Summary
A quarter of all seasonal influenza cases are caused by type B influenza virus (IBV) that also dominates periodically. Here, we investigated a recombinant adenovirus vaccine carrying a synthetic HA2 representing the consensus sequence of all IBV hemagglutinins. The vaccine fully protected mice from lethal challenges by IBV of both genetic lineages, demonstrating its breadth of protection. The protection was not mediated by neutralizing antibodies but robust antibody-dependent cellular cytotoxicity and cell-mediated immune responses. Complete protection of the animals required the entire codon-optimized HA2 sequence that elicited a balanced immune response, whereas truncated vaccines without either the fusion peptide or the transmembrane domain reduced the efficacy of protection. Finally, the vaccines did not demonstrate any sign of disease exacerbation following lung pathology and morbidity monitoring. Collectively, these data suggest that it could be worth further exploring this prototype universal vaccine because of its considerable efficacy, safety, and breadth of protection.
Subject areas: Immunology, immune response, Virology
Graphical abstract

Highlights
-
•
The evolution of influenza B viruses necessitates development of universal vaccines
-
•
Consensus sequence of all viral hemagglutinins induces broad protection
-
•
Efficacy and safety of the vaccine are underpinned by balanced immune responses
-
•
Truncated vaccines facilitate mechanistic investigation of vaccine-elicited protection
Immunology; Immune response; Virology
Introduction
Influenza viruses known to infect humans are classified into three types based on their genetic makeup: A, B, and C. The first two are known to cause seasonal epidemics, whereas influenza C causes a mild respiratory illness without causing epidemics. The influenza A viruses (IAV) are divided into subtypes based on their two surface proteins namely, hemagglutinin and neuraminidase, whereas influenza B viruses (IBV) are classified based on their genetic lineages, i.e., B/Victoria and B/Yamagata (Zambon, 1999). IBV cases constitute ∼25% of all influenza cases annually, but it is not rare for IBV to dominate in a given season. For example, data from Europe in the 2017–2018 season revealed IBV cases were responsible for ∼65% of influenza cases (European Center for Disease Prevention and Control, 2018; Sun et al., 2019); however, even in IAV-dominated seasons, IBV has been shown to cause a second wave of infection as cases of IAV infection decline (Epperson et al., 2014; Garten et al., 2018). The severity of the IBV is also notable, with co-circulation of the two genetic lineages contributing significantly to the ∼290,000–650,000 annual influenza-attributed mortality reported globally (Iuliano et al., 2018).
Although the two IBV genetic lineages derived from a common ancestor, they are now antigenically different (Rota et al., 1990). For at least the last two decades, they have been co-circulating worldwide (Chen and Holmes, 2008; Shaw et al., 2002). Moreover, since 2015, both genetic lineages have displayed remarkable epidemic activity likely resulting from selective sweeps across the genome, followed by alternate evolutionary trajectories (Virk et al., 2020). Notably, although the evolution of the IBV viruses involve multiple viral proteins (Virk et al., 2020), the viral hemagglutinin (HA) remains of particular interest, given its critical impact on host immune response and annual vaccine preparations.
The hemagglutinin of IBV consists of two subunits, i.e., HA1 and HA2. Genetic analyses have revealed four major domains on the IBV HA1 protein; these epitopes are mainly responsible for the antigenic differences between influenza B strains. The 4 domains, 120-loop, 150- loop, 160-loop, and 190-helix, and their respective surrounding regions have demonstrated a high degree of structural flexibility. Antigenic drift generally does not significantly compromise the structural integrity; yet, mutations in these domains could be sufficient to allow the virus to evade neutralizing antibodies (Berton et al., 1984; Hovanec and Air, 1984; Koel et al., 2013; Kordyukova et al., 2011; Ni et al., 2013; Rivera et al., 1995).
Given the unpredictable nature of the switch in predominance, the inclusion of antigenic components from both IBV lineages in quadrivalent vaccine formulations is currently recommended, but a good antigenic match is still necessary (Ambrose and Levin, 2012; McCullers et al., 2004; Reed et al., 2012). Indeed, markedly high epidemic activity in recent years is of concern, with an increasing likelihood of diversification of IBV into multiple genetically distinct lineages (Virk et al., 2020). Broadly protective or universal influenza virus vaccines which could abolish the need for annual reformulation and re-administration of seasonal influenza virus vaccines are essential (Krammer and Palese, 2015; Wang et al., 2010).
A previous study showed that a peptide conjugate vaccine based on the maturational cleavage site of the IBV hemagglutinin precursor induced protection against lethal challenge of IBVs from both genetic lineages (Bianchi et al., 2005). Sequential immunization of chimeric HA proteins comprised of IBV stalk and the head of HA of exotic IAV provided protection against diverse strains of the IBV viruses (Ermler et al., 2017), while similar observations were also made on a mosaic IBV HA with major antigenic sites replaced with those from an exotic IAV counterpart (Sun et al., 2019). These findings are consistent with studies on the conserved protective epitopes in the stalk of the IBV HA (Dreyfus et al., 2012). In this communication, we report a recombinant adenovirus expressing the consensus sequence of the HA2 subunit that affords full protection to mice against lethal challenge with IBVs of either genetic lineage. Furthermore, dissection of the synthetic sequence allowed us to shed light on the mechanisms underlying immune protection as well as pulmonary pathology.
Results
Consensus sequence of HA2 affords full protection against IBV of both genetic lineages
All known hemagglutinin sequences of influenza B viruses from NCBI were downloaded and identical sequences removed to create a non-redundant dataset. The sequences were subjected to multiple alignments using CLUSTALW-MPI on a 64-processor Linux cluster, followed by the extraction of the target region from the full-gene alignment. The Shannon entropy for each amino acid position of the identified consensus sequences was then calculated to determine the degree of variation. The consensus sequence represents the highest degree of conservation of all HA2 sequences (Figure S1, supplemental information). Specifically, all positions in the reference remain the consensus, most represented residue at or near 100% (Figure S1).
Recombinant adenoviruses constructs (rAds) were designed to express a trimeric, secreted form of the consensus HA2 (Figure S2) (for details see STAR Methods). In brief, it includes 23 amino acids from the human tyrosinase signal peptide as previously described (Hashem et al., 2014) as well, two separate fragments from the HA1 subunit of influenza B/Florida/04/06, (amino acids 16–69 and 306–361), joined by a GSGSG linker were added to the N-terminus of the HA2 consensus sequence. A 27-amino acid fragment from the bacteriophage T4 fibritin trimerization motif was also added to the C-terminus of the HA2 sequence (rAd-HA2F), preceded by another GSGSG linker. Protein expression by the adenovirus construct was confirmed by Western blot (Figure S3, supplemental information).
To evaluate the efficacy of the vaccine in animals, we first compared the route of administration and prime/boost immunization regimen (Figure 1A). As shown in Figures 1B and 1C, subcutaneous administration of the vaccine failed to protect the mice from a lethal dose of IBV, whereas intranasal administration resulted in 60% survival. Using intranasal administration with a prime/boost dosing regimen resulted in full protection of the animals. These results were consistent with the changes in body weight (Figures 1D and 1E). Although the difference in survival rate and morbidity was drastically different between the two routes of administration, the HA2-specific antibody titers were similar between the two routes of administration (Figures 1F and 1G). Moreover, the antibody titers against HA2 were similar following either the single (Day 21) or the prime/boost (Day 49) immunization (Figures 1H and 1I), suggesting that the quality of immune responses is drastically different following the two routes of immunization. Notably, in contrast to the strong antibody responses to the HA2 stalk (Figures 1H and 1I), neither of the two fragments derived from HA1 (a.a. 16–69; 306–361), used as the linkers in the vaccine, induced detectable antibody responses (Figure S5), even though the two sequence fragments are also very conserved, with overall conservation rates of >99.7% except for four a.a. substitutions (Table S1, in supplemental information). These findings suggest that the HA1-derived fragments are unlikely to contribute to antibody-mediated immune responses (see below for more discussion).
Figure 1.
Intranasal immunization with a prime/boost regimen provides the best protection against lethal IBV challenge compared to subcutaneous immunization
(A–I) (A) Schematic diagram of the immunization, viral challenge, and necropsy timeline. DBA2 mice were subcutaneously (B, D, F, and H) or intranasally (C, E, G, and I) vaccinated once or twice before an intranasal challenge with B/Victoria/02/87. Survival (B, C; n = 10), weight (D and E; n = 10), Day 21 anti-HA2 IgG endpoint titer (F and G; n = 3–5), and Day 49 anti-HA2 IgG endpoint titer (H and I; n = 8–10) are shown.
