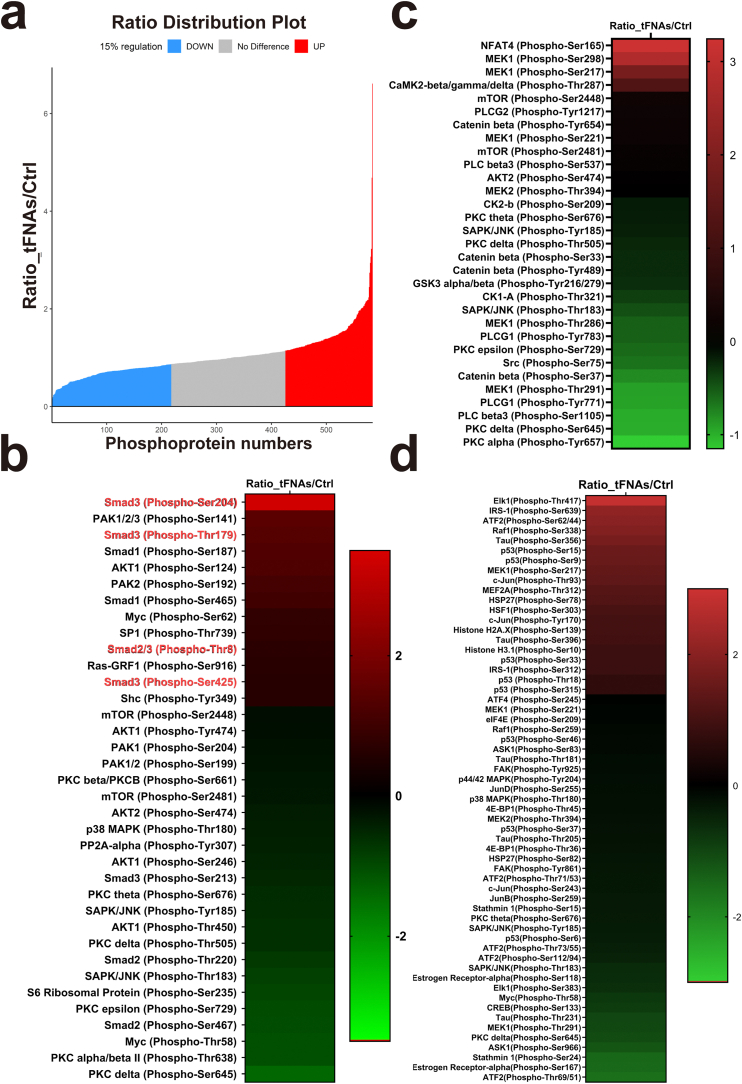
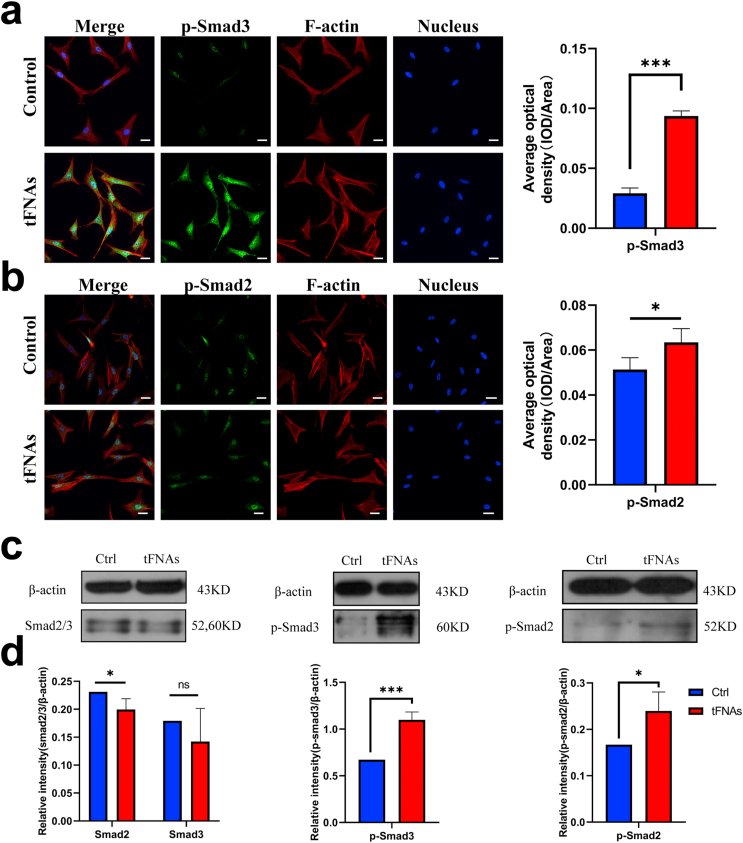
To investigate the molecular basis of the effect of tFNAs on SMSC chondrogenic differentiation, we performed a comprehensive proteomic screen using phosphorylated antibody arrays to identify proteins that were differentially regulated before and after tFNA treatment at the working concentration. The microarray slide images generated are shown in Supplementary Fig. S2. When the regulatory value was 15%, the cells treated with tFNAs phosphorylated 160 proteins compared with the control cells (Fig. 6a). We then focused on the phosphorylated proteins in three pathways, the TGFβ, Wnt and MAPK pathways (Fig. 6b–d), and found that phosphorylation of Smad2/3, the most important downstream target in the TGF pathway, was significantly enhanced. Then, we discovered that in the 250 nM tFNA-treated cells, Smad2/3 (phospho-Thr8), Smad3 (phospho-Ser 204), Smad3 (phospho-Thr179), and Smad3 (phospho-Ser 425) levels were enhanced by 1.18, 2.07, 1.43, and 1.15 times, respectively, compared to those of the control group (Supplementary Fig. S3). Furthermore, as shown in Fig. 7a–d, the results of immunofluorescence and Western blot analysis confirmed the increased phosphorylation of Smad2/3. Therefore, we concluded that tFNAs could improve the chondrogenic differentiation of SMSCs by regulating the TGF/Smad2/3 signaling pathway.
