Abstract
Cellular immunotherapeutic approaches such as chimeric antigen receptor (CAR) T-cell therapy in chronic lymphocytic leukemia (CLL) thus far have not met the high expectations. Therefore it is essential to better understand the molecular mechanisms of CLLinduced T-cell dysfunction. Even though a significant number of studies are available on T-cell function and dysfunction in CLL patients, none examine dysfunction at the epigenomic level. In non-malignant T-cell research, epigenomics is widely employed to define the differentiation pathway into T-cell exhaustion. Additionally, metabolic restrictions in the tumor microenvironment that cause T-cell dysfunction are often mediated by epigenetic changes. With this review paper we argue that understanding the epigenetic (dys)regulation in T cells of CLL patients should be leveled to the knowledge we currently have of the neoplastic B cells themselves. This will permit a complete understanding of how these immune cell interactions regulate T- and B-cell function. Here we relate the cellular and phenotypic characteristics of CLL-induced T-cell dysfunction to epigenetic studies of T-cell regulation emerging from chronic viral infection and tumor models. This paper proposes a framework for future studies into the epigenetic regulation of CLL-induced Tcell dysfunction, knowledge that will help to guide improvements in the utility of autologous T-cell based therapies in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is a B-cell malignancy characterized by the accumulation of CD5+ B cells in the bone marrow, spleen, lymph nodes (LN) and peripheral blood (PB).1 The disease course is heterogeneous; some patients require early treatment for progressive disease whilst others never require clinical intervention. Mutational status of the variable region of the immunoglobulin heavy chain (IGHV) gene reflects the stage of B-cell maturation from which the CLL cells originate and is a powerful prognostic and predictive biomarker.2,3 These immunogenetic studies are part of a great effort to identify the cell-of-origin of CLL subsets and thereby trace the differentiation path from normal to neoplastic B cells.4 A range of therapies are currently available for CLL. Whilst a proportion of patients can be successfully treated with chemo-immunotherapy and many achieve meaningful remissions during treatment with Ibrutinib and Venetoclax, patients eventually develop resistance.5,6
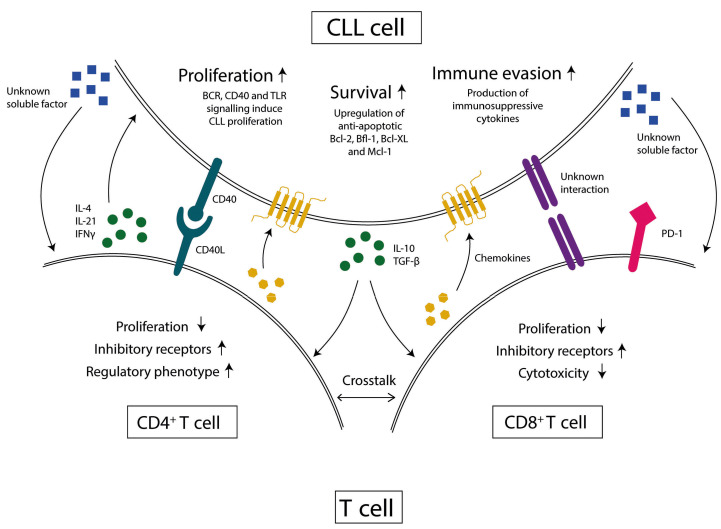
CLL cells actively migrate to the LN to receive survival signals and interact with other cell types, including stromal cells, monocyte-derived macrophages and T cells,7 that provide soluble factors and cell-cell interactions. Within this tumor microenvironment (TME), CD40/CD40L interactions between CLL cells and Thelper cells (Th) induce tumor cell proliferation and upregulation of anti-apoptotic proteins (Figure 1).8 The T-cell derived cytokines interleukin-4 (IL-4), IL-21 and interferon-gamma (IFNγ), also promote CLL cell proliferation and survival.9–11 In addition, CLL cells enforce immunosuppression by the production of IL-10. As described below, T cells from CLL patients develop a dysfunctional phenotype.
Figure 1.
The chronic lymphocytic leukemia microenvironment. In the tumor microenvironment (TME) of chronic lymphocytic leukemia (CLL) patients, T cells and B cells interact with each other. The CLL cells provide soluble factors and cell-cell interactions to suppress T-cell function and the T cells provide soluble factors and cell-cell interactions to sustain CLL cells. BCR: B-cell receptor, TLR: Toll-like receptor, Bcl-2: B-cell lymphoma-2, Bfl-1: Bcl2 related protein A1, Bcl-XL: B-cell lymphomaextra large, Mcl-1: induced myeloid leukemia cell differentiation protein, IL-4: interleukin-4, IL-21: interleukin-21, IFNγ: interferon-gamma, IL-10: interleukin-10, TBG- β: transforming growth factor-beta, PD-1: programmed death protein-1
Over the last decade, the mobilization of host T-cell responses to combat their cancer pathogenesis has revolutionized therapy. Strategies to reinforce an autologous immune response based on cellular cytotoxicity include immune checkpoint blockade (ICB), such as the programmed cell death protein-1 (PD-1) targeting agents, chimeric antigen receptor (CAR) T-cell therapeutic approaches and bi-specific antibody-derived constructs.12 These strategies direct T cells to the malignant cells and have been effective in a variety of neoplasms including hematological malignancies.13,14 Despite high expectations in CLL, results of clinical trials have been disappointing. For example, effective PD-1 blockade was only observed for patients with Richter’s transformation;15 demonstrating that targeting the PD-1/PDL-1 axis is not sufficient to rescue T-cell dysfunction in CLL patients and that other mechanisms are involved. It has been suggested that there is involvement of a novel metabolic immune checkpoint, IL4I1, in resistance to ICB, as IL4I1 expressed on CLL cells suppresses the adaptive immune responses in a murine CLL model.16 CAR T-cell therapies have been developed against the CD19 antigen, expressed by CLL cells as well as healthy B cells.1 CD19-CAR T cells have induced durable response albeit in a small fraction of CLL patients.14,17 Since CAR T cells are generated from autologous T cells, it is likely that CLL-induced T-cell dysfunction may contribute to suboptimal results in patients. The generation of CAR T cells provides a window-of-opportunity to optimize these cells, either by specific culture conditions or additional genetic editing. In order for this to succeed, it is essential to understand the underlying mechanisms of dysfunction.