Data shown is mean ± SEM; ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (Mann-Whitney test).
In light of these findings, we employed the intranasal administration with a prime/boost regimen in all subsequent experiments. We found that all vaccinated animals survived the lethal challenge by IBV from both lineages (Figures 2A and 2B), whereas unvaccinated animals were not protected as all animals died of B/Victoria challenge and only 20% survived following B/Florida challenge (Figures 2A and 2B). Again, the survival rates were consistent with the change of body weight (Figures 2C and 2D). Moreover, the protection induced by the vaccines was confirmed by pathological examination of the lung tissues (Figures 2E and 2F) and the assessment of viral burden in the lungs (Figures 2G and 2H). These results indicate that intranasal administration of the vaccine affords better protection of the animals from IBV of both genetic lineages, with a prime/boost regimen being the most effective.
Figure 2.
Intranasal immunization with rAd-HA2F provides effective cross-protection against a lethal challenge with IBV from both lineages without disease enhancement
DBA2 mice were intranasally vaccinated twice before an intranasal challenge with B/Victoria/02/87 (A, C, E, and G) or B/Florida/04/06 from the Yamagata lineage (B, D, F, and H). Mice were euthanized 3 days post-challenge for viral titer and histology analysis or monitored for 14 days post-challenge for survival. Survival (A and B), weight (C and D), average pathological score (E and F), and lung viral titer (G and H) are shown. Data shown is mean ± SEM representative of 2 independent experiments; In each experiment, n = 10 per group for survival and weight change, and n = 4 per group for histology and lung viral titer; ∗p < 0.05 (Mann-Whitney test).
Recombinant adenovirus-vectored vaccine induces higher levels of antigen-specific antibodies and antibody-dependent cellular cytotoxicity (ADCC)
Having observed the protective efficacy of the vaccine, we next investigated the immunological correlates. We observed HA2-specific IgG1 antibodies were detectable in the sera 21 days after a prime immunization, but the level of IgG1 appeared to decrease slightly following the boost (Figure 3A). In sharp contrast, antigen-specific IgG2a and IgG2b antibodies, known to be potent IgG subclasses in terms of ADCC induction (Collins, A.M. 2016), were observed to have a sustained high level of expression in the serum (Figures 3B and 3C). Mucosal antibody responses were also detected with high antigen-specific IgG and IgA levels in the bronchoalveolar lavage (BAL) fluid (Figures 3D and 3E).
Figure 3.
rAd-HA2F elicits robust HA2-specific antibody responses with high ADCC activity against multiple IBV strains
Serum from DBA2 mice collected 21 days after prime vaccination (Day 21) and 21 days after boost vaccination (Day 49) was used to determine anti-HA2 endpoint titer. IgG1 (A), IgG2a (B), and IgG2b (C) titers are shown. Bronchoalveolar lavage fluid (BALF) was collected on Day 49 to determine anti-HA2 IgG (D) and IgA (E) endpoint titers. Day 49 serum was used to determine the ADCC activity against B/Victoria/2/87 (F), B/Florida/04/06 (G; Yamagata lineage), B/Brisbane/60/08 (H; Victoria lineage), and B/Yamagata/16/88 (I). Data shown is mean ± SEM; n = 6 per group in each experiment; ∗∗p < 0.01 (Mann-Whitney test).
We tested if sera from vaccinated animals could have neutralizing activities. To this end, we incubated the vaccine-elicited sera with the virus and tested their neutralizing activities. We found no detectable neutralizing antibodies were induced in the vaccinated animals (Figure S6, supplemental information).
We then investigated their ADCC potential, given that IgG2a and IgG2b, not IgG1, are known to be prime inducers of this important antiviral effector function in mice (Collins, A.M. 2016). To this end, we evaluated sera isolated 49 days after prime vaccination. As shown in Figures 3F–3I, the antibodies demonstrated strong ADCC activities against viruses from both Yamagata and Victoria lineages, an observation which may explain the lethal challenge experiments (Figure 2).
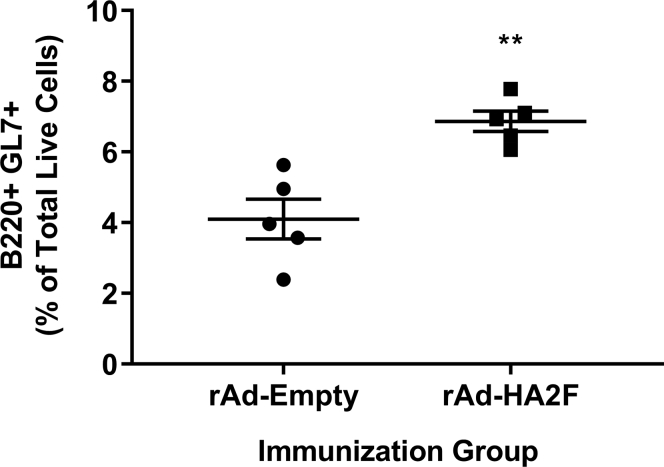
Moreover, we analyzed germinal center (GC) B cells (B220+ GL7+) following vaccination, as higher levels of GC B cells are correlated with enhanced affinity maturation to the antigen. We isolated cells from the draining lymph nodes of immunized mice on Day 49 and stained the cells with corresponding antibodies for flow cytometry analysis (Figure S4). As shown in Figure 4, a modest but significant increase of B220+ GL7+ cells was observed in the vaccinated group compared to the vector control.
Figure 4.
Immunization with rAd-HA2F increases the number of germinal center B cells
Cells isolated from draining lymph nodes of immunized mice at Day 49 were stained for flow cytometry analysis. Percentage of germinal center B cells (B220 + GL7+) among total live cells is shown. Data shown is mean ± SEM; n = 5 per group; ∗∗p < 0.01 (Mann-Whitney test).
Immunization with truncated HA2 vaccines reduces efficacy
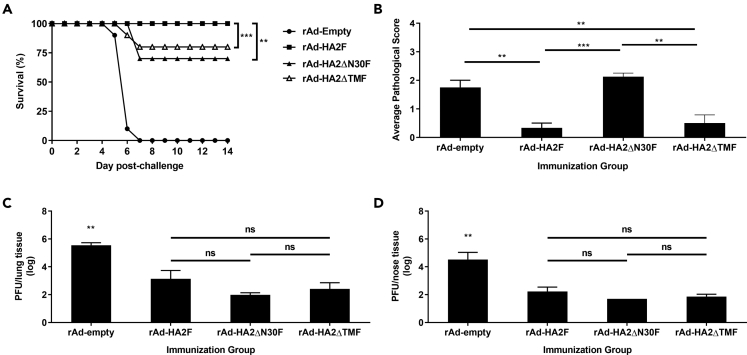
As the full length HA2 had shown robust protection, we next investigated if two regions known to be of critical importance for the virus were involved in inducing protection. Specifically, the N-terminus of the HA2 and the transmembrane domain were deleted from the vaccine construct. The N-terminus of the HA2 contains the only universally conserved linear sequence (fusion peptide) (Chun et al., 2008) and is indispensable for virus entry (Skehel and Wiley, 2000). The transmembrane domain is not only indispensable for the IBV virus but also has high immunogenicity (Muralidharan et al., 2018; Zhang et al., 2017). These two truncated vaccines, designated as rAd-HA2ΔN30F and rAd-HA2ΔTMF, respectively, were compared head-to-head with the full-length rAd-HA2F.
As shown in Figure 5, both truncated forms of the vaccine had modest but significant reductions in protecting the mice from lethal challenge, i.e., a loss of protection rate by 30% (rAd-HA2ΔN30F) and 20% (rAd-HA2ΔTMF), respectively (Figure 5A), whereas, interestingly, no significant difference in lung and nasal virus burdens were found between them and the full length HA2 vaccine (Figures 5C and 5D). Notably, pathological examination of the lung tissues revealed higher scores of vessel mural inflammation and septal thickening because of increased cellularity and congestion in the vector control and rAd-HA2ΔN30F (Figure 5B). These results indicate that the full length of HA2 is needed for complete protection and the removal of the N-terminal 30 a.a. sequence and the transmembrane domain reduces but does not abolish this protective effect. In addition, the inability of the rAd-HA2ΔN30F to effectively reduce pulmonary inflammation (Figure 5B) may be because of immunopathology associated with this vaccine (see below for more discussion).
Figure 5.