Fig. 6.
The results of the phospho-antibody array and regulation of proteins associated with chondrogenic differentiation of SMSCs by tFNAs. a. Statistical diagram of the phosphorylation proteins produced by tFNAs in the phospho-antibody array. Phosphoproteins whose levels increased or decreased by more than 15% were labeled red and blue. b. Heat map representing that the fold change in phosphorylation expression of TGF-β pathway-related proteins. c. Heat map representing that the fold change in phosphorylation expression of Wnt pathway-related proteins. d. Heat map representing that the fold change in phosphorylation expression of MAPK pathway-related proteins.
Fig. 7.
tFNAs promoted the chondrogenic differentiation of SMSCs by up-regulating the phosphorylation expression of Smad2/3. a-b. Immunofluorescence detection of p-Smad3 and p-Smad2. (F-actin: red, nucleus: blue, and protein: green). Scale bars are 25 μm. Quantitative analysis of the average optical density of fluorescence images. Data are presented as the mean ± SD (n = 3). Statistical analysis: *p < 0.05, ***p < 0.001. c. WB analysis of the expression levels of Smad 2/3, p-Smad2 and p-Smad3 (the internal control was β-actin). d. Quantitative analysis of the expression levels of Smad 2/3, p-Smad2 and p-Smad3. Data are presented as the mean ± SD (n = 3). Statistical analysis: *p < 0.05, ***p < 0.001, n.s. represents no significant difference.
3.7. tFNAs promoted cartilage regeneration in vivo
3.7.1. Clinical evaluation of the regeneration of AC
To investigate whether tFNAs can promote articular cartilage in situ regeneration, we established a rabbit model with a cylindrical cartilage defect, and then, the same amount of saline or 250 nM tFNAs was injected into the articular cavity. As shown in Fig. 8a, the cartilage defect in the control group was still obvious at 6 weeks after the operation, the repaired tissue was mostly white granulation tissue, and the new tissue still had an obvious boundary with the surrounding cartilage. Although the tFNA group also showed discontinuity with the normal cartilage boundary at 6 weeks, the degree of repair was significantly better than that of the control group. At 12 weeks after the operation, cartilage defects in the control group were nearly filled. However, the surface was uneven, and the color difference between the new tissue and the surrounding normal cartilage was obvious. In contrast, the new cartilage in the tFNA group was more integrated, looked more like normal cartilage and had more continuous connections to the surrounding normal cartilage. The ICRS macroscopic score of the tFNA group was also better than that of the control group at both 6 weeks and 12 weeks (Fig. 8b and c).
Fig. 8.
Macroscopic and micro-CT evaluation of repaired knees. a. Representative macroscopic analysis of repaired tissues at 6 and 12 weeks post-operation. Red circles show the defect area. b. ICRS score for macroscopic assessment. Data are presented as the mean ± SD (n = 8). Statistical analysis: *p < 0.05, ***p < 0.001. c. Heat map of variables for the ICRS scoring. d. Micro-CT images contain 2D and 3D reconstruction of repaired cartilage at 6 and 12 weeks post-operation. e-f. Quantitative analysis of BMD and BV/TV in the regenerated area. Data are presented as the mean ± SD (n = 8). Statistical analysis: *p < 0.05, ***p < 0.001.
To compare the regenerative effect of tFNAs with that of the control, we performed micro-CT analysis. The femur samples at two time points were reconstructed by two-dimensional and three-dimensional images, and then, the two important indexes (BV/TV and BMD) were analyzed. Reconstruction images showed that the repaired tissue in the tFNA group was much greater than that in the control group at both 6 and 12 weeks after the operation (Fig. 8d). The BV/TV value also confirmed this finding (Fig. 8f). In addition, the quantitative assessment of BMD suggested that the BMD values of the tFNA group were substantially increased at both 6 weeks and 12 weeks after the operation (Fig. 8e).