Epigenetic mechanisms regulate cell differentiation and function by modulating gene expression patterns (Table 1). A disrupted and unstable epigenome is often described in cancer18 and has received widespread attention in virtually all types of malignancies, including CLL,19,20 and technological advances allow epigenomic profiling on a genome-wide scale from low cell numbers. In addition, bioinformatic tools facilitate easier integration across different omics platforms. In CLL, this has led to studies of DNA methylation, histone modifications and chromatin accessibility for the purpose of understanding clinical behavior and cellular origin.19,21 Similarly, in non-malignant T-cell research these same technologies have been harnessed to study cellular differentiation.22 Advances in immunotherapy are accompanied by an increasing need to understand the epigenome of the dysfunctional and exhausted T cell,23 a research area that is unexplored in CLL T cells.24,25
The aim of this review is to examine the literature on epigenetic mechanisms of T-cell function and dysfunction, thereby posing a number of critical questions on the future research direction for the epigenetic regulation of CLL-induced T-cell dysfunction. The epigenetic mechanisms discussed here are DNA methylation, histone modification and chromatin structure; transcriptional regulation by non-coding RNA is outside the scope of this paper. We outline the features of CLL-induced T-cell dysfunction, the molecular mechanisms of functional and dysfunctional T cells and the epigenetic regulation of T-cell differentiation. Finally, we argue that the analysis of malignant B cells needs to be complemented with similar approaches in T cells of CLL patients in the future to fully understand how these immune cells affect each other in CLL. With this we intend to provide a framework for future studies into CLL-induced T-cell dysfunction.
Chronic lymphocytic leukemia-induced T-cell dysfunction
In a healthy T-cell response, presentation of antigen combined with the appropriate cytokine costimulation induces an expansion of effector T cells, followed by a contraction phase under antigen clearance. However, in the case of prolonged exposure to antigen, such as in chronic viral infection and cancer, T cells upregulate inhibitory receptors and gradually become dysfunctional. 26,27 Due to interactions with CLL cells, T cells acquire immune dysfunction that worsens during disease development. 28,29 It is tempting to suggest that T-cell dysfunction contributes to an increased risk for infection and a diminished capacity to control CLL progression12,30 although formal proof is currently lacking.
CD4+ T cells have been reported to have at least two tumor-promoting roles in CLL.31,32 Firstly, Th cells provide survival signals to CLL cells in the LN through contactdependent interaction, e.g., CD40/CD40L, and by secreting soluble factors such as IL-4, IFNγ and IL-21 (Figure 1). These signals promote both the upregulation of anti-apoptotic proteins such as B-cell lymphoma-2 (BCL-2) family members8,33 and tumor proliferation.9 Secondly, regulatory T cells (Tregs) mediate immune tolerance and these cells are found in higher frequency in CLL patients, leading to a dampened immune response.28,34Whether there is Th1 or Th2 skewing in CLL is still under debate as studies have resulted in contradictory findings.31,35,36
Cytotoxic T cells in CLL patients show increased expression of inhibitory receptors. In addition, functional defects such as reduced synapse formation, proliferative capacity and cytotoxicity are reported.37 These features are indicative of T-cell dysfunction, though cytokine production appears to be intact or even enhanced.24
Both CD4+ and CD8+ T cells from CLL patients display an altered subset distribution, with a reduced proportion of naive T cells and skewing towards differentiated effector cells.38,39 Whilst this may partially explain the reduced immune response with autologous T-cell-based therapies in CLL,12,25 it remains unknown whether this is the result of prolonged exposure to CLL antigens as the prevalence of CLL antigen-specific T cells is unknown.
Several studies have demonstrated higher expression inhibitory receptors such as PD-1, CD160 and CD244 on CD4+ and CD8+ cells on CLL T cells.24,36,40,41 Persistent PD-1 expression is a hallmark characteristic of T-cell exhaustion in both tumor and chronic infection models42 and is often co-expressed with other inhibitory markers such as T-cellimmunoglobulin and mucin-domain containing-3 (TIM-3) and cytotoxic T-lymphocyte-associated protein-4 (CTLA4) on CD8+ T cells, and CTLA4 and T-cell immunoreceptor with Ig and ITIM domains (TIGIT) on CD4+ cells.32,36 All of these surface proteins exert an inhibitory effect on T cells when engaged with their specific ligand, and as such represent an opportunity for ICB therapy.
The presence of inhibitory markers indicates diminished T-cell function which is substantiated by direct functional studies. Research shows that CLL T cells have impaired synapse formation.35,43 Proliferation is also affected as Riches et al.24 showed a reduced proliferative index of CD8+ T cells after a αCD3/αCD28 stimulation when comparing CLL patients to age-matched healthy donors. Additionally, even though these CD8+ T cells retained the capacity to degranulate in a killing assay, killing of target cells was defective in CLL T cells.24 In contrast, others described reduced degranulation in CLL CD8+ T cells compared to healthy donors.25,37 These contradictory findings could reflect a difference in experimental design or highlight different stages of dysfunction between patients.
CLL cells and Tregs produce the anti-inflammatory cytokines IL-10 and transforming growth factor-b (TGF-b) and thereby create an immunosuppressive environment.44 Since Tregs are present at higher frequency in CLL, it is likely that these cytokines play a role in the T-cell dysfunction. Additionally, Jadidi-Niaragh et al.45 demonstrated that the expansion of Tregs coincided with a decrease in IL-17 producing cells during CLL progression. IFNγ, TNF and granzyme B production was not reduced in CLL CD8+ T cells compared to age-matched healthy donors.46,47 Riches et al.24 even reported enhanced expression of IFNγ and TNF. These findings seem to reflect a heightened proinflammatory state of CD8+ T cells in CLL .
The majority of aforementioned studies describing CLLinduced T-cell dysfunction were derived from the analysis of PB rather than the LN, that more accurately reflects the physiological CLL TME. T cells from the PB and LN compartments exhibit phenotypic differences, with LNderived T cells exhibiting signatures that reflect recent TME interactions. The dysfunctional T-cell phenotype is more severe in the LN,10,28 suggesting the analysis of PBderived T cells is compromised. In addition, bulk analyzes lead to a mixed phenotype. Analyzing paired LN and PB samples with single-cell approaches such as high-dimensional flow cytometry or single-cell transcriptional and epigenetic sequencing technologies would reveal mechanisms of dysfunction that are not detectable in bulk PB samples.
In conclusion, T-cell abnormalities in CLL patients include altered cytokine secretion profiles, an exhausted phenotype and compromised cytotoxicity which might be more prominent in the CLL TME. In-depth characterization of both LN and PB compartments will hopefully enhance our understanding of the interactions between CLL cells and T cells that induce T-cell dysfunction.
Transcriptional control of T-cell function and dysfunction
Cancer-induced T-cell exhaustion and dysfunction is mainly caused by the interactions with malignant cells within the TME,26 through ongoing engagement of inhibitory receptors with their ligands, excretion of immunosuppressive cytokines by the tumor cells and persistent antigen exposure to the T cells.48 Together this results in reduced effector function and a dysfunctional transcriptional program, the latter of which is driven by differential transcription factor activation and dictates cell fate and function.49,50 Research into T-cell exhaustion and dysfunction, which is almost exclusively focused on CD8+ T cells,26,51 led to identification of transcription-factor dynamics that determine these aberrant T-cell phenotypes.