Deletion of the extended fusion peptide (ΔN30) or the transmembrane domain (ΔTM) attenuates protection
DBA2 mice were intranasally immunized twice before an intranasal challenge with B/Victoria/02/87. Mice were euthanized 3 days post-challenge for viral titer and histology analysis or monitored for 14 days post-challenge for survival. Survival (A), average pathological score (B), lung viral titer (C), and nose viral titer (D) are shown. Data shown is mean ± SEM; n = 10 per group for survival and n = 4 per group for histology, lung and nose viral titer; ∗∗p < 0.01, ∗∗∗p < 0.001 (Mann-Whitney test or one-way ANOVA with Tukey's post-test); ns: not significant.
Potent IgG effector functions require the N-terminal sequence of the HA stalk
As IgG2a is a more potent ADCC mediating IgG subclass than IgG1 in mice (Collins, A.M. 2016), we investigated the effect of removing the N-terminal and transmembrane domain regions on the IgG1/IgG2a ratio and ADCC activity of the antibodies induced by vaccination. As shown in Figure 6A, the full-length vaccine (rAd-HA2F) had the lowest level of IgG1 antibodies compared to the two truncated vaccines, after a prime/boost immunization, with rAd-HA2ΔN30F displaying the highest level. rAd-HA2ΔN30F vaccination also induced a higher IgG1/IgG2a ratio than immunization with rAd-HA2F (Figure 6B). Consistent with the observed higher level of IgG2a (Figure 6C), rAd-HA2F was found to induce the highest level of ADCC activity, with the lowest activity by rAd-HA2FΔN30F (Figure 6D). The ADCC activity induced by the vaccine devoid of the transmembrane domain (rAd-HA2ΔTMF) was lower than the full-length vaccine but not statistically significant. Taken together, deletion of the fusion peptide containing N-terminal 30 a.a. sequence led to higher IgG1/IgG2a ratio and a reduction in ADCC activities.
Figure 6.
Truncated vaccines elicit higher Th2 antibody isotypes than rAd-HA2F affecting ADCC activity
Serum from DBA2 mice collected 21 days after boost vaccination (Day 49) was used to determine anti-HA2 endpoint titer. IgG1 (A), and IgG2a (C) endpoint titers, and IgG1:IgG2a ratios (B) are shown. (D) Day 49 serum was used to determine the ADCC activity against B/Victoria/2/87. Fold induction over ‘no antibody’ controls is shown. Data shown is mean ± SEM; n = 6 per group in each experiment; ∗p < 0.05, ∗∗p < 0.01 (one-way ANOVA with Tukey's post-test in panels (A–C) and two-way ANOVA with Tukey's post-test in panel (D)). In (D), rAd-HA2ΔN30F is significantly different from rAd-HA2F at the first 2 points (dilution (log) = 1.48 and 1.95).
Truncated forms of the vaccine induce altered cytokine profiles
We next investigated antigen-specific cytokine levels following vaccination by the three vaccines. To this end, splenocytes from vaccinated animals were stimulated with peptides identified to be immunogenic for the quantitation of cytokines (Muralidharan et al., 2018). Specifically, P1 (FFGAIAGFL), located in the fusion peptide, was used to stimulate the splenocytes from animals vaccinated with rAd-HA2ΔN30F, in comparison with the full-length rAd-HA2F, to assess the role of the fusion peptide in one set of experiments. In another set of experiments, P2 (YYSTAASSL), situated within the transmembrane domain, was used to stimulate splenocytes from animals vaccinated with rAd-HA2ΔTMF, in comparison with the full-length rAd-HA2F. In both experiments, splenocytes from mice vaccinated with the empty vector were included as a control and the levels of secreted cytokines were measured in the cell supernatants.
As shown in Figure 7A, rAd-HA2ΔN30F vaccine resulted in reduced levels of IL-2, IL-9, IL-17, and GM-CSF. Although IL-2 is a strong proinflammatory cytokine secreted by Th1 cells, IL-9 was shown to be involved in suppressing influenza immunopathology (Murphy-Schafer and Paust, 2021). Moreover, the drastically reduced IL-17 level in the rAd-HA2ΔN30F group is noteworthy, as IL-17 plays an important role in mucosal immunity against pathogens (Flórido et al., 2018; Khader et al., 2009; Kumar et al., 2013).The rAd-HA2ΔN30F vaccination also resulted in decreased level of GM-CSF, known to improve vaccine efficacy against influenza (Huang et al., 2011; Littauer et al., 2018). In addition, the immunodominant epitope in the N-terminus did not appear to effectively stimulate IFN-γ response as neither IFN-γ and IL-18 (an IFN-γ inducer) (Nakanishi, 2018; Okamura et al., 1995) were elevated by vaccinations with rAd-HA2F or rAd-HA2ΔN30F.
Figure 7.
Truncated vaccines diminish antigen-specific cytokine responses. Cells were isolated from the spleen of twice-immunized DBA2 mice at Day 49
(A) Cells were stimulated ex vivo with P1 (FFGAIAGFL), an epitope missing in rAd-HA2ΔN30F.
(B) Cells were stimulated with P2 (YYSTAASSL), an epitope missing in rAd-HA2ΔTMF. Following 48-h stimulation, the supernatant was used with the Luminex system to quantify cytokines. pg/ml of IL-2, IFN-γ, IL-9, IL-17, IL-18, and GM-CSF is shown.
Data shown is mean ± SEM; n = 4 per group in each experiment; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (one-way ANOVA with Tukey's post-test).
The antigen-specific cytokine profile following vaccination with rAd-HA2ΔTMF was slightly different from that obtained with rAd-HA2ΔN30F. As shown in Figure 7B, although deletion of the transmembrane domain (rAd-HA2ΔTMF) also resulted in decreased levels of IL-2, IL-17, and GM-CSF, similar to what was observed with rAd-HA2ΔN30F (Figure 7A), IFN-γ and IL-18 (IFN-γ inducer) were also greatly reduced, an observation different from that achieved with rAd-HA2ΔN30F vaccination.
We did not find any significant differences in other cytokines (IL-1β, IL-4, TNF-α, or IL-6), which could be due to the limitation of the cytokine assays or the different signaling pathways induced by these cytokines. Nonetheless, these results collectively indicate that both epitopes in the N-terminus and transmembrane domain contribute to the robust antiviral activities against IBV infection.
Discussion
IBV infections are a serious public health issue and disease severity is comparable to that caused by IAV in both adults (Gaglani et al., 2021; Hu et al., 2021; Su et al., 2014; Xie et al., 2021) and children (Chi et al., 2008). Furthermore, although IBV cases usually constitute about ¼ of all cases, IBV viruses are predominant in some seasons (Ambrose and Levin, 2012; McCullers et al., 2004; Reed et al., 2012; Sun et al., 2019). The current vaccines against influenza are not optimal in that the vaccine seeds must be selected to produce the vaccines several months before the next flu season starts. Mismatches between the vaccine seeds used for vaccine production and the viruses circulating in the upcoming season could substantially weaken the vaccine effectiveness. Indeed, the success rate in the prediction of the correct B strain to be included in the vaccine was less than 50% in the US from 2000 to 2010 (Reed et al., 2012). Although the quadrivalent influenza vaccine containing viral proteins from both genetic lineages would help improve vaccine coverage against IBV (Heikkinen et al., 2014), the rapid and divergent trajectories of the IBV viruses could potentially result in the emergence of 3 or more distinct IBVs, underlining the urgency for developing universal influenza vaccines (Virk et al., 2020). Although numerous studies have been conducted to develop vaccines capable of inducing broadly reactive immune responses against IAV (Chen et al., 2016; Fan et al., 2015; Hai et al., 2012; Hashem et al., 2010; Impagliazzo et al., 2015; Krammer et al., 2013, 2014; Laursen et al., 2018; Lu et al., 2014; Sui et al., 2009; Ueda et al., 2020), much fewer studies on universal vaccines against IBV have been reported (Bianchi et al., 2005; Ermler et al., 2017; Sun et al., 2019).
In this communication, we reported that a recombinant adenovirus expressing a consensus sequence of the IBV HA2 domain afforded complete protection in mice against IBVs from both genetic lineages. Several lines of evidence prompted us to conduct the current study on the IBV HA stalk-based vaccine. First, the effects of different administration routes on the IBV stalk-induced protection remains poorly understood (Sun et al., 2019), particularly for virally vectored vaccines. Second, the protective epitopes in the stalk of IBV remain to be better delineated although the targets of a monoclonal antibody (CR9114) (Dreyfus et al., 2012) and a monospecific antibody (Uni-1) capable of binding to both IAV and IVB have been located in the stalk (Chun et al., 2008). Third, the immunological mechanisms underlying the stalk-induced protection require further elucidation. Fourth, disease exacerbation has been associated with antibodies against the stalk (Khurana et al., 2013) and intranasal delivery of inactivated and split whole virus vaccine adjuvanted with strong TLR4 agonist (Maroof et al., 2014), respectively. To this end, we constructed a recombinant adenovirus expressing the full length or truncated forms of the HA2 stalk and investigated some specific questions related to the aforementioned knowledge gaps with animal studies.