3.7.2. Histomorphologicalevaluation
Histomorphological staining was used to show the properties of the repaired AC tissue. Consistent with the results of the macroscopic evaluation, the results of H&E staining (Fig. 9a) showed that the repaired area was still not well integrated with normal cartilage at 6 weeks in the tFNA group, partial collapse was detected, and the staining was shallow. However, at 12 weeks, the repaired cartilage area was well integrated with normal cartilage, and chondrocytes in the repaired area were clearly aligned. In the control group, the regenerated area was largely unstained at 6 weeks, and tissue collapse showed no sign of repair. In contrast, at 12 weeks in the control group, the tissue surface was uneven and accompanied by flocculated fibrous tissue, and the staining was shallow compared to that of the tFNA group.
Fig. 9.
Histomorphological evaluation of repaired tissue. a. Histological (H&E, safranin O, and Sirius red staining) and immunohistochemical staining (collagen II) of the defect area. Solid arrows indicate the repair interface. (N: normal cartilage; R: repaired cartilage). Scale bars are 500 μm. b. Modified O'Driscoll scores for histological evaluation of cartilage repair after 6 weeks and 12 weeks. Data are presented as the mean ± SD (n = 4). Statistical analysis: *p < 0.05, **p < 0.01, ***p < 0.001. c. Statistical results of Young's modulus of regenerated tissue in the defect area after repair. Data are presented as the mean ± SD (n = 4). Statistical analysis: *p < 0.05, ***p < 0.001.
Safranin O can dye polysaccharides in cartilage red, and Fast Green can dye bone tissue green (Fig. 9a). In the tFNA group, all tissues after repair were stained red at 6 weeks and 12 weeks, but at 6 weeks, the repaired tissue was clearly demarcated from the normal tissue with irregular surfaces. At 12 weeks, the repaired tissue was closely connected to the normal tissue, and the arrangement of chondrocytes in the repaired area was more similar to that in the normal tissue. At 6 weeks, there was almost no repaired tissue and no polysaccharides in the control group. At 12 weeks, the control group showed red staining, but the surface of the repaired tissue was irregular, and flocculent fibrous tissue was observed. Compared with the normal cells, the repaired chondrocytes showed slight abnormalities in morphology and arrangement.
The total collagen content of repaired tissues was verified by Sirius red staining (Fig. 9a). At 6 weeks, birefringent collagen fibers were observed in the repaired area of the tFNA group, and these molecules were tightly and regularly arranged but had gaps with the normal tissue. At 12 weeks, the repaired areas were more regular and orderly. At 6 weeks in the control group, the repaired area was still defective, and there were basically no regenerated collagen fibers. At 12 weeks, there were birefringent collagen fibers, which were closely arranged and regular in shape, but flocculent fibrous tissue was present on the surface.
The collagen II content was verified by immunohistochemical analysis (Fig. 9a). At 6 weeks, immunohistochemical analysis showed that the regenerated cartilage in the tFNA group had more obvious coloration than that in the control group but was lighter than that of the surrounding normal cartilage, indicating that the expression of collagen II was lower than that of normal AC. At 12 weeks, the coloration of the new cartilage in the tFNA group was close to the color of the surrounding normal AC, demonstrating that the collagen II content was similar to that of normal AC. In the control group, less collagen II staining than that in the tFNA group was observed at 12 weeks, with flocculent fibrous tissue on the surface.
Consistent with the staining results, the results of the O'Driscoll scoring system showed that the histological scores were higher in the tFNA group than in the control group at both 6 weeks and 12 weeks (Fig. 9b). These results indicated that tFNAs can promote articular cartilage in situ regeneration.
3.7.3. Biomechanical evaluation
The mechanical properties of regenerated AC were evaluated by biomechanical testing. As shown in Fig. 9c, the results revealed that the Young's modulus in the tFNA group was significantly higher than that in the control group at both 6 weeks and 12 weeks, indicating that the mechanical properties of regenerated AC in the tFNA group were better. However, compared with those of the sham group (normal cartilage), the mechanical properties of the tFNA group were still poorer.
