Highlights
-
•
Runx2 loss in hypertrophic chondrocytes (Runx2HC/HC) causes limb dwarfism in mice.
-
•
Decreased apoptosis leads to doubling of the zone of hypertrophic chondrocytes.
-
•
Runx2HC/HC hypertrophic chondrocytes barely expresses Mmps and aggrecanase.
-
•
Calcified cartilage is accumulated in the long bones of Runx2HC/HC mice.
-
•
Runx2 regulates the expression of Rankl and Opg in hypertrophic chondrocytes.
Keywords: Dwarfism, Apoptosis, Chondroclast/osteoclast, Matrix-metalloproteinase, Aggrecanase
Abbreviations: SOX9, SRY box transcription factor; RUNX2, Runt related transcription factor 2; PTHRP, Parathyroid hormone-related peptide; ACAN, Aggrecan; COL2, Collagen type II; IHH, Indian hedgehog; COL10, Collagen type X; MMP, Matrix metalloproteinase; VEGFA, Vascular endothelial growth factor a; BAC, Bacterial artificial chromosome; CDK1, Cyclin-dependent kinase 1; RANKL, Receptor activator of nuclear factor Kappa B ligand; OPG, Osteoprotegerin; Wnt/PCP, Wnt/planar cell polarity; PCNA, Proliferating cell nuclear antigen; CCND1, Cyclin D1; TRAP, Tartrate-resistant acid phosphatase; TNAP, Tissue-nonspecific alkaline phosphatase
Abstract
The RUNX2 transcription factor is a key regulator for the development of cartilage and bone. Global or resting chondrocyte-specific deletion of the Runx2 gene results in failure of chondrocyte hypertrophy, endochondral ossification, and perinatal lethality. The terminally mature hypertrophic chondrocyte regulates critical steps of endochondral ossification. Importantly, expression of the Runx2 gene starts in the resting chondrocyte and increases progressively, reaching the maximum level in hypertrophic chondrocytes. However, the RUNX2 role after chondrocyte hypertrophy remains unknown. To answer this question, we deleted the Runx2 gene specifically in hypertrophic chondrocytes using the Col10-Cre line. Mice lacking the Runx2 gene in hypertrophic chondrocytes (Runx2HC/HC) survive but exhibit limb dwarfism. Interestingly, the length of the hypertrophic chondrocyte zone is doubled in the growth plate of Runx2HC/HC mice. Expression of pro-apoptotic Bax decreased significantly while anti-apoptotic Bcl2 remains unchanged leading to a four-fold increase in the Bcl2/Bax ratio in mutant mice. In line with this, a significant reduction in apoptosis of Runx2HC/HC hypertrophic chondrocyte is noted. A large amount of cartilage matrix is present in the long bones that extend toward the diaphyseal region of Runx2HC/HC mice. This is not due to enhanced synthesis of the cartilage matrix as the expression of both collagen type 2 and aggrecan were comparable among Runx2HC/HC and WT littermates. Our qPCR analysis demonstrates the increased amount of cartilage matrix is due to impaired expression of cartilage degrading enzymes such as metalloproteinase and aggrecanase as well as tissue inhibitor of metalloproteinases. Moreover, a significant decrease of TRAP positive chondroclasts was noted along the cartilage islands in Runx2HC/HC mice. Consistently, qPCR data showed an 81% reduction in the Rankl/Opg ratio in Runx2HC/HC littermates, which is inhibitory for chondroclast differentiation. Finally, we assess if increase cartilage matrix in Runx2HC/HC mice serves as a template for bone and mineral deposition using micro-CT and Von Kossa. The mutant mice exhibit a significant increase in trabecular bone mass compared to littermates. In summary, our findings have uncovered a novel role of Runx2 in apoptosis of hypertrophic chondrocytes and degradation of cartilage matrix during endochondral ossification.
Introduction
The majority of the bones in the mammalian skeleton are generated through the process of endochondral ossification. This sequential process of bone formation starts with the condensation of mesenchyme during early embryonic development [1]. The SRY box transcription factor (SOX9) is required for the commitment of undifferentiated cells in the condensed mesenchyme to chondrocytes [2], [3]. The immature chondrocytes residing within the central regions of the hyaline cartilage primordia undergo proliferation and secrete the initial matrix which contributes to the growth of the cartilage template [4]. The hyaline cartilage is degraded by enzymes released from the hypertrophic chondrocytes and is eventually replaced with mineralized bone produced by osteoblasts [5], [6].
During endochondral ossification, chondrocytes sequentially pass through resting, proliferative, prehypertrophic, and hypertrophic stages before undergoing apoptosis [7], [8]. Chondrocytes in each stage are defined by unique morphology, arrangement, gene expression, and location within the growth plate [9]. The small resting chondrocyte expresses SOX9, parathyroid hormone-related peptide (PTHRP), collagen type II (COL2), and aggrecan (ACAN) [3], [9], [10], [11]. During cartilage growth, resting chondrocytes undergo mitotic duplication with one daughter cell moving underneath the other, thereby resulting in the characteristic columnar arrangement of proliferative chondrocytes. The Cyclin-dependent kinase 1 (CDK1) and Wnt/planar cell polarity (Wnt/PCP) pathway regulate the development of proliferative chondrocytes characterized by the expression of cyclin d1 (CCND1) and Proliferating cell nuclear antigen (PCNA) [12], [13], [14]. The post-mitotic columnar chondrocyte transition to the prehypertrophic chondrocyte is marked by the expression of Indian hedgehog (IHH), PTHRP receptor (PTHRPR), and Runt related transcription factor 2 (RUNX2) [15], [16], [17]. Hypertrophic chondrocytes are the final stage in chondrocyte differentiation with a conspicuous expression of collagen type X (Col10), matrix metalloproteinase (MMP), aggrecanases (ADAMTS), and vascular endothelial growth factor-a (VEGFA) [17], [18], [19]. The sequential transition from the resting, proliferative to the hypertrophic stage is also regulated by extracellular signaling from BMP/TGFβ, FGF, NOTCH, and WNT/β-Catenin pathways [5], [20], [21], [22], [23].
The hypertrophic chondrocyte is the terminally mature and largest cell in the growth plate. Hypertrophic chondrocytes control angiogenesis, degradation of cartilage matrix, cartilage calcification, and differentiation of perichondral cells [4], [8]. The VEGFA produced by the hypertrophic chondrocytes in response to hypoxia is an essential mediator of growth plate angiogenesis [18], [24]. The vascular invasion facilitates the recruitment of progenitors for cartilage degrading chondroclasts and bone synthesizing osteoblasts [25]. Hypertrophic chondrocytes secrete aggrecanase, collagenase, and gelatinase that directly promote the degradation of the cartilage matrix [26], [27]. Chondrocytes also express tissue inhibitor of matrix metalloproteinase (TIMPs) that inhibits the activity of these enzymes [28], [29], [30], [31]. In addition, hypertrophic chondrocytes control the differentiation of the cartilage resorbing chondroclasts by releasing receptor activator of nuclear factor Kappa B ligand (RANKL) and osteoprotegerin (OPG) [32]. Deletion of Rankl gene specifically from hypertrophic chondrocytes leads to retention of calcified cartilage [33]. Hypertrophic chondrocytes regulate cartilage calcification through membrane bound enzymes and secretion of matrix vesicles [34], [35]. The chondrocyte secreted vesicles are enriched in tissue-nonspecific alkaline phosphatase (TNAP), inorganic phosphate, calcium ions, and several calcium binding proteins. Secreted or membrane bound TNAP hydrolyzes pyrophosphate into inorganic phosphate to induce calcification. Hypertrophic chondrocytes also induce differentiation of perichondral cells to osteoblasts and the development of bone collar through secretion of the IHH and BMP3 [15], [36]. Thus, the hypertrophic chondrocyte plays a central role in initiating both cartilage degradation and bone formation during endochondral ossification.
The RUNX2 transcription factor is essential for skeletal development. Global deletion of the Runx2 gene results in failure of both osteoblast and chondrocyte differentiation and lack of intramembranous and endochondral ossification [37], [38], [39]. Although Runx2 expression starts in the early chondrocyte, its role in chondrocyte hypertrophic differentiation is most appreciated. To test the spatiotemporal role of Runx2 during endochondral ossification, many laboratories employed transgenic approaches using mouse models to overexpress Runx2 in resting and hypertrophic chondrocytes. Overexpression of Runx2 in resting chondrocytes accelerates hypertrophic maturation, endochondral ossification, and ectopic mineralization of permanent cartilage [40], [41]. In contrast, blocking endogenous Runx2 function in resting chondrocytes by dominant negative Runx2 results in delayed chondrocyte hypertrophy and endochondral ossification [40]. Transient overexpression of Runx2 in resting chondrocytes also causes accelerated hypertrophy, endochondral ossification, and apoptosis of chondrocytes [42]. Interestingly, overexpression of Runx2 in hypertrophic chondrocytes delays chondrocyte hypertrophy, reduces mineralization and decreases chondrocyte apoptosis [43]. Overexpression of Runx2 in resting chondrocytes lead to perinatal lethality but mice overexpressing Runx2 in hypertrophic chondrocytes survive [40], [43]. Thus, overexpression models have failed to reveal the spatiotemporal role of the Runx2 gene during endochondral ossification.
Both global and resting chondrocyte-specific Runx2 null models show a complete lack of chondrocyte hypertrophy and endochondral ossification and lethality at birth [17], [37], [44]. The failed endochondral ossification in these models is attributed to the absence of chondrocyte hypertrophy. It is important to note that during chondrogenesis, expression of the Runx2 gene starts in resting chondrocytes and increases progressively, reaching the maximum level in hypertrophic chondrocytes. However, the physiologic role of the endogenous Runx2 gene after chondrocyte hypertrophy remains unknown. We address this question by deleting the Runx2 gene in hypertrophic chondrocytes using the BAC-Col10a-Cre transgenic model [45].
Results
Runx2 deletion in hypertrophic chondrocytes results in poorly mineralized extremities and limb dwarfism
To study the regulatory role of RUNX2 beyond chondrocyte hypertrophy, the Runx2 gene was deleted in hypertrophic chondrocytes by mating a Runx2F/F mouse with BAC-Col10-Cre transgenic mouse. We confirmed that Col10-Cre activity is specific to the hypertrophic chondrocytes by using Ai9-tdTomato reporter mice (Fig. 1A). Histologic analysis of femurs from newborn mice showed tdTomato positive cells appearing in the last two rows of the hypertrophic zone and extending into the calcified cartilage (Fig. 1A). These data are consistent with the previously reported pattern of BAC-Col10-Cre gene expression only in hypertrophic chondrocytes [45].
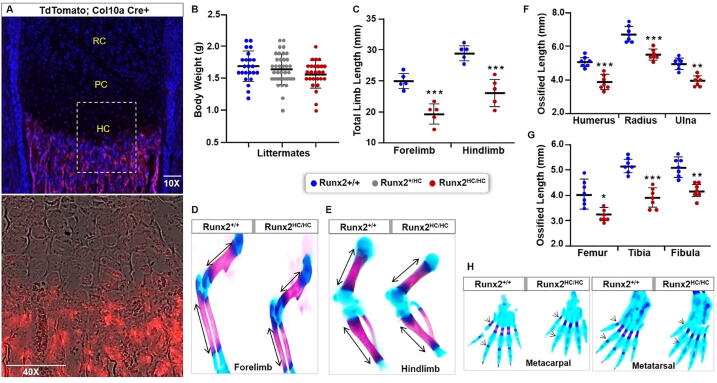
Fig. 1.
Deletion of Runx2 in hypertrophic chondrocytes results in decreased mineralization of limbs and extremities. (A) Cryo-section of the femur from a newborn pup containing BAC-Col10-Cre and ROSA26-tdTomato transgenes. A representative image with DAPI/TRITC overlay is shown at 10x magnification. The boxed region captured at 40X magnification is shown as phase contrast with TRITC overlay. RC: resting chondrocytes; PC: proliferative chondrocytes; HC: hypertrophic chondrocytes. Scale bar: 100 μm. (B) The whole-body weight of 3-days old littermates from multiple pregnancies are presented in a scatter plot. (Runx2+/+ n = 23, Runx2+/HC n = 39, Runx2HC/HC n = 25). (C) The total length of intact forelimbs and hindlimbs from 3-days old Runx2+/+ and Runx2HC/HC littermates is presented in the scatterplot plot (n = 5). (D, E) A representative image of alizarin red and alcian blue stained forelimbs and hindlimbs from Runx2+/+ and Runx2HC/HC littermates. (F, G) The length of ossified regions in each element of the forelimb (humerus, radius, ulna) and the hindlimb (femur, tibia, and fibula) was measured digitally and presented in the scatter plot (n = 7). (H) A representative image of the hand and paw stained with alizarin red and alcian blue. Arrowheads indicate unmineralized digits in the metacarpal and metatarsal of the Runx2HC/HC mice. (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, homozygous mice (Runx2HC/HC) are born alive with the expected Mendelian ratio and have comparable bodyweight to heterozygous (Runx2+/HC) and wild-type (Runx2+/+) littermates (Fig. 1B). Whole mount alizarin red and alcian blue staining revealed an overall well-formed skeleton, indicating the activity of the Runx2 gene in the hypertrophic chondrocyte is not essential for embryonic skeletogenesis (data not shown). However, the total length of intact forelimb and hindlimb are significantly shorter in homozygous mice (Fig. 1C). We assessed contributions of the proximal, intermediate, and distal elements toward limb dwarfism by measuring the alizarin red stained regions among littermates (Fig. 1D, E). Quantification of the ossified region from seven littermates showed a 20% shorter length of the humerus and a 15% shortening of both radius and ulna in homozygous mice (Fig. 1F). Similarly, the length of the ossified regions in the femur, tibia, and fibula is reduced by 15% and 14% respectively. (Fig. 1G). Moreover, the distal elements (metacarpal and metatarsal) were unmineralized and the phalanges were poorly developed in Runx2HC/HC mice (Fig. 1H). Together these results demonstrate that Runx2 loss in hypertrophic chondrocytes leads to limb dwarfism.
Runx2 deficiency in the hypertrophic chondrocyte leads to significant expansion of the zone of hypertrophy in the growth plate
To understand the effect of Runx2 loss in hypertrophic chondrocytes during endochondral ossification, we performed histological analysis. H&E staining revealed that the overall length of the growth plate is increased significantly in the Runx2HC/HC mice (Fig. 2A). Quantification of growth plate length confirmed an increase of 14% at the proximal end and 12% at the distal end of the tibia (Fig. 2C). Surprisingly, the hypertrophic chondrocyte zone is enlarged in both the proximal and distal growth plate of the Runx2HC/HC littermates (Fig. 2A-B). Quantification of the length of hypertrophic chondrocyte zone showed an increase of 89% in proximal and 87% in the distal growth plate (Fig. 2D). The significant lengthening of the hypertrophic zone is also noted in the growth plate of the femur, metacarpal, and metatarsal of the Runx2HC/HC mice (data not shown). In contrast, the length of the bone region between the proximal and distal growth plate in the tibia is decreased by 32% in the Runx2HC/HC littermates (Fig. 2A, E).
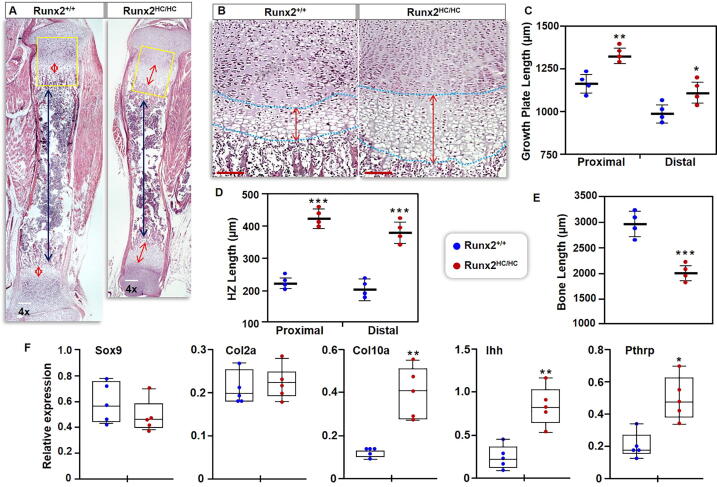
Fig. 2.
The length of the hypertrophic zone is doubled in Runx2HC/HC mice. (A) Hindlimbs from 3-days old littermates were processed for histology. Representative images of the whole tibia stained with H&E are shown at 4x magnification. Double headed red arrows indicate the length of the hypertrophic zone and black arrows show the length of the ossified region. Scale bar: 100 μm. (B) Box regions are shown at 10x magnifications. Scale bar: 200 μm. (C) Length of the overall growth plate from the articular surface to the end of the hypertrophic zone, (D) length of the zone of chondrocyte hypertrophy (HZ), and (E) length of the ossified region was quantified from four different tibias (n = 4) obtained from Runx2+/+ and Runx2HC/HC littermates. Data are presented in a scatter plot showing mean and SDM. (F) Growth plates from the enzymatically cleared hindlimbs of 3-days old Runx2+/+ and Runx2HC/HC littermates (n = 5) were dissected, and flash frozen for RNA extraction. Relative expression of Ihh, Col10a, Pthrp, Sox9, and Col2 genes normalized with Gapdh is presented in box and whisker plot. (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To explore the molecular basis for changes in the growth plate, the expression of chondrocyte marker genes was assessed by qPCR analysis. Consistent with the enlarged hypertrophic zone, a 3.5 fold increase in expression of the pre-hypertrophic marker (Ihh) and hypertrophic chondrocyte marker (Col10) was noted in Runx2HC/HC littermates (Fig. 2F). Interestingly, Pthrp, which is expressed by resting chondrocytes, also showed a 2.5-fold increased expression in Runx2HC/HC mice (Fig. 2F). However, the mRNA levels of the essential chondrocyte transcription factor Sox9 and the major collagen in the hyaline cartilage (Col2) are comparable among the littermates. (Fig. 2F). Thus, loss of the Runx2 gene from hypertrophic chondrocyte results in significant changes in both the growth plate and the ossified zone, leading to limb dwarfism.
Runx2 promotes apoptosis of hypertrophic chondrocytes
The enlarged zone of hypertrophic chondrocytes in Runx2HC/HC mice could be due to an increase in chondrocyte proliferation and subsequent hypertrophic differentiation or a decrease in apoptosis of hypertrophic chondrocytes. To assess these possibilities, cell death and proliferation assays were performed. We evaluated apoptosis of chondrocytes in paraffin-embedded sections of the hindlimb. A significant reduction in TUNEL positive hypertrophic chondrocytes is noted in both the distal femur and proximal tibial growth plates of Runx2HC/HC littermates (Fig. 3A). Quantification of TUNEL positive hypertrophic chondrocytes revealed a 25% and 30% decrease in the femur and tibia growth plates respectively (Fig. 3B). To independently confirm changes in chondrocyte apoptosis, we monitored the expression of pro-apoptotic and anti-apoptotic genes in the epiphyseal region of the tibia and femur from 3-days old littermates. The expression of the pro-apoptotic Bax gene was drastically reduced in Runx2HC/HC mice. Quantification of Bax mRNA from five littermates showed an 81% decrease in the Runx2HC/HC mice (Fig. 3C). Interestingly, the expression levels of the anti-apoptotic Bcl2 gene showed no significant difference among littermates (Fig. 3C). The ratio between the expression level of an anti-apoptotic and pro-apoptotic gene determines a cells resistance to apoptosis. The Bcl2 to Bax ratio is increased by 4-fold in the Runx2HC/HC mice, indicating resistance to apoptosis (Fig. 3C). These results strongly suggest that RUNX2 regulates apoptosis of hypertrophic chondrocytes during endochondral ossification.
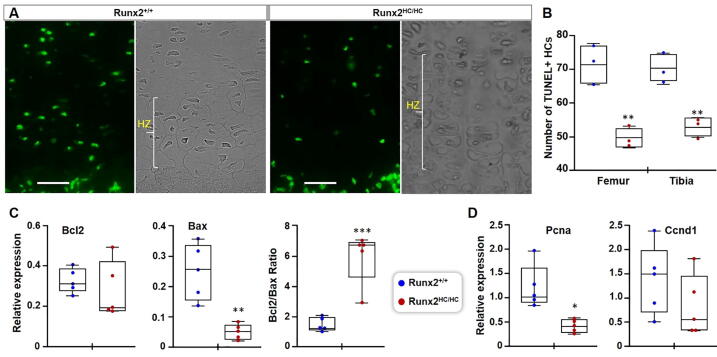
Fig. 3.
Decrease apoptosis of hypertrophic chondrocytes contributes to the lengthening of the zone of hypertrophy. (A) Hindlimbs from 3-days old littermates were processed for histology. Representative images from tibial growth plates are shown at 40x magnification. Scale bar: 50 μm. (B) TUNEL positive hypertrophic chondrocytes counted from proximal and distal growth plates of tibia and femur from three littermates (n = 3) are presented in box and whisker plot. (C, D) Enzymatically cleared distal femur and proximal tibia epiphyseal growth plates from 3-days old Runx2+/+ and Runx2HC/HC littermates (n = 5) were used for RNA extraction. Relative expression of (C) Bax and Bcl2, (D) Pcna, and Ccnd1 genes normalized with Gapdh are shown in the box and whisker plot. (*P < 0.05, **P < 0.01, ***P < 0.001).
To test if chondrocyte proliferation contributes to an enlarged hypertrophic zone, the expression of proliferation marker genes was compared among littermates (Fig. 3D). Runx2HC/HC mice showed a significant reduction in the expression of proliferating cell nuclear antigen (Pcna). However, expression of cyclin D1(Ccnd1), another marker of cell proliferation, showed no change among littermates (Fig. 3D). Taken together these data indicate that the lengthening of the zone of hypertrophy in Runx2HC/HC is likely due to a decrease in the apoptosis of hypertrophic chondrocytes.
Deletion of Runx2 in hypertrophic chondrocytes leads to impaired resorption of cartilage matrix
The decreased ossified region in the long bones of Runx2HC/HC mice prompted the assessment of the cartilage matrix in the developing limbs. Alcian blue stained tibia sections showed the presence of a significant amount of cartilage matrix in both the proximal and distal metaphysis region in Runx2HC/HC mice (Fig. 4A). The cartilage erosion zone is significantly expanded and the remnants of hypertrophic cartilage are present up to the mid-diaphysis region of the tibia (Fig. 4B). Quantification of hypertrophic cartilage length indicates a 56% increase in both the proximal and the distal growth plate of homozygous mice (Fig. 4C). A significant increase in the cartilage erosion zone is also noted in the femur of the Runx2HC/HC mice (data not shown). The cartilage accumulation in the mice may be due to an increased synthesis or decreased resorption of the cartilage matrix. To distinguish these possibilities, we analyzed the expression of the most abundant proteoglycan (Acan) and collagen (Col2) produced by chondrocytes. As noted earlier the expression of Col2 is comparable among littermates (Fig. 2F). To our surprise, the expression of Acan is decreased by 20% in the Runx2HC/HC littermates (Fig. 4D). These results indicate that the increased amount of cartilage in the Runx2 mutant mice is not due to enhanced cartilage synthesis.
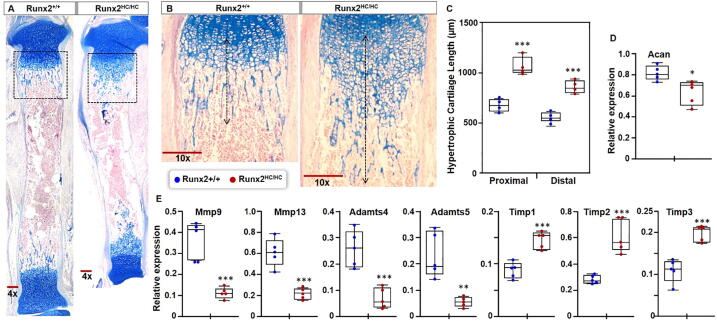
Fig. 4.
Accumulation of hypertrophic cartilage in Runx2HC/HC mice is associated with decreased expression of matrix metalloproteinase. (A) Representative images of alcian blue stained whole tibia from 3-days old littermates. (B) The boxed region is shown at 10x magnification. Double headed arrows indicate the length of hypertrophic cartilage. Scale bar: 200 μm. (C) The length of hypertrophic cartilage measured from the start of the hypertrophic zone to the end of cartilage remnant in littermates (n = 4) is presented in a box and whisker plot. (D, E) RNA isolated from the enzymatically cleared distal femur and proximal tibia epiphyseal growth plates from Runx2+/+ and Runx2HC/HC littermates (n = 5) subjected to qPCR. Relative expression of Acan, Mmp9, Mmp13, Adamts4, Adamts5, Timp1, Timp2 and Timp3 normalized with Gapdh is shown in the box and whisker plot. (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The turnover of the calcified cartilage in the developing limb is regulated by several matrix degrading enzymes primarily produced by hypertrophic chondrocytes. Therefore, we evaluated the expression of collagenase-3 (Mmp13), gelatinase-B (Mmp9), and aggrecanases (Adamts4, Adamts5) among littermates. Expression of both collagenase-3 and gelatinase-B is decreased by 65% in Runx2HC/HC compared to littermates (Fig. 4E). Expression of Adamts4 and Adamts5 was significantly decreased by 75% and 81% respectively in Runx2HC/HC littermates (Fig. 4E). We also examined gene expression of tissue inhibitors of metalloproteinases (Timps), that are known to regulate the activity of collagenases and aggrecanases. Interestingly, expression of Timp1, Timp2 and Timp3 was increased by 65%, 120% and 80% respectively in Runx2HC/HC littermates. These results suggests that cartilage accumulation is likely linked with the decreased expression and activity of cartilage degrading enzymes. Together our data indicate that RUNX2 activity in hypertrophic chondrocytes is required for the expression of the enzymes responsible for the degradation of the calcified cartilage matrix.
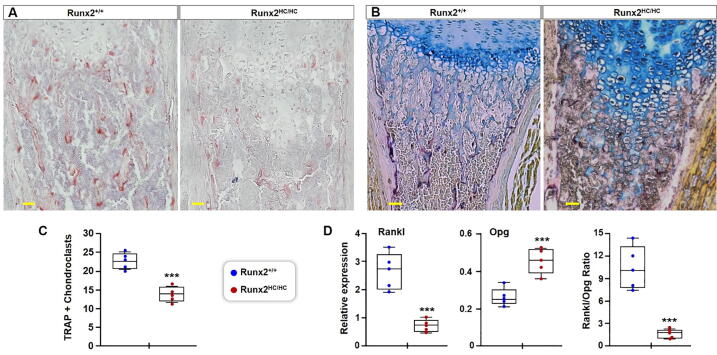
Increased calcified cartilage in Runx2HC/HC mice is associated with a decrease in the number of chondroclasts
In addition to the matrix degrading enzymes secreted by hypertrophic chondrocytes, turnover of the hypertrophic cartilage is regulated by chondroclasts. TRAP staining of the tibial section showed a marked decrease in the number of TRAP positive cells in the cartilage erosion zone of Runx2HC/HC littermates (Fig. 5A). To better quantify the difference in cartilage resorbing cells, the paraffin sections from hindlimb were double stained with TRAP and alcian blue. Consistent with the earlier observation, Runx2HC/HC bones showed a significant decrease in the number of TRAP + chondroclasts lining the cartilage island within the erosion zone (Fig. 5B). Quantification from five littermates revealed a 35% decrease in the number of TRAP + chondroclast in Runx2HC/HC mice (Fig. 5C). Next, we performed qPCR analysis to evaluate the expression of genes regulating the differentiation of chondroclasts. Interestingly, expression of Rankl is decreased by 73% in the growth plate chondrocyte of Runx2HC/HC mice. In sharp contrast expression of Opg is increased by 75% in the Runx2HC/HC littermates. This differential gene expression in the Runx2HC/HC mice resulted in a decreased ratio of Rankl/Opg, which is inhibitory for chondroclast differentiation (Fig. 5D). Together our results suggest that Runx2 deficiency in the hypertrophic chondrocytes leads to impaired differentiation of chondroclasts and cartilage resorption in the developing bones.
Fig. 5.
Runx2HC/HC mice show a decreased number of TRAP positive chondroclasts in the cartilage erosion zone. (A) Hindlimbs from 3-days old littermates were processed for histology and stained with TRAP. A representative image of TRAP-stained tibia from Runx2+/+ and Runx2HC/HC littermates is shown at 10x magnification. (B) TRAP and alcian blue double staining performed to reveal the cartilage associated with TRAP + chondroclasts. Representative images are presented at 10x magnification. Scale bar: 100 μm. (C) The number of TRAP + chondroclasts counted from five littermates (n = 5) is presented in a box and whisker plot. (D) RNA isolated from enzymatically cleared epiphyseal growth plates from indicated littermates was used for qPCR performed with four replicates. Relative Opg and Rankl expression normalized with Gapdh is shown in the box and whisker plot. The calculated Rankl/Opg ratio for each sample is presented in a box and whisker plot. (***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
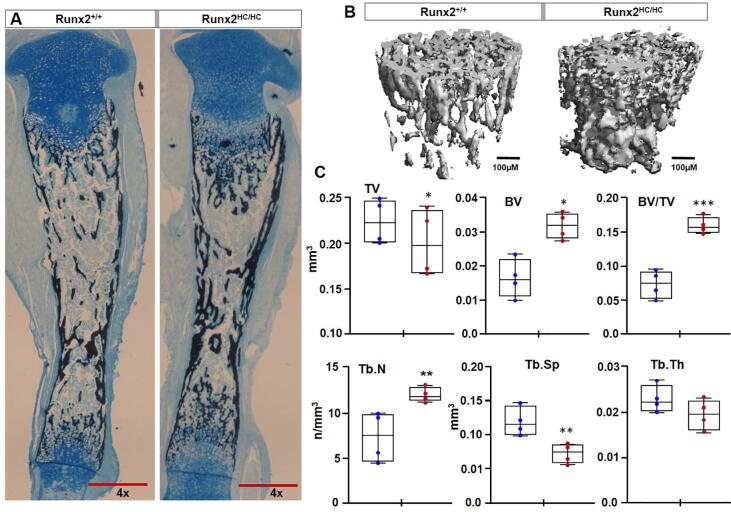
Runx2HC/HC mice exhibits increased trabecular bone mass
During endochondral ossification, hypertrophic cartilage provides the template for bone synthesis. Therefore, we performed Von Kossa staining to assess if an increased amount of hypertrophic cartilage in Runx2HC/HC mice affects mineral deposition. A marked increase in mineral deposition was evident at both the proximal and distal growth plates of the tibia of Runx2HC/HC littermates (Fig. 6A). Hindlimbs used to evaluate bone mass by micro-CT analysis confirmed an increase in trabecular bone (Fig. 6B). Quantification from four littermates showed a 2-fold increase in the BV/TV ratio of the trabecular bone in Runx2HC/HC mice (Fig. 6C). Importantly, a 50% increase in the trabecular number was coupled with a 30% decrease in trabecular spacing in Runx2HC/HC mice (Fig. 6C). The trabecular thickness and bone surface, however, remain unchanged among littermates (Fig. 6C and data not shown). The cortical bone was comparable among littermates and bone parameters determined at the mid-diaphysis region such as BV/TV, cortical thickness, bone surface and porosity were comparable between Runx2+/+ and Runx2HC/HC littermates (data not shown). Together our data demonstrate that increase mineral deposition in the trabecular bone is associated with retention of hypertrophic cartilage.
Fig. 6.
Increased trabecular mineralization in Runx2 mutant mice. (A) Paraffin sections of the tibia from 3-days old littermates were stained with Von Kossa followed by alcian blue. Representative images of the tibia taken at 4x magnification are shown. Scale bar: 500 μm. (B) Representative 3D-image of trabecular bone from the femur of 3-days old Runx2+/+ and Runx2HC/HC littermates. Scale bar: 100 μm. (C) Trabecular bone parameters were analyzed from a 2.4 mm region immediately beneath the growth plate. Quantified data of bone parameters from Runx2+/+ and Runx2HC/HC littermates (n = 4) is presented in a box and whisker plot. BV; bone volume, TV; total volume, Tb.N; trabecular number, Tb.Th; trabecular thickness, Tb.Sp; trabecular space. (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
In the current study, the Runx2 gene is deleted in hypertrophic chondrocytes to assess its physiologic role after chondrocyte hypertrophy. Homozygous mice have comparable body weights and survive to adulthood. Runx2HC/HC mice exhibit limb dwarfism and unmineralized metacarpals and metatarsals. Runx2 gene ablation in the hypertrophic chondrocyte does not affect the resting or proliferative zone in the growth plate but the zone of chondrocyte hypertrophy is nearly doubled. The expansion of the hypertrophic zone in growth plates is mainly due to a significant reduction in apoptosis of the hypertrophic chondrocytes. Decreased expression of the matrix degrading enzymes by Runx2-deficient hypertrophic chondrocytes leads to accumulation of calcified cartilage in the limb bones. Moreover, Runx2HC/HC mice showed a significant reduction in TRAP + chondroclasts and an increase in trabecular bone mass. Thus, RUNX2 continues to function after chondrocyte hypertrophy to control the turnover of hypertrophic cartilage during endochondral ossification.
Impairments in the chondrocyte differentiation and endochondral ossification process are central to the dwarfism phenotype. Runx2HC/HC mice have shorter limbs and a decreased length of the ossified region. The reason for the shortening of the long bone could be a decrease in the proliferation of chondrocytes. IHH produced by hypertrophic chondrocyte regulates chondrocyte proliferation by inducing expression of Pthrp in the resting chondrocyte [46]. Deletion of either Ihh or Pthrp, therefore, leads to proliferative defects and dwarfism [15], [47]. RUNX2 directly regulates the expression of both Ihh and Pthrp [17], [48], [49]. Deletion of Runx2 in resting chondrocytes and the resultant decrease in Ihh expression and chondrocyte proliferation leads to dwarfism [17]. However, Runx2 deletion in hypertrophic chondrocyte did not show any change in chondrocyte proliferation as the length of the proliferative zone and Ccnd1 expression were comparable among wild-type and homozygous littermates. Our data are consistent with the recent report showing no change in chondrocyte proliferation upon deletion of the Runx2 gene in hypertrophic chondrocytes [50]. The increased levels of Ihh and Pthrp noted in Runx2HC/HC mice likely reflect an increase in the number of the pre-hypertrophic/hypertrophic chondrocytes within the enlarged growth plate. These results indicate that dwarfism in Runx2HC/HC mice is not due to chondrocyte proliferation.
A surprising finding in our study is the doubling of the zone of hypertrophic chondrocytes in Runx2HC/HC littermates. Accelerated maturation or decreased apoptosis of hypertrophic chondrocytes can lead to the lengthening of the hypertrophic zone. For example, activation of β-catenin signaling in resting chondrocytes leads to a significant expansion of the hypertrophic zone due to accelerated maturation and shortened proliferative zone, but without affecting chondrocyte apoptosis [51]. Our histological analysis reveals that the overall length of the growth plate is significantly increased but the length of the resting and proliferating zone is comparable among wild-type and homozygous littermates. Moreover, the expression of the proliferative marker is either unchanged (Ccnd1) or significantly decreased (Pcna) in Runx2HC/HC mice. Our results are consistent with no change in proliferation demonstrated by BrdU labeling in mice where the Runx2 gene is deleted in hypertrophic chondrocytes [50]. Thus, the accelerated maturation is unlikely a contributing factor for the doubling of the zone of chondrocyte hypertrophy in Runx2HC/HC mice.
A significant decrease in apoptosis of hypertrophic chondrocytes is associated with enlargement of the zone of hypertrophy [26]. BCL2 and BAX proteins promote cell survival and apoptosis, respectively. Expression of the anti-apoptotic Bcl2 gene is highest in the proliferative chondrocytes but is nearly absent in hypertrophic chondrocytes [52]. Deletion of the Bcl2 gene results in the shortened hypertrophic zone due to increased apoptosis of hypertrophic chondrocytes [52]. In sharp contrast, expression of the pro-apoptotic BAX protein increases progressively during chondrocyte differentiation with the maximum levels noted in hypertrophic chondrocyte [52]. Deletion of the Bax gene results in decreased apoptosis of chondrocytes [53]. In line with the decreased apoptosis of hypertrophic chondrocytes in Runx2HC/HC mice, our qPCR analysis showed no change in Bcl2 mRNA but a significant decrease in the expression of the Bax gene. This differential expression leads to a 4-fold increase in the Bcl2/Bax ratio, which normally inhibits cellular apoptosis. Therefore, the expansion of the hypertrophic zone in Runx2HC/HC mice is likely due to a decrease in apoptosis of hypertrophic chondrocytes. Importantly, Runx2 binds and regulates the activity of Bcl2 and Bax promoters [54], [55]. In stark contrast to our data, a recent report shows that Runx2 deletion in hypertrophic chondrocytes promotes apoptosis of hypertrophic chondrocytes without changing the length of the hypertrophic zone. Surprisingly, increase apoptosis of hypertrophic chondrocytes was noted despite no change in the expression of the pro-apoptotic Bax gene and a significant increase in the mRNA of the anti-apoptotic Bcl2 gene [50]. The underlying reasons for these contrasting findings need further investigation.
During endochondral ossification, degradation of cartilage and formation of bone are coupled. Compared to wild-type littermates, a large amount of calcified cartilage was present in the Runx2HC/HC mice. The cartilage islands were noted well beyond the metaphyseal regions of the endochondral bones. The production of ACAN and COL2 mainly determines the amount of cartilage matrix in the developing endochondral bones [56], [57]. A similar expression level of Acan and Col2 mRNA among littermates rules out that increased cartilage synthesis is contributing to the accumulation of cartilage matrix in Runx2HC/HC mice. Enzymes produced by hypertrophic chondrocytes such as aggrecanase, gelatinase, and collagenase are critical for the degradation of the calcified cartilage [58]. Deletion of gelatinase (Mmp9) or collagenase (Mmp13) genes in mice disrupts cartilage degradation, leading to accumulation of hypertrophic cartilage in the long bone [26], [27]. In humans, a mutation in the gelatinase or collagenase genes results in metaphyseal dysplasia characterized by defective growth and modeling of the spine and long bones [59], [60].
ADAMTS4 and ADAMTS5 are the major aggrecanase involved in degradation of aggrecan [61]. Global deletion of Adamts4 or Adamts5 gene in mice does not impair embryonic skeletogenesis [62], [63]. However, Adamts5 is required for development of trabeculated bone in the mandibular condyle [64]. Interestingly, Adamts5 deficiency prevents cartilage degradation in both inflammatory and surgically induced murine model of osteoarthritis [62], [63]. In humans, single nucleotide polymorphism in ADAMTS5 gene is associated with lumbar degenerative disc disease [65]. It is important to note that RUNX2 directly regulates the expression of gelatinase, collagenase, and aggrecanase [66], [67], [68], [69]. We find Runx2 deletion in hypertrophic chondrocytes results in a significant reduction in expression of Adamts4, Adamts5, Mmp9, and Mmp13. The reported decrease in Mmp13 expression due to deletion of Runx2 in hypertrophic chondrocytes at the embryonic stage is consistent with our data, but expression of Mmp9, Adamts4 or Adamts5 is not described in this paper [47]. Thus, RUNX2 activity in hypertrophic chondrocytes is required to produce enzymes that are responsible for the degradation of the calcified cartilage.
During development, the four members of the tissue inhibitors of metalloproteinases gene family (Timp1-4), are ubiquitously expressed in several tissues and cancers [70]. TIMPs inhibits activity of MMPs and ADAMTSs by forming an irreversible complex with the enzyme in a 1:1 ratio [71]. During cartilage development, Timp1, Timp2 and Timp3 are expressed by chondrocytes [28], [29], [30], [31]. Due to the redundant functions, no skeletal phenotype is noted when Timp genes are individually deleted in mice [72]. However, combined deletion of Timp1-4 leads to enhanced degradation of cartilage and significant shortening of long bones [72]. The proximal promoters of Timp1, 2 and 3 contain a Runx binding motif and RUNX2 overexpression induce activity of Timp1 promoter in fibroblasts [73], [74]. We noted a significant increase in expression of Timp1, Timp2 and Timp3 in Runx2HC/HC mice. Therefore, the accumulation of hypertrophic cartilage in Runx2HC/HC mice is likely due to a decrease in expression and activity of cartilage degrading enzymes.
Another surprising finding of our study is the significant increase of trabecular bone mass in Runx2HC/HC mice. The high bone mass is not due to increased bone synthesis, as the expression of osteoblast markers such as collagen type I and osteocalcin are comparable among the littermates (unreported observation). Chondroclasts are essential for the turnover of hypertrophic cartilage, which serves as a template for mineral deposition during endochondral ossification. A 35% decrease in the number of chondroclasts noted in the cartilage erosion zone/trabecular bone region of the Runx2HC/HC mice is likely responsible for the retention of hypertrophic cartilage. Hypertrophic chondrocytes produced RANKL and OPG is central for the differentiation of chondroclasts/osteoclasts [33], [75]. During endochondral ossification, RUNX2 regulates the expression of both Rankl and Opg in chondrocytes and the Rankl/Opg ratio is critical for chondroclast/osteoclast differentiation [76], [77]. We find a significant increase in Opg mRNA and a significant decrease of Rankl expression in Runx2HC/HC mice. This differential expression leads to 81% decrease in the Rankl/Opg ratio, which is inhibitory for chondroclast/osteoclast differentiation. We have previously reported that deletion of the Runx2 gene in resting chondrocytes blocks chondrocyte hypertrophy, decreases expression of Rankl/Opg and results in an absence of TRAP + chondroclasts [17]. Together, our results demonstrate RUNX2 controlled Rankl and Opg expression in hypertrophic chondrocytes is required for chondroclast/osteoclast differentiation. In contrast to our finding, a significant increase in the number of TRAP + osteoclasts is reported in E16.5 Runx2 homozygous null mice but without any change in the Rankl/Opg ratio. Interestingly, a significant decrease in the Rankl/Opg ratio is noted a day earlier in this model but neither the TRAP staining nor the quantification of TRAP + osteoclast is presented at E15.5 [50]. Further investigation is needed to reconcile the discrepancies in the cartilage and trabecular bone phenotype between the two Runx2 mouse models.
Conclusion
In conclusion, our findings uncover a novel role of RUNX2 in regulating apoptosis of hypertrophic chondrocytes and degradation of cartilage matrix during endochondral ossification. Regulation of Rankl and Opg expression by RUNX2 in hypertrophic chondrocytes is critical for chondroclast/osteoclast differentiation and resorption of cartilage and bone matrix.
Experimental procedures
Generation and genotyping of Runx2HC/HC mouse
Generation of exon 8 Runx2-floxed (Runx2F/F) mice is reported previously [17]. To delete Runx2 specifically in hypertrophic chondrocytes, Runx2F/F mice were crossed with BAC-collagen type10-Cre mice [45]. The resulting heterozygous (Runx2+/HC) mice were intercrossed to obtain Runx2HC/HC, Runx2+/HC, and Runx2+/+ mice. The Ai9-tdTomato reporter mouse was used to confirm the specificity of Cre-expression [78]. All genotypes were determined using DNA from the tail biopsy with Direct PCR lysis reagent (Viagen biotech, Cat#102-T). The Runx2 wild type and floxed alleles, ROSA26-Tdtomato and Cre transgene were detected by PCR using the specific primers in Table 1. All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham and conformed to relevant federal and state guidelines and regulations.
Table 1.
Primer sequence used for genotyping of mice.
| Name | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| Runx2 | atc agt tcc caa tgg tac ccg | gca aga tca tga cta ggg att g |
| Col10-Cre | ttt aga gca tta ttt caa ggc agt ttc | agg caa att ttg gtg tac gg |
| tdTomato | ctg ttc ctg tac ggc atg g | ggc att aaa gca gcg tat cc |
Skeletal staining and histology
For skeletal staining with alizarin red and alcian blue, littermates of all genotypes were collected at 3-days of age. We follow previously published protocol for staining of newborn pups with some modification [17]. Hindlimbs and forelimbs were dissected and de-skinned, and muscles were partially removed. Limbs were fixed for 3 days in 95% ethanol (EtOH) and then placed in acetone for 2 days to remove fat tissues. Skeletons were then placed in alizarin red and alcian blue staining solution 0.015% alcian blue (Sigma-Aldrich, Cat#A5268), 0.005% alizarin red (MP Biomedicals, Cat# 100375), and 5% Acetic acid in 70% ethanol for 3 days with gentle shaking. The soft tissues from the stained skeleton were cleared with 0.5% KOH for 2 days with gentle shaking and the fresh solution was used every day. The further clearing was carried out with 0.5% KOH and sequential concentrations of glycerol (20%, 50%, and 80%) for 3 days for each with gentle shaking. Stained skeleton preparations were stored in 0.5% KOH and 80% glycerol for complete cleaning. Skeletal elements of both WT and Runx2HC/HC were captured in the same frame with Nikon Cool PIX S10 digital camera (Nikon, Cat#2555B).
Tissue embedding for histological sectioning and staining was performed essentially as previously published [17], [79]. For histological analysis, hindlimbs from 3-days old littermates were fixed overnight in 4% paraformaldehyde (Sigma-Aldrich, Cat#P6148). Tibia and femur were dehydrated in gradient ethanol, embedded in paraffin, and sectioned at 7 μm thickness. Tissue sections were dewaxed in xylene, rehydrated in gradient ethanol, and processed for hematoxylin and eosin, alcian blue, and TRAP staining as described elsewhere [79].
TRAP and alcian blue double staining
The tissue sections were double stained with TRAP and alcian blue to reveal the cartilage associated chondroclasts. TRAP staining was performed by incubating slides in the naphthol AS-BI phosphate solution for 2 hrs at room temperature using acid phosphatase leucocyte kit (Sigma-Aldrich, Cat#387A). The slides were washed in PBS for 5 min and then stained with 0.2% alcian blue solution in 3% acetic acid. The slides were washed with three changes of water for 5 min each.
Micro-CT analysis
Trabecular and cortical bone structure and mineral density were analyzed in 3-days old mice by µCT analysis with the help of UAB Small Animal Imaging and Analysis Core. Undecalcified femurs were scanned by a cone beam microcomputed tomography system µCT40 to produce 3D images (Scanco Medical AG, Brüttisellen, Switzerland). Images were analyzed using µCT 3D Viewer V3.8 software. Quantification of bone parameters for cortical bone were derived from a 0.6 mm region of the femoral mid-shaft and trabecular bone from a 2.4 mm region immediately beneath the growth plate. Bone parameters analyzed were; bone volume (BV), total volume (TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular space (Tb.Sp).
RNA isolation and quantitative real time PCR
Analysis of the gene expression by qPCR was performed as previously published [80]. Briefly, total RNA was extracted from the distal femur and proximal tibia of hindlimbs of 3-days old littermates. Deskinned hindlimbs were completely cleared of skeletal muscle by digestion initially with pronase (Roche Diagnostic, Cat#10165921001) and collagenase-D (Roche Diagnostic, Cat#11088882001) for 1-hr and subsequently with collagenase-D for 2 hrs at 37 °C. The epiphysial growth plates were harvested under dissecting microscope and flash frozen in liquid nitrogen and homogenized in PBS containing DEPC. The growth plate homogenates were sonicated in TRIZOL reagent (Life Technologies, Cat# 15596018) to extract total RNA. The cDNA was prepared from 1.5 µg of total RNA using a cDNA synthesis kit (BioRad, Cat# 170-8891). Quantitative real time PCR with four replicates was performed using iQ SYBR Green Supermix (BioRad, Cat# 1725124) with specific primer pairs detailed in Table 2. Gene expression values were normalized with Gapdh, used as an internal control. Relative gene expression was obtained using the 2^-ΔCT method.
Table 2.
Sequence of primers used for qPCR.
| Name | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| Acan | ccc tcc ggc aga aga aag at | cgc ttc tgt agc ctg tgc ttg |
| Col2 | tcc tct gcg atg aca tta tct | gat cat ctc tgg gtc ctt gtt |
| Sox9 | agg aag ctg gca gac cag ta | tgt aat cgg ggt ggt ctt tc |
| Col10a | gca gca tta cga ccca aag at | ctt gaa gcc tga tcc agg ta |
| Ihh | acg tgc att gct ctg tca ag | ctc gat gac ctg gaa agc tc |
| Pthrp | ttc agc agt gga gtg tcc tg | agc tct gat ttc ggc tgt gt |
| Adamts4 | caa gca gtc ggg ctc ctt | gatcgtgaccacatcgctgta |
| Adamts5 | cca gtt gta caa aga tta tcg gaa cct | gtt gct cct tca ggg atc ct |
| Mmp9 | gat ccc cag agc gtc att | cca cct tgt tca cct cat tt |
| Mmp13 | tgt ttg cag agc act act tga a | cag tca cct cta agc caa aga aa |
| Bax | tgc aga gga tga ttg ctg ac | gat cag ctc ggg cac ttt ag |
| Bcl2 | ctg gca tct tct cct tcc ag | gac ggt agc gac gag aga ag |
| Cyclin d1 | gcg tac cct gac acc aat ctc | ctc ctc ttc gca ctt ctg ctc |
| Pcna | ttt gag gca cgc ctg atc c | gga gacgtg aga cga gtc cat |
| Rankl | act tgg gat ttt gat gct ggt t | tgg gcc aag atc tct aac atg a |
| Opg | atg aac aag tgg ctg tg | cct cac tgt gca gtg ctg tt |
| Timp1 | agg tgg tct cgt tga ttt ct | gta agg cct gta gct gtg cc |
| Timp2 | gaa tcc tct tga tgg ggt tg | cgt ttt gca atg cag acg ta |
| Timp3 | ctt ctg caa ctc cga cat cgt | ggg gca tct tac tga agc ctc |
| Gapdh | ccg cct gga gaa acc tgc caa g | gga tag ggc ctc tct tgc tca g |
TUNEL assay
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed using fluorescein in situ cell death detection kit (Roche Applied Science, Cat# 11684795910). Paraffin sections from hindlimbs of 3-day old littermates were processed for TUNEL labeling according to the manufacturer’s directions. The tissue was permeabilized with 0.1% Triton-X 100 in 0.1% sodium citrate for 20 min at room temperature. Slides were placed in PBS for 10 mins and the TUNEL labeling reaction carried for 1.5 hrs at room temperature. Slides were washed twice with PBS for 10 mins each prior to mounting. Images of growth plate were captured using BZ-X8000 Keyence Fluorescence microscope. TUNEL positive nuclei within hypertrophic chondrocyte zone from both distal femur and proximal tibia were quantified.
Statistical analysis
Mean values and standard deviation of the means (SDM) were calculated using GraphPad Prism 9.0.0. Statistical significance was determined between Runx2+/+ and Runx2HC/HC using an unpaired Student’s t test. Data with P < 0.05 were considered statistically significant and are indicated with an asterisk in the figures. Data are presented either in a Scatter plot or a Box-whisker plot showing maximum to minimum values, mean, and SDM.
CRediT authorship contribution statement
Harunur Rashid: Conceptualized the experiments, Data curation, Validation, Visualization, Performed formal analysis, Funding acquisition, Writing of the original draft, Review and editing. Haiyan Chen: Conceptualized the experiments, Data curation, Validation, Visualization. Amjad Javed: Conceptualized the experiments, Performed formal analysis, Funding acquisition, Writing of the original draft, Review and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and National Institute on Aging of the National Institutes of Health under Award Number R01AR062091 and R56AG065129. H.R was supported by NIDCR training grant number, T-90DE022736, and a pilot grant from UAB GC-CODED. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contribution
H.R, H.C and A.J outlined and designed experiments. H.R and H.C performed most of the experiments. H.R and A.J performed analysis, interpretation of data and writing of the manuscript.
References
- 1.Berendsen A.D., Olsen B.R. Bone development. Bone. 2015;80:14–18. doi: 10.1016/j.bone.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;6(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Crombrugghe B., Lefebvre V., Behringer R.R., Bi W., Murakami S., Huang W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000;19(5):389–394. doi: 10.1016/s0945-053x(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 4.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 5.F. Long, D.M. Ornitz, Development of the endochondral skeleton, Cold Spring Harb Perspect Biol 5 (2013) a008334. [DOI] [PMC free article] [PubMed]
- 6.Kozhemyakina E., Lassar A.B., Zelzer E. A pathway to bone: signalingmolecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallett S.A., Ono W., Ono N. Growth plate chondrocytes: skeletal development, growth and beyond. Int. J. Mol. Sci. 2019;20:6009. doi: 10.3390/ijms20236009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hojo H., McMahon A.P., Ohba S. An emerging regulatory landscape for skeletal development. Trends Genet. 2016;32(12):774–787. doi: 10.1016/j.tig.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Y., Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol. Cell. Biol. 2008;28(16):4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuhashi K., Ono W., Matsushita Y., Sakagami N., Takahashi A., Saunders T.L., Nagasawa T., Kronenberg H.M., Ono N. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563(7730):254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Q.i., Eberspaecher H., Lefebvre V., de Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 1997;209(4):377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 12.Saito M., Mulati M., Talib S.Z., Kaldis P., Takeda S., Okawa A., Inose H. The indispensable role of cyclin-dependent kinase 1 in skeletal development. Sci. Rep. 2016;6:20622. doi: 10.1038/srep20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato K., Bhattaram P., Penzo‐Méndez A., Gadi A., Lefebvre V. SOXC transcription factors induce cartilage growth plate formation in mouse embryos by promoting noncanonical Wnt signaling. J. Bone Miner. Res. 2015;30(9):1560–1571. doi: 10.1002/jbmr.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyngaarden L.A., Vogeli K.M., Ciruna B.G., Wells M., Hadjantonakis A.K., Hopyan S. Oriented cell motility and division underlie early limb bud morphogenesis. Development. 2010;137(15):2551–2558. doi: 10.1242/dev.046987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St-Jacques B., Hammerschmidt M., McMahon A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Eerden B.C., Karperien M., Gevers E.F., Löwik C.W., Wit J.M. Expression of Indian hedgehog, parathyroid hormone-related protein, and their receptors in the postnatal growth plate of the rat: evidence for a locally acting growth restraining feedback loop after birth. J. Bone Miner. Res. 2000;15(6):1045–1055. doi: 10.1359/jbmr.2000.15.6.1045. [DOI] [PubMed] [Google Scholar]
- 17.Chen H., Ghori-Javed F.Y., Rashid H., Adhami M.D., Serra R., Gutierrez S.E., Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J. Bone Miner. Res. 2014;29(12):2653–2665. doi: 10.1002/jbmr.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zelzer E., Mamluk R., Ferrara N., Johnson R.S., Schipani E., Olsen B.R. VEGFA is necessary for chondrocyte survival during bone development. Development. 2004;131(9):2161–2171. doi: 10.1242/dev.01053. [DOI] [PubMed] [Google Scholar]
- 19.Inada M., Wang Y., Byrne M.H., Rahman M.U., Miyaura C., Lopez-Otin C., Krane S.M. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. PNAS. 2004;101(49):17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Retting K.N., Song B., Yoon B.S., Lyons K.M. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136(7):1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill T.P., Später D., Taketo M.M., Birchmeier W., Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell. 2005;8(5):727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Naski M.C., Colvin J.S., Coffin J.D., Ornitz D.M. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125(24):4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- 23.Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H.M., Teitelbaum S.L., Ross F.P., Kopan R., Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14(3):306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schipani E., Ryan H.E., Didrickson S., Kobayashi T., Knight M., Johnson R.S. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15(21):2865–2876. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maes C., Kobayashi T., Selig M.K., Torrekens S., Roth S.I., Mackem S., Carmeliet G., Kronenberg H.M. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu T.H., Shipley J.M., Bergers G., Berger J.E., Helms J.A., Hanahan D., Shapiro S.D., Senior R.M., Werb Z. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93(3):411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stickens D., Behonick D.J., Ortega N., Heyer B., Hartenstein B., Yu Y., Fosang A.J., Schorpp-Kistner M., Angel P., Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131(23):5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bord S., Horner A., Beeton C.A., Hembry R.M., Compston J.E. Tissue inhibitor of matrix metalloproteinase-1 (TIMP-1) distribution in normal and pathological human bone. Bone. 1999;24(3):229–235. doi: 10.1016/s8756-3282(98)00174-4. [DOI] [PubMed] [Google Scholar]
- 29.Apte S.S., Fukai N., Beier D.R., Olsen B.R. The matrix metalloproteinase-14 (MMP-14) gene is structurally distinct from other MMP genes and is co-expressed with the TIMP-2 gene during mouse embryogenesis. J. Biol. Chem. 1997;272(41):25511–25517. doi: 10.1074/jbc.272.41.25511. [DOI] [PubMed] [Google Scholar]
- 30.Apte S.S., Hayashi K., Seldin M.F., Mattei M.-G., Hayashi M., Olsen B.R. Gene encoding a novel murine tissue inhibitor of metalloproteinases (TIMP), TIMP-3, is expressed in developing mouse epithelia, cartilage, and muscle, and is located on mouse chromosome 10. Dev. Dyn. 1994;200(3):177–197. doi: 10.1002/aja.1002000302. [DOI] [PubMed] [Google Scholar]
- 31.Poulet B., Liu K.e., Plumb D., Vo P., Shah M., Staines K., Sampson A., Nakamura H., Nagase H., Carriero A., Shefelbine S., Pitsillides A.A., Bou-Gharios G., Zhang C. Overexpression of TIMP-3 in chondrocytes produces transient reduction in growth plate length but permanently reduces adult bone quality and quantity. PLoS ONE. 2016;11(12):e0167971. doi: 10.1371/journal.pone.0167971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuyama R., Stockmans I., Torrekens S., Van Looveren R., Maes C., Carmeliet P., Bouillon R., Carmeliet G. Vitamin D receptor in chondrocytes promotes osteoclastogenesis and regulates FGF23 production in osteoblasts. J Clin Invest. 2006;116(12):3150–3159. doi: 10.1172/JCI29463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong J., Onal M., Jilka R.L., Weinstein R.S., Manolagas S.C., O'Brien C.A. Matrix-embedded cells control osteoclast formation. Nat. Med. 2011;17(10):1235–1241. doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang K.Y., Chan D., Cheah K.S. Fate of growth plate hypertrophic chondrocytes: death or lineage extension? Dev. Growth Differ. 2015;57:179–192. doi: 10.1111/dgd.12203. [DOI] [PubMed] [Google Scholar]
- 35.Amizuka N., Hasegawa T., Oda K., Luiz de Freitas P.H., Hoshi K., Li M., Ozawa H. Histology of epiphyseal cartilage calcification and endochondral ossification. Front. Biosci. (Elite Ed) 2012;4:2085–2100. doi: 10.2741/e526. [DOI] [PubMed] [Google Scholar]
- 36.Colnot C., de la Fuente L., Huang S., Hu D., Lu C., St-Jacques B., Helms J.A. Indian hedgehog synchronizes skeletal angiogenesis and perichondrial maturation with cartilage development. Development. 2005;132(5):1057–1067. doi: 10.1242/dev.01649. [DOI] [PubMed] [Google Scholar]
- 37.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.-H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 38.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R., Selby P.B., Owen M.J. Cbfa1, a candidate gene for cleidocranial dysplaysia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 39.Choi J.Y., Pratap J., Javed A., Zaidi S.K., Xing L., Balint E., Dalamangas S., Boyce B., van Wijnen A.J., Lian J.B., Stein J.L., Jones S.N., Stein G.S. Subnuclear targeting of RunxyCbfayAML factors is essential for tissue-specific differentiation during embryonic development. PNAS. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueta C., Iwamoto M., Kanatani N., Yoshida C., Liu Y., Enomoto-Iwamoto M., Ohmori T., Enomoto H., Nakata K., Takada K., Kurisu K., Komori T. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J. Cell Biol. 2001;153(1):87–100. doi: 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda S., Bonnamy J.P., Owen M.J., Ducy P., Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15(4):467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Catheline S.E., Hoak D., Chang M., Ketz J.P., Hilton M.J., Zuscik M.J., Jonason J.H. Chondrocyte-Specific RUNX2 overexpression accelerates post-traumatic osteoarthritis progression in adult mice. J. Bone Miner. Res. 2019;34(9):1676–1689. doi: 10.1002/jbmr.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding M., Lu Y., Abbassi S., Li F., Li X., Song Y., Geoffroy V., Im H.-J., Zheng Q. Targeting Runx2 expression in hypertrophic chondrocytes impairs endochondral ossification during early skeletal development. J. Cell. Physiol. 2012;227(10):3446–3456. doi: 10.1002/jcp.24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takarada T., Hinoi E., Nakazato R., Ochi H., Xu C., Tsuchikane A., Takeda S., Karsenty G., Abe T., Kiyonari H., Yoneda Y. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J. Bone Miner. Res. 2013;28(10):2064–2069. doi: 10.1002/jbmr.1945. [DOI] [PubMed] [Google Scholar]
- 45.Gebhard S., Hattori T., Bauer E., Schlund B., Bösl M.R., de Crombrugghe B., von der Mark K. Specific expression of Cre recombinase in hypertrophic cartilage under the control of a BAC-Col10a1 promoter. Matrix Biol. 2008;27(8):693–699. doi: 10.1016/j.matbio.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H.M. Kronenberg, PTHrP and skeletal development, Ann. N.Y. Acad. Sci., 1068 (2006) 1-13. [DOI] [PubMed]
- 47.Karaplis A.C., Luz A., Glowacki J., Bronson R.T., Tybulewicz V.L., Kronenberg H.M., Mulligan R.C. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994;8(3):277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- 48.Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., Ochi T., Endo N., Kitamura Y., Kishimoto T., Komori T. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 1999;214(4):279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 49.Pratap J., Wixted J.J., Gaur T., Zaidi S.K., Dobson J., Gokul K.D., Hussain S., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68(19):7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin X., Jiang Q., Nagano K., Moriishi T., Miyazaki T., Komori H., Ito K., Mark K.V.D., Sakane C., Kaneko H., Komori T. Runx2 is essential for the transdifferentiation of chondrocytes into osteoblasts. PLoS Genet. 2020;16(11):e1009169. doi: 10.1371/journal.pgen.1009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dao D.Y., Jonason J.H., Zhang Y., Hsu W., Chen D.i., Hilton M.J., O'Keefe R.J. Cartilage-specific β-catenin signaling regulates chondrocyte maturation, generation of ossification centers, and perichondrial bone formation during skeletal development. J. Bone Miner. Res. 2012;27(8):1680–1694. doi: 10.1002/jbmr.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amling M., Neff L., Tanaka S., Inoue D., Kuida K., Weir E., Philbrick W.M., Broadus A.E., Baron R. Bcl-2 lies downstream of parathyroid hormone-related peptide in a signaling pathway that regulates chondrocyte maturation during skeletal development. J. Cell Biol. 1997;136(1):205–213. doi: 10.1083/jcb.136.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaman F., Chrysis D., Huntjens K., Fadeel B., Sävendahl L., Bobé P. Ablation of the pro-apoptotic protein Bax protects mice from glucocorticoid-induced bone growth impairment. PLoS ONE. 2012;7(3):e33168. doi: 10.1371/journal.pone.0033168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eliseev R.A., Dong Y.-F., Sampson E., Zuscik M.J., Schwarz E.M., O'Keefe R.J., Rosier R.N., Drissi M.H. Runx2-mediated activation of the Bax gene increases osteosarcoma cell sensitivity to apoptosis. Oncogene. 2008;27(25):3605–3614. doi: 10.1038/sj.onc.1211020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Browne G., Nesbitt H., Ming L., Stein G.S., Lian J.B., McKeown S.R., Worthington J. Bicalutamide-induced hypoxia potentiates RUNX2-mediated Bcl-2 expression resulting in apoptosis resistance. Br. J. Cancer. 2012;107(10):1714–1721. doi: 10.1038/bjc.2012.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe H., Kimata K., Line S., Strong D., Gao L.Y., Kozak C.A., Yamada Y. Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat. Genet. 1994;7(2):154–157. doi: 10.1038/ng0694-154. [DOI] [PubMed] [Google Scholar]
- 57.Li S.W., Prockop D.J., Helminen H., Fässler R., Lapveteläinen T., Kiraly K., Peltarri A., Arokoski J., Lui H., Arita M., et al. Transgenic mice with targeted inactivation of the Col2 alpha 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes Dev. 1995;9(22):2821–2830. doi: 10.1101/gad.9.22.2821. [DOI] [PubMed] [Google Scholar]
- 58.Ortega N., Behonick D.J., Werb Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004;14(2):86–93. doi: 10.1016/j.tcb.2003.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lausch E., Keppler R., Hilbert K., Cormier-Daire V., Nikkel S., Nishimura G., Unger S., Spranger J., Superti-Furga A., Zabel B. Mutations in MMP9 and MMP13 determine the mode of inheritance and the clinical spectrum of metaphyseal anadysplasia. Am. J. Hum. Genet. 2009;85(2):168–178. doi: 10.1016/j.ajhg.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy A.M., Inada M., Krane S.M., Christie P.T., Harding B., López-Otín C., Sánchez L.M., Pannett A.A., Dearlove A., Hartley C., Byrne M.H., Reed A.A., Nesbit M.A., Whyte M.P., Thakker R.V. MMP13 mutation causes spondyloepimetaphyseal dysplasia, Missouri type (SEMD(MO) J Clin Invest. 2005;115(10):2832–2842. doi: 10.1172/JCI22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma P., Dalal K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J. Cell. Biochem. 2011;12(12):3507–3514. doi: 10.1002/jcb.23298. [DOI] [PubMed] [Google Scholar]
- 62.Stanton H., Rogerson F.M., East C.J., Golub S.B., Lawlor K.E., Meeker C.T., Little C.B., Last K., Farmer P.J., Campbell I.K., Fourie A.M., Fosang A.J. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434(7033):648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 63.Glasson S.S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H.L., Flannery C.R., Peluso D., Kanki K., Yang Z., Majumdar M.K., Morris E.A. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434(7033):644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 64.Rogers-DeCotes A.W., Porto S.C., Dupuis L.E., Kern C.B. ADAMTS5 is required for normal trabeculated bone development in the mandibular condyle. Osteoarthritis Cartilage. 2021;2(4):547–557. doi: 10.1016/j.joca.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajasekaran S., Kanna R.M., Senthil N., Raveendran M., Ranjani V., Cheung K.M., Chan D., Kao P.K., Yee A., Shetty A.P. Genetic susceptibility of lumbar degenerative disc disease in young Indian adults. Eur. Spine J. 2015;24(9):1969–1975. doi: 10.1007/s00586-014-3687-y. [DOI] [PubMed] [Google Scholar]
- 66.Hirata M., Kugimiya F., Fukai A., Saito T., Yano F., Ikeda T., Mabuchi A., Sapkota B.R., Akune T., Nishida N., Yoshimura N., Nakagawa T., Tokunaga K., Nakamura K., Chung U.I., Kawaguchi H. C/EBPbeta and RUNX2 cooperate to degrade cartilage with MMP-13 as the target and HIF- 2alpha as the inducer in chondrocytes. Hum. Mol. Genet. 2012;21(5):1111–1123. doi: 10.1093/hmg/ddr540. [DOI] [PubMed] [Google Scholar]
- 67.Pratap J., Javed A., Languino L.R., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol. Cell. Biol. 2005;25(19):8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thirunavukkarasu K., Pei Y., Moore T.L., Wang H., Yu X.P., Geiser A.G., Chandrasekhar S. Regulation of the human ADAMTS-4 promoter by transcription factors and cytokines. Biochem. Biophys. Res. Commun. 2006;345(1):197–204. doi: 10.1016/j.bbrc.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 69.Thirunavukkarasu K., Pei Y., Wei T. Characterization of the human ADAMTS-5 (aggrecanase-2) gene promoter. Mol. Biol. Rep. 2007;34(4):225–231. doi: 10.1007/s11033-006-9037-3. [DOI] [PubMed] [Google Scholar]
- 70.Jackson H.W., Defamie V., Waterhouse P., Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat. Rev. Cancer. 2017 Jan;17(1):38–53. doi: 10.1038/nrc.2016.115. Epub 2016 Dec 9 PMID: 27932800. [DOI] [PubMed] [Google Scholar]
- 71.Cawston T.E., Murphy G., Mercer E., Galloway W.A., Hazleman B.L., Reynolds J.J. The interaction of purified rabbit bone collagenase with purified rabbit bone metalloproteinase inhibitor. Biochem. J. 1983;211(2):313–318. doi: 10.1042/bj2110313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saw S., Aiken A., Fang H., McKee T.D., Bregant S., Sanchez O., Chen Y., Weiss A., Dickson B.C., Czarny B., Sinha A., Fosang A., Dive V., Waterhouse P.D., Kislinger T., Khokha R. Metalloprotease inhibitor TIMP proteins control FGF-2 bioavailability and regulate skeletal growth. J. Cell Biol. 2019;218(9):3134–3152. doi: 10.1083/jcb.201906059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark I.M., Swingler T.E., Sampieri C.L., Edwards D.R. The regulation of matrix metalloproteinases and their inhibitors. Int. J. Biochem. Cell Biol. 2008;40(6–7):1362–1378. doi: 10.1016/j.biocel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Bertrand-Philippe M., Ruddell R.G., Arthur M.J., Thomas J., Mungalsingh N., Mann D.A. Regulation of tissue inhibitor of metalloproteinase 1 gene transcription by Runx1 and Runx2. J. Biol. Chem. 2008;279(23):24530–24539. doi: 10.1074/jbc.M311804200. [DOI] [PubMed] [Google Scholar]
- 75.Kishimoto K., Kitazawa R., Kurosaka M., Maeda S., Kitazawa S. Expression profile of genes related to osteoclastogenesis in mouse growth plate and articular cartilage. Histochem. Cell Biol. 2006;125(5):593–602. doi: 10.1007/s00418-005-0103-z. [DOI] [PubMed] [Google Scholar]
- 76.Kearns A.E., Khosla S., Kostenuik P.J. Receptor activator of nuclear factor kappaB ligand and osteoprotegerin regulation of bone remodeling in health and disease. Endocr. Rev. 2008;29(2):155–192. doi: 10.1210/er.2007-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Enomoto H., Shiojiri S., Hoshi K., Furuichi T., Fukuyama R., Yoshida C.A., Kanatani N., Nakamura R., Mizuno A., Zanma A., Yano K., Yasuda H., Higashio K., Takada K., Komori T. Induction of osteoclast differentiation by Runx2 through receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin regulation and partial rescue of osteoclastogenesis in Runx2-/- mice by RANKL transgene. J. Biol. Chem. 2003;278(26):23971–23977. doi: 10.1074/jbc.M302457200. [DOI] [PubMed] [Google Scholar]
- 78.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.J., Lein E.S., Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adhami M.D., Rashid H., Chen H., Clarke J.C., Yang Y., Javed A. Loss of Runx2 in committed osteoblasts impairs postnatal skeletogenesis. J. Bone Miner. Res. 2015;30(1):71–82. doi: 10.1002/jbmr.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.H. Rashid, H. Chen, Q. Hassan, A. Javed, Dwarfism in homozygous Agc1CreERT mice is associated with decreased expression of aggrecan, Genesis 55(10) (2017) 10.1002. [DOI] [PMC free article] [PubMed]