We found a sharp distinction depending on the route of vaccine administration; namely, although subcutaneous administration of the vaccine failed to provide protection, intranasal administration afforded full protection of the animals from a lethal dose (Figure 1). The complete absence of protection by subcutaneous administration was rather surprising, given that both routes of administration resulted in robust and comparable levels of HA2-specific antibodies. Although additional investigations focused on elucidating the specific mechanisms underlying this vaccine failure are needed, we elected instead to focus on better understanding the effects of the intranasal route of administration. In sharp contrast to that with the subcutaneous route, nasal administration afforded full protection against lethal challenges of viruses from both genetic lineages. Even though the nasal administration is absolutely required, the breadth of the protection is because of the synthetic sequence representing the highest degree of conservation of all HA2 sequences. All positions in the reference (synthetic gene) remain the consensus, most represented residue at or near 100% (Figure S1).
The intranasal administered vaccine did not induce disease enhancement whereas bronchiolar epithelial apoptotic necrosis and bronchiolitis were notable in the vector control group (Figure 2). It is also of note that intranasal administration of adenoviral-vectored IAV nucleocapsid protein did not cause injury to pulmonary tissue (Dhakal et al., 2021).The vaccine induced higher levels of antigen-specific IgG2a and IgG2b antibodies than IgG1. Interestingly, IgG2a/2b displayed different dynamics from IgG1, with the titer of the latter declining slightly (not statistically significant) even after the boost (Figure 3). Importantly, vaccine-induced antibodies had no neutralizing activities but demonstrated strong ADCC effects against two IBV strains from each of the genetic lineages. These results were consistent with the more potent characteristics of IgG2a and IgG2b for the FcγR-mediated effector functions (Bachmann and Jennings, 2010; Collins, 2016; Ermler et al., 2017; Nimmerjahn et al., 2010; Sun et al., 2019). The efficacy of the vaccine was also demonstrated by an increased pool of GC B cells in the draining lymph nodes (Figure 4), an important process for the generation of long-lived antibody secreting plasma cells and memory B cells (Stebegg et al., 2018).
Our characterization of the two truncated forms of the vaccine yielded additional interesting findings. Consistent with our recent immunoinformatics analyses (Muralidharan et al., 2018), deletion of either the N-terminus or transmembrane domain reduced the vaccine efficacy by ∼ 30% and ∼20%, respectively (Figure 5). The partial reduction instead of a complete abolition of protection by the two truncated vaccines is not surprising, as the vaccine-induced protective immunity does not depend on neutralizing antibodies of which the potency could be drastically reduced or even abolished if a few key amino acids are mutated or substituted. As such, vaccines inducing protection independently of neutralizing activities could be advantageous in this regard.
The role of the N-terminus in inducing protection is noteworthy, as truncation of the N-terminus encompassing the fusion peptide (rAd-HA2ΔN30F) was associated with a higher IgG1/IgG2a ratio, in agreement with reduced ADCC activities (Figure 6). However, the exact mechanisms underlying the reduction of the ADCC remain to be understood. Interestingly, the 30 a.a. sequence at the N-terminus of the HA2 is extremely conserved (close to 99.96%), whether missing this region would affect the exposure of ADCC epitopes would need further investigation in future studies. Moreover, it also remains to be understood as to how this region could be involved in reducing lung inflammation, which was shown by the rAd-HA2ΔN30F vaccine resulting in higher pulmonary pathology score equal to the vector control (Figure 5B). A human CD4+ T cell epitope has also been reported in this region (Babon et al., 2012), whereas antibodies against this region could inhibit the entry of the virus (Hashem et al., 2010). Finally, the transmembrane domain appeared to play a more minimal role in terms of affecting the IgG1/IgG2a ratio, compared with the full-length vaccine (rAd-HA2F), with a slight but not statistically significant reduction in ADCC activities.
Cytokine profiling shed some light on the mechanisms underlying the N-terminal fusion peptide sequences and the transmembrane domains (Figure 7). It is worth noting that the epitope (YYSTAASSL) in the transmembrane domain of the HA2 is a very potent stimulator for IFNγ and its inducer IL-18 (Nakanishi, 2018; Okamura et al., 1995) as deletion of the transmembrane domain resulted in a drastic reduction of both cytokines. Finally, both N-terminus and transmembrane domain regions were found to be important in inducing IL-2, IL-17 and GM-CSF. As mentioned earlier, IL-2 and GM-CSF are well known to be a strong, secreted, Th1 proinflammatory cytokine and molecular adjuvant for influenza vaccine, respectively (Huang et al., 2011; Littauer et al., 2018). The drastically decreased levels of IL-17 with both truncated vaccines are of particular interest as IL-17 has been suggested to be involved in strengthening mucosal immunity against pathogens (Counoupas et al., 2020; Flórido et al., 2018; Khader et al., 2009; Kumar et al., 2013). Clearly, both the epitopes in the N-terminus and transmembrane domain may contribute to the mobilization of mucosal immunity. Interestingly, mice vaccinated with inactivated and detergent-split influenza vaccines adjuvanted with TLR agonist induced transient weight loss through Th17-mediated immunity (Maroof et al., 2014), an observation made in a different experimental condition from ours. Indeed, we did not observe any transiently exacerbated inflammation and weight loss with any of the vectored vaccines, in agreement with the other study using adenoviral vector for intranasal administration of IAV nucleocapsid protein (Dhakal et al., 2021). Nonetheless, induction of a balanced immune response is of critical importance in the development of safe and efficacious vaccine against influenza. With that said, the vectored vaccine described here merits further exploration as a potent universal vaccine due to its efficacy, safety and breadth of protection against virtually all influenza B viruses.
Limitations of the study
We were unable to quantitatively determine the resident T cells in the lungs because of technical difficulties. We have not conducted investigation of the longevity of the induced immune responses and protection of aged animals in this study either. Although we succeeded in identifying important epitopes in the N terminus and transmembrane domains, we did not conduct dissection of more domains. For example, although the two HA-1 derived fragments have been found to be less likely to be involved in inducing antibody-mediated protection, we did not conduct experiments to investigate if they are able to induce protective cell mediated immune responses. Nonetheless, given the high degree of conservation rates of these viral sequences, the inclusion of these two viral sequences is likely to be beneficial rather than detrimental. It should be mentioned that we only tested adenovirus 5 as the vector as it is a well characterized viral vector. Future studies should be considered to include different vectors or delivery systems, along with better understanding of structures of the consensus stalk vaccine and the deletion mutants. These studies are currently ongoing in our laboratories.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mouse influenza B HA | Sino Biological Inc. | 11053-RP01 |
| HRP-conjugated anti-mouse IgG1, IgG2a, IgG2b | Jackson Immunoresearch Laboratories | 115-035-205, 115-035-206, 115-035-207 |
| HRP-conjugated anti-mouse IgG | Cytiva Lifescience | NA9311ML |
| anti-mouse IgA | Life Technologies | LS626720 |
| Tetramethylbenzidine (TMB) substrate | Cell Signaling Technology | 7004 |
| Rabbit polyclonal anti-influenza B NP | Genetex | GTX128538 |
| HRP-conjugated anti-rabbit IgG antibody | Cytiva | NA9341ML |
| Fixable Viability Dye eFluor 506 | eBioscience | 65-0866-14 |
| anti-mouse CD16/CD32 | eBioscience | 16-0161-86 |
| AF647-conjugated anti-mouse Ly77/GL7 (clone GL7) | BD Biosciences | 561529 |
| PE-Cy7-conjugated anti-mouse CD45R/B220 (clone RA3-6B2) | BD Biosciences | 552772 |
| Bacterial and virus strains | ||
| B/Victoria/2/87, | Icahn School of Medicine at Mount Sinai (Ermler et al., 2017 | N/A |