4. Discussion
Due to a poor regenerative ability and lack of appropriate treatment, damaged AC can eventually develop into osteoarthritis, substantially reducing patient quality of life [1,37]. In recent years, MSCs-based tissue engineering therapies have been promising approaches for AC regeneration, and major progress has been made in these strategies [7]. However, the lack of MSCs sources and the decreased potential of stem cell culture in vitro restrict its further application [9,19]. Therefore, more studies have focused on improving the in situ regeneration of AC based on endogenous MSCs to overcome the above limitations [17,18]. In particular, joint-resident MSCs are the driving factor for in situ regeneration of AC, among which SMSCs play a dominant role in adult cartilage repair due to their good chondrogenic potential and proximity to AC [19]. Nevertheless, an ideal regenerative microenvironment for endogenous stem cells to promote regeneration-relevant biological functions such as proliferation, migration and chondrogenic differentiation is one of the main challenges [20]. The most common method to improve the regenerative microenvironment is the use of chemoattractants. Traditional chemoattractants include growth factors and chemokines, such as bone morphology protein-2, transforming growth factor-β3 and stromal cell derived factor-1. In addition to the existence of expensive shortcomings, large doses of treatment often cause adverse pathological changes, such as hypertrophic synovium [17]. Furthermore, the function of most chemokines is relatively simple and does not comprehensively promote the biological function of MSCs. Due to the ability to create a biomimetic cellular environment that promotes cartilage regeneration, the application of nanoengineering systems in AC tissue engineering has attracted extensive attention [[38], [39], [40]]. The role and prospects of tFNAs, promising nanomaterials, in the field of biomedicine have been widely demonstrated [24]. tFNAs are not only able to provide a unique cellular environment for chondroblastic growth and phenotype maintenance [31] but also promote the proliferation and osteogenic differentiation of MSCs [30]. Herein, we sought to explore whether tFNAs could have positive effects on the relevant microenvironment for AC in situ regeneration.
Previous studies have shown that tFNAs can enter cells through caveolin-mediated endocytosis and are stable in the cell for a certain period of time, while high levels of ssDNAs cannot be absorbed [41]. Furthermore, the key to the positive effects of tFNAs on the AC regeneration-relevant microenvironment is cellular uptake. Thus, we not only successfully synthesized tFNAs with a stable size and zeta potential but also labeled the tFNAs with Cy5 to investigate the uptake of SMSCs. The results revealed that tFNAs can be absorbed specifically by SMSCs and that ssDNAs cannot enter SMSCs in abundance.
The proliferation, migration and chondrogenic differentiation of MSCs are essential biological functions in cartilage repair. In this study, based on our experimental data, we discovered that tFNAs can also significantly improve SMSC proliferation in a concentration-dependent manner by regulating the cell cycle, and tFNAs at a concentration of 250 nM provided an ideal cellular environment for SMSC proliferation. In addition, we proved that SMSCs exposed to tFNAs showed enhanced proliferation via enhancement of the classic Wnt signaling pathway, similar to previous research. As an important positive mediator in the classic Wnt pathway, β-catenin is transported to the nucleus when it accumulates in the cytoplasm and then interacts with Lef-1 and other proteins to produce a series of biological behaviors, including the expression of cyclin D1. The primary function of cyclin D1 is to promote cell proliferation by regulating cyclin-dependent kinases, and in particular, this molecule is essential for the transition from the G1 to the S phase in mitosis. These results are consistent with the results of cell proliferation and cell cycle experiments.
In addition to proliferation, MSC migration plays a decisive role in cartilage repair. For the in vitro migration experiment, we used a bidirectional wound healing assay to explore the effects of tFNAs on the horizontal migration of SMSCs. As a widely used experiment in the study of cell migration, the wound healing assay is often used to study horizontal cell migration in a two-dimensional plane. However, this assay cannot simulate the formation of gradient chemokines, which is quite different from the migration of MSCs in vivo. To further investigate the effects of tFNAs on SMSC migration in three dimensions, we performed Transwell chamber assays. We used 5% FBS in the lower chamber as a chemokine, and SMSCs were seeded in the upper chamber. Then, we applied a rabbit articular cartilage defect model to explore the effect of tFNAs on the migration of SMSCs in vivo. In fact, when cartilage is injured, some chemokines are produced in the articular cavity to promote the migration of MSCs to the injured site [18]. However, due to insufficient signals, the number of migrating MSCs is usually limited, which leads to difficulty in cartilage regeneration [17]. After the injection of tFNAs into the articular cavity, the migratory abilities of peripheral MSCs, especially SMSCs, were significantly improved, which has been confirmed in vitro. Therefore, when our in vivo cartilage defect model was established, the effect of the released chemotaxis signal was also significantly improved due to the enhancement of MSC migration, which may be the main reason why we observed more cells in the regenerative tissue after 7 days of tFNA injection.
