Abstract
Spinal muscular atrophy (SMA) is a severe neuromuscular disease affecting children, due to mutation/deletion of survival motor neuron 1 (SMN1) gene. The lack of functional protein SMN determines motor neuron (MN) degeneration and skeletal muscle atrophy, leading to premature death due to respiratory failure. Nowadays, the Food and Drug Administration approved the administration of three drugs, aiming at increasing the SMN production: although assuring noteworthy results, all these therapies show some non-negligible limitations, making essential the identification of alternative/synergistic therapeutic strategies. To offer a valuable in vitro experimental model for easily performing preliminary screenings of alternative promising treatments, we optimized an organotypic spinal cord culture (derived from murine spinal cord slices), which well recapitulates the pathogenetic features of SMA. Then, to validate the model, we tested the effects of human mesenchymal stem cells (hMSCs) or murine C2C12 cells (a mouse skeletal myoblast cell line) conditioned media: 1/3 of conditioned medium (obtained from either hMSCs or C2C12 cells) was added to the conventional medium of the organotypic culture and maintained for 7 days. Then the slices were fixed and immunoreacted to evaluate the MN survival. In particular we observed that the C2C12 and hMSCs conditioned media positively influenced the MN soma size and the axonal length respectively, without modulating the glial activation. These data suggest that trophic factors released by MSCs or muscular cells can exert beneficial effects, by acting on different targets, and confirm the reliability of the model. Overall, we propose the organotypic spinal cord culture as an excellent tool to preliminarily screen molecules and drugs before moving to in vivo models, in this way partly reducing the use of animals and the costs.
Key words: 3D cell culture, motor neuron disease, drug screening
Introduction
Spinal muscular atrophy (SMA) is a progressive neurodegenerative disorder and one of the main causes of infantile mortality. It is characterized by the degeneration and loss of lower motor neurons (MNs), determining a progressive muscle atrophy, respiratory failure and, in most severe cases, premature death.1 SMA is due to the homozygous deletion or mutation of the Survival Motor Neuron 1 (SMN1) gene, resulting in the lack of the full-length SMN (FL-SMN) protein. Humans also carry an almost identical gene called SMN2 which in the 90% of cases undergoes an alternative splicing, mainly producing a trunked (lacking exon 7, SMNΔ7) and not-functional protein. The SMN2 copy number inversely correlates with the disease severity.2
Since the genetic causes of SMA are well known, the current available treatments (Spinraza, Zolgensma and Evrysdi) consist in SMN-dependent approaches: however, albeit effective, they present several limitations, such as high costs, still unknown long-term effects, uncomfortable administration route (Spinraza) and some side effects;3,4 moreover they disregard SMN-independent targets, although additional cellular and molecular mechanisms have been demonstrated contributing to MN death (in particular autophagy and apoptosis dysregulation)5,6 and muscular atrophy/innervation (neurofilament accumulation, denervation and neuromuscular junction immaturity).7 Therefore, the identification of alternative/synergistic therapeutic strategies is still a hot topic in the SMA field.
Most clinical trials based on experimental results fail to obtain significant effects on human patients, therefore, it is necessary to improve the preclinical model reliability, possibly replacing/reducing animal studies with valid in vitro models. A good compromise between 2D cell cultures and experimental animal models is represented by 3D cultures, as organotypic cell cultures. This method allows easy and repeated manipulation while maintaining intact morphology and local synaptic connections in vitro (reminiscent of the in vivo situation). Indeed, organotypic slice cultures are very useful to study the tissue cytoarchitecture, thanks to long-term survival, the preservation of regional differentiation, neuroanatomic organization and cell-to-cell interaction.8 Moreover, compared to animal models, organotypic cultures require fewer animals, allow real time monitoring and are less expensive.9
In this scenario, to easily perform a preliminary screening of promising treatments,10 we developed a 3D organotypic spinal cord culture (by adapting the most used method, i.e. the ‘membrane technique’ or interface method11), which well recapitulated the pathogenetic features of SMA: in particular, to assess the neurodegeneration, we evaluated the MN soma size and axonal length, two parameters previously reported as altered in SMA.12,13 To verify the reliability of the model, we tested on SMA slices the paracrine effects of human mesenchymal stem cells (hMSCs) or murine C2C12 cells (a mouse skeletal myoblast cell line). The secretome of these cells (especially MSCs) has been extensively studied and proposed as therapeutic tool for many neurodegenerative and traumatic pathologies: indeed, the broad range of secreted factors can exert beneficial effects as inhibiting cell death and oxidative stress, enhancing immunomodulation and angiogenesis, stimulating neurogenesis, supporting regeneration, therefore counteracting many SMN-independent altered pathways in SMA.14-19
This work represents a proof-of-concept study, demonstrating the possibility to use the 3D organotypic spinal cord culture as an effective therapeutic screening tool; moreover, even if the results here presented refer to SMA, this model could be suitable for all the other MN and neuromuscular diseases.