| B/Yamagata/16/88 | Icahn School of Medicine at Mount Sinai (Ermler et al., 2017 | N/A |
| B/Brisbane/60/08 | Icahn School of Medicine at Mount Sinai (Ermler et al., 2017 | N/A |
| mouse-adapted B/Florida/4/06 | Icahn School of Medicine at Mount Sinai (Ermler et al., 2017 | N/A |
| Biological samples | ||
| Tissue samples from DBA/2J mice used in this study; the animals were purchased from Jackson lab | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| FFGAIAGFL | New England Peptides | P1 |
| YYSTAASSL | New England Peptides | P2 |
| DRICTGITSSNSPHVVKTATQGEVNVTGVIPLTTTP TKSYFANLKGTRTRGKLC |
New England Peptides | HA1 linker (N-term |
| EADCLHEKYGGLNKSKPYYTGEHAKAIGNCPIWVK TPLKLANGTKYRPPAKLLKER |
New England Peptides | HA1 linker (C-term |
| B/Florida/4/06 HA2 protein | Sinobiologcals | Cat: 11053-V01H2 |
| Critical commercial assays | ||
| Promega ADCC Reporter Bioassay | Promega | M1212 |
| Bio-GloTM luciferase assay | Promega | G7941 |
| Adeno-X Rapid Titer Kit | Takara Bio USA Inc | 632250 |
| Gateway®-adapted ViraPowerTM adenoviral expression system | Life Technologies | V49320 |
| ProcartaPlex Multiplex Immunoassay kit | Life Technologies | EPX170-26087-901 |
| Deposited data | ||
| Division of Regulatory Research (CG-IBV data) | Health Canada, Ottawa, Canada | N/A |
| Experimental models: Cell lines | ||
| FcγRIV effector cell | Promega | M1212 |
| MDCK | ATCC | CCL-34 |
| HeLa | ATCC | CCL-2 |
| HEK293A | Qbiogene | AES2044 |
| Experimental models: Organisms/strains | ||
| DBA/2J mice | (The Jackson Laboratory) | 000671 |
| Recombinant DNA | ||
| pBluescript II SK+ vector | Bio Basic Canada Inc. | N/A |
| pENTRTM/SD/D-TOPO vector | Life Technologies | K242020 |
| pAd/CMV/V5-DEST vector | Life Technologies | V493-20 |
| Software and algorithms | ||
| Geneious Prime | Geneious. | Geneious Prime |
| GraphPad Prism 8 | GraphPad Software Inc. | GraphPad Prism 8 |
| BD FACSDIVA 6.2 | BD Biosciences | BD FACSDIVA 6.2 |
| MILLIPLEX Analyst version 5.1 | Merck Millipore | MILLIPLEX Analyst version 5.1 |
| Original Code for Bioinformatics Analysis | Zenodo | https://doi.org/10.5281/zenodo.5573271 (https://zenodo.org/record/5573271) |
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Xuguang Li (sean.li@canada.ca).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Design of universal influenza B HA2 sequence and generation of recombinant adenoviruses
A bioinformatics approach was used to identify a universally conserved consensus sequence for the HA2 subunit of influenza B, as described before (Chun et al., 2008). Briefly, generation of consensus sequence was accomplished by first downloading two sets of Influenza B HA sequences from the NCBI Flu Database with the following filters: Flu B, Human, All Countries/Regions, HA, Any Subtype, Full-length only. For the first set the additional filter of "Collapse identical sequences" was also applied. Set 1 included 3699 HA sequences and Set 2 16011. Sequences with ambiguous codons (BJOUX) were filtered out using perl scripts prior to alignment against the generated peptide sequence. Alignment was accomplished with MAFFT using the --add-fragments and --keeplength options to restrict the alignment length to that of the peptide sequence. Per position Shannon entropy and the consensus sequence were calculated using perl scripts and the EMBOSS toolkit respectively. Result and figure presentation were finalized in R. This analysis revealed only 4 positions where variability exceeded 5% of the total dataset and 2 positions with ∼5% variability. At the four positions of higher variability the dataset has an approximately even distribution between one of two residues (minor allele frequency from 46-49%). The reference remains the consensus in two of the positions (158 N/D, 208 I/V) for both sets. A slight shift is seen at positions 132(ref E 49.5%--> D 50.5%) and 204(ref L 46%--> I 53.7%) in set 1 (collapsed identical sequences) and only at position 132(ref E 47.4%--> D 52.6%) in set 2 (all available sequences considered).
Recombinant adenoviruses constructs (rAds) were designed to express a trimeric, secreted form of the consensus HA2 by including 23 amino acids from the human tyrosinase signal peptide (; GenBank accession #AH003020) at the N-terminus as described (Hashem et al., 2014). Two separate fragments from the HA1 subunit of influenza B/Florida/04/06, corresponding to amino acids 16-69 and 306-361 and joined by a GSGSG linker, were added to the N-terminus of the HA2 consensus sequence, to stabilize its conformation in a similar approach described for the trimeric influenza A (H1N1, A/California/ 05/2009) hemagglutinin stem proteins (Lu et al., 2014). A 27 amino acid fragment from the bacteriophage T4 fibritin trimerization motif (YIPEAPRDGQAYVRKDGEWVLLSTFLG) was also added to the C-terminus of the HA2 sequence (rAd-HA2F), preceded by another GSGSG linker. Adenovirus constructs containing truncated HA2 genes were also generated, by deleting the extended fusion peptide region (aa 362-392) (rAd-HA2ΔN30F) and the transmembrane domain (aa 549-574) (rAd-HA2ΔTMF), as well as an empty vector control (rAd-empty). rAds were generated using the Gateway®-adapted ViraPowerTM adenoviral expression system (Life Technologies), following the manufacturer’s instructions.
Briefly, the HA2F, HA2ΔN30F and HA2ΔTMF sequences were codon-optimized for mouse, synthesized and cloned in a pBluescript II SK+ vector by Bio Basic Canada Inc. The genes were isolated from the pBluescript II SK+ plasmid by PCR. The PCR products were cloned into a pENTRTM/SD/D-TOPO vector (Life Technologies) and transformed into E. coli as per manufacturer’s instructions. Following isolation of the plasmid, the genes were transferred into a pAd/CMV/V5-DEST vector (Life Technologies) by a recombination reaction. Sequencing was performed at every step of the subcloning procedure to confirm the integrity of the gene sequences. Following digestion with PacI restriction enzyme, the resulting pAd-DEST vectors were purified by phenol-chloroform extraction/ethanol precipitation before transfection into HEK-293A cells for packaging of the recombinant adenoviruses. Cells and supernatant were harvested once the cytopathic effect reached 80%. Cell lysates obtained by freeze-thawing were combined with the supernatant and rAds were purified by ultracentrifugation with a 30% sucrose cushion. rAd stocks were titrated using the Adeno-X Rapid Titer Kit (Takara Bio USA Inc.).
Animal studies (Mice)
All animal procedures were approved by the Health Canada Ottawa Animal Care Committee and performed in accordance with institutional guidelines. Six-week-old female DBA/2J mice (The Jackson Laboratory) were used for all animal experiments. Mice were immunized subcutaneously or intranasally with 109 PFU of each rAd construct. Subcutaneous and intranasal inoculations were given in 200ul and 25ul, respectively. Mice were first immunized on day 0 and received a boost vaccination on day 28, when applicable. Serum was collected from each animal 3 weeks after the prime and boost immunizations for measurement of antibody levels. Spleens and axillary lymph nodes were harvested from some of the vaccinated animals on day 49 and single cell suspensions were prepared for flow cytometry and cytokines analysis. Bronchoalveolar lavage fluid (BALF) was also collected from some mice 3 weeks following the second immunization for mucosal antibodies analysis. Mice were challenged intranasally, 4 weeks after the boost immunization, with 3.75x105 PFU of the B/Victoria/2/87 or 1.66x104 PFU of the B/Florida/4/06 virus, in 25ul. Following the challenge, mice were weighed daily and monitored for signs of illness for 14 days. 3 days post challenge, lung and nasal tissues were collected from 4 mice per group for viral load determination and pathology analysis. Survival data is presented as the percentage of surviving animals at each time point, compared to the initial number of animals in each group. Weight loss data is presented as the percentage of animal weight relative to their initial body weight on the day of challenge.
Methods details
In vitro protein expression
Generated rAds were used to infect confluent HeLa cells at a multiplicity of infection (MOI) of 200. After 48 hours, infected cells were washed twice with PBS, lysed with RIPA buffer and slightly sonicated. Cell lysate proteins were separated using 4-15% gradient SDS-PAGE and transferred to a PVDF membrane. Protein expression was confirmed by Western blot using a polyclonal rabbit antibody against influenza B HA (Sino Biological Inc.).
ELISA
The end-point titers of serum and BALF anti-HA2 antibodies were determined by ELISA as described previously, with minor modifications (Hashem et al., 2014). Briefly, 96-well plates were coated with 100ul/well of 0.5ug/ml of recombinant influenza B/Florida/4/06 HA2 protein (Sino Biological Inc.), overnight at 4°C. Plates were washed with PBS containing 0.05% Tween 20 (PBS-0.05T) and blocked for 2 hours at 37°C with 3% BSA in PBS-0.05T. Following a wash step, two-fold serial dilutions of the mouse serum or BALF in blocking buffer were added for 1 hour at 37°C. After washing, HRP-conjugated anti-mouse IgG (Cytiva), anti-mouse IgG1, IgG2a, IgG2b (Jackson Immunoresearch Laboratories) or anti-mouse IgA (Life Technologies) were added at a 1:2000 dilution, for 1 hour at 37°C. After additional washing, Tetramethylbenzidine (TMB) substrate (Cell Signaling Technology) was added. The reaction was stopped by addition of 0.16M sulfuric acid and absorbance was read at 450nm. End-point antibody titers were expressed as the reciprocals of the final detectable dilution, with a cut-off defined as the mean of rAd-Empty samples plus 3 standard deviations.
Lung and nose viral titer
The right lobe of the lungs and nasal tissues were harvested 3 days post challenge and viral titers were measured by plaque assay. Briefly, tissues were collected and flash frozen in liquid nitrogen prior to analysis. Following thawing on ice, lungs and nasal tissues were homogenized in 300μl and 250μl of PBS, respectively. The homogenates were clarified by centrifugation and filtered using a 0.45um syringe filter. Ten-fold serial dilutions of the homogenates were prepared in serum-free complete DMEM medium supplemented with 25mM Hepes, 0.2%BSA and 2ug/ml TPCK-treated trypsin and incubated on confluent MDCK cells for 2 hours, at 35°C. The inoculum was removed, cells were washed with dilution media, and overlaid with complete DMEM medium containing 25mM Hepes, 0.2% BSA, 2ug/ml TPCK-treated trypsin and 0.8% agarose. After incubation for 4 days at 35°C/5% CO2, the overlay was removed and cell monolayers were stained with crystal violet for plaque counting. Results are expressed as PFU/lung tissue or PFU/nasal tissue.
Histopathology
The lungs were collected from DBA/2J mice 3 days after challenge with influenza B viruses and were fixed in 10% neutral buffered formalin. They were then trimmed to produce sections from different areas of the lobe, routinely processed, and embedded in paraffin. Four-micron sections were cut and stained with Hematoxylin and Eosin (H and E), for evaluation. Scoring was done by a veterinary pathologist who was blinded to the experimental design. The lung lesions were classified using the Society of Toxicologic Pathology’s International Harmonization and Diagnostic Criteria (INHAND) for the respiratory tract publication, by Renne et al. (Renne et al., 2009). A numeric lesion severity score was assigned to each lesion where 0 means no lesion, 1 means a minimal degree of severity, 2 means mild, 3 means moderate, 4 signifies marked and 5 means severe. An average of individual scores are shown.
Microneutralization
Microneutralization assay was carried out as described in the 2011 version of the WHO Manual for the laboratory diagnosis and virological surveillance of influenza (WHO, 2011). Briefly, serum samples were treated with receptor-destroying enzymes (RDE) (Denka Seiken Co.) for 18 hours at 37°C, followed by a 30-minute incubation at 56°C. One hundred TCID50 of the tested influenza B viruses were mixed with equal volume of 2-fold serial dilutions of the treated samples in a 96-well tissue culture plate and were incubated for 1 hour at 37°C. Pelleted MDCK cells were washed twice and resuspended with serum-free complete DMEM medium supplemented with 25mM Hepes and 1%BSA and were added to the plate at a density of 1.5x104 cells/well, following the 1-hour incubation. The plate was then incubated for 20 hours, at 37°C/5% CO2. After the incubation, the media was removed, cells were washed twice with PBS and were fixed with cold 80% acetone, for 10 minutes. Following a wash with PBS containing 0.1% Tween 20 (PBS-0.1T), the viral NP antigen was detected by indirect ELISA with a rabbit polyclonal antibody to influenza B virus NP (GeneTex), diluted to a concentration of 0.5ug/ml, in PBS-0.1T containing 5% skim milk, and incubated at 37°C for 1 hour. Cells were then washed again with PBS-0.1T and incubated with a 1:2000 dilution of an HRP-conjugated anti-rabbit IgG antibody (Cytiva) for 1 hour, at 37°C. After additional washes with PBS-0.1T, TMB substrate (Cell Signaling Technology) was added. The reaction was stopped by addition of 0.16M sulfuric acid after 10 minutes and absorbance was read at 450nm. Inhibition was calculated as a percentage of the average absorbance of 5 biological replicates from virus-antibody mixture relative to no antibody control.
Secreted cytokines
H-2Kd-restricted MHC class I binding and immunogenicity predictions were performed using syfpeithi. de, NeTMHC3.4, and Immune Epitope Database (IEDB) Analysis Resource (Kim et al., 2012; Lundegaard et al., 2008; Nielsen et al., 2003; Peters and Sette, 2005; Rammensee et al., 1999; Sidney et al., 2008). The selected peptides (FFGAIAGFL (P1) and YYSTAASSL (P2)), where P1 is absent from rAd-HA2ΔN30F and P2 is absent from rAd-HA2ΔTMF, were synthesized with over 95% purity (New England Peptides). Four million spleen cells were then stimulated with 10 ug/ml of each of the selected peptides. Following a 48-hour incubation, secreted cytokines levels in the supernatant were measured using a ProcartaPlex Multiplex Immunoassay kit (Life Technologies). The plates were read on a Luminex 200 system (MilliporeSigma). Data analysis was performed using MILLIPLEX Analyst version 5.1 software for determining pg/ml of each cytokine.
Flow cytometry
Axillary lymph nodes were collected from immunized mice 21 days after receiving a second immunization and single cell suspensions were prepared. Cell suspensions were washed with FACS wash buffer (PBS with 1% BSA and 0.05% sodium azide) and were first stained with Fixable Viability Dye eFluor 506 (eBioscience) for 15 minutes, at 4°C. Following another wash with FACS wash buffer, cells were incubated with purified anti-mouse CD16/CD32 (eBioscience) as a Fc block, for 5 minutes. Lymph node cells were then stained for 30 minutes, at 4 °C, with AF647-conjugated anti-mouse Ly77/GL7 (clone GL7) and PE-Cy7-conjugated anti-mouse CD45R/B220 (clone RA3-6B2) (BD Biosciences). After washing the cells with FACS wash buffer, cells were resuspended with PBS containing 1% formaldehyde and 0.02% sodium azide. Single-stained/FMO controls and compensation beads were used where appropriate to correct for spectral overlap. Using a BD LSRF Fortessa flow cytometer, 50,000 singlet events were recorded. Data analysis was performed using BD FACSDIVA 6.2 software.
Antibody-dependent cellular cytotoxicity (ADCC)
The ADCC function of the serum antibodies was measured with the Promega ADCC Reporter Bioassay, according to the manufacturer’s instructions. Briefly, 50,000 MDCK cells were seeded in a white clear bottom 96-well plate and grown overnight. Cells were infected with influenza B viruses at a MOI of 5 for 20-24 hours. The following day, the serum samples were heat-inactivated for 30 minutes, at 56°C, serially diluted, and added to the infected cells. Mouse FcγRIV effector cells (Promega) were then added to each well (100,000 cells/well) and the plate was incubated at 37°C/5% CO2 for 5 hours. Bio-GloTM luciferase assay substrate (Promega) was then added and luminescence values were read in relative luminescence units (RLU) after 5 minutes. ADCC activity is expressed as fold induction, relative to a ‘no antibody’ control.
Quantification and statistical analysis
Statistical analysis was conducted using Mann-Whitney test, one-way ANOVA or two-way ANOVA when appropriate. Tukey’s post-test was used to adjust for multiple comparisons between different test groups. Tests were done at 5% significance level. All statistical analyses were performed using GraphPad Prism 8 software.
Acknowledgments
We thank Dr. Roger Tam and Dr. Neda Nasheri for commenting on the manuscript, Marsha Russell and Bozena Jaentschke for technical assistance, Dr. Martha Navarro and her team at Health Canada's Animal Care Facility for the animal studies, and Dr. Don Caldwell for pathological examination of the lung tissues. This work is supported by the Government of Canada (Intramural funding from Health Canada).
Author contributions
AZ, GVD, AH, AF, HL, SS, MRM and XL led the conceptualization of the project and acquisition of initial funding. CG, AM, AD, AZ, AP, WZ, LT, DS, JC, and GVD were involved in conducting experiments and acquisition of data. CG, AM, AD, WZ, DS, JC, FK, LW, SS, MRM, GVD, and XL analyzed data and wrote the manuscript.
Declaration of interests
AZ, GVD and XL are listed as inventors in an application for provisional patents filed by the Government of Canada.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103328.
Contributor Information
Gary Van Domselaar, Email: gary.van-domselaar@canada.ca.
Xuguang Li, Email: sean.li@canada.ca.
Supplemental information
Data and code availability
-
•
All data have been deposited in the Government of Canada (Health Canada) data deposition site and will be available to any interested party upon request.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. The interested party should contact the lead contact Xuguang Li (XL) (sean.li@canada.ca).
References
- Ambrose C.S., Levin M.J. The rationale for quadrivalent influenza vaccines. Hum. Vaccin. Immunother. 2012;8:81–88. doi: 10.4161/hv.8.1.17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babon J.A.B., Cruz J., Ennis F.A., Yin L., Terajima M. A human CD4+ T cell epitope in the influenza hemagglutinin is cross-reactive to influenza A virus subtypes and to influenza B virus. J. Virol. 2012;86:9233–9243. doi: 10.1128/JVI.06325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M.F., Jennings G.T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- Berton M.T., Naeve C.W., Webster R.G. Antigenic structure of the influenza B virus hemagglutinin: nucleotide sequence analysis of antigenic variants selected with monoclonal antibodies. J. Virol. 1984;52:919–927. doi: 10.1128/jvi.52.3.919-927.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., Liang X., Ingallinella P., Finotto M., Chastain M.A., Fan J., Fu T.-M., Song H.C., Horton M.S., Freed D.C., et al. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J. Virol. 2005;79:7380–7388. doi: 10.1128/JVI.79.12.7380-7388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Holmes E.C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008;66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-J., Ermler M.E., Tan G.S., Krammer F., Palese P., Hai R. Influenza A viruses expressing intra- or intergroup chimeric hemagglutinins. J. Virol. 2016;90:3789–3793. doi: 10.1128/JVI.03060-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C.Y., Wang S.M., Lin C.C., Wang H.C., Wang J.R., Su I.J., Liu C.C. Clinical features of children infected with different strains of influenza B in southern taiwan. Pediatr. Infect. Dis. J. 2008;27:640–645. doi: 10.1097/INF.0b013e31816be008. [DOI] [PubMed] [Google Scholar]
- Chun S., Li C., Van Domselaar G., Wang J., Farnsworth A., Cui X., Rode H., Cyr T.D., He R., Li X. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine. 2008;26:6068–6076. doi: 10.1016/j.vaccine.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Collins A.M. IgG subclass co-expression brings harmony to the quartet model of murine IgG function. Immunol. Cell Biol. 2016;94:949–954. doi: 10.1038/icb.2016.65. [DOI] [PubMed] [Google Scholar]
- Counoupas C., Ferrell K.C., Ashhurst A., Bhattacharyya N.D., Nagalingam G., Stewart E.L., Feng C.G., Petrovsky N., Britton W.J., Triccas J.A. Mucosal delivery of a multistage subunit vaccine promotes development of lung-resident memory T cells and affords interleukin-17-dependent protection against pulmonary tuberculosis. Npj Vaccin. 2020;5:105. doi: 10.1038/s41541-020-00255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal S., Loube J., Misplon J.A., Lo C.-Y., Creisher P.S., Mulka K.R., Deshpande S., Mitzner W., Klein S.L., Epstein S.L. Effect of an adenovirus-vectored universal influenza virus vaccine on pulmonary pathophysiology in a mouse model. J. Virol. 2021;95 doi: 10.1128/JVI.02359-20. e02359–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V., Jongeneelen M., et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson S., Blanton L., Kniss K., Mustaquim D., Steffens C., Wallis T., Dhara R., Leon M., Perez A., Chaves S.S., et al. Influenza activity - United States, 2013-14 season and composition of the 2014-15 influenza vaccines. MMWR. Morb. Mortal. Wkly. Rep. 2014;63:483–490. [PMC free article] [PubMed] [Google Scholar]
- Ermler M.E., Kirkpatrick E., Sun W., Hai R., Amanat F., Chromikova V., Palese P., Krammer F. Chimeric hemagglutinin constructs induce broad protection against influenza B virus challenge in the mouse model. J. Virol. 2017;91 doi: 10.1128/JVI.00286-17. e00286–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . European Centre for Disease Prevention and Control, An agency of the European Union; 2018. Influenza in Europe, Summary of the Season 2017–18. [Google Scholar]
- Fan X., Hashem A.M., Chen Z., Li C., Doyle T., Zhang Y., Yi Y., Farnsworth A., Xu K., Li Z., et al. Targeting the HA2 subunit of influenza A virus hemagglutinin via CD40L provides universal protection against diverse subtypes. Mucosal Immunol. 2015;8:211–220. doi: 10.1038/mi.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flórido M., Muflihah H., Lin L.C.W., Xia Y., Sierro F., Palendira M., Feng C.G., Bertolino P., Stambas J., Triccas J.A., et al. Pulmonary immunization with a recombinant influenza A virus vaccine induces lung-resident CD4 + memory T cells that are associated with protection against tuberculosis. Mucosal Immunol. 2018;11:1743–1752. doi: 10.1038/s41385-018-0065-9. [DOI] [PubMed] [Google Scholar]
- Gaglani M., Vasudevan A., Raiyani C., Murthy K., Chen W., Reis M., Belongia E.A., McLean H.Q., Jackson M.L., Jackson L.A., et al. Effectiveness of trivalent and quadrivalent inactivated vaccines against influenza B in the United States, 2011-2012 to 2016-2017. Clin. Infect. Dis. 2021;72:1147–1157. doi: 10.1093/cid/ciaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten R., Blanton L., Elal A.I.A., Alabi N., Barnes J., Biggerstaff M., Brammer L., Budd A.P., Burns E., Cummings C.N., et al. Update: influenza activity in the United States during the 2017–18 season and composition of the 2018–19 influenza vaccine. MMWR. Morb. Mortal. Wkly. Rep. 2018;67:634–642. doi: 10.15585/mmwr.mm6722a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai R., Krammer F., Tan G.S., Pica N., Eggink D., Maamary J., Margine I., Albrecht R.A., Palese P. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J. Virol. 2012;86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem A.M., Van Domselaar G., Li C., Wang J., She Y.M., Cyr T.D., Sui J., He R., Marasco W.A., Li Xuguang X. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem. Biophys. Res. Commun. 2010;403:247–251. doi: 10.1016/j.bbrc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- Hashem A.M., Gravel C., Chen Z., Yi Y., Tocchi M., Jaentschke B., Fan X., Li C., Rosu-Myles M., Pereboev A., et al. CD40 ligand preferentially modulates immune response and enhances protection against influenza virus. J. Immunol. 2014;193:722–734. doi: 10.4049/jimmunol.1300093. [DOI] [PubMed] [Google Scholar]
- Heikkinen T., Ikonen N., Ziegler T. Impact of influenza B lineage-level mismatch between trivalent seasonal influenza vaccines and circulating viruses, 1999-2012. Clin. Infect. Dis. 2014;59:1519–1524. doi: 10.1093/cid/ciu664. [DOI] [PubMed] [Google Scholar]
- Hovanec D.L., Air G.M. Antigenic structure of the hemagglutinin of influenza virus B/Hong Kong/8/73 as determined from gene sequence analysis of variants selected with monoclonal antibodies. Virology. 1984;139:384–392. doi: 10.1016/0042-6822(84)90384-2. [DOI] [PubMed] [Google Scholar]
- Hu W., Gruner W.E., DeMarcus L.S., Thervil J.W., Kwaah B., Fries A.C., Sjoberg P.A., Robbins A.S. Influenza surveillance trends and influenza vaccine effectiveness among department of defense beneficiaries during the 2019-2020 influenza season. Med. Surveill. Mon. Rep. 2021;28:2–8. [PubMed] [Google Scholar]
- Huang F.F., Barnes P.F., Feng Y., Donis R., Chroneos Z.C., Idell S., Allen T., Perez D.R., Whitsett J.A., Dunussi-Joannopoulos K., et al. GM-CSF in the lung protects against lethal influenza infection. Am. J. Respir. Crit. Care Med. 2011;184:259–268. doi: 10.1164/rccm.201012-2036OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A., Milder F., Kuipers H., Wagner M.V., Zhu X., Hoffman R.M.B., Van Meersbergen R., Huizingh J., Wanningen P., Verspuij J., et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader S.A., Gaffen S.L., Kolls J.K. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Loving C.L., Manischewitz J., King L.R., Gauger P.C., Henningson J., Vincent A.L., Golding H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013;5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E., et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel B.F., Burke D.F., Bestebroer T.M., Van Der Vliet S., Zondag G.C.M., Vervaet G., Skepner E., Lewis N.S., Spronken M.I.J., Russell C.A., et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Kordyukova L.V., Serebryakova M.V., Polyansky A.A., Kropotkina E.A., Alexeevski A.V., Veit M., Efremov R.G., Filippova I.Y., Baratova L.A. Linker and/or transmembrane regions of influenza A/Group-1, A/Group-2, and type B virus hemagglutinins are packed differently within trimers. Biochim. Biophys. Acta Biomembr. 2011;1808:1843–1854. doi: 10.1016/j.bbamem.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Krammer F., Palese P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- Krammer F., Pica N., Hai R., Margine I., Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013;87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Margine I., Hai R., Flood A., Hirsh A., Tsvetnitsky V., Chen D., Palese P. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J. Virol. 2014;88:2340–2343. doi: 10.1128/JVI.03183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.P., Sridhar R., Banurekha V.V., Jawahar M.S., Fay M.P., Nutman T.B., Babu S. Type 2 diabetes mellitus coincident with pulmonary tuberculosis is associated with heightened systemic type 1, type 17, and other proinflammatory cytokines. Ann. Am. Thorac. Soc. 2013;10:441–449. doi: 10.1513/AnnalsATS.201305-112OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen N.S., Friesen R.H.E., Zhu X., Jongeneelen M., Blokland S., Vermond J., Van Eijgen A., Tang C., Van Diepen H., Obmolova G., et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362:598–602. doi: 10.1126/science.aaq0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littauer E.Q., Mills L.K., Brock N., Esser E.S., Romanyuk A., Pulit-Penaloza J.A., Vassilieva E.V., Beaver J.T., Antao O., Krammer F., et al. Stable incorporation of GM-CSF into dissolvable microneedle patch improves skin vaccination against influenza. J. Control Release. 2018;276:1–16. doi: 10.1016/j.jconrel.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Welsh J.P., Swartz J.R. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc. Natl. Acad. Sci. U S A. 2014;111:125–130. doi: 10.1073/pnas.1308701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof A., Yorgensen Y.M., Li Y., Evans J.T. Intranasal vaccination promotes detrimental Th17-mediated immunity against influenza infection. PLoS Pathog. 2014;10:e1003875. doi: 10.1371/journal.ppat.1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J.A., Saito T., Iverson A.R. Multiple genotypes of influenza B virus circulated between 1979 and 2003. J. Virol. 2004;78:12817–12828. doi: 10.1128/JVI.78.23.12817-12828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan A., Gravel C., Duran A., Larocque L., Li C., Zetner A., Van Domselaar G., Wang L., Li X. Identification of immunodominant CD8 epitope in the stalk domain of influenza B viral hemagglutinin. Biochem. Biophys. Res. Commun. 2018;502:226–231. doi: 10.1016/j.bbrc.2018.05.148. [DOI] [PubMed] [Google Scholar]
- Murphy-Schafer A.R., Paust S. Divergent mast cell responses modulate antiviral immunity during influenza virus infection. Front. Cell. Infect. Microbiol. 2021;11:580679. doi: 10.3389/fcimb.2021.580679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K. Unique action of Interleukin-18 on T cells and other immune cells. Front. Immunol. 2018;9:763. doi: 10.3389/fimmu.2018.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni F., Kondrashkina E., Wang Q. Structural basis for the divergent evolution of influenza B virus hemagglutinin. Virology. 2013;446:112–122. doi: 10.1016/j.virol.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Lux A., Albert H., Woigk M., Lehmann C., Dudziak D., Smith P., Ravetch J.V. FcYγRIV deletion reveals its central role for IgG2a and IgG2b activity in vivo. Proc. Natl. Acad. Sci. U S A. 2010;107:19396–19401. doi: 10.1073/pnas.1014515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H., Tsutsul H., Komatsu T., Yutsudo M., Tanimoto T., Torigoe K., Okura T., Nukada Y., Hattori K., Akita K., et al. Cloning of a new cytokine that induces IFN-γ production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinforma. 2005;61:1–9. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H.-G., Bachmann J., Emmerich N.P.N., Bachor O.A., Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenet. 1999;503:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Reed C., Meltzer M.I., Finelli L., Fiore A. Public health impact of including two lineages of influenza B in a quadrivalent seasonal influenza vaccine. Vaccine. 2012;30:1993–1998. doi: 10.1016/j.vaccine.2011.12.098. [DOI] [PubMed] [Google Scholar]
- Renne R., Brix A., Harkema J., Herbert R., Kittel B., Lewis D., March T., Nagano K., Pino M., Rittinghausen S., et al. Proliferative and Nonproliferative lesions of the Rat and mouse respiratory tract. Toxicol. Pathol. 2009;37:5S–73S. doi: 10.1177/0192623309353423. [DOI] [PubMed] [Google Scholar]
- Rivera K., Thomas H., Zhang H., Bossart-Whitaker P., Wei X., Air G.M. Probing the structure of influenza B hemagglutinin using site-directed mutagenesis. Virology. 1995;206:787–795. doi: 10.1006/viro.1995.1001. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Wallis T.R., Harmon M.W., Rota J.S., Kendal A.P., Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- Shaw M.W., Xu X., Li Y., Normand S., Ueki R.T., Kunimoto G.Y., Hall H., Klimov A., Cox N.J., Subbarao K. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology. 2002;303:1–8. doi: 10.1006/viro.2002.1719. [DOI] [PubMed] [Google Scholar]
- Sidney J., Assarsson E., Moore C., Ngo S., Pinilla C., Sette A., Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;41:1–14. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Stebegg M., Kumar S.D., Silva-Cayetano A., Fonseca V.R., Linterman M.A., Graca L. Regulation of the germinal center response. Front. Immunol. 2018;9:2469. doi: 10.3389/fimmu.2018.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Chaves S.S., Perez A., D’Mello T., Kirley P.D., Yousey-Hindes K., Farley M.M., Harris M., Sharangpani R., Lynfield R., et al. Comparing clinical characteristics between hospitalized adults with laboratory-confirmed influenza A and B virus infection. Clin. Infect. Dis. 2014;59:252–255. doi: 10.1093/cid/ciu269. [DOI] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M., et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Kirkpatrick E., Ermler M., Nachbagauer R., Broecker F., Krammer F., Palese P. Development of influenza B universal vaccine candidates using the “mosaic” hemagglutinin approach. J. Virol. 2019;93::e00333–19. doi: 10.1128/JVI.00333-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda G., Antanasijevic A., Fallas J.A., Sheffler W., Copps J., Ellis D., Hutchinson G.B., Moyer A., Yasmeen A., Tsybovsky Y., et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife. 2020;9:1–30. doi: 10.7554/eLife.57659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk R.K., Jayakumar J., Mendenhall I.H., Moorthy M., Lam P., Linster M., Lim J., Lin C., Oon L.L.E., Lee H.K., et al. Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity. Proc. Natl. Acad. Sci. U S A. 2020;117:619–628. doi: 10.1073/pnas.1916585116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.T., Tan G.S., Hai R., Pica N., Ngai L., Ekiert D.C., Wilson I.A., García-Sastre A., Moran T.M., Palese P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U S A. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2011. WHO Global Influenza Surveillance Network: Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. [Google Scholar]
- Xie H., Xiang R., Wan H.J., Plant E.P., Radvak P., Kosikova M., Li X., Zoueva O., Ye Z., Wan X.F. Reduced influenza B-specific postvaccination antibody cross-reactivity in the B/Victoria lineage-predominant 2019/20 season. Clin. Infect. Dis. 2021;72:E776–E783. doi: 10.1093/cid/ciaa1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon M.C. Epidemiology and pathogenesis of influenza. J. Antimicrob. Chemother. 1999;44:3–9. doi: 10.1093/jac/44.suppl_2.3. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wei Y., Liu K., Huang M., Li R., Wang Y., Liu Q., Zheng J., Xue C., Cao Y. Recombinant influenza H9N2 virus with a substitution of H3 hemagglutinin transmembrane domain showed enhanced immunogenicity in mice and chicken. Sci. Rep. 2017;7:17923. doi: 10.1038/s41598-017-18054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data have been deposited in the Government of Canada (Health Canada) data deposition site and will be available to any interested party upon request.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. The interested party should contact the lead contact Xuguang Li (XL) (sean.li@canada.ca).