As a key factor in the process of articular cartilage regeneration, we investigated the role of tFNAs in SMSC chondrogenic differentiation, which has never been studied before. In our study, we used a pellet culture system to evaluate the chondrogenic differentiation of SMSCs exposed to 250 nM tFNAs. The results of histological staining, qPCR and WB revealed that the chondrogenic inductive environment with 250 nM tFNAs is more suitable for chondrogenic differentiation. To further confirm the important role of tFNAs in promoting SMSC chondrogenic differentiation, we used phospho-antibody array analysis to investigate the potential mechanism by which tFNAs enhance SMSC chondrogenic differentiation. In fact, chondrogenic differentiation of SMSCs is regulated by crosstalk of multiple signaling pathways [42,43]. For example, TGFβ-Smad2/3 can activate the transcription factor SOX9 to initiate and maintain chondrogenesis, which is the classic chondrogenic pathway [44]. In addition, phosphorylation of the MAPK family members p38, ERK-1 and JNK can stimulate the expression of Sox9 [43,45]. Furthermore, some studies have shown that the Wnt signaling pathway is associated with the chondrogenic differentiation of MSCs [46]. Therefore, we used phospho-antibody array analysis to conduct a comprehensive screen. The microarray results showed a significant increase in Smad2/3 phosphorylation in the tFNA-treated group, which was also confirmed by subsequent WB and immunofluorescence. These results suggested that the promotion of SMSC chondrogenic differentiation after treatment with 250 nM tFNAs was related to the enhancement of Smad2/3 phosphorylation.
To investigate the effect of tFNAs on AC regeneration in situ, we used New Zealand white rabbits to establish in vivo cartilage defect models. After strict disinfection, 250 nM tFNAs were injected into the joint cavity of the rabbits, while saline was injected into the control rabbits. When treated with 250 nM tFNAs, the rabbits not only showed more rapid cartilage regeneration than those in the control group but also had less fibrous tissue after the defects repaired. The results of histomorphological staining showed that the application of tFNAs led to a better alignment of chondrocytes in the regenerated tissue and higher expression of collagen II. Furthermore, the mechanical properties of regenerated cartilage in situ have also been greatly improved by the treatment of tFNAs.
According to our findings, we confirmed that tFNAs had positive effects on the proliferation, migration, and chondrogenic differentiation of SMSCs as well as the in situ regeneration of damaged AC in rabbits. Compared with traditional chemoattractants such as bone morphology protein-2, transforming growth factor-β3 and stromal cell derived factor-1, tFNAs have some obvious advantages, including simple preparation, low cost, comprehensive functions and fewer side effects. However, there are still some limitations and challenges for the application of tFNAs in AC regeneration. First, although tFNAs are more stable due to their particular structure than double-stranded DNAs, studies have shown that tFNAs will degrade rapidly in vivo, especially in the blood. Therefore, in this study, we used continuous joint cavity injection, which will not only cause pain to patients but will also result in the risk of joint infection. Furthermore, compared with scaffold sustained release factors, intraarticular injection of tFNAs results in uneven biological distribution, which may reduce therapeutic efficacy. Therefore, to more effectively use tFNAs to promote cartilage regeneration, our next work will focus on solving the above problems.
5. Conclusion
In conclusion, our study demonstrated that tFNAs, novel biological nanophase materials, could enhance both the proliferation and migration of SMSCs, and 250 nM was identified as the best working concentration. We also found that 250 nM tFNAs could promote chondrogenic differentiation of SMSCs by increasing the phosphorylation level of Smad2/3. In addition, in rabbit models of AC defects, tFNAs significantly increased the number of migrated MSCs in the defect area and enhanced AC in situ regeneration. To the best of our knowledge, this study is the first to report the treatment of cartilage defects using tFNAs, indicating that tFNAs have great potential for clinical articular cartilage repair and represent a promising therapeutic for AC regeneration in the future.
CRediT authorship contribution statement
Liwei Fu: Writing – original draft, Investigation, Data curation, Formal analysis, Visualization. Pinxue Li: Writing – original draft, Investigation, Data curation, Formal analysis, Visualization. Junyao Zhu: Investigation, Data curation, Formal analysis. Zhiyao Liao: Investigation, Data curation. Cangjian Gao: Investigation, Formal analysis. Hao Li: Data curation, Formal analysis. Zhen Yang: Data curation. Tianyuan Zhao: Formal analysis. Wei Chen: Methodology. Yu Peng: Software. Fuyang Cao: Investigation. Chao Ning: Methodology. Xiang Sui: Resources. Quanyi Guo: Supervision. Yunfeng Lin: Supervision. Shuyun Liu: Conceptualization, Supervision.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Key R&D Program of China (2019YFA0110600).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.07.028.
Contributor Information
Quanyi Guo, Email: doctorguo_301@163.com.
Yunfeng Lin, Email: yunfenglin@scu.edu.cn.
Shuyun Liu, Email: clear_ann@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Armiento A.R., Alini M., Stoddart M.J. Articular fibrocartilage - why does hyaline cartilage fail to repair. Adv. Drug Deliv. Rev. 2019;146:289–305. doi: 10.1016/j.addr.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Huey D.J., Hu J.C., Athanasiou K.A. Unlike bone, cartilage regeneration remains elusive. Science. 2012;338(6109):917–921. doi: 10.1126/science.1222454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon H., Brown W.E., Lee C.A., Wang D., Paschos N., Hu J.C., Athanasiou K.A. Surgical and tissue engineering strategies for articular cartilage and meniscus repair. Nat. Rev. Rheumatol. 2019;15(9):550–570. doi: 10.1038/s41584-019-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saris T., de Windt T.S., Kester E.C., Vonk L.A., Custers R., Saris D. Five-year outcome of 1-stage cell-based cartilage repair using recycled autologous chondrons and allogenic mesenchymal stromal cells: a first-in-human clinical trial. Am. J. Sports Med. 2021;49(4):941–947. doi: 10.1177/0363546520988069. [DOI] [PubMed] [Google Scholar]
- 5.Gilat R., Haunschild E.D., Huddleston H.P., Tauro T.M., Patel S., Wolfson T.S., Parvaresh K.C., Yanke A.B., Cole B.J. Osteochondral allograft transplant for focal cartilage defects of the femoral condyles: clinically significant outcomes, failures, and survival at a minimum 5-year follow-up. Am. J. Sports Med. 2021;49(2):467–475. doi: 10.1177/0363546520980087. [DOI] [PubMed] [Google Scholar]
- 6.Jung M., Karampinos D.C., Holwein C., Suchowierski J., Diallo T.D., Gersing A.S., Bamberg F., Baumann F.A., Ruschke S., Jungmann P.M. Quantitative 3-T magnetic resonance imaging after matrix-associated autologous chondrocyte implantation with autologous bone grafting of the knee: the importance of subchondral bone parameters. Am. J. Sports Med. 2021;49(2):476–486. doi: 10.1177/0363546520980134. [DOI] [PubMed] [Google Scholar]
- 7.Huang B.J., Hu J.C., Athanasiou K.A. Cell-based tissue engineering strategies used in the clinical repair of articular cartilage. Biomaterials. 2016;98:1–22. doi: 10.1016/j.biomaterials.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeira C., Santhagunam A., Salgueiro J.B., Cabral J.M. Advanced cell therapies for articular cartilage regeneration. Trends Biotechnol. 2015;33(1):35–42. doi: 10.1016/j.tibtech.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Szychlinska M.A., Stoddart M.J., D'Amora U., Ambrosio L., Alini M., Musumeci G. Mesenchymal stem cell-based cartilage regeneration approach and cell senescence: can we manipulate cell aging and function. Tissue Eng. B Rev. 2017;23(6):529–539. doi: 10.1089/ten.TEB.2017.0083. [DOI] [PubMed] [Google Scholar]
- 10.Song N., Scholtemeijer M., Shah K. Mesenchymal stem cell immunomodulation: mechanisms and therapeutic potential. Trends Pharmacol. Sci. 2020;41(9):653–664. doi: 10.1016/j.tips.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Rosa M., Castrogiovanni P., Musumeci G. The synovium theory: can exercise prevent knee osteoarthritis? The role of “mechanokines”, A possible biological key. J Funct Morphol Kinesiol. 2019;4(1):11. doi: 10.3390/jfmk4010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castrogiovanni P., Di Rosa M., Ravalli S., Castorina A., Guglielmino C., Imbesi R., Vecchio M., Drago F., Szychlinska M.A., Musumeci G. Moderate physical activity as a prevention method for knee osteoarthritis and the role of synoviocytes as biological key. Int. J. Mol. Sci. 2019;20(3):511. doi: 10.3390/ijms20030511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones B.A., Pei M. Synovium-derived stem cells: a tissue-specific stem cell for cartilage engineering and regeneration. Tissue Eng. B Rev. 2012;18(4):301–311. doi: 10.1089/ten.TEB.2012.0002. [DOI] [PubMed] [Google Scholar]
- 14.Fan J., Varshney R.R., Ren L., Cai D., Wang D.A. Synovium-derived mesenchymal stem cells: a new cell source for musculoskeletal regeneration. Tissue Eng. B Rev. 2009;15(1):75–86. doi: 10.1089/ten.teb.2008.0586. [DOI] [PubMed] [Google Scholar]