Materials and Methods
Experimental model
The original breeding pairs of SMA mice were purchased from Jackson Laboratory (stock number 005025; Jackson Lab, Bar Harbor, ME, USA). To maintain the colony, carrier mice (SMN2+/+; SMNΔ7+/+; Smn+/-) were interbred and the offspring was genotyped by PCR assays as previously described:20 briefly, animals were genotyped for the presence of the two human transgenes (SMN2 and SMNΔ7) and the three possible genotype variants of the mouse Smn locus (Smn+/+, Smn+/− and Smn−/−). WT (Smn+/+) and SMA (Smn−/−) animals were used to perform organotypic cell cultures.
Animals had free access to food and water. All experimental procedures on live animals were performed according with national (DL n. 116, G.U., Supp. 40, February 18, 1992; permit number 17/2010-B, June 30, 2010) and European Community Council guidelines (Council Directive 86/609/EEC). Additionally, an ad hoc Ethical Committee of the University of Turin specifically approved this study. Particular care was taken to minimize the number of animals, their discomfort and pain.
Organotypic cell cultures
At postnatal day 8 [P8, identified from pilot experiments (here not shown) as the most suitable time point to assure good cell survival and slice morphology], SMA (n=15) and WT animals (n=16) were decapitated and the T13-L5 spinal cord segments were rapidly dissected. After removal, the samples were cut into 350 μmthick sections (about 10 per dissected segment) with a mechanical tissue chopper (McIlwain) and the slices were immediately transferred in ice-cold 6% glucose saline solution. Organotypic cultures were prepared according to the standard membrane interface method.11 The isolated slices were transferred onto a porous translucent membrane (Millicell®-CM:PICM03050, Millipore, Billerica, MA, USA) and placed in a petri dish with 1 mL of medium. i.e., a mixture of 50% MEM (Invitrogen-Gibco, Waltham, MA, USA) with 25 mM HEPES (0.625 ml in 25 mL; Sigma- Aldrich, St. Louis, MO, USA), 25% horse serum, 25% HBSS with glucose 25.76 mg/mL 1% glutamine, 1% penicillin/streptomycin.21 The slices were maintained in the interface between culture medium, in a humidified atmosphere of 95% air with 5% CO2 at 37°C. The medium was changed at day 2 and 5 after seeding, and the culture was maintained for 7 days. During the whole period in culture, the slices were observed and monitored with a Leica 0 550FW inverted phase contrast microscope, equipped with an Euromex digital camera.
To evaluate the efficiency of protocol and exclude artifacts, immediately after the tissue chopper cutting some organotypic slices were fixed with 4% paraformaldehyde (PFA) in 0.1M phosphate buffered saline (PBS) for 4 h at 4°C, washed with PBS and then immunoreacted as described in the following paragraph.
Cell cultures and collection of conditioned media
C2C12 cells (mouse skeletal myoblast cell line) and human mesenchymal stem cells (hMSCs) were prepared and expanded as follows (respectively according to22,23). C2C12 cells from ATCC [ATCC-LGC Standards S.r.l., Sesto San Giovanni (MI), Italy] were cultured in DMEM high glucose (Sigma-Aldrich) with 10% fetal bovine serum (FBS; Sigma-Aldrich), containing 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen-Gibco). Cells were maintained in a humidified atmosphere of 95% air with 5% CO2 at 37°C. In order to allow C2C12 myoblasts to differentiate into myotubes, the serum concentration was reduced to 3% (myotube formation took about 5 days). On day 5 post-differentiation, the surnatant was collected.
Concerning hMSCs, bone marrow was aspirated from the iliac crest of healthy donors after informed consensus in accordance with the Declaration of Helsinki and the City of Health & Science of Turin - Ordine Mauriziano Hospital Ethics Committee. Cells were plated at 8×105 cells/cm2 in T flasks, and cultivated in Eagle’s alpha minimum essential medium (a-MEM; Cambrex Bioscience, East Rutherford, NJ, USA), containing 10% FBS, 2 mM L-glutamine (Invitrogen-Gibco), 100 U⁄mL penicillin and 100 μg⁄mL streptomycin (Invitrogen-Gibco). Cells were maintained in a humidified atmosphere of 95% air with 5% CO2 at 37°C. After 3 days free floating cells were removed and replaced with fresh culture medium. The cells were analyzed for their viability and for their immunophenotype by flow cytometry (data not shown), as reported by Mareschi et al.23 At 4-5th passage, when at about 80% confluence, we collected the surnatant after approximately 24-48 h after refreshing medium.
After collection, both conditioned media were centrifuged at 2000 RPM for 5 min, to remove detached cells and stored at −80°C until further use.
Treatment with conditioned media
At day 7 in vitro (7 DIV) in standard conditions, SMA (n=9) and WT (n=8) organotypic cultures underwent a treatment with conditioned media for additional 7 days (replaced once during the week), in order to study their potential neuroprotective effects and to validate the model. As controls, other organotypic cultures were maintained in standard medium (SMA untreated n=6; WT untreated n=8).
In detail, the WT and SMA organotypic cultures were incubated with 2/3 of conventional medium and 1/3 of conditioned medium obtained by either a culture of C2C12 cells or a culture of hMSCs. Accordingly, the different experimental conditions will be referred as WT hMSCs (n=4), WT C2C12 (n=4), SMA hMSCs (n=4) and SMA C2C12 (n=5), depending on the genotype and the treatment.
Altogether after 14 DIV, both treated and control organotypic SMA and WT slices were fixed with 4% PFA in 0.1M PBS for 4 h at 4°C, and then extensively washed with PBS.
Immunofluorescence reactions
For the immunofluorescence (IF) reactions, SMA (n=23) and WT (n=24) sections were washed three times (5 min each) with PBS. To allow cell permeabilization, the samples were eventually incubated for 20 min with PBS-Triton 0.3% at room temperature (RT) and then washed with PBS. After the incubation for 30 min in a blocking solution containing 10% Normal Donkey Serum (NDS) and PBS-Triton 0.3%, the samples were incubated with the following primary antibodies, at 4°C for 48 h: monoclonal anti-microtubule- associated protein 2 (MAP2) (anti-mouse; 1:200; Chemicon®, Sigma-Aldrich); polyclonal anti-glial fibrillary acidic protein (GFAP) (anti-rabbit; 1:500; Dako Cytomation, Glostrup, Denmark); monoclonal anti-neurofilament SMI32 (anti-mouse, 1:1000; Covance, Princeton, NJ, USA); monoclonal anti-Ki67 (anti-mouse, 1:400; Novocastra Laboratories, Ltd., Newcastle upon Tyne, UK); polyclonal anti-ionized calcium binding adaptor molecule 1 (IBA1) (anti-rabbit, 1:500; Wako). Afterwards the samples were washed in PBS and incubated for 1 at RT with 1:200 anti-rabbit or anti-mouse cyanine 3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), as appropriate. Then the slices were incubated with 4’-6- Diamidino-2-phenylindole (DAPI) 1:50 in PBS, for labelling the cell nuclei. Finally, coverslips were mounted on the slices with a drop of PBS-glycerol 1:1.
Morphometric analyses
The slices were analyzed with a Nikon Eclipse 90i epifluorescence microscope and photographed by a Nikon DS-5Mc digital camera. For checking double staining and making 3D reconstructions, some preparations were examined also with a Leica TCS SP5 confocal laser scanning microscope.
For evaluating the effects of conditioned media, the analysis of cell bodies and axons was performed with the Neurolucida software (extension module AutoNeuron) and the associated data analysis software NeuroExplorer (MicroBrightField): for the morphometric analysis, only SMI32-positive neurons located in the ventral horns and presenting a diameter not smaller than 10 μm were considered alpha MNs (according to6,12) and were drawn and measured (WT untreated n=32; WT hMSCs n=6; WT C2C12 n=15; SMA untreated n=19; SMA hMSCs n=5; SMA C2C12 n=18).
Statistical analysis
Data are shown as mean ± standard error of mean (SEM) and inter-group differences were statistically compared: unpaired t-test two tails was used for morphological analysis (MN soma size and axonal length) between SMA and WT organotypic slices, while one way ANOVA for comparing the treatment effects in WT or SMA conditions. Differences were considered to be significant when p≤0.05. Statistical analysis was performed using GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA, USA).
Results
The organotypic spinal cord culture model
We have optimized a protocol -starting from the Stoppini’s interface method10- able to ensure excellent tissue preservation and cell survival (in terms of well-preserved morphology, structure and dorso-ventral orientation of the slices) during the whole observation period of the organotypic cultures, although a physiological limited degradation of the slices was progressively observed at the edges (starting from 4 DIV) (Figure 1A). During the whole observation period (14 days), the spinal cord slices appeared intact, with the dorsal and ventral horns well recognizable. Pyknotic nuclei (identifying dying cells) were not observed (data not shown).
Immediately after tissue chopper cutting, neither signs of alteration nor astrogliosis (identified by GFAP immunolabeling) were detected (Figure 1B), while it tended to appear during the following days (Figure 1C), both in WT and SMA slices. Moreover, SMI32-positive MNs were easily detectable, in good conditions and correctly placed in the ventral horns both immediately after cutting (Figure 1D) and until 14 DIV in WT animals (Figure 1E).
In absence of any treatments, at 14 DIV we measured the cell body size and the axon length of the MNs by the Neurolucida software (Figure 2): we observed significant differences in the soma area between WT and SMA samples (WT 655.21 μm2 ± 53.33; SMA 476.78 μm2 ± 50.65; unpaired t-test p≤0.05), and a trend in the axonal length reduction (WT 564.08 μm ± 63.43; SMA 381.02 μm ± 56.14; unpaired t-test p>0.05). These analyses confirmed the occurring atrophy and neurodegeneration affecting SMA MNs compared to the WT ones, and demonstrated the reliability of the organotypic culture as a valid model to study SMA hallmarks and to preliminary test the treatment efficacy.
Organotypic culture treatment
After 7 DIV, the standard medium was replaced for WT (n=11) and SMA (n=12) slices with hMSCs or C2C12 cell conditioned medium and maintained for additional 7 days, before PFA-fixation. Control slices were maintained in standard medium for the whole observation period.
We first evaluated the effect of the conditioned media on WT cells. MN soma areas were not influenced by any treatment (WT untreated 655.21 μm2 ± 53.33; WT hMSCs 635.87 μm2 ± 146.32; WT C2C12 652.85 μm2 ± 57.98; one way ANOVA p>0.05). However, the axonal length was slightly increased by the presence of C2C12 conditioned medium (WT untreated 564.08 μm ± 63.43; WT hMSCs 439.22 μm ± 92.89; WT C2C12 660.12 μm ± 112.95; one way ANOVA p>0.05), even if these differences were not significantly different (Figure 3).
Concerning the SMA organotypic cultures, both treatments induced an effect, although influencing different parameters. The administration of C2C12 conditioned medium (Figure 4) significantly affected the MN area (SMA untreated 476.78 μm2 ± 50.65; SMA C2C12 659.58 μm2 ± 46.24; one way ANOVA p≤0.05), but not the axonal length (SMA untreated 381.02 μm ± 56.14; SMA C2C12 359.99 μm ± 38.82; one way ANOVA p>0.05). On the contrary, in presence of hMSCs conditioned medium (Figure 4), the axonal length was significantly increased in SMA treated MNs compared to untreated and C2C12 treated conditions (SMA untreated 381.02 μm ± 56.14; SMA hMSCs 717.38 μm ± 228.05; SMA C2C12 359.99 μm ± 38.82; one way ANOVA p≤0.05); as regards the soma size, it was not affected by the hMSC secretome (SMA untreated 476.78 μm2 ± 50.65; SMA hMSCs 433.67 μm2 ± 110.57; one way ANOVA p>0.05).
Moreover, independently from the genotype and the group, a certain degree of astrocyte activation appeared during the days: however, the treatment with either hMSC or C2C12 conditioned media did not seem to modulate the astrogliosis, although these observations were only qualitative (Figure 5A).
Figure 1.
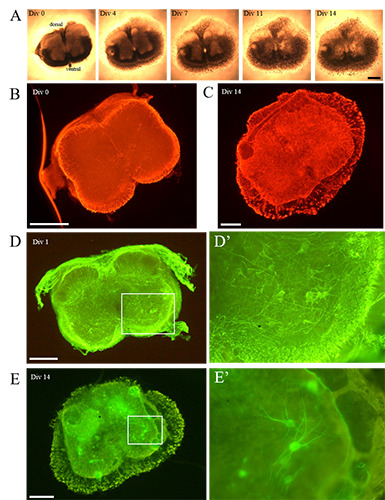
A) The time lapse analysis shows the gradual modifications occurring to a representative organotypic spinal cord SMA slices, during the 14 DIV: although a progressive thinning of the section was evident (from 4 DIV), overall the slice morphology appeared well preserved during the whole period of observation. B) Immediately after the tissue chopper cutting, no signs of astrogliosis (GFAP) were evident (here showed a SMA slice), whereas C) it increased over time (as visible in this representative WT slice). D) Concerning the cell preservation, at 1 DIV the WT MNs (SMI32- positive cells) were in good conditions and showed numerous neurites (well visible in the inset D’). E) At 14 DIV, the WT MN survival was still good: the cells were correctly located in the ventral horns and several extensions were detectable (a detail is appreciable in the inset E’). Scale bar: A-E) 500 μm; D’, E’) 250 μm.
Figure 2.
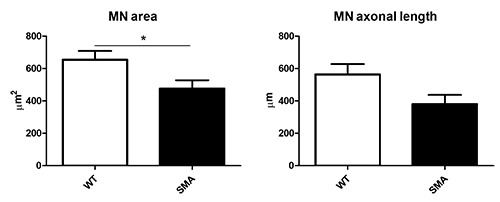
Morphological analysis highlighted the soma atrophy and axonal degeneration occurring in SMA MNs compared to WT MNs (unpaired t-test *p≤0.05).
Figure 3.
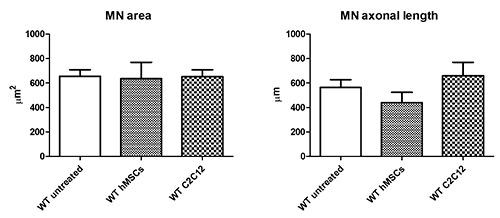
hMSCs and C2C12 conditioned media, administered to WT organotypic slices, did not induce evident modification of the MN soma area. A slight increase of MN axonal length was observed in presence of C2C12 conditioned medium.
Figure 4.
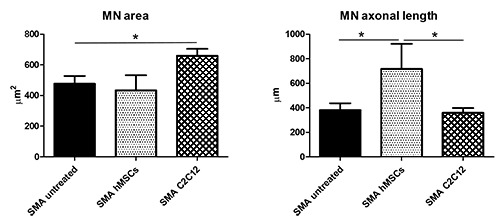
The administration of C2C12 conditioned medium significantly increased the cell body area of SMA MNs, whereas the hMSCs conditioned medium significantly affected the SMA MN axonal length (one way ANOVA *p≤0.05).
Finally, as previously stated, during the 14 DIV of observation a physiological limited degradation of the slices is progressively observed at the edges (involving the white matter), characterized by a progressive centrifugal migration of the cells: indeed, during the days, the slice borders tended to become progressively thinner, and the cells appeared as arranged in a monolayer in proximity to the white matter, seemingly moving away from the slice edges: this progressive flattening has been already reported by other authors.24 In case of C2C12 conditioned treatment, it appeared slightly more pronounced (Figure 5B). Among the involved cells, in particularly we observed GFAP-positive and IBA1-positive cells (Figure 5 C,D). Rare MNs (SMI32-positive) were observed among the migrating cells (data not shown).
In conclusion, these data suggest that the SMA spinal cord organotypic culture is a reliable model and, as a validation, we demonstrated that different treatments can effectively influence different MN morphology-related features.
Discussion
The study aimed to develop a reliable 3D in vitro model of SMA, using organotypic culture technique. This method allowed maintaining the tissue slices alive until 14 DIV after their cutting, preserving quite intact the in vivo cytoarchitecture and highly simplifying the experimental conditions. This is a pivotal approach i) to study pathogenetic mechanisms in a rather complex cellular environment; ii) to preliminary screen and select hit compounds and drugs, using a 3D model (composed by different cell types, interacting each other); iii) to discard ineffective molecules before going to in vivo experiments; and consequently iv) to highly reduce the use of experimental animals and the therapeutic failure of clinical trials.
Figure 5.
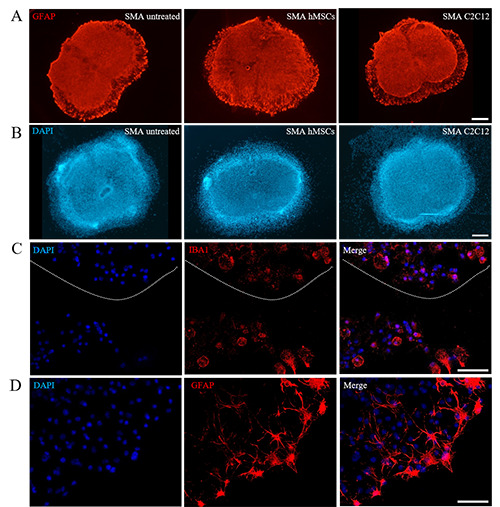
A) Neither treatment was able to modulate astrogliosis, B) but C2C12 conditioned medium administration increased the dispersion of cells from the slice edges (as demonstrated by the numerous DAPI positive nuclei located over the edge of the slice): C) among the migrating cells, we mainly observed microglial (IBA1 positive) and D) astrocytes (GFAP positive). DAPI was used to label the cell nuclei. Scale bar: A,B) 500 μm; C,D) 50 μm.
The protocol employed is based on the Stoppini’s interface method11, but has been optimized for this work. Stoppini’s technique was conceived in particular for brain organotypic cultures. We placed the spinal cord slices at the air-medium interface on millicell semiporous membranes, maintaining them stationary during the entire culturing process: in this way, they received oxygen from above and medium from below.25 With this procedure the cells can potentially survive more than 2 months, maintaining their metabolic and structural characteristics and allowing both electrophysiological and morphological experiments.11 In our experimental design, we decided to stop the cultures after 14 DIV, according to the average lifespan of the employed SMA model.
Organotypic cultures show evident advantages compared to standard in vitro models. Indeed, they assure a complex 3D architecture and maintain local neuronal circuits, although the pathways are largely disconnected and the axotomy can induce neuronal cell death.10,26 However in our optimized protocol we did not detect massive apoptosis. Moreover, the organotypic cultures can also maintain the interactions between neurons and glial cells, allowing to understand their crosstalk (largely involved in the pathogenesis of several neurological diseases27). Finally, compared to other 3D cell cultures (as organoids), organotypic cultures are better standardized, more homogenous and cheaper.28 Moreover, compared to in vivo animal models, organotypic cultures require fewer animals, allow real time monitoring and are less expensive.9
Here, we developed a 3D organotypic spinal cord culture, which well recapitulates the pathogenetic features of SMA and can be used to preliminary screen therapeutic molecules. Indeed, we compared the morphology of SMA and WT MNs, and we observed a significant reduction of cell body area and a remarkable (albeit not significant) axonal length decrease in the SMA samples. These data highlight the reliability of the model, since both the decrease of MN soma size12 and the reduction in axon length13 have been previously described in vivo in SMNΔ7 MNs. Therefore, these features confirm the occurring SMA-related neurodegeneration. However, as concerns the neuroinflammation, at 14 DIV we observed a certain degree of astrogliosis both in WT and SMA slices, suggesting that it was more due to a technical concern than to the disease per se.
As a proof-of-concept study, we tested the effect of paracrine effects of hMSCs and C2C12 conditioned media. As known, the secretome of MSCs consists in several bioactive molecules (including neurotrophins, microRNAs, hormones, cytokines and interleukins) with remarkable neuroprotective, anti-inflammatory, immunoregulatory, angiogenic and trophic effects:29 indeed their positive and supportive effects have been demonstrated in several neurodegenerative and traumatic disease models.15,30,31 On the other hand, the C2C12 cells are known to secrete the so-called “myokines”, molecules able to influence metabolic regulation, inflammatory processes, angiogenesis, and myogenesis.32 Therefore we evaluated their effect on WT and SMA organotypic spinal cord cultures. In the WT we only observed a slight increase (not statistically significant) in the MN axonal length triggered by C2C12 conditioned medium, whereas the soma size did not vary in any conditions. It is likely that for the WT slices (i.e., in healthy conditions) the conditioned media supplement did not exert particular additive effects. Instead, in the SMA samples, we observed a different effect induced by hMSCs and C2C12 cells, which respectively determined an increase in the axonal length and in the cell body area. These effects probably reflect the different mixture of cell type secretome. Indeed, on one side, MSC paracrine activity effectively prevented the axon length reduction due to SMN lack,33 supporting axonal elongation thanks to several factors and miRNAs (as BDNF and miRNA-17-92 cluster) presented in the MSC-exosomes.34-36 On the other side, the neurotrophic factors (as FGF21, IGF1 and TGFb1) produced by C2C1232 and contained in their conditioned medium have been able to counteract the MN soma atrophy, a feature already observed in SMA mice and probably antecedent to the MN cell death.12 Moreover in presence of C2C12 conditioned media, we observed a progressive centrifugal migration of the spinal cord cells, possibly in the attempt to arrange in a monolayer in search to a better access to nutrients present in the C2C12-medium, compared to the multilayer disposition in the slice. Lastly, we cannot exclude that the presence of FBS in the culture medium could partially affect the morphological changes we observed37 as well as its eventual deprivation:38 however, the significant differences between the treated and untreated samples confirm the prevailing and effective action of the conditioned media.
Finally, although it is well known the anti-inflammatory effect exerted by these cells, we did not notice a reduction of astrocyte activation in neither condition: however, as previously stated, a certain degree of astrogliosis is always present at 14 DIV (also in WT slices), therefore the conditioned media could be not sufficient to counteract such phenomenon.
To conclude, it is evident that the 3D organotypic spinal cord model proposed is valid and useful, since in presence of two different treatments we appreciated a number of varied effects. In particular we suggest the use of this model to unravel cell death mechanisms or cell-glia interactions, and to preliminary screen neuroprotective molecules. In addition, it could be also possible to setup a co-culture (with organotypic spinal cord slices and muscular cells), to study the neuron-muscle interplay and the innervation mechanisms (known to be altered in SMA7), and test therapies to support these phenomena.
Funding Statement
Funding: This work was supported by funds from Girotondo/ONLUS and from Smarathon-ONLUS associations.
References
- 1.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 1999;96:6307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995;80:155-65. [DOI] [PubMed] [Google Scholar]
- 3.Chen TH. New and developing therapies in spinal muscular atrophy: From genotype to phenotype to treatment and where do we stand? Int J Mol Sci 2020;21:3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menduti G, Rasà DM, Stanga S, Boido M. Drug screening and drug repositioning as promising therapeutic approaches for spinal muscular atrophy treatment. Front Pharmacol 2020;11:592234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piras A, Schiaffino L, Boido M, Valsecchi V, Guglielmotto M, De Amicis E, et al. Inhibition of autophagy delays motoneuron degeneration and extends lifespan in a mouse model of spinal muscular atrophy. Cell Death Dis 2017;8:3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellino R, Boido M, Borsello T, Vercelli A. Pharmacological c-Jun NH2-terminal kinase (JNK) pathway inhibition reduces severity of spinal muscular atrophy disease in mice. Front Mol Neurosci 2018;11:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boido M, Vercelli A. Neuromuscular junctions as key contributors and therapeutic targets in spinal muscular atrophy. Front Neuroanat 2016;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stavridis SI, Dehghani F, Korf HW, Hailer NP. Characterisation of transverse slice culture preparations of postnatal rat spinal cord: preservation of defined neuronal populations. Histochem Cell Biol 2005;123:377-92. [DOI] [PubMed] [Google Scholar]
- 9.Pineau H, Sim V. POSCAbilities: The application of the prion organotypic slice culture assay to neurodegenerative disease research. Biomolecules 2020;10:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandamooz S, Nabiuni M, Miyan J, Ahmadiani A, Dargahi L. Organotypic spinal cord culture: a proper platform for the functional screening. Mol Neurobiol 2016;53:4659-74. [DOI] [PubMed] [Google Scholar]
- 11.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods 1991;37:173-82. [DOI] [PubMed] [Google Scholar]
- 12.d'Errico P, Boido M, Piras A, Valsecchi V, De Amicis E, Locatelli D, et al. Selective vulnerability of spinal and cortical motor neuron subpopulations in delta7 SMA mice. PLoS One 2013;8:e82654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alrafiah A, Karyka E, Coldicott I, Iremonger K, Lewis KE, Ning K, et al. Plastin 3 promotes motor neuron axonal growth and extends survival in a mouse model of spinal muscular atrophy. Mol Ther Methods Clin Dev 2018;9:81-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henningsen J, Rigbolt KT, Blagoev B, Pedersen BK, Kratchmarova I. Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol Cell Proteomics 2010;9:2482-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boido M, Piras A, Valsecchi V, Spigolon G, Mareschi K, Ferrero I, et al. Human mesenchymal stromal cell transplantation modulates neuroinflammatory milieu in a mouse model of amyotrophic lateral sclerosis. Cytotherapy 2014;16:1059-72. [DOI] [PubMed] [Google Scholar]
- 16.Bonafede R, Mariotti R. ALS pathogenesis and therapeutic approaches: The role of mesenchymal stem cells and extracellular vesicles. Front Cell Neurosci 2017;11:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boido M, Ghibaudi M, Gentile P, Favaro E, Fusaro R, Tonda-Turo C. Chitosan-based hydrogel to support the paracrine activity of mesenchymal stem cells in spinal cord injury treatment. Sci Rep 2019;9:6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cofano F, Boido M, Monticelli M, Zenga F, Ducati A, Vercelli A, et al. Mesenchymal stem cells for spinal cord injury: Current options, limitations, and future of cell therapy. Int J Mol Sci 2019;20:2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohseni R, Hamidieh AA, Shoae-Hassani A, Ghahvechi-Akbari M, Majma A, Mohammadi M, et al. An open-label phase 1 clinical trial of the allogeneic side population adiposederived mesenchymal stem cells in SMA type 1 patients. Neurol Sci 2021. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 20.Boido M, De Amicis E, Valsecchi V, Trevisan M, Ala U, Ruegg MA, et al. Increasing agrin function antagonizes muscle atrophy and motor impairment in spinal muscular atrophy. Front Cell Neurosci 2018;12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corse AM, Rothstein JD. Organotypic spinal cord cultures and a model of chronic glutamate-mediated motor neuron degeneration. In: Ohnishi ST, Ohnishi T, editors. Central nervous system trauma: research techniques. CRC: Boca Raton; 1995. p. 341–51. [Google Scholar]
- 22.Gunetti M, Tomasi S, Giammò A, Boido M, Rustichelli D, Mareschi K, et al. Myogenic potential of whole bone marrow mesenchymal stem cells in vitro and in vivo for usage in urinary incontinence. PLoS One 2012;7:e45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mareschi K, Ferrero I, Rustichelli D, Aschero S, Gammaitoni L, Aglietta M, et al. Expansion of mesenchymal stem cells isolated from pediatric and adult donor bone marrow. J Cell Biochem 2006;97:744-54. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz N, Uysal B, Welzer M, Bahr JC, Layer N, Löffler H, et al. Long-term adult human brain slice cultures as a model system to study human CNS circuitry and disease. Elife 2019;8:e48417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends Neurosci 1997;20:471-7. [DOI] [PubMed] [Google Scholar]
- 26.Humpel C. Organotypic brain slice cultures: A review. Neuroscience 2015;305:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernaus A, Blanco S, Sevilla A. Glia crosstalk in neuroinflammatory diseases. Front Cell Neurosci 2020;14:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol 2020;21:571-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baez-Jurado E, Hidalgo-Lanussa O, Barrera-Bailón B, Sahebkar A, Ashraf GM, Echeverria V, et al. Secretome of mesenchymal stem cells and its potential protective effects on brain pathologies. Mol Neurobiol 2019;56:6902-27. [DOI] [PubMed] [Google Scholar]
- 30.Boido M, Garbossa D, Fontanella M, Ducati A, Vercelli A. Mesenchymal stem cell transplantation reduces glial cyst and improves functional outcome after spinal cord compression. World Neurosurg 2014;81:183-90. [DOI] [PubMed] [Google Scholar]
- 31.Yao P, Zhou L, Zhu L, Zhou B, Yu Q. Mesenchymal stem cells: A potential therapeutic strategy for neurodegenerative diseases. Eur Neurol 2020;83:235-41. [DOI] [PubMed] [Google Scholar]
- 32.Deshmukh AS, Cox J, Jensen LJ, Meissner F, Mann M. Secretome analysis of lipid-induced insulin resistance in skeletal muscle cells by a combined experimental and bioinformatics workflow. J Proteome Res 2015;14:4885-95. [DOI] [PubMed] [Google Scholar]
- 33.Pletto D, Capra S, Finardi A, Colciaghi F, Nobili P, Battaglia GS, et al. Axon outgrowth and neuronal differentiