Summary
Tight coordination between transcription and translation has long been recognized as the hallmark of gene expression in bacteria. In Escherichia coli cells, disruption of the transcription-translation coordination leads to the loss of transcription processivity via triggering Rho-mediated premature transcription termination. Here we quantitatively characterize the transcription and translation kinetics in Gram-positive model bacterium Bacillus subtilis. We found that the speed of transcription elongation is much faster than that of translation elongation in B. subtilis under various growth conditions. Moreover, a Rho-independent loss of transcription processivity occurs constitutively in several genes/operons but is not subject to translational control. When the transcription elongation is decelerated under poor nutrients, low temperature, or nucleotide depletion, the loss of transcription processivity is strongly enhanced, suggesting that its degree is modulated by the speed of transcription elongation. Our study reveals distinct design principles of gene expression in E. coli and B. subtilis.
Subject areas: Biochemistry, Bacteriology, Synthetic biology
Graphical abstract
Highlights
-
•
Asynchronous Transcription-Translation in B. subtilis under various growth conditions
-
•
Loss of transcription processivity occurs constitutively in several genes/operons
-
•
Lack of Rho-mediated translation control of transcription kinetics in B. subtilis
-
•
Transcription processivity is affected by the transcription elongation speed
Biochemistry; Bacteriology; Synthetic biology
Introduction
It is widely accepted that, in E. coli cells, transcription and translation process are tightly coordinated where the lead ribosome closely follows RNA polymerase (RNAP) during gene expression (McGary and Nudler, 2013; Miller et al., 1970). Specially, the speeds of transcription and translation elongation are synchronized in bacterial cells under various nutrient conditions, enabling the full processivity of transcription (Iyer et al., 2018; Proshkin et al., 2010; Vogel and Jensen, 1994b; Zhu et al., 2019). A series of recent biochemical and structural studies have provided evidences that such coordination is mediated by the physical coupling between RNAP and the lead ribosome(Burmann et al., 2010; Demo et al., 2017; Kohler et al., 2017; O'Reilly et al., 2020; Saxena et al., 2018; Wang et al., 2020; Webster et al., 2020). Alternative models such as the stochastic coupling model have also been proposed (Chen and Fredrick, 2018; Rui et al., 2016). In addition, it has been found that alternative factors such as (p)ppGpp assist the kinetic coordination between transcription and translation (Iyer et al., 2018; Kingston et al., 1981; Vogel and Jensen, 1994a; Vogel et al., 1992; Zhu et al., 2019).
It has been well established that disruption of transcription-translation coordination, by, e.g., nonsense mutation or antibiotic treatment, leads to Rho-mediated premature transcription termination (PTT), a loss of transcription processivity (Adhya and Gottesman, 1978; Newton et al., 1965; Zhu et al., 2019). Such mechanism suppresses persuasive transcription during transcription-translation dissociation so that the cellular investments on transcription and translation are coordinated (Richardson, 1991, 2003). It could also function in limiting the expression of deleterious foreign DNA (e.g., phage-related or xenogenic DNA) (Aleksandra et al., 2016). Rho-mediated control of transcription processivity could significantly affect the gene expression process (exemplified in the polarity phenomenon) (Adhya and Gottesman, 1978; Newton et al., 1965) and fundamental physiological processes of bacterial cells (Aleksandra et al., 2016).
The notion of Rho-mediated loss of transcription processivity during transcription-translation dissociation, although firmly established in E. coli, is not necessarily applicable to other bacterial species. For example, previous studies have shown that Rho factor has a diminished role in the Gram-positive bacterium B. subtilis, as reflected by (1) Rho factor is dispensable in B. subtilis, whereas it is essential in E. coli (Ingham et al., 1999; Quirk et al., 1993; Vladimir et al., 2017; Yakhnin et al., 2001); (2) the majority of terminators in B. subtilis are intrinsic terminators, and Rho-dependent terminators are rare (De Hoon et al., 2005; Naville and Gautheret, 2009). A very recent study has reported that, in contrast to the tight transcription-translation coordination in E. coli, RNAP outpaces the lead ribosomes in B. subtilis cells grown in rich medium, providing a functional explanation on the diminished role of Rho-dependent transcription termination in B. subtilis (Johnson et al., 2020).
Since Rho-mediated loss of transcription processivity is a consequence of the transcription-translation dissociation in E. coli, the observation of transcription-translation dissociation of B. subtilis in rich medium poses several important questions: What is the status of transcription processivity in B. subtilis? How is it regulated, and is it also subject to Rho-mediated translational control? Moreover, considering that B. subtilis cells frequently encounter various stressful conditions in their natural niches (soil) (Gray et al., 2019; Van Dijl and Hecker, 2013), it remains undetermined whether and how transcription-translation interplay and transcription processivity are affected by stress. Here we quantitatively characterize the transcription and translation kinetics in B. subtilis under a broad range of growth conditions. We confirm the dissociation status between translation and transcription under various growth conditions in B. subtilis. In particular, we identify that the loss of transcription processivity occurs constitutively during transcription-translation dissociation. In addition, it is Rho independent and is not subject to translational control and instead is mainly affected by the elongation speed of RNAP.
Results
Asynchronous transcription and translation in B. subtilis
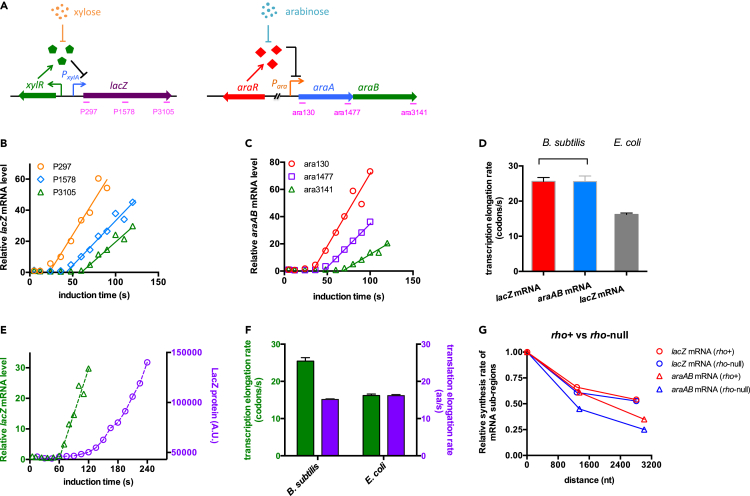
We first focused on the B. subtilis 168 strain exponentially growing in glycerol plus casamino acid (gly + cAA) medium (a rich medium supporting a doubling time of 29 min). To quantitatively measure the transcription kinetics, we applied our recently established multiple-probes qRT-PCR method based on the lacZ induction system of E. coli (Zhu et al., 2019). To adapt this approach to B. subtilis, we integrated a xylose-inducible xylR-PxylA-lacZ cassette into the chromosome of B. subtilis (Figure 1A, left). In this case, the transcription of lacZ mRNA in B. subtilis could be induced by xylose and be further measured by qRT-PCR. Since lacZ is an exogenous gene for B. subtilis, we also focused on the transcription of the native araAB mRNA that is inducible by arabinose (Figure 1A, right) (Sá-Nogueira et al., 1997).
Figure 1.
Characterization of transcription and translation kinetics of Bacillus subtilis
Cells were grown in glycerol plus casamino acid (gly + cAA) medium.
(A) Two inducible systems used in this study. A xylR-PxylA-lacZ cassette was integrated into the lacA locus of B. subtilis genome. The expression of lacZ gene is controlled by the XylR repressor and thus is induced by xylose. The other inducible system is the native ara operon of B. subtilis, which is controlled by AraR and is induced by arabinose. We focused on the araAB region of the ara operon. Regions detected by three qRT-PCR primers are labeled in pink.
(B) The induction kinetics of the lacZ mRNA. Three pairs of primers were used to detect the abundances of lacZ mRNA sub-regions. The rising part of each induction curve was fitted to a linear line.
(C) The induction kinetics of araAB mRNA detected by three pairs of qRT-PCR primers, being similar to (B)
(D) The transcription elongation rates of both B. subtilis and E. coli. Data of E. coli lacZ mRNA are from Zhu et al (2019). Data are represented as mean ± SD.
(E) The induction curve of the intact lacZ mRNA (detected by P3105, green triangles) is plotted together with the induction curve of LacZ protein (purple circles).
(F) The transcription and translation elongation rates of B. subtilis and E. coli. Data of E. coli cells are from Zhu et al (2019). Data are represented as mean ± SD.
(G) The transcription processivity of lacZ mRNA and araAB mRNA in both rho+ (red symbols) and rho-null strains (blue symbols) of B. subtilis. The relative accumulation rates of mRNA sub-regions (slope of the linear induction curve in B and C) are plotted against the hybridization locations of primers in lacZ or araAB mRNA. The detection positions of the 5' primer (P297 for lacZ mRNA and ara130 for araAB mRNA) were set as location “zero” (x axis). The accumulation rates of the 5′ mRNA sub-regions detected by P297 or ara130 were set as “1” (y axis).
For both lacZ and araAB mRNA, we used three pairs of qRT-PCR primers to detect the induction kinetics of different mRNA sub-regions, the head (detected by P297 for lacZ; ara130 for araAB), the middle (detected by P1578 for lacZ; ara1477 for araAB), and the tail (detected by P3105 for lacZ; ara3141 for araAB) (Figure 1A, Figure S1). As exemplified in Figures 1B and 1C, both the lacZ and araAB mRNA abundance are linearly correlated with the time following the addition of inducers. From the linear induction curve of each pair of primers, we could deduce the transcription time of each mRNA sub-regions (Thead, Tmid, Ttail) and further obtain the transcription elongation rate of RNAP (see STAR method section). We found that the transcription elongation rates of lacZ mRNA and araAB mRNA in B. subtilis were similar to each other, at a value of ∼25–26 codons/s (75–80 nt/s) (Figure 1D) with a high reproducibility (Figure S2). This value is much higher than that of E. coli cells, ∼16 codons/s (Zhu et al., 2019) and is consistent with the report of Johnson et al. (2020).
We next measured the induction kinetics of LacZ protein in order to investigate the potential interplay between transcription and translation. As shown in Figure 1E, the time required to transcribe the complete lacZ mRNA was significantly shorter than that required to translate the complete LacZ protein, indicating that transcription is naturally dissociated from translation in B. subtilis. This result is consistent with the recent report of Johnson et al., which was also done in nutrient-rich medium (Johnson et al., 2020). Quantitatively, the speed of transcription elongation (∼25 codon/s) is 65% higher than that of translation elongation (∼15 aa/s) in B. subtilis, whereas these two values are equal in E. coli (Figure 1F) (Zhu et al., 2019).
Rho-independent loss of transcription processivity in B. subtilis
An important parameter of transcription kinetics is the transcription processivity, which is affected by the degree of PTT (Iyer et al., 2016). A loss of transcription processivity means that an elongating RNA polymerase fails to reach the 3′ end of an operon to generate a full-length transcript (Iyer et al., 2016). It could mechanistically originate from various mechanisms such as the Rho-mediated form occurring when transcription-translation coordination is disrupted in E. coli (Iyer et al., 2018; Zhu et al., 2019) or a permanent arrest of transcription in cases such as treating cells with a transcription blocker such as actinomycin D (Levinthal and Higa, 1962; Miller, 1987). Since translation is naturally dissociated from transcription in B. subtilis, we wondered the status of transcription processivity in such cases. The slope of the linear transcriptional induction curve (Figures 1B and 1C) denotes the relative accumulation rate of each mRNA sub-region (head, middle and tail regions) (Iyer et al., 2016; Zhu et al., 2019). Comparing the slope of the head and tail region is a reliable and standard approach to quantify the processivity of RNAP during transcription elongation given that the stabilities of different mRNA sub-regions are almost the same as each other (Figure S3) (Iyer et al., 2016, 2018; Vogel et al., 1992; Zhu et al., 2019). As shown in Figure 1G, we found that the mRNA accumulation rate significantly decreased from 5′ to 3′ direction for both lacZ and araAB mRNA (red circles and triangles), suggesting a significant loss of transcription processivity. Quantitatively, the mRNA accumulation rates of lacZ and araAB dropped by ∼40% and ∼65% from the head region to tail region, respectively. The loss of transcription processivity was also observed in two additional native operons, mtlAFD and xynPB operons (Figure S4). To investigate the potential origin of the loss of transcription processivity, we next measured the transcription kinetics of rho-null mutant (Figure S5). If the loss of transcription processivity originates from Rho-dependent PTT in B. subtilis, it should be substantially alleviated in the rho-null strain. The growth rate of the rho-null mutant was mildly lower than that of wild-type cells (Figure S6), being consistent with previous reports that Rho is dispensable for B. subtilis (Ingham et al., 1999; Vladimir et al., 2017). However, the loss of transcription processivity was not alleviated in the rho-null strain relative to rho+ strain (Figure 1G), indicating that it is Rho independent.
Lack of translational control on the transcription processivity in B. subtilis
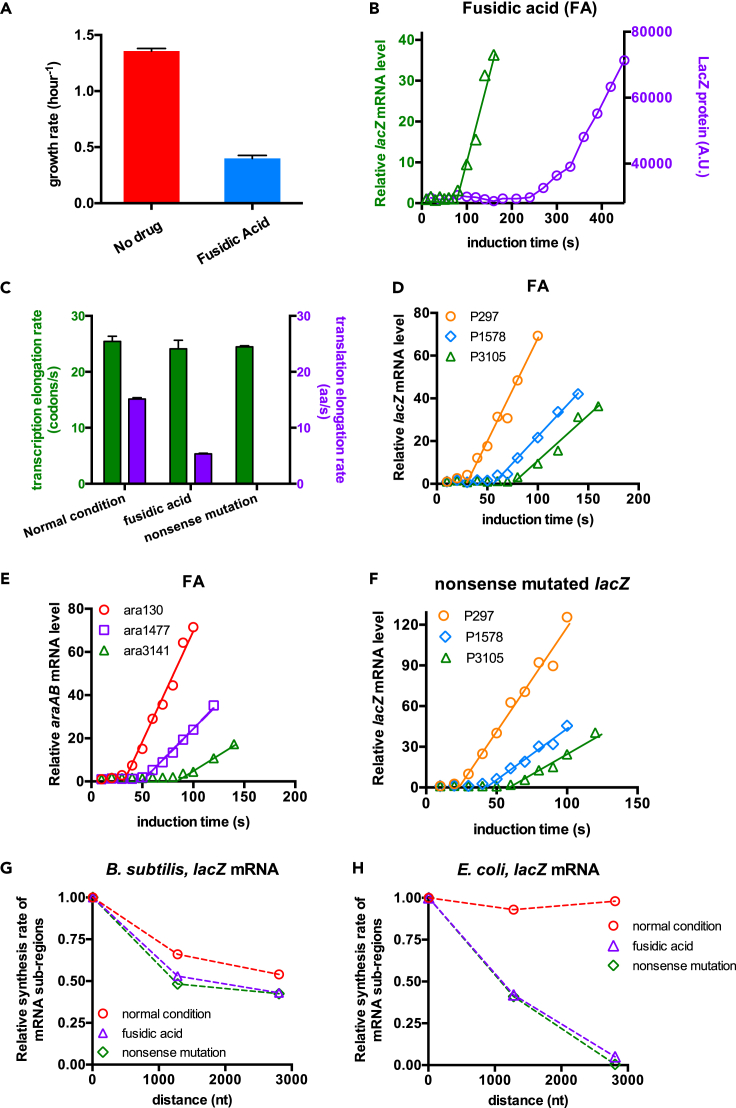
We next sought to explore the potential effect of translation on transcription processivity in B. subtilis. We first applied fusidic acid (FA), an antibiotic that targets ribosome translocation via inhibiting EF-G recycling, to perturb the translation elongation process (Bennett and Maaloe, 1974; Seo et al., 2006). The growth rate of B. subtilis in gly + cAA medium dropped by 70% during treatment with 0.2 μg/mL FA (Figure 2A, DT: ∼100 min). Concomitantly, the translation time of the complete LacZ protein under FA treatment became much longer than that under normal condition (purple circles in Figures 2B and 1E), suggesting a much slower translation elongation rate. As shown in Figure 2C (purple bar), the translation elongation rate of B. subtilis under FA treatment was only ∼5 aa/s, being approximately one-third of the normal value (∼15 aa/s). In contrast, the transcription elongation rate remained almost unaffected under FA treatment (Figure 2C, green bar).
Figure 2.
Transcription and translation kinetics of B. subtilis following fusidic acid (FA) treatment and nonsense mutation
Cells were grown in gly + cAA medium.
(A) THE growth rates of B. subtilis in gly + cAA medium supplemented with/without 0.2 μg/mL FA. Data are represented as mean ± SD.
(B) The induction curve of the complete lacZ mRNA (green triangles) and LacZ protein (purple circles) for B. subtilis grown in gly + cAA medium supplemented with 0.2 μg/mL FA.
(C) Summary of the transcription and translation elongation rates of lacZ mRNA for B. subtilis0020 under normal condition, FA treatment, and nonsense mutation. Data are represented as mean ± SD.
(D) The induction kinetics of the lacZ mRNA for B. subtilis under FA treatment.
(E) The induction kinetics of the araAB mRNA for B. subtilis under FA treatment.
(F) The induction kinetics of a nonsense-mutated lacZ mRNA. The coding sequence of the 154th amino acid residue of the native lacZ, “TGG,” was mutated to “TAA” stop codon.
(G) The transcription processivity of lacZ mRNA in B. subtilis under normal condition, FA treatment, and nonsense mutation. The same as Figure 1G, the relative accumulation rates of mRNA sub-regions (slope of the linear induction curve) are plotted against the hybridization locations of primers for lacZ mRNA. (H) The transcription processivity of lacZ mRNA in E. coli under normal condition, FA treatment, and nonsense mutation. Data originate from Zhu et al (2019).
Introducing nonsense mutation is another approach to investigate the effect of translation on transcription (Newton et al., 1965). We introduced a “TAA” stop codon in the upper part of lacZ gene and repeated the transcription kinetics of lacZ mRNA. Being similar to FA treatment, nonsense mutation did not affect the transcription elongation rate as well (Figure 2C, green bar). Therefore, both FA treatment and nonsense mutation significantly amplify the degree of transcription-translation dissociation in B. subtilis. However, analysis of the transcription kinetics (Figures 2D–2F) showed that transcription processivity was only weakly affected by FA treatment and nonsense mutation (Figures 2G and S7). In contrast, our recent quantitative study shows that FA treatment and nonsense mutation, which both disrupt the transcription-translation coordination, cause a severe loss of transcription processivity during lacZ mRNA induction in E. coli cells (Figure 2H) (Zhu et al., 2019). In summary, the degree of transcription-translation dissociation, which is known to strongly modulate the Rho-mediated PTT in E. coli cells, is not an important determinant of the loss of transcription processivity here observed in B. subtilis.
Loss of transcription processivity is more severe under poor nutrients and low temperatures
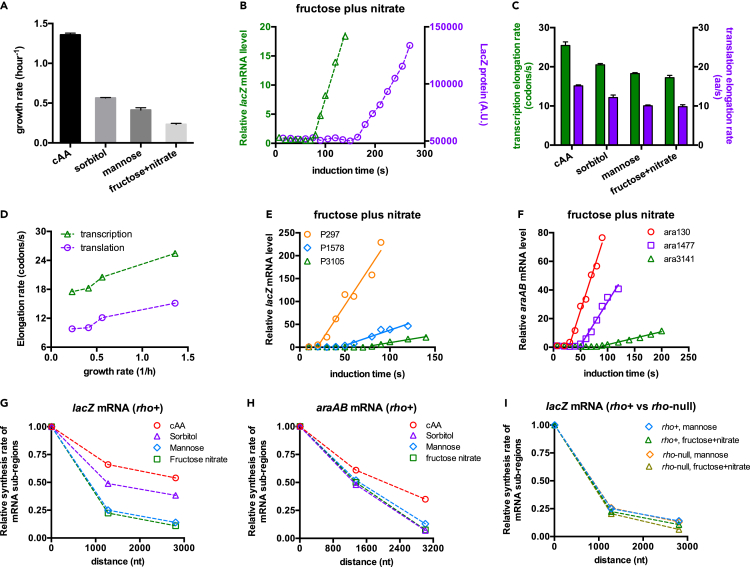
B. subtilis frequently encounters a variety of environment stressors in their natural niches (soil), including poor nutrients and suboptimal temperatures (Gray et al., 2019; Janna et al., 2005; Van Dijl and Hecker, 2013). We therefore extended the kinetics study to poor nutrient conditions and low temperatures in order to gain an insight into the impact of stress on the transcription-translation interplay. The nutrient-limited minimal medium supports much lower growth rates than gly + cAA medium (Figure 3A). Again, we found that translation remains dissociated from transcription in poor nutrients (Figures 3B and S8). However, compared with rich medium, both transcription and translation elongation rates became lower under poor nutrients (Figure 3C) and were positively correlated with growth rate (Figure 3D). We further quantified the transcription processivity by analyzing the transcription induction kinetics (Figures 3E, 3F, and S9). Strikingly, transcription processivity drops more substantially for cells under poor nutrients than their counterparts growing under rich medium (Figures 3G and 3H). In addition, the loss of transcription processivity in poor nutrient conditions was still Rho independent since its level was not reduced in the rho-null strains (Figures 3I and S10).
Figure 3.
Transcription and translation kinetics of B. subtilis under poor nutrient conditions
(A) The growth rates of B. subtilis under different nutrient conditions. Data are represented as mean ± SD.
(B) The induction curves of the intact lacZ mRNA (green triangles) and LacZ protein (purple circles) for B. subtilis grown in a poor nutrient condition, fructose plus nitrate medium, which supports a doubling time of ∼3 h.
(C) Summary of transcription (green bar) and translation (purple bar) elongation rates of B. subtilis under different nutrient conditions. Data are represented as mean ± SD.
(D) Growth rate-dependent transcription and translation elongation rates under different nutrient conditions. Data of growth rates are from (A) Data of transcription and translation elongation rates are from (C)
(E) The induction kinetics of lacZ mRNA in B. subtilis grown in fructose plus nitrate medium.
(F) The induction kinetics of araAB mRNA in B. subtilis grown in fructose plus nitrate medium.
(G) The transcription processivity of lacZ mRNA in B. subtilis (rho+) under different nutrient conditions.
(H) The transcription processivity of araAB mRNA in B. subtilis (rho+) under different nutrient conditions.
(I) The transcription processivity of lacZ mRNA in both rho+ and rho-null strains of B. subtilis under mannose medium and fructose plus nitrate medium.
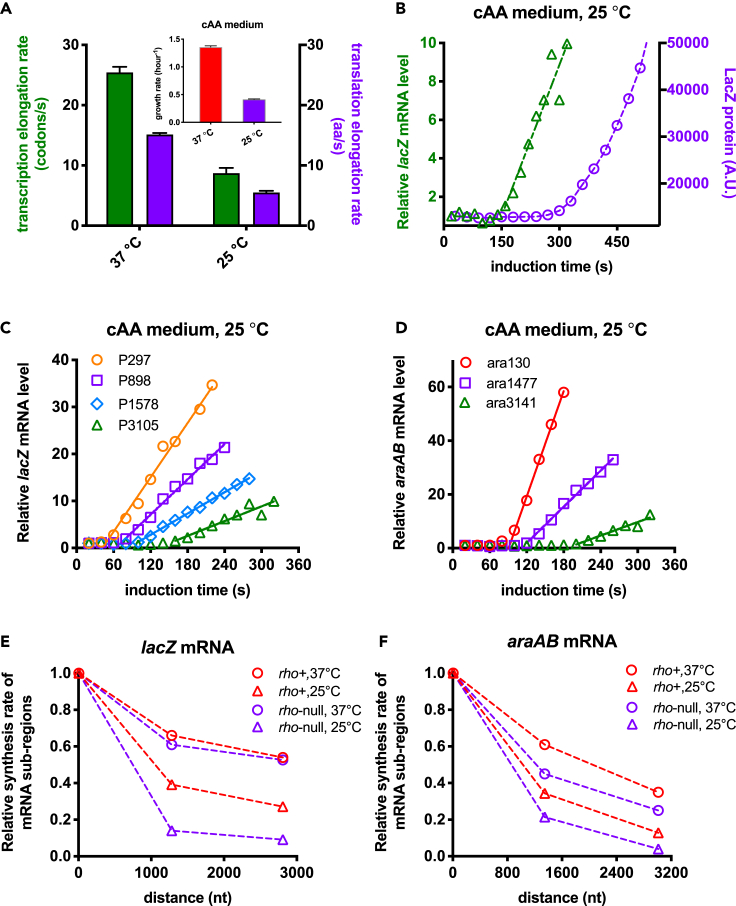
The growth rate of B. subtilis at a low temperature, 25°C, decreased to only ∼30% of that at 37°C (subpanel in Figure 4A). Strikingly, both transcription and translation elongation rates dropped by a factor of ∼3 at 25°C (Figure 4A), and therefore, transcription and translation were still dissociated from each other (Figure 4B). From the induction kinetics of both lacZ and araAB mRNA (Figures 4C and 4D), we found that the loss of transcription processivity of both rho+ and rho-null strains at 25°C became more marked than that at 37°C (compare circles and triangles in Figures 4E and 4F), being similar to the case of nutrient limitation. Moreover, the rho-null mutant exhibited an even stronger loss of transcription processivity than rho+ strain at 25°C (compare red and purple triangles in Figures 4E and 4F; also see induction kinetics in Figure S11). In summary, being different from the cases of fusidic acid and nonsense mutation (Figure 2), the status of transcription processivity depends strongly on the nutrient quality and temperature, implying that it is controlled by factors other than transcription-translation dissociation.
Figure 4.
Transcription and translation kinetics of B. subtilis at 25°C
B. subtilis cells were grown in gly + cAA medium.
(A) The transcription and translation elongation rates of B. subtilis under 37°C and 25°C. Data of the growth rate are shown in the subpanel. Data are represented as mean ± SD.
(B) The induction curves of the intact lacZ mRNA and LacZ protein for B. subtilis at 25°C.
(C) The induction kinetics of lacZ mRNA in B. subtilis at 25°C.
(D) The induction kinetics of araAB mRNA in B. subtilis at 25°C.
(E) The transcription processivity of lacZ mRNA for both rho+ and rho-null strains of B. subtilis at two temperatures.
(F) The transcription processivity of araAB mRNA for both rho+ and rho-null strains of B. subtilis at two temperatures.
Loss of transcription processivity is more severe during deceleration of transcription elongation
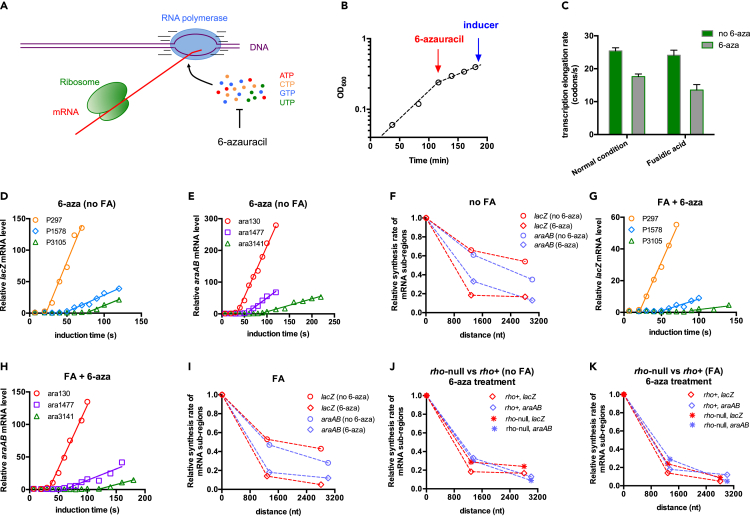
Given that the transcription elongation rate decreases under both poor nutrients and low temperatures, the more substantially compromised transcription processivity could simply be due to a further slowdown of RNAP movement as increased pauses of RNAP could create more windows of time for the occurrence of an RNAP drop-off and transcriptional termination event, further resulting in the loss of processivity (Ray-Soni et al., 2016). To test this hypothesis, we sought to investigate whether directly perturbing the transcription elongation rate could affect the transcription processivity in rich medium. We applied 6-azauracil (6-aza), a drug that depletes intracellular nucleotide pools (especially UTP and GTP) of microorganisms (Exinger and Lacroute, 1992; Ishiguro et al., 2000; Lopez et al., 1979), to inhibit the transcription elongation of RNAP (illustrated on Figure 5A). As expected, addition of 6-aza caused rapid slowdown of cell growth and decelerated the transcription elongation process (Figures 5B and 5C). Strikingly, the loss of transcription processivity is indeed enhanced by 6-aza treatment (Figures 5D and 5E), as quantitively displayed in Figure 5F. To remove any potential interference from translation, we also repeated the 6-aza experiment in medium supplementing FA (Figure S12), where the ribosome is far away behind RNAP (Figures 2B and 2C). The loss of transcription processivity also became much more marked by 6-aza treatment in this case, as shown in Figure 5I (also see induction kinetics in Figures 5G and 5H). In addition, the stronger loss of transcription processivity under 6-aza treatment was also not related to Rho factor (Figures 5J, 5K, and S13). Taken together, the above results support that the elongation rate of RNAP is an important determinant of the transcription processivity in B. subtilis.
Figure 5.
Transcription kinetics of B. subtilis treated with 6-azauracil (6-aza)
B. subtilis cells were grown in gly + cAA medium.
(A) 6-Aza was used to perturb the transcription elongation process of B. subtilis through depleting the cellular nucleotides pools.
(B) The growth of B. subtilis treated with 6-aza. Cells were first exponentially grown in gly + cAA medium to OD600∼0.25. 6-Aza, 500 μg/mL, was then added to deplete the cellular nucleotides pools. The cell culture was further incubated for 1 h before measuring transcription kinetics.
(C) The effect of 6-aza on the transcription elongation rate of B. subtilis. Cells were grown in gly + cAA medium supplemented with/without 0.2 μg/mL FA. Data are represented as mean ± SD.
(D) and (E) The induction kinetics of lacZ mRNA and araAB mRNA in B. subtilis treated with 6-aza. FA was not supplemented in this case.
(F) Comparison of the transcription processivities of lacZ (red) and araAB (light purple) mRNA in B. subtilis (rho+ strain) with (diamond)/without (circle) 6-aza treatment. FA was not supplemented in this case.
(G) and (H) The induction kinetics of lacZ mRNA and araAB mRNA in B. subtilis treated with 6-aza. Being different from D and E, cells were grown with 0.2 μg/mL FA. The growth curve is shown in Figure S12.
(I) Comparison of the transcription processivities of lacZ (red) and araAB (light purple) mRNA in B. subtilis (rho+ strain) with (diamond)/without (circle) 6-aza treatment. Cells were grown with 0.2 μg/mL FA.
(J) and (K) Comparison of the transcription processivities of rho+ strain and rho-null strain during 6-aza treatment.
Discussion
In E. coli, the tight transcription-translation coordination effectively suppresses Rho-mediated premature transcription termination, guaranteeing the full processivity of transcription in cells under different nutrient conditions (Iyer et al., 2018; Zhu et al., 2019). Rho-mediated premature transcription termination (PTT) strongly occurs under special conditions such as nonsense mutation or treating cells with translation-blocking antibiotics (Adhya and Gottesman, 1978; Zhu et al., 2019), further substantially compromising the transcription processivity. Therefore, the transcription processivity of E. coli is strongly affected by the degree of transcription-translation dissociation. Being different from the scenario of E. coli, here we show that translation and transcription are naturally uncoordinated in B. subtilis under various growth conditions. Of importance, notable loss of transcription processivity occurs constitutively even in nutrient-rich medium. However, in contrast to the case of E. coli, it is Rho independent and does not subject to translational control and likely depends on the elongation speed of RNAP. The loss of transcription processivity is naturally attributed to the occurrence of PTT of RNAP. Given that the way of transcription termination in bacteria falls into two major categories: Rho-dependent termination and Rho-independent intrinsic termination (Aleksandra et al., 2016), the loss of transcription processivity could thus arise from intragenic Rho-independent intrinsic termination. The elongation process of transcription in vivo (nucleotide incorporation) is actually competitive with the process of termination (Greive and von Hippel, 2005). Studies have also shown that intrinsic termination could occur even in the absence of termination signals in which the paused/halted RNAP elongation complex could dissociate via a general forward-translocation mechanism (Santangelo and Roberts, 2004; Zhou et al., 2007). Therefore, transcription termination could occur prematurely within a gene/operon given that pausing is frequent in vivo (Larson et al., 2014) and can serve to halt RNAP. Recent systematic studies have also shown that there are many intrinsic NusA- or NusG-dependent pause sites within the genome of B. subtilis (Mondal et al., 2016; Yakhnin et al., 2020). Following this logic, a slowdown of transcription elongation (increased frequency of pausing) under harsh environments could amplify the degree of loss of transcription processivity (Davenport et al., 2000; Yang et al., 2021; Zhou et al., 2007).
In addition, alternative factors could also affect the transcription processivity. We tested two potential candidate proteins, Rnase J and GreA since they have been shown to participate in rescuing and liberating stalled/backtracked RNAP elongation complex in cases of severe transcription stalling in B. subtilis (Kusuya et al., 2011; Sikova et al., 2020). We found that notable loss of transcription processivity still occurs in rnjA-null mutant and greA-null mutant (Figure S14), suggesting that those two proteins do not significantly affect transcription processivity. However, an unexpected finding is that the transcription elongation speed of rnjA-null mutant is only half that of wild-type cells (Figure S14E), suggesting that Rnase J might be an important regulator of the transcription elongation process. This finding may provide meaningful clues for future exploration of the origin of the intriguing fast elongation speed of RNAP in B. subtilis.
The loss of transcription processivity could be viewed as a form of transcription attenuation by nature. In E. coli cells, ribosome translation plays a key role in controlling transcription attenuation, as well exemplified in the case of trp, pyr operon, and Rho-mediated polarity phenomenon (Adhya and Gottesman, 1978; Merino and Yanofsky, 2005; Turnbough, 2019). Instead, the control of transcription attenuation in B. subtilis mainly falls into two categories: (1) mediated by RNA-binding proteins such as TRAP and PyrR (Turnbough, 2019), as found in e.g., typ and pyr operons; (2) mediated by riboswitches (Winkler and Breaker, 2005), including tRNA-binding riboswitches (e.g., tyrS operon) and small-molecule binding riboswitches (e.g., rib operon in B. subtilis) (Turnbough, 2019). Given that the loss of transcription processivity here also occurred at the exogeneous lacZ mRNA, and therefore might not be related to those known mechanisms of transcription attenuation.
Mechanistically, the lack of Rho-mediated control of transcription processivity during transcription-translation dissociation in B. subtilis may be intriguing given previous reports that Rho factors from E. coli and B. subtilis share similar RNA-activated ATPase activity and in vitro termination efficiency on the λtR1 terminator (Ingham et al., 1999). One possible origin could lie at the Rho specificity on rut site. The Rho factor in E. coli cells does not require a specific consensus rut site, and its activity could be stimulated on various artificial C-rich RNA sequences and even non-C-rich RNA sequences (Hart and Roberts, 1992; Mitra and Nagaraja, 2012; Ray-Soni et al., 2016). It is possible that the activity of Rho factor in B. subtilis is highly sequence specific, thus being limited to only a few operons (Johnson et al., 2020).
Limitations of the study
Our results of transcription processivity are derived from a few operons. The quantitative behavior of transcription processivity could in principle be affected by both trans-acting protein factors that affect transcription pausing/termination and cis-acting DNA elements such as intragenic anti-terminators. We expect future systematic genome-wide studies to further elucidate the status of transcription processivity among various operons in B. subtilis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Bacillus subtilis 168 | Professor Jianhua Zhang | N/A |
| Bacillus subtilis rho-null strain | This study | N/A |
| Bacillus subtilis rnjA-null strain | This study | N/A |
| Bacillus subtilis greA-null strain | This study | N/A |
| Bacillus subtilis PxylA-lacZ strain | This study | N/A |
| Bacillus subtilis PxylA-lacZ-TAA strain | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| KH2PO4 | Aladdin (Shanghai) | CAS: 7778-77-0 |
| K2HPO4 | Aladdin (Shanghai) | CAS: 7758-11-4 |
| MnSO4·4H2O | Macklin | CAS: 10101-68-5 |
| MgSO4·7H2O | Aladdin (Shanghai) | CAS: 10034-99-8 |
| ZnCl2 | Macklin | CAS: 7646-85-7 |
| Tryptophan | Aladdin (Shanghai) | CAS: 73-22-3 |
| ferric ammonium citrate | Aladdin (Shanghai) | CAS: 1185-57-5 |
| NH4Cl | Sigma | G5767 |
| Glucose | Sigma | 213330 |
| Glycerol | Aladdin (Shanghai) | CAS: 56-81-5 |
| Fructose | Aladdin (Shanghai) | CAS: 57-48-7 |
| Mannose | Aladdin (Shanghai) | CAS: 3458-28-4 |
| casamino acid | Aladdin (Shanghai) | CAS: 65072-00-6 |
| LB broth | Coolaber (Beijing) | PM0010 |
| KNO3 | Aladdin (Shanghai) | CAS: 7757-79-1 |
| fusidic acid | Aladdin (Shanghai) | F134821 |
| 6-azauracil | Aladdin (Shanghai) | A124269 |
| Actinomycin D | GLPBIO | GC16866 |
| 4-methylumbelliferyl-D-galactopyranoside (MUG) | Aladdin (Shanghai) | CAS: 6160-78-7 |
| Chloramphenicol | Coolaber (Beijing) | CC3451 |
| Erythromycin | Coolaber (Beijing) | CE5091 |
| Spectinomycin | Coolaber (Beijing) | CS10421 |
| Critical commercial assays | ||
| Golden Green high-fidelity PCR mix | Tsingke Biotech | TSE101 |
| T3 super PCR mix | Tsingke Biotech | TSE030 |
| Total RNA extraction kit | TianGen | DP430 |
| First-strand cDNA synthesis reverse transcriptase kit | TianGen | KR118 |
| Plasmid extraction kit | TianGen | DP103 |
| Bacterial genome extraction kit | TianGen | DP302 |
| DNA purification kit | TianGen | DP209 |
| PowerUp SYBR green Master mix | Thermo Fisher | Lot # 01000432 |
| Super-premix SYBR green Plus kit | Yeasen Biotech | 11201ES08 |
| Oligonucleotides | ||
| P297-F/R | Tsingke Biotech | N/A |
| P898-F/R | Tsingke Biotech | N/A |
| P1578-F/R | Tsingke Biotech | N/A |
| P3105-F/R | Tsingke Biotech | N/A |
| Ara130-F/R | Tsingke Biotech | N/A |
| Ara1477-F/R | Tsingke Biotech | N/A |
| Ara3141-F/R | Tsingke Biotech | N/A |
| Mtl117-F/R | Tsingke Biotech | N/A |
| Mtl2926-F/R | Tsingke Biotech | N/A |
| Xyn162-F/R | Tsingke Biotech | N/A |
| Xyn2727-F/R | Tsingke Biotech | N/A |
| Recombinant DNA | ||
| pAX01 integration vector | Professor Zheming Zhou | N/A |
| pDG1730 integration vector | Professor Ming Sun | N/A |
Resource availability
Lead contact
Further information and requests for materials should be directed to and will be fulfilled by the lead contact, Xiongfeng Dai (daixiongfeng@mail.ccnu.edu.cn) upon reasonable request.
Material availability
This study did not generate unique reagents.
Experimental model and subject details
Wild type Bacillus subtilis 168 strain and its derivatives were used in this study. For all related experiments, we always prepared fresh LB plates of B. subtilis cells (from the glycerol stock of −80°C freezer) as the starting material.
Methods details
Strains
All the strains used in this study were derivatives of Bacillus subtilis 168 strain (Burkholder and Giles, 1947).
To construct an inducible lacZ reporter strain of B. subtilis, the lacZ gene from E. coli K-12 strain was inserted into the BamHI site of the pAX01 vector (with erythromycin resistance marker, ermR) (Härtl et al., 2001), which bears the xylose-inducible xylR-PxylA cassette, generating pAX01-lacZ vector. The pAX01-lacZ vector was directly transformed into B. subtilis 168 strain so that the xylA-PxylA-lacZ cassette was integrated into the B. subtilis genome. In the case of nonsense mutation, the coding sequence of the 154th residues of lacZ, “TGG”, was mutated to a stop codon “TAA” by PCR-mediated point mutation using pAX01-lacZ as the template (Golden Green high-fidelity PCR mix from Tsingke Biotech, Beijing). The resultant vector, pAX01-lacZ-null, was also transformed into B. subtilis 168 strain to obtain the nonsense-mutated B. subtilis lacZ-null strain.
To make a rho-null strain, a ∼600 bp intragenic region of rho gene of B. subtilis was inserted into a pUC19-derived vector (with chloramphenicol resistance marker, catR) and integrated into the rho locus of B. subtilis 168 strain by Campbell single-crossover integration. In such case, the ORF of the native rho gene was disrupted by the insertion of the vector. To make the rnjA-null and greA-null strains, the flanking regions (including both upstream and downstream part) of rnjA and greA in the genome of B. subtilis were PCR amplified and placed at the flanking regions of the spectinomycin resistance marker (specR) in the integration vector, pDG1730(Guérout-Fleury et al., 1996), to replace its original amyE regions and further transformed into the B. subtilis so that the native rnjA and greA locus were replaced by specR.
Medium
Growth media used in this study were based on C-minimal medium with slight modifications (Commichau et al., 2008). The medium contained 4 g/L KH2PO4, 16 g/L K2HPO4, 2.32 mg/L MnSO4·4H2O, 0.123 g/L MgSO4·7H2O, 12.5 μM ZnCl2, 50 mg/L tryptophan, and 22 mg/L ferric ammonium citrate. 20 mM NH4Cl was used as the nitrogen source, with exception to fructose plus nitrate medium, where 20 mM KNO3 was used as the nitrogen source. Carbon sources included 0.4% (v/v) glycerol, 0.4% (w/v) sorbitol, 0.2% (w/v) mannose, and 0.4% fructose. In addition, glycerol plus casamino acid medium (gly + cAA) contains 0.3% (w/v) casamino acids. Fusidic acid and 6-azauracil were used at a concentration of 0.2 μg/mL and 500 μg/mL, respectively.
Cell growth
Cell growth was performed in a 37°C air bath shaker shaking at 200 rpm, with exception to the case of low temperature in which 25°C was used. A standard procedure of culturing B. subtilis contained three steps: seed culture, pre-culture, and the final experimental culture. Cells from a fresh colony were first inoculated into LB broth and grew for several hours, as the seed culture. The seed culture was then transferred to the minimal medium (identical to the final experimental culture) and grew overnight as pre-culture. On the next day, precultures was transferred to a fresh minimal medium at an initial OD600∼0.02, and this was the final experimental culture. 5-8 OD600 data points during exponential stage (usually at the OD600 range of 0.08–0.6) were measured by a Thermo Sci Genesys 30 spectrophotometer to obtain an exponential growth curve from which the growth rate was calculated. For the fusidic acid study, 0.2 μg/mL fusidic acid was supplemented to the final experimental culture at OD600 ∼0.05. The cell culture was adapted for additional two generations before reaching a stable exponential growth stage. For the 6-azauracil (6-aza) study, 500 μg/mL 6-aza was supplemented to the exponential growing culture at OD600∼0.25. The cell culture was further incubated for 1 h before performing transcription kinetics measurement.
Measurement of transcription kinetics
The protocol of transcription kinetics measurement was modified from the qRT-PCR method that established before (Johnson et al., 2020; Zhu et al., 2019). 1% xylose and 1% arabinose were supplemented to the exponentially growing B. subtilis lacZ reporter strain (OD600∼0.4) to induce the expression of PxylA-lacZ and the native araAB operon, respectively. 1% mannitol and 1% xylose were used to induce the expression of the native mtlAFD and xynPB operon, respectively. Immediately after inducer addition, 0.8 mL cell culture was withdrawn at a 10-20 s interval (depends on the exact growth condition) and transferred to 0.9 mL pre-chilled stop solution (60% ethanol, 2% phenol and 10 mM EDTA). Considering the thick cell wall structure of gram-positive bacteria, the stop solution also contained 10 μg/mL actinomycin D (GLPBIO), a strong blocker of transcription elongation, to ensure the immediate arrest of cellular RNA transcription(Levinthal and Higa, 1962; Pollock, 1963). Cells were pelleted at 4°C at 12000 rpm for 2 min and flash frozen by liquid nitrogen (if not immediately used for RNA extraction). Cell pellets could be stored at −80°C for a few days before RNA extraction. For RNA extraction, cell pellets were first lysed by 10 mg/mL lysozymes for 10 min at room temperature. Total cellular RNA was then extracted using a bacterial total RNA extraction kit (TianGen Biotech, Beijing). Note that during RNA extraction, the cellular DNA was removed by Dnase I. For cDNA synthesis (cDNA), 0.5 to 2 μg total cellular RNA was used with a first-strand cDNA synthesis reverse transcriptase kit (TianGen). During cDNA synthesis, a second type of Dnase, gDnase (genomic deoxyribonuclease), was added to ensure the complete elimination of genome contamination. The real time qRT-PCR was performed using either the PowerUp SYBR green Master mix (Thermo fisher) or Super-premix SYBR green Plus kit (Yeasen Biotech) with the ABI QuantStudio 3 real-time Thermocycler. The PCR reaction procedure for thermo fisher reagents was as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The specificity of the primers is confirmed by single-peak melting curve of qPCR process. The mRNA abundance of a sample taken immediately after inducer addition (referred to as ‘basal sample’), M(0), was set as ‘1’. The relative mRNA abundance at each time point, M(t), equals to 2Ct(0)−Ct(t). The mRNA abundance was plotted with time to obtain the transcription kinetics of each mRNA sub-regions, including head region, middle region and tail region (detected by different pairs of qRT-PCR primers). For data of each pair of primers, the linear range of the transcription kinetics data was fit with a linear line, being expressed as y = a × x – b, where “a” (the slope of the linear line) denotes the relative mRNA accumulation rate of each sub-regions. The transcription time of each mRNA sub-region, namely Thead, Tmid, Ttail, equals to (1 + b)/a since “1” is the basal line value before mRNA induction. The transcription elongation rates of lacZ and araAB mRNA equaled to 2808/(Ttail-Thead) and 3011/(Ttail-Thead), respectively, where 2808 and 3011 denote the distances (nt) between the tail region and head region of lacZ and araAB mRNA, respectively.
Measurement of mRNA degradation kinetics
B. subtilis lacZ reporter strain was first exponentially cultured to OD600∼0.4 in cAA medium or fructose plus nitrate medium (supplemented with 0.5% xylose or 0.5% arabinose). At time zero, 20 μg/mL actinomycin D (GLPBIO) were supplemented to completely block the cellular transcription process of B. subtilis(Levinthal and Higa, 1962). 0.8 mL cell culture was then withdrawn at a 1 min interval and transferred to 0.9 mL pre-chilled stop solution (60% ethanol, 2% phenol and 10 mM EDTA). Cell samples were then subject to RNA extraction and qRT-PCR as detailed in the section of measurement of transcription kinetics. Different sets of qRT-PCR probes were used to detect different sub-regions of lacZ and araAB mRNA. The relative abundances of lacZ and araAB mRNA were plotted with time to obtain the exponential mRNA degradation curve (see Figure S3) from which mRNA decay rates and half-lives could be calculated.
Measurement of translation kinetics
Measurement of the translation elongation rate was based on the classical LacZ induction assay, initially developed by Schelif et al(Schleif et al., 1973) and extensively used in recent studies(Dai et al., 2016; Iyer et al., 2018; Johnson et al., 2020; Zhu et al., 2019). 1% xylose was supplemented to the exponentially growing B. subtilis lacZ reporter strain (OD600∼0.4) to induce the expression of PxylA-lacZ. Immediately after induction, at 15-s to 30-s intervals (depending on the growth condition), aliquots of 400-μL culture were transferred into pre-chilled microfuge tubes (Nest Biotech) containing 10-μL chloramphenicol (34 mg/mL) for a total of 18–20 time points. Samples were flash frozen at liquid nitrogen and stored at −80°C prior to LacZ assay. Measurement of LacZ activity was based on a sensitive fluorescence substrate 4-methylumbelliferyl-D-galactopyranoside (MUG). Briefly, 100-μL cell sample was added to 400-μL Z-buffer and warmed at 37°C for 10 min 50 μL of 2 mg/mL MUG was added and the reaction mixtures were incubated for 0.5–1 h. The reaction was stopped by 250 μL of 1 M Na2CO3. The fluorescence intensity was measured with a micro-plate reader (360 nm excitation filter, 460 nm emission filters). LacZ induction curve was made by plotting the LacZ activity against the induction time and further analyzed using a square root plot (Schleif plot) to obtain the lag time for the synthesis of the first LacZ molecule (Tfirst). The translation elongation rate equals to 1024/(Tfirst-Tini), where Tini denotes the time cost of initiation steps including the penetration of xylose into cells, XylR repressor depression and transcription initiation. Tini was obtained by analyzing the induction kinetics of head region of lacZ mRNA. Tini was relatively constant at ∼20 s and ∼45 s under 37°C and 25°C, respectively.
Quantification and statistical analysis
The slope of the linear mRNA induction curve was analyzed by GraphPad Prism 8 software using linear fit.
Acknowledgment
We thank Professor Jianhua Zhang (Shanghai Jiao Tong University), Professor Zhemin Zhou (Jiang Nan University), and Professor Ming Sun (Central China Agricultural University) for kindly providing the B. subtilis 168 strain and pAX01 and pDG1730 vectors, respectively. We also greatly acknowledge Gene-Wei Li and Kurt Fredrick for useful discussions and critical reading of the manuscript during various stages of the work. This work was supported by the National Natural Science Foundation of China (31970027, 31870028, and 32022001) and by self-determined research funds of CCNU from the colleges' basic research and operation of MOE (CCNU19TS028 and CCNU20TS023).
Author contributions
Conceptualization, M.Z. and X.D.; methodology, M.Z. and X.D.; investigation, M.Z., X.D., H.M., F.H., and Q.W.; writing-original draft, M.Z. and X.D.; writing-review & editing, M.Z. and X.D.; funding acquisition, M.Z. and X.D.; resources, M.Z., H.M., and X.D.; supervision, M.Z. and X.D.
Declaration of interests
The authors declare no conflict of interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103333.
Contributor Information
Manlu Zhu, Email: zhumanlu@mail.ccnu.edu.cn.
Xiongfeng Dai, Email: daixiongfeng@mail.ccnu.edu.cn.
Supplemental information
Data and code availability
All related data are provided in the article and any further details is available from the lead contact upon reasonable request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- Adhya S., Gottesman M. Control of transcription termination. Annu. Rev. Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Aleksandra G.-M., Vladimir B., Jacek B., Elena B. Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology. 2016;162:433–447. doi: 10.1099/mic.0.000244. [DOI] [PubMed] [Google Scholar]
- Bennett P.M., Maaloe O. The effects of fusidic acid on growth, ribosome synthesis and RNA metabolism in Escherichia coli. J. Mol. Biol. 1974;90:541–561. doi: 10.1016/0022-2836(74)90234-4. [DOI] [PubMed] [Google Scholar]
- Burkholder P.R., Giles N.H. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 1947;34:345–348. [PubMed] [Google Scholar]
- Burmann B.M., Schweimer K., Luo X., Wahl M.C., Stitt B.L., Gottesman M.E., Rosch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- Chen M., Fredrick K. Measures of single- versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation. Proc. Natl. Acad. Sci. USA. 2018;115:10774–10779. doi: 10.1073/pnas.1812940115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau F.M., Gunka K., Landmann J.J., Stulke J. Glutamate metabolism in Bacillus subtills: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 2008;190:3557–3564. doi: 10.1128/JB.00099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhu M., Warren M., Balakrishnan R., Patsalo V., Okano H., Williamson J.R., Fredrick K., Wang Y.P., Hwa T. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2016;2:16231. doi: 10.1038/nmicrobiol.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R.J., Wuite G.J., Landick R., Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- De Hoon M.J.L., Makita Y., Nakai K., Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:212–221. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demo G., Rasouly A., Vasilyev N., Svetlov V., Loveland A.B., Diaz-Avalos R., Grigorieff N., Nudler E., Korostelev A.A. Structure of RNA polymerase bound to ribosomal 30S subunit. Elife. 2017;6:e28560. doi: 10.7554/eLife.28560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F., Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 1992;22:9. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Gray D.A., Dugar G., Gamba P., Strahl H., Jonker M.J., Hamoen L.W. Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat. Commun. 2019;10:890. doi: 10.1038/s41467-019-08719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greive S.J., von Hippel P.H. Thinking quantitatively about transcriptional regulation. Nat. Rev. Mol. Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- Guérout-Fleury A., Frandsen N., Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hart C.M., Roberts J.W. Rho-dependent transcription termination. Characterization of the requirement for cytidine in the nascent transcript. J. Biol. Chem. 1992;266:24140–24148. [PubMed] [Google Scholar]
- Härtl B., Wehrl W., Wiegert T., Homuth G., Schumann W. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J. Bacteriol. 2001;183:2696. doi: 10.1128/JB.183.8.2696-2699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham C.J., Dennis J., Furneaux P.A. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- Ishiguro A., Nogi Y., Hisatake K., Muramatsu M., Ishihama A. The Rpb6 subunit of fission yeast RNA polymerase II is a contact target of the transcription elongation factor TFIIS. Mol. Cell Biol. 2000;20:1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S., Le D., Park B.R., Kim M. Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 2018;3:741. doi: 10.1038/s41564-018-0161-3. [DOI] [PubMed] [Google Scholar]
- Iyer S., Park B.R., Kim M. Absolute quantitative measurement of transcriptional kinetic parameters in vivo. Nucleic Acids Res. 2016;44:e142. doi: 10.1093/nar/gkw596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janna P., Marie P., Erland B. Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 2005:49–58. doi: 10.1016/j.femsec.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Johnson G.E., Lalanne J.B., Peters M.L., Li G.W. Functionally uncoupled transcription–translation in Bacillus subtilis. Nature. 2020;585:124–128. doi: 10.1038/s41586-020-2638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R.E., Nierman W.C., Chamberlin M.J. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J. Biol. Chem. 1981;256:2787–2797. [PubMed] [Google Scholar]
- Kohler R., Mooney R.A., Mills D.J., Landick R., Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356:194–197. doi: 10.1126/science.aal3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuya Y., Kurokawa K., Ishikawa S., Ogasawara N., Oshima T. Transcription factor GreA contributes to resolving promoter-proximal pausing of RNA polymerase in Bacillus subtilis cells. J. Bacteriol. 2011;193:3090–3099. doi: 10.1128/JB.00086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal C., Higa K.A. Messenger RNA turnover and protein synthesis in B. subtilis inhibited by actinomycin D. Proc. Natl. Acad. Sci. USA. 1962;48:1631–1638. doi: 10.1073/pnas.48.9.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.M., Marks C.L., Freese E. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta. 1979;587:238–252. doi: 10.1016/0304-4165(79)90357-x. [DOI] [PubMed] [Google Scholar]
- McGary K., Nudler E. RNA polymerase and the ribosome: the close relationship. Curr. Opin. Microbiol. 2013;16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E., Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Miller M.J. Sensitivity of RNA synthesis to actinomycin D inhibition is dependent on the frequency of transcription: a mathematical model. J. Theor. Biol. 1987;129:289–299. doi: 10.1016/s0022-5193(87)80003-6. [DOI] [PubMed] [Google Scholar]
- Miller O.L., Jr., Hamkalo B.A., Thomas C.A., Jr. Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Mitra A., Nagaraja V. Under-representation of intrinsic terminators across bacterial genomic islands: rho as a principal regulator of xenogenic DNA expression. Gene. 2012;508:221–228. doi: 10.1016/j.gene.2012.07.064. [DOI] [PubMed] [Google Scholar]
- Mondal S., Yakhnin A.V., Sebastian A., Albert I., Babitzke P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 2016;1:15007. doi: 10.1038/nmicrobiol.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M., Gautheret D. Transcription attenuation in bacteria: theme and variations. Brief Funct. Genom. Proteom. 2009;8:482–492. doi: 10.1093/bfgp/elp025. [DOI] [PubMed] [Google Scholar]
- Newton W.A., Beckwith J.R., Zipser D., Brenner S. Nonsense mutants and polarity in the lac operon of Escherichia coli. J. Mol. Biol. 1965;14:290–296. doi: 10.1016/s0022-2836(65)80250-9. [DOI] [PubMed] [Google Scholar]
- O’Reilly F.J., Xue L., Graziadei A., Sinn L., Lenz S., Tegunov D., Blötz C., Singh N., Hagen W.J.H., Cramer P., et al. In-cell architecture of an actively transcribing-translating expressome. Science. 2020;369:554–557. doi: 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock M.R. The differential effect of actinomycin D on the biosynthesis of enzymes in Bacillus subtilis and Bacillus cereus. Biochim. Biophys. Acta. 1963;76:80–93. [PubMed] [Google Scholar]
- Proshkin S., Rahmouni A.R., Mironov A., Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk P.G., Dunkley E.A., Lee P., Krulwich T.A. Identification of a putative Bacillus subtilis rho gene. J. Bacteriol. 1993;175:8053. doi: 10.1128/jb.175.24.8053.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A., Bellecourt M.J., Landick R. Mechanisms of bacterial transcription termination: all good things must end. Annu. Rev. Biochem. 2016;85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- Richardson J.P. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64:1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- Richardson J.P. Loading Rho to terminate transcription. Cell. 2003;114:157–159. doi: 10.1016/s0092-8674(03)00554-3. [DOI] [PubMed] [Google Scholar]
- Rui L., Qing Z., Junbai L., Hualin S. Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator. Nucleic Acids Res. 2016:2554–2563. doi: 10.1093/nar/gkv1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Nogueira I., Nogueira T.V., Soares S., Lencastre H.D. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology. 1997;143(Pt 3):957–969. doi: 10.1099/00221287-143-3-957. [DOI] [PubMed] [Google Scholar]
- Santangelo T.J., Roberts J.W. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- Saxena S., Myka K.K., Washburn R., Costantino N., Court D.L., Gottesman M.E. Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 2018;108:495–504. doi: 10.1111/mmi.13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R., Hess W., Finkelstein S., Ellis D. Induction kinetics of the L-arabinose operon of Escherichia coli. J. Bacteriol. 1973;115:9–14. doi: 10.1128/jb.115.1.9-14.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Abedin S., Kamp D., Wilson D.N., Nierhaus K.H., Cooperman B.S. EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- Sikova M., Wiedermannova J., Prevorovsky M., Barvik I., Sudzinova P., Kofronova O., Benada O., Sanderova H., Condon C., Krasny L. The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription complexes. EMBO J. 2020;39:e102500. doi: 10.15252/embj.2019102500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough C.L. Regulation of bacterial gene expression by transcription attenuation. Microbiol. Mol. Biol. Rev. 2019;83 doi: 10.1128/MMBR.00019-19. e00019–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijl J.M., Hecker M. Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb. Cell Fact. 2013;12:3. doi: 10.1186/1475-2859-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimir B., Pierre N., Aleksandra G.M., Olivier D., Sandrine A., Anne A., Cyprien G., Francis R., Jacek B., Stéphane A. Termination factor Rho: from the control of pervasive transcription to cell fate determination in Bacillus subtilis. PLoS Genet. 2017;13:e1006909. doi: 10.1371/journal.pgen.1006909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Jensen K.F. Effects of guanosine 3',5'-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J. Biol. Chem. 1994;269:16236–16241. [PubMed] [Google Scholar]
- Vogel U., Jensen K.F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Sorensen M., Pedersen S., Jensen K.F., Kilstrup M. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol. Microbiol. 1992;6:2191–2200. doi: 10.1111/j.1365-2958.1992.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Wang C., Molodtsov V., Firlar E., Kaelber J.T., Blaha G., Su M., Ebright R.H. Structural basis of transcription-translation coupling. Science. 2020;369:1359–1365. doi: 10.1126/science.abb5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.W., Takacs M., Zhu C., Vidmar V., Eduljee A., Abdelkareem M.m., Weixlbaumer A. Structural basis of transcription-translation coupling and collision in bacteria. Science. 2020;369:1355–1359. doi: 10.1126/science.abb5036. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Breaker R.R. Regulation of bacterial gene expression by riboswitches. Ann. Rev. Microbiol. 2005;59:487. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Yakhnin A.V., FitzGerald P.C., McIntosh C., Yakhnin H., Kireeva M., Turek-Herman J., Mandell Z.F., Kashlev M., Babitzke P. NusG controls transcription pausing and RNA polymerase translocation throughout the Bacillus subtilis genome. Proc. Natl. Acad. Sci. U S A. 2020;117:21628–21636. doi: 10.1073/pnas.2006873117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H., Babiarz J.E., Yakhnin A.V., Babitzke P. Expression of the Bacillus subtilis trpEDCFBA operon is Influenced by translational coupling and rho termination factor. J. Bacteriol. 2001;183:5918–5926. doi: 10.1128/JB.183.20.5918-5926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Han Y.H., Im J., Seo S.W. Synthetic protein quality control to enhance full-length translation in bacteria. Nat. Chem. Biol. 2021;17:421–427. doi: 10.1038/s41589-021-00736-3. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Navaroli D.M., Enuameh M.S., Martin C.T. Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:10352–10357. doi: 10.1073/pnas.0606306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Mori M., Hwa T., Dai X. Disruption of transcription-translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 2019;4:2347–2356. doi: 10.1038/s41564-019-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All related data are provided in the article and any further details is available from the lead contact upon reasonable request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.