Development of an exhausted and dysfunctional state allows T cells to stay alive, possibly retaining some residual function, although incapable of effectively eradicating the tumor or pathogen.42 Since studies have shown that early cancer-induced T-cell dysfunction is still a plastic differentiation state,27 reversing or preventing the dysfunctional state is the ‘holy grail’ for immunotherapy. Although, transcriptional dynamics of CLL-induced T-cell dysfunction have rarely been studied so far, we argue that several of these mechanisms can be extrapolated to CLL T-cells.
T-cell factor-1 (TCF-1) is a fate-defining transcription factor,52 important for self-renewal and therefore crucial in naive and memory T-cell subsets. It functions as a transcription factor and histone deacetylase,53 meaning that it can both activate and suppress gene expression. Chen et al.52 showed that early in CD8+ T-cell differentiation after chronic infection, TCF-1 initiates development of precursor exhausted T cells by antagonizing effector T-cell differentiation. Loss of TCF-1 in stem-cell like precursor exhausted cells is irreversible in vivo and these cells differentiate into terminally exhausted and dysfunctional cells. In human cancer studies, TCF-1 expression on tumor-infiltrating lymphocytes (TIL) is positively correlated to ICB responses,54,55 as TCF-1+ T cells tend to be more persistent than those that have lost TCF-1 expression; a key concept relevant for CAR T-cell production.
T-cell exhaustion is accompanied by global chromatin remodeling, details of which are discussed in the section Epigenetics as regulator of T-cell function. Until recently, the molecular cause of chromatin remodeling was unclear, however, several groups have shown an essential role for the transcription factor thymocyte selection-associated high mobility group box protein (TOX) in terminal T-cell exhaustion.56–60 As is often the case with transcription factors, TOX may play a more versatile and species-specific role than initially thought.61 A recent study showed that TOX+CD8+ T cells exist in the circulation of most humans,62 especially effector memory cells tend to express TOX. Still, human studies also confirmed the involvement of TOX in T-cell exhaustion, as HIV- and HCV-specific CD8+ T cells show high levels of TOX58,62 as well as human TIL.56 Dynamic assessment of TOX expression in T cells during CLL progression or the production of CAR T-cells could indicate the level of exhaustion in these cells.
The tight relationship between two T-box transcription factors, T-bet and Eomes, has been linked to specific checkpoints in T-cell differentiation.63,64 T-bet is well-known for its role in CD4+-Th1 differentiation by establishing IFNγ expression. In CD8+ T cells, T-bet is similarly involved in effector functions. In addition, T-bet regulates PDCD1, the gene encoding PD-1, expression through an exhaustionspecific enhancer.65 However, T-bet expression is not sustained in terminally exhausted T cells; upon loss of TCF-1 expression, a transition from T-bet to Eomes expression takes place in the final stages of differentiation.52,66
Eomes is known for its role in CD8+ T-cell differentiation and effector function,50 and is involved in early memory formation in CD8+ cells.67 For these reasons, Eomes is an essential transcription factor for anti-tumor immunity.68 In exhausted cells, Eomes often co-expresses with inhibitory receptors such as PD-1 and TIM-3,69 and is elevated in TIL.68,70 Loss of Eomes also leads to increased TCF-1 expression and reduced expression of TOX,68 suggesting this could be a target to improve stemness of T cells and prevent terminal exhaustion.
Whilst expression patterns of TCF-1 and TOX are largely unexplored in T cells from CLL patients, we can speculate on their role. The T-cell compartment of CLL patients displays a skewing towards differentiated effector cells and this is likely reflected by a reduction in TCF-1 expression because of its prominent role in self-renewal of naive and memory cells.53,71 A clue to whether TCF-1 plays a role in CLL T cells comes from the TCL1 mouse model; a widely accepted model for human CLL including the Tcell dysfunction.69,72,73 Targeting SLAMF6, a surrogate marker for TCF-1, resulted in a reduced leukemic burden and a reduction in exhausted T cells.74 The role of TCF-1 in self-renewal is an important factor for CAR T-cell development as persistence and stemness are beneficial to outcome. 75 Pre-selection for TCF-1+ cells prior to CAR T-cell infusion, or expansion conditions that induce TCF-1 expression, could be a strategy to improve outcome.
Currently, no studies have found differential TOX expression in CLL T cells neither in patient samples nor in the TCL-1 model.35,69,76 It is likely that CLL T cells have not reached the terminally exhausted state that is associated with high, TOX expression,58 which is also evident by their residual cytokine production. Especially loss of IFNγ expression is known to occur very late in development of exhaustion.42 Possibly, a small subset of CLL T cells are terminally exhausted but cannot be picked up by bulk RNA sequencing methods. High-dimensional flow cytometry and single-cell sequencing technologies on both LN and PB compartments could provide new insights into this matter
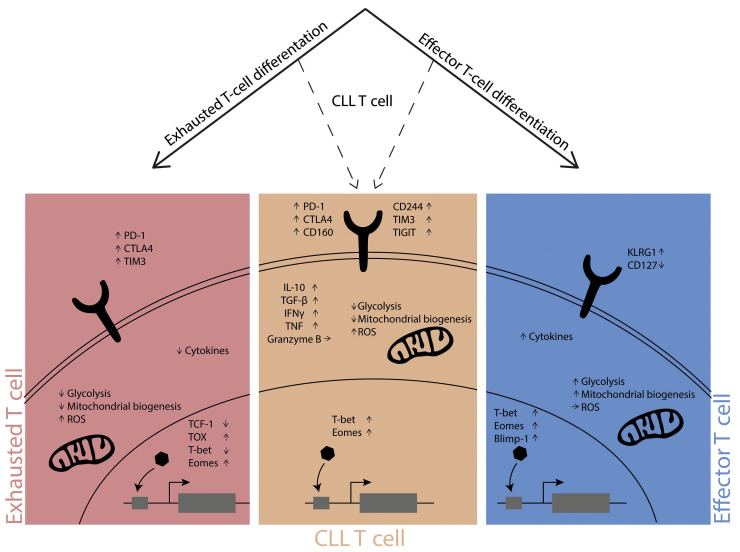
High expression of T-bet has been reported in CD8+ T cells of CLL patients, which coincided with an elevated IFNγ expression.24 Since T-bet expression decreases upon differentiation into terminal T-cell exhaustion, there is likely effector skewing rather than true terminal T-cell exhaustion in CLL (Figure 2). With regards to T-bet expression in CD4+ T cells, Roessner et al.31 found increased levels in CLL patients compared to healthy controls. The accumulation of T-bet expressing CD4+ T cells was also confirmed in the TCL1 mouse model, however, reducing their number had no impact on CLL development. Thus, the mechanistic role of T-bet in this model is not clear yet. Eomes has been studied in the TCL1 mouse model as well. Knock-out of Eomes in CD8+ and CD4+ T cells resulted in impaired tumor control, concluding that this transcription factor seems essential for adaptive immunity against CLL cells.77 However, this needs to be confirmed in human CLL studies.
Overall, whilst T-bet and Eomes expression patterns have been studied in CLL T cells, the patterns of TCF-1 and TOX expression are relatively unknown in CLL T cells, especially in CD4+ T cells. Since T-cell function arises from a dynamic interplay between transcription factors, it is clear that, rather than an analysis of a single transcription factor, the study of CLL T cells requires an integrated network analysis of genome-wide data. Integrating the level of transcription factor expression with the set of genes they control, could help uncover mechanisms that lead to T-cell dysfunction in CLL. Interestingly, recent studies demonstrated recovery of T-cell function upon treatment with Venetoclax and Ibrutinib,28,78 indicating a plasticity of T-cell phenotypes by therapeutic eradication of the tumor cells. Whether this recovery is due to selective survival of functional T cells or true reversibility of the dysfunctional phenotype, due to return to normal homeostasis, is currently unknown. We believe analyzing epigenetic profiles of these T cells could provide answers since epigenetic regulation can represent an interface between a changing environment and T-cell functionality.
Figure 2.
T-cell differentiation. The differentiation pathway of exhausted T cells (left panel) is distinct from that of effector T cells (right panel), and these cells show clear phenotypic and functional differences. The differentiation pathway of dysfunctional CLL T cells (middle panel) is currently unknown and these cells show features of exhausted as well as effector T cells. The dotted lines represent the possibility of CLL T cells to be on the path towards exhaustion or effector cell. The phenotypic characteristics depicted here are cell-surface markers, metabolic features and transcription factor profiles. PD-1: programmed death protein-1, CTLA4: cytotoxic T-lymphocyte-associated protein-4, TIM3: T-cell immunoglobulin and mucin-domain containing-3, TIGIT: T cell immunoreceptor with Ig and ITIM domains, KLRG1: killer cell lectin-like receptor subfamily G member-1, ROS: reactive oxygen species, IL-10: interleukin-10, TBG-b: transforming growth factor-beta, IFNγ: interferongamma, TNF: tumor necrosis factor, TCF-1: T cell factor-1, TOX: thymocyte selection-associated high mobility group box protein, T-bet: T-box expressed in T cells, Eomes: eomesodermin, Blimp-1: B-lymphocyte-induced maturation protein-1.
Epigenetics as regulator of T-cell function
Tight regulation of epigenetic mechanisms is essential for immune cell differentiation (Table 1)22,79 and the function of transcription factors depends on the epigenetic landscape in which they operate. Instructed by intra- or extracellular cues, cells alter their epigenetic profile and differentiate, a process that is hierarchical; cells generally do not de-differentiate in vivo. Naive T cells are characterized by high methylation and closed chromatin of effector genes and, upon activation, an accessible and gene activating epigenetic profile is established.79 Promoters and enhancers often retain their permissive chromatin state, allowing for rapid re-activation of effector function upon re-encounter with antigen.79 This epigenetic memory helps to understand the cell-of-origin and lineage history of dysfunctional T cells found in cancer.
The CD4+ T cells in the CLL TME produce a set of cytokines that implicates their cellular identity and epigenomic profile. The production of IFNγ, a typical Th1 cytokine, is strongly regulated by histone acetylation and DNA methylation. T-bet establishes these activating histone marks on the IFNG locus63 and Th1 cells are characterized by a demethylated IFNG promoter, allowing expression. Normally, Th1 cells retain highly methylated promoters of other lineage-specific cytokine genes, for example IL4 and IL17, preventing expression of these genes.80 This epigenetic programming allows differentiation into a specific lineage and concomitant exclusion of the opposite fate.
IL-4 and IL-21 in the CLL TME are likely produced by Tfollicular- helper (Tfh) cells, a distinct subset of CD4+ cells that localize to the LN and expresses PD-1, CXCR5 and CD40 on its cell surface. Tfh cells have overlapping functions with other Th cells. The chromatin of its key transcription factor BCL-6, shows to be accessible in multiple Th subsets,81 likely accounting for plasticity between Tfh and other Th cells.82 However, Tfh cells also display unique epigenetic features such as enhancer activity on a specific conserved noncoding sequences (CNS) of the IL4 locus, which distinguishes them from Th2 cells.
Tregs, which contribute to immunosuppression in CLL, highly express forkhead box P3 (FOXP3) and are characterized by a demethylated region within the FOXP3 gene.79 The core epigenetic program of thymic-derived Tregs is relatively stable but non-Treg cells can also acquire a Treg phenotype after leaving the thymus, known as inducible Tregs (iTregs). Conversion of Treg to Th-17 and vice versa can occur due to inflammatory signals or metabolic alterations of the cellular microenvironment. 82 Thymic-derived Tregs can be distinguished from iTregs by the epigenetic program on specific CNS within the FOXP3 gene.
Currently, the CAR T-cell production process for CD4+ and CD8+ T cells is identical but as described above, these cells can have opposing roles in CLL. Monitoring the CAR T-cell phenotype before and after infusion is highly relevant. Tfh cells, that support CLL cell proliferation in the LN, and Tregs, that provide immune suppression, are particularly detrimental and profiling epigenetic marks such as the ones described above can help to understand the behavior and plasticity of CAR T cells upon infusion and interaction with their target cells.
As we have discussed previously, CD8+ T cells of CLL patients show signs of exhaustion especially by the presence of inhibitory receptors. Exhausted CD8+ T cells are recognized as a separate lineage within CD8+ differentiation, 51 and although similarities exist to effector and memory cells, many studies have shown extensive epigenetic remodeling in exhausted T-cells.65,66 The key characteristic of T-cell exhaustion, persistent PD-1 expression, is tightly regulated by DNA methylation and chromatin structure. 83,84 Within these PD-1 expressing cells we can define several intermediate states with different expression patterns and functional capacities. Beltra et al.66 describe four subsets within exhausted CD8+ T cells and show that robust chromatin remodeling occurred during differentiation into terminal exhaustion, enforcing different levels of functionality in the subsets. Due to the ability of TOX to bind and recruit a diverse set of chromatin remodeling complexes,57 this transcription factor plays a crucial role in establishing the chromatin landscape of exhausted T cells. Since LN and PB compartments are accessible in the TCL1 murine model, it would be interesting to apply Ly108 and CD69 phenotyping and investigate the exhausted T-cell subsets proposed by Beltra et al.66 in this model of CLL.
Progressive loss of T-cell function during differentiation towards terminal exhaustion has implications for immunotherapy. Pauken et al.23 showed that upon PD-1 blockade therapy, T cells regained some effector function but retained their exhausted epigenetic profile. For this reason, researchers are exploring combinations of epigenetic drugs with ICB to reverse the exhausted epigenetic program and achieve a more potent and long-lasting immune response against the tumor cells;85 a strategy that might have potential for CLL treatment especially since ICB has not been successful in CLL patients yet.
Metabolism has recently received widespread attention in studying anti-tumor immunity, and chromatin-modifying enzymes are highly dependent on metabolites and metabolic cofactors to manage the epigenetic program within a cell.86Metabolic alterations in the TME can therefore hamper an adequate immune response through epigenetic mechanisms.87 When T cells encounter their cognate antigen, they switch from oxidative phosphorylation (OXPHOS) to a more glycolysis-based metabolism. A key player in the switch to glycolysis is Sirtuin 6 (SIRT6)88 which belongs to a subclass of histone deacetylases (HDAC) that depend on NAD+ for removal of an acetylgroup. 86 It functions as a gene repressor together with hypoxia-inducible factor 1-α (HIF1α), a transcription factor that activates glycolytic genes such as glucose transporter- 1 and hexokinase-2;89 genes that CLL T cells fail to upregulate after activation.25,90
In addition to diminished glucose metabolism of T cells in the TME, mitochondrial function and fitness of tumorinfiltrating and exhausted T cells is also impaired.91,92 Scharping et al.91 showed that loss of mitochondrial function is due to a defect in mitochondrial biogenesis, which is regulated by peroxisome proliferator-activated receptor- γ co-activator-1α (PGC1α). PGC1α is a transcriptional coactivator and it recruits both histone acetylases and the SWI/SNF chromatin remodeling complex to the genome to activate gene expression.93 Expression of PGC1α itself was shown to be reduced by PD-1 signaling in a chronic infection model, which could explain the relationship between T-cell exhaustion and reduced mitochondrial biogenesis.94 Altogether, PGC1α presents a potential target to improve metabolic function and anti-tumor immunity of T cells.95
Diminished oxygen availability (hypoxia) in the TME arises from an imbalance between increased oxygen consumption and inadequate oxygen supply, and leads to elevated levels of reactive oxygen species (ROS). A recent study showed that the phosphatase PAC1 (also known as DUSP2) mediated ROS-induced CD8+ T-cell exhaustion and reduced anti-tumor immunity. PAC1 recruits the Mi- 2b nucleosome-remodeling-deacetylase complex, an ATPdependent chromatin remodeling complex, to the chromatin of T-cell effector genes. This results in a global reduction in chromatin accessibility and consequently decreased expression of these genes, establishing the dysfunctional T-cell state.96
Even though metabolic rewiring is a hallmark of cancer, CLL does not necessarily follow the same paradigm. Due to circulation in and out of the LN and contact with oxygenated blood, the CLL TME might not be nutrient-deprived or hypoxic, although studies on this are limited. Research shows heightened mitochondrial respiration and increased levels of ROS within CLL cells while glycolysis and the production of lactate is not enhanced.97 Immunosuppression in CLL is most prominent in the LN28 and CD8+ T-cell dysfunction in CLL seems to be instructed in part by metabolic dysregulation. 25,90 However, it is not clear if the metabolic dysregulation is initiated in the TME and whether it mediates dysfunction through epigenetic mechanisms.
The required metabolic switch from OXPHOS to glycolysis is impaired in CLL T cells.90 The HDAC SIRT6 regulates glucose homeostasis as co-repressor with HIF1α. Even though it is unknown whether SIRT6 is increased in CLL T-cells, it might explain why higher levels of HIF1α levels do not coincide with increased glycolysis in CD8+ T cells from CLL patients.25 These cells also show reduced expression of PGC1α and impaired mitochondrial biogenesis upon T-cell activation.25 Whether PGC1α plays a causal role in the observed dysfunction or is a consequence of increased PD-1 expression remains unclear, however overexpression of PGC1α could be a strategy to improve the metabolic fitness of CAR T cells.91
Besides inducing elevated levels of ROS, hypoxia also mediates immunosuppression through induction of CD39 and CD73, two ectonucleotidases that metabolize ATP to adenosine. In the CLL TME, high adenosine levels are detected and CLL cells express CD39 and CD73.98 Serra et al.99 showed an anti-apoptotic effect of adenosine on CLL cells, whilst the effect of adenosine on T cells was 2-fold, firstly it reduced IFNγ production and proliferation of effector CD8+ T cells and it then induced proliferation of Tregs.100 The adenosine signaling axis has not been studied yet in T cells of CLL patients, and we hypothesize that it could play an important role. This is strengthened by a study in the TCL1 mouse model. Targeting the A2A extracellular adenosine receptor led to restoration of immune competence towards the CLL cells by increased degranulation and cytokine release, and decreased Treg expansion.101 These data indicate the potential of targeting the adenosinergic axis for immunotherapeutic approaches in CLL.
Taken together, the epigenetic profiles of CD4+ and CD8+ T cells can demonstrate the differentiation they have undergone which will be helpful in determining the level of exhaustion and potential reversibility of the dysfunctional phenotype of CLL T cells. In addition, metabolic alterations in the CLL TME likely cause T-cell aberrations through epigenetic mechanisms.
Future considerations
There are several outstanding questions in the field of CLL-induced T-cell dysfunction (Table 2) and given the rapid development of technologies available to study the epigenome, the possibilities seem ample. Study design will be critical to permit accurate interpretation and integration with large multi-omics datasets. It is essential to choose the correct ‘normal’ counterpart for comparison; in early CLL studies biological inference was occasionally predicated on the selection of an inaccurate control.102 In general, T cells from CLL patients are compared to agematched healthy controls but a more detailed characterization of normal T-cell states for accurate comparisons is required. Perhaps machine learning approaches could assist in making these decisions.103
Table 1.
Epigenetic mechanisms.
Table 2.
Outstanding questions.
Studying single-cell epigenomic and transcriptomic profiles have gained attention in research on CLL cells and non-malignant T cells, however no such studies are available on CLL T cells yet. In a similar approach like determining the cell-of-origin of CLL,4 lineage tracing might address whether a hierarchical pathway of differentiation towards dysfunction exists in CLL T cells. If such a hierarchical pathway exists, it could assist in disease monitoring of patients. Currently, treatment decisions are made based on CLL characteristics but possibly the development of Tcell dysfunction over time provides clues on disease progression and the need for treatment in the future.
Single-cell approaches also allow increased resolution for individual populations of cells with a distinct functional or dysfunctional phenotype. These single-cell maps are becoming available for many different T-cell states in a healthy and malignant setting104 and comparison to CLL T cells might allow better understanding of the states of these cells. An unanswered question in the field is to what extent CLL T cells are truly exhausted, because cytokine expression seems intact. This residual function could indicate a progenitor exhaustive state that is likely more reversible than a terminally exhaustive state. A combined analysis of key exhaustion-related features such as expression of inhibitory receptors, transcription factors such as TOX and TCF-1, and chromatin remodeling, could help answering this question.
High-dimensional flow or mass cytometry is also an excellent means to study single-cell phenotypes. Hartmann et al.105 described a method to characterize the metabolic regulome of CD8+ T cells with mass cytometry and their method also allowed integration with imaging to study spatial organization of different metabolic programs in the tissue. Although it is not easy to obtain human CLL LN, this technology would permit detailed characterization of the metabolic program within the CLL TME and potentially answer questions on the role of T cells in that environment. It would also allow simultaneous study of the CLL B cells and the T cells within the same patient. We believe that is essential for determining how these cellular interactions regulate T- and B-cell function in CLL.
As mentioned in the introduction, understanding the molecular mechanisms of CLL-induced T-cell dysfunction is essential to improve CAR T-cell therapy, and the in vitro generation of CAR T cells provides a window-of-opportunity to optimize these cells. Additional genetic editing of targets such as TCF-1, PGC1α and PAC1 might improve the stemness and memory phenotype, mitochondrial fitness and the response to hypoxia. In addition, the differential roles of CD4+ and CD8+ T cells in CLL might call for a different approach for CAR generation of the two subsets. Although both subsets display features of exhaustion, for CD4+ cells the prevention of Treg expansion is important whilst for the CD8+ the cytotoxic capacity needs to be improved. Current approaches do not distinguish between CD4+ and CD8+ cells during the expansion phase and the CAR construct is the same, it might be worthwhile to explore a subset-specific approach in CLL.
Concluding remarks
In summary, we have described the features of T-cell dysfunction and can conclude that CD4+ and CD8+ T cells have both overlapping as well as distinct roles in CLL. Both show features of exhaustion and whilst CD4+ T cells possibly play a tumor-supportive role in the TME, CD8+ T cells have reduced cytotoxic capacity. Transcriptional control of T cells is executed by several key transcription factors that are known for their role in effector T-cell function but, depending on the context, could also steer cells into dysfunction and exhaustion. Epigenetic marks and their regulators play a major role in determining the context in which transcription factors operate and it is likely that T-cell dysfunction caused by metabolic perturbations in the TME is epigenetically mediated. The dynamics and plasticity of these transcriptional profiles of dysfunctional T cells are still an active topic of debate in healthy as well as CLL T cells. For CLL, plasticity of T-cell function and phenotype is important for immunotherapeutic approaches such as CAR T-cell therapy. Although in vitro generation of CAR T cells can be optimized, the signals they receive once transferred back into the patients and entering the CLL TME are important to take into account. Therefore, integrating molecular studies of CLL cells and T cells is necessary to unravel factors leading to T-cell dysfunction, an understanding that can improve re-invigoration of T cells by autologous T-cell-based immunotherapy.
Supplementary Material
References
- 1.Kipps TJ, Stevenson FK, Wu CJ, et al. Chronic lymphocytic leukaemia. Nat Rev Dis Primers. 2017;3(1):1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840-1847. [PubMed] [Google Scholar]
- 3.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848-1854. [PubMed] [Google Scholar]
- 4.Gaiti F, Chaligne R, Gu H, et al. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature. 2019;569 (7757):576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kater AP, Wu JQ, Kipps T, et al. Venetoclax plus rituximab in relapsed chronic lymphocytic leukemia: 4-year results and evaluation of impact of genomic complexity and gene mutations from the MURANO Phase III Study. J Clin Oncol. 2020;38(34):4042-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burger JA. Treatment of chronic lymphocytic leukemia. N Engl J Med. 2020;383(5):460-473. [DOI] [PubMed] [Google Scholar]
- 7.Attekum MH van, Eldering E, Kater AP. Chronic lymphocytic leukemia cells are active participants in microenvironmental cross-talk. Haematologica. 2017;102(9): 1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haselager MV, Kielbassa K, Ter Burg J, et al. Changes in Bcl-2 members in response to ibrutinib or venetoclax uncover functional hierarchy in determining resistance to venetoclax in CLL. Blood. 2020;136(25):2918-2926. [DOI] [PubMed] [Google Scholar]
- 9.Pascutti MF, M J, Jm T, et al. IL-21 and CD40L signals from autologous T cells can induce antigen-independent proliferation of CLL cells. Blood. 2013;122(17):3010-3019. [DOI] [PubMed] [Google Scholar]
- 10.Hanna BS, Roessner PM, Yazdanparast H, et al. Control of chronic lymphocytic leukemia development by clonally-expanded CD8 + T-cells that undergo functional exhaustion in secondary lymphoid tissues. Leukemia. 2019;33(3):625-637. [DOI] [PubMed] [Google Scholar]
- 11.Haselager MV, Kater AP, Eldering E. Proliferative signals in chronic lymphocytic leukemia; What are we missing? Front Oncol. 2020;10:592205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofland T, Eldering E, Kater AP, Tonino SH. Engaging cytotoxic T and NK cells for immunotherapy in chronic lymphocytic leukemia. Int J Mol Sci. 2019;20(17):4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viardot A, Hess G, Bargou RC, et al. Durability of complete response after blinatumomab therapy for relapsed/refractory diffuse large B-cell lymphoma. Leuk Lymphoma. 2020;61(11):2767-2770. [DOI] [PubMed] [Google Scholar]
- 14.Cao J-X, Gao W-J, You J, Wu L-H, Liu J-L, Wang Z-X. The efficacy of anti-CD19 chimeric antigen receptor T cells for B-cell malignancies. Cytotherapy. 2019;21(7):769-781. [DOI] [PubMed] [Google Scholar]
- 15.Ding W, LaPlant BR, Call TG, et al. Pembrolizumab in patients with CLL and Richter transformation or with relapsed CLL. Blood. 2017;129(26):3419-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadik A, Patterson LFS, Öztürk S, et al. IL4I1 Is a metabolic immune checkpoint that activates the AHR and promotes tumor progression. Cell. 2020;182(5):1252-1270.e34. [DOI] [PubMed] [Google Scholar]
- 17.Kochenderfer JN, Dudley ME, Feldman SA, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119(12):2709-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14(Suppl 1):R65-R76. [DOI] [PubMed] [Google Scholar]
- 19.Xanthopoulos C, Kostareli E. Advances in epigenetics and epigenomics in chronic lymphocyticlLeukemia. Curr Genet Med Rep. 2019;7(4):214-226. [Google Scholar]
- 20.Guièze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126(4):445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulis M, Merkel A, Heath S, et al. Wholegenome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47(7):746-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suarez-Alvarez B, Rodriguez RM, Fraga MF, López-Larrea C. DNA methylation: a promising landscape for immune system-related diseases. Trends Genet. 2012;28(10):506-514. [DOI] [PubMed] [Google Scholar]
- 23.Pauken KE, Sammons MA, Odorizzi PM, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riches JC, Davies JK, McClanahan F, et al. T cells from CLL patients exhibit features of Tcell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Bruggen JAC, Martens AWJ, Fraietta JA, et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8+ T cells and impede CAR T-cell efficacy. Blood. 2019;134(1):44-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell. 2018;33(4): 547-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schietinger A, Philip M, Krisnawan VE, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45(2):389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Weerdt I, Hofland T, de Boer R, et al. Distinct immune composition in lymph node and peripheral blood of CLL patients is reshaped during venetoclax treatment. Blood Adv. 2019;3(17):2642-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riches JC, Gribben JG. Understanding the immunodeficiency in chronic lymphocytic leukemia: potential clinical implications. Hematol Oncol Clin North Am. 2013;27(2):207-235. [DOI] [PubMed] [Google Scholar]
- 30.Man S, Henley P. Chronic lymphocytic leukaemia: the role of T cells in a B cell disease. Br J Haematol. 2019;186(2):220-233. [DOI] [PubMed] [Google Scholar]
- 31.Roessner PM, Hanna BS, Öztürk S, et al. TBET-expressing Th1 CD4+ T cells accumulate in chronic lymphocytic leukaemia without affecting disease progression in Eμ- TCL1 mice. Br J Haematol. 2020;189(1):133-145. [DOI] [PubMed] [Google Scholar]
- 32.Catakovic K, Gassner FJ, Ratswohl C, et al. TIGIT expressing CD4+T cells represent a tumor-supportive T cell subset in chronic lymphocytic leukemia. OncoImmunology. 2018;7(1):e1371399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tromp JM, Tonino SH, Elias JA, et al. Dichotomy in NF-κB signaling and chemoresistance in immunoglobulin variable heavy-chain-mutated versus unmutated CLL cells upon CD40/TLR9 triggering. Oncogene. 2010;29(36):5071-5082. [DOI] [PubMed] [Google Scholar]
- 34.Jak M, Mous R, Remmerswaal EBM, et al. Enhanced formation and survival of CD4+ CD25hi Foxp3+ T-cells in chronic lymphocytic leukemia. Leuk Lymphoma. 2009;50(5):788-801. [DOI] [PubMed] [Google Scholar]
- 35.Görgün G, Holderried TAW, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115(7):1797-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palma M, Gentilcore G, Heimersson K, et al. T cells in chronic lymphocytic leukemia display dysregulated expression of immune checkpoints and activation markers. Haematologica. 2017;102(3):562-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taghiloo S, Allahmoradi E, Tehrani M, et al. Frequency and functional characterization of exhausted CD8+ T cells in chronic lymphocytic leukemia. Eur J Haematol. 2017;98(6):622-631. [DOI] [PubMed] [Google Scholar]
- 38.Tinhofer I, Weiss L, Gassner F, Rubenzer G, Holler C, Greil R. Difference in the relative distribution of CD4+ T-cell subsets in B-CLL with mutated and unmutated immunoglobulin (Ig) VH genes: implication for the course of disease. J Immunother. 2009;32(3):302-309. [DOI] [PubMed] [Google Scholar]
- 39.Mackus WJM, Frakking FNJ, Grummels A, et al. Expansion of CMV-specific CD8+CD45RA+CD27- T cells in B-cell chronic lymphocytic leukemia. Blood. 2003;102(3):1057-1063. [DOI] [PubMed] [Google Scholar]
- 40.Brusa D, Serra S, Coscia M, et al. The PD- 1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia. Haematologica. 2013;98(6):953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.te Raa GD, Pascutti MF, García-Vallejo JJ, et al. CMV-specific CD8+ T-cell function is not impaired in chronic lymphocytic leukemia. Blood. 2014;123(5):717-724. [DOI] [PubMed] [Google Scholar]
- 42.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492-499. [DOI] [PubMed] [Google Scholar]
- 43.Ramsay AG, Gribben JG. Immune dysfunction in chronic lymphocytic leukemia T cells and lenalidomide as an immunomodulatory drug. Haematologica. 2009;94(9):1198-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiLillo DJ, Weinberg JB, Yoshizaki A, et al. Chronic lymphocytic leukemia and regulatory B cells share IL-10 competence and immunosuppressive function. Leukemia. 2013;27(1):170-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jadidi-Niaragh F, Ghalamfarsa G, Memarian A, et al. Downregulation of IL-17-producing T cells is associated with regulatory T cell expansion and disease progression in chronic lymphocytic leukemia. Tumour Biol. 2013;34(2):929-940. [DOI] [PubMed] [Google Scholar]
- 46.Lee B-N, Gao H, Cohen EN, et al. Treatment with lenalidomide modulates T-cell immunophenotype and cytokine production in patients with chronic lymphocytic leukemia. Cancer. 2011;117(17):3999-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens AWJ, Janssen SR, Derks IAM, et al. CD3xCD19 DART molecule treatment induces non-apoptotic killing and is efficient against high-risk chemotherapy and venetoclax- resistant chronic lymphocytic leukemia cells. J Immunother Cancer. 2020;8(1): e000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kallies A, Good-Jacobson KL. Transcription factor T-bet orchestrates lineage development and function in the immune system. Trends Immunol. 2017;38(4):287-297. [DOI] [PubMed] [Google Scholar]
- 50.Pearce EL, Mullen AC, Martins GA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302(5647):1041-1043. [DOI] [PubMed] [Google Scholar]
- 51.Blank CU, Haining WN, Held W, et al. Defining “T cell exhaustion.” Nat Rev Immunol. 2019;19(11):665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Ji Z, Ngiow SF, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51(5):840-855.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raghu D, Xue H-H, Mielke LA. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 2019;40(12):1149-1162. [DOI] [PubMed] [Google Scholar]
- 54.Brummelman J, Mazza EMC, Alvisi G, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J Exp Med. 2018;215(10):2520-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175(4):998-1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott AC, Dündar F, Zumbo P, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571(7764):270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan O, Giles JR, McDonald S, et al. TOX transcriptionally and epigenetically programs CD8 + T cell exhaustion. Nature. 2019;571(7764):211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alfei F, Kanev K, Hofmann M, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571(7764):265-269. [DOI] [PubMed] [Google Scholar]
- 59.Yao C, Sun H-W, Lacey NE, et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8 + T cell persistence in chronic infection. Nat Immunol. 2019;20(7):890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seo H, Chen J, González-Avalos E, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc Natl Acad Sci U S A. 2019;116(25):12410-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Utzschneider DT, Kallies A. Human effector T cells express TOX—Not so “TOX”ic after all. Sci Immunol. 2020;5(49):eabc8272. [DOI] [PubMed] [Google Scholar]
- 62.Sekine T, Perez-Potti A, Nguyen S, et al. TOX is expressed by exhausted and polyfunctional human effector memory CD8+ T cells. Sci Immunol. 2020;5(49):eaba7918. [DOI] [PubMed] [Google Scholar]
- 63.Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126(3):299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Istaces N, Splittgerber M, Lima Silva V, et al. EOMES interacts with RUNX3 and BRG1 to promote innate memory cell formation through epigenetic reprogramming. Nat Commun. 2019;10(1):3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sen DR, Kaminski J, Barnitz RA, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beltra J-C, Manne S, Abdel-Hakeem MS, et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity. 2020;52(5):825-841.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kavazović I, Han H, Balzaretti G, et al. Eomes broadens the scope of CD8 T-cell memory by inhibiting apoptosis in cells of low affinity. PLoS Biol. 2020;18(3):e3000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, He Y, Hao J, Ni L, Dong C. High levels of Eomes promote exhaustion of anti-tumor CD8+ T cells. Front Immunol. 2018;9:2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Llaó Cid L, Hanna BS, Iskar M, et al. CD8+ T-cells of CLL-bearing mice acquire a transcriptional program of T-cell activation and exhaustion. Leuk Lymphoma. 2020;61(2): 351-356. [DOI] [PubMed] [Google Scholar]