- 15.De Bari C., Dell F., Accio, Tylzanowski P., Luyten F.P. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44(8):1928–1942. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Tan A.R., Hung C.T. Concise review: mesenchymal stem cells for functional cartilage tissue engineering: taking cues from chondrocyte-based constructs. Stem Cells Transl Med. 2017;6(4):1295–1303. doi: 10.1002/sctm.16-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z., Li H., Yuan Z., Fu L., Jiang S., Gao C., Wang F., Zha K., Tian G., Sun Z., Huang B., Wei F., Cao F., Sui X., Peng J., Lu S., Guo W., Liu S., Guo Q. Endogenous cell recruitment strategy for articular cartilage regeneration. Acta Biomater. 2020;114:31–52. doi: 10.1016/j.actbio.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Hu H., Liu W., Sun C., Wang Q., Yang W., Zhang Z., Xia Z., Shao Z., Wang B. Endogenous repair and regeneration of injured articular cartilage: a challenging but promising therapeutic strategy. Aging Dis. 2021;12(3):886–901. doi: 10.14336/AD.2020.0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGonagle D., Baboolal T.G., Jones E. Native joint-resident mesenchymal stem cells for cartilage repair in osteoarthritis. Nat. Rev. Rheumatol. 2017;13(12):719–730. doi: 10.1038/nrrheum.2017.182. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S., Hu B., Liu W., Wang P., Lv X., Chen S., Liu H., Shao Z. Articular cartilage regeneration: the role of endogenous mesenchymal stem/progenitor cell recruitment and migration. Semin. Arthritis Rheum. 2020;50(2):198–208. doi: 10.1016/j.semarthrit.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Sun X., Yin H., Wang Y., Lu J., Shen X., Lu C., Tang H., Meng H., Yang S., Yu W., Zhu Y., Guo Q., Wang A., Xu W., Liu S., Lu S., Wang X., Peng J. In situ articular cartilage regeneration through endogenous reparative cell homing using a functional bone marrow-specific scaffolding system. ACS Appl. Mater. Interfaces. 2018;10(45):38715–38728. doi: 10.1021/acsami.8b11687. [DOI] [PubMed] [Google Scholar]
- 22.Szychlinska M.A., Calabrese G., Ravalli S., Parrinello N.L., Forte S., Castrogiovanni P., Pricoco E., Imbesi R., Castorina S., Leonardi R., Di Rosa M., Musumeci G. Cycloastragenol as an exogenous enhancer of chondrogenic differentiation of human adipose-derived mesenchymal stem cells. A Morphological Study, Cells. 2020;9(2):347. doi: 10.3390/cells9020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szychlinska M.A., Calabrese G., Ravalli S., Dolcimascolo A., Castrogiovanni P., Fabbi C., Puglisi C., Lauretta G., Di Rosa M., Castorina A., Parenti R., Musumeci G. Evaluation of a cell-free collagen type I-based scaffold for articular cartilage regeneration in an orthotopic rat model. Materials. 2020;13(10):2369. doi: 10.3390/ma13102369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Tian T., Zhang T., Cai X., Lin Y. Advances in biological applications of self-assembled DNA tetrahedral nanostructures. Mater. Today. 2019;24:57–68. [Google Scholar]
- 25.Shao X., Cui W., Xie X., Ma W., Zhan Y., Lin Y. Treatment of Alzheimer’s disease with framework nucleic acids. Cell Prolif. 2020;53(4) doi: 10.1111/cpr.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M., Zhu J., Qin X., Zhou M., Zhang X., Gao Y., Zhang T., Xiao D., Cui W., Cai X. Cardioprotection of tetrahedral DNA nanostructures in myocardial ischemia-reperfusion injury. ACS Appl. Mater. Interfaces. 2019;11(34):30631–30639. doi: 10.1021/acsami.9b10645. [DOI] [PubMed] [Google Scholar]
- 27.Zhao D., Liu M., Li Q., Zhang X., Xue C., Lin Y., Cai X. Tetrahedral DNA nanostructure promotes endothelial cell proliferation, migration, and angiogenesis via notch signaling pathway. ACS Appl. Mater. Interfaces. 2018;10(44):37911–37918. doi: 10.1021/acsami.8b16518. [DOI] [PubMed] [Google Scholar]
- 28.Shi S., Lin S., Li Y., Zhang T., Shao X., Tian T., Zhou T., Li Q., Lin Y. Effects of tetrahedral DNA nanostructures on autophagy in chondrocytes. Chem. Commun. (Camb.) 2018;54(11):1327–1330. doi: 10.1039/c7cc09397g. [DOI] [PubMed] [Google Scholar]
- 29.Zhou M., Gao S., Zhang X., Zhang T., Zhang T., Tian T., Li S., Lin Y., Cai X. The protective effect of tetrahedral framework nucleic acids on periodontium under inflammatory conditions. Bioact Mater. 2021;6(6):1676–1688. doi: 10.1016/j.bioactmat.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao X.R., Lin S.Y., Peng Q., Shi S.R., Li X.L., Zhang T., Lin Y.F. Effect of tetrahedral DNA nanostructures on osteogenic differentiation of mesenchymal stem cells via activation of the Wnt/β-catenin signaling pathway. Nanomedicine. 2017;13(5):1809–1819. doi: 10.1016/j.nano.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Shao X., Lin S., Peng Q., Shi S., Wei X., Zhang T., Lin Y. Tetrahedral DNA nanostructure: a potential promoter for cartilage tissue regeneration via regulating chondrocyte phenotype and proliferation. Small. 2017;13(12) doi: 10.1002/smll.201602770. [DOI] [PubMed] [Google Scholar]
- 32.Zhang T., Tian T., Zhou R., Li S., Ma W., Zhang Y., Liu N., Shi S., Li Q., Xie X., Ge Y., Liu M., Zhang Q., Lin S., Cai X., Lin Y. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat. Protoc. 2020;15(8):2728–2757. doi: 10.1038/s41596-020-0355-z. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Wu N., Cheng J., Sun M., Yang P., Zhao F., Zhang J., Duan X., Fu X., Zhang J., Hu X., Chen H., Ao Y. Biomechanically, structurally and functionally meticulously tailored polycaprolactone/silk fibroin scaffold for meniscus regeneration. Theranostics. 2020;10(11):5090–5106. doi: 10.7150/thno.44270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Liao Z., Yang Z., Gao C., Fu L., Li P., Zhao T., Cao F., Chen W., Yuan Z., Sui X., Liu S., Guo Q. 3D printed poly(ε-caprolactone)/meniscus extracellular matrix composite scaffold functionalized with kartogenin-releasing PLGA microspheres for meniscus tissue engineering. Front Bioeng Biotechnol. 2021;9:662381. doi: 10.3389/fbioe.2021.662381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan H., Zhang C., Li J., Bi L., Qin L., Wu H., Hu Y. Gelatin microspheres containing TGF-beta 3 enhance the chondrogenesis of mesenchymal stem cells in modified pellet culture. Biomacromolecules. 2008;9(3):927–934. doi: 10.1021/bm7013203. [DOI] [PubMed] [Google Scholar]
- 36.Smith G.D., Taylor J., Almqvist K.F., Erggelet C., Knutsen G., Garcia Portabella M., Smith T., Richardson J.B. Arthroscopic assessment of cartilage repair: a validation study of 2 scoring systems. Arthroscopy. 2005;21(12):1462–1467. doi: 10.1016/j.arthro.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Loeser R.F., Collins J.A., Diekman B.O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12(7):412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdollahiyan P., Oroojalian F., Mokhtarzadeh A. The triad of nanotechnology, cell signalling, and scaffold implantation for the successful repair of damaged organs: an overview on soft-tissue engineering. J. Contr. Release. 2021;332:460–492. doi: 10.1016/j.jconrel.2021.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Saraswat R., Ratnayake I., Perez E.C., Schutz W.M., Zhu Z., Ahrenkiel S.P., Wood S.T. Micropatterned biphasic nanocomposite platform for maintaining chondrocyte morphology. ACS Appl. Mater. Interfaces. 2020;12(13):14814–14824. doi: 10.1021/acsami.9b22596. [DOI] [PubMed] [Google Scholar]
- 40.Deng C., Xu C., Zhou Q., Cheng Y. Advances of nanotechnology in osteochondral regeneration. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11(6):e1576. doi: 10.1002/wnan.1576. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J., Zhang M., Gao Y., Qin X., Zhang T., Cui W., Mao C., Xiao D., Lin Y. Tetrahedral framework nucleic acids promote scarless healing of cutaneous wounds via the AKT-signaling pathway. Signal Transduct Target Ther. 2020;5(1):120. doi: 10.1038/s41392-020-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu M., Chen G., Li Y.P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong L., Huang X., Karperien M., Post J.N. The regulatory role of signaling crosstalk in hypertrophy of MSCs and human articular chondrocytes. Int. J. Mol. Sci. 2015;16(8):19225–19247. doi: 10.3390/ijms160819225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budi E.H., Duan D., Derynck R. Transforming growth factor-β receptors and smads: regulatory complexity and functional versatility. Trends Cell Biol. 2017;27(9):658–672. doi: 10.1016/j.tcb.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 45.van Gastel N., Stegen S., Eelen G., Schoors S., Carlier A., Daniëls V.W., Baryawno N., Przybylski D., Depypere M., Stiers P.J., Lambrechts D., Van Looveren R., Torrekens S., Sharda A., Agostinis P., Lambrechts D., Maes F., Swinnen J.V., Geris L., Van Oosterwyck H., Thienpont B., Carmeliet P., Scadden D.T., Carmeliet G. Lipid availability determines fate of skeletal progenitor cells via SOX9. Nature. 2020;579(7797):111–117. doi: 10.1038/s41586-020-2050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z., Zou Y., Guo X.M., Tan H.S., Denslin V., Yeow C.H., Ren X.F., Liu T.M., Hui J.H., Lee E.H. Temporal activation of β-catenin signaling in the chondrogenic process of mesenchymal stem cells affects the phenotype of the cartilage generated. Stem Cell. Dev. 2012;21(11):1966–1976. doi: 10.1089/scd.2011.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.