- 70.Park J, Kwon M, Kim KH, et al. Immune Checkpoint inhibitor-induced reinvigoration of tumor-infiltrating CD8+ T cells is determined by their differentiation status in glioblastoma. Clin Cancer Res. 2019;25(8):2549-2559. [DOI] [PubMed] [Google Scholar]
- 71.Hofland T, Martens AWJ, van Bruggen JAC, et al. Human CXCR5+PD-1+ CD8 T cells in healthy individuals and patients with hematologic malignancies. Eur J Immunol. 2021;51(3):703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bresin A, D’Abundo L, Narducci MG, et al. TCL1 transgenic mouse model as a tool for the study of therapeutic targets and microenvironment in human B-cell chronic lymphocytic leukemia. Cell Death Dis. 2016;7(1):e2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hofbauer JP, Heyder C, Denk U, et al. Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia. 2011;25(9):1452-1458. [DOI] [PubMed] [Google Scholar]
- 74.Yigit B, Wang N, Hacken E ten, et al. SLAMF6 as a regulator of exhausted CD8+ T cells in cancer. Cancer Immunol Res. 2019;7(9):1485-1496. [DOI] [PubMed] [Google Scholar]
- 75.Huang R, Li X, He Y, et al. Recent advances in CAR-T cell engineering. J Hematol Oncol. 2020;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kiaii S, Kokhaei P, Mozaffari F, et al. T cells from indolent CLL patients prevent apoptosis of leukemic B cells in vitro and have altered gene expression profile. Cancer Immunol Immunother. 2013;62(1):51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roessner PM, Cid LL, Lupar E, et al. EOMES and IL-10 regulate anti-tumor activity of PD- 1+ CD4+ T-cells in B-cell non-Hodgkin lymphoma. bioRxiv. 2020;2020.03.09.983098. [Google Scholar]
- 78.Solman IG, Blum LK, Hoh HY, et al. Ibrutinib restores immune cell numbers and function in first-line and relapsed/refractory chronic lymphocytic leukemia. Leuk Res. 2020;97:106432. [DOI] [PubMed] [Google Scholar]
- 79.Schmidl C, Delacher M, Huehn J, Feuerer M. Epigenetic mechanisms regulating T-cell responses. J Allergy Clin Immunol. 2018;142(3):728-743. [DOI] [PubMed] [Google Scholar]
- 80.Janson PCJ, Linton LB, Bergman EA, et al. Profiling of CD4+ T cells with epigenetic immune lineage analysis. J Immunol. 2011;186(1):92-102. [DOI] [PubMed] [Google Scholar]
- 81.Shih H-Y, Sciumè G, Poholek AC, et al. Transcriptional and epigenetic networks that drive helper T cell identities. Immunol Rev. 2014;261(1):23-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Youngblood B, Oestreich KJ, Ha S-J, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen- specific CD8(+) T cells. Immunity. 2011;35(3):400-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boer K, de Wit LEA, Peters FS, et al. Variations in DNA methylation of interferon gamma and programmed death 1 in allograft rejection after kidney transplantation. Clin Epigenetics. 2016;8:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zebley CC, Gottschalk S, Youngblood B. Rewriting history: epigenetic reprogramming of CD8+ T cell differentiation to enhance immunotherapy. Trends Immunol. 2020;41(8):665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haws SA, Leech CM, Denu JM. Metabolism and the epigenome: a dynamic relationship. Trends Biochem Sci. 2020;45(9):731-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. 2020;78(6):1019-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong L, D’Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finlay DK, Rosenzweig E, Sinclair LV, et al. PDK1 regulation of mTOR and hypoxiainducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209(13):2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siska PJ, van der Windt GJW, Kishton RJ, et al. Suppression of Glut1 and glucose metabolism by decreased Akt/mTORC1 signaling drives T cell impairment in B cell leukemia. J Immunol. 2016;197(6):2532-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scharping NE, Menk AV, Moreci RS, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity. 2016;45(2):374-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Siska PJ, Rathmell JC. T cell metabolic fitness in antitumor immunity. Trends Immunol. 2015;36(4):257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krämer AI, Handschin C. How Epigenetic modifications drive the expression and mediate the action of PGC-1α in the regulation of metabolism. Int J Mol Sci. 2019;20(21):5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bengsch B, Johnson AL, Kurachi M, et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity. 2016;45(2):358-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scharping NE, Rivadeneira DB, Menk AV, et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat Immunol. 2021;22(2):205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dan Lu, Liu L, Sun Y, et al. The phosphatase PAC1 acts as a T cell suppressor and attenuates host antitumor immunity. Nat Immunol. 2020;21(3):287-297. [DOI] [PubMed] [Google Scholar]
- 97.Galicia-Vázquez G, Aloyz R. Metabolic rewiring beyond Warburg in chronic lymphocytic leukemia: how much do we actually know? Crit Rev Oncol Hematol. 2019;134:65-70. [DOI] [PubMed] [Google Scholar]
- 98.Serra S, Vaisitti T, Audrito V, et al. Adenosine signaling mediates hypoxic responses in the chronic lymphocytic leukemia microenvironment. Blood Adv. 2016;1(1):47-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Serra S, Horenstein AL, Vaisitti T, et al. CD73-generated extracellular adenosine in chronic lymphocytic leukemia creates local conditions counteracting drug-induced cell death. Blood. 2011;118(23):6141-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leone RD, Powell JD. Metabolism of immune cells in cancer. Nat Rev Cancer. 2020;20(9):516-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arruga F, Serra S, Vitale N, et al. Targeting of the A2A adenosine receptor counteracts immunosuppression in vivo in a mouse model of chronic lymphocytic leukemia. Haematologica. 2021;106(5):1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mansouri L, Wierzbinska JA, Plass C, Rosenquist R. Epigenetic deregulation in chronic lymphocytic leukemia: clinical and biological impact. Semin Cancer Biol. 2018;51:1-11. [DOI] [PubMed] [Google Scholar]
- 103.Jabbari P, Rezaei N. Artificial intelligence and immunotherapy. Exp Rev Clin Immunol. 2019;15(7):689-691. [DOI] [PubMed] [Google Scholar]
- 104.Nieto P, Elosua-Bayes M, Trincado JL, et al. A single-cell tumor immune atlas for precision oncology. bioRxiv.;2020.10.26.354829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hartmann FJ, Mrdjen D, McCaffrey E, et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat Biotechnol. 2021;30(2):186-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soshnev AA, Josefowicz SZ, Allis CD. Greater than the sum of parts: complexity of the dynamic epigenome. Mol Cell. 2016;62(5):681-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17(11):661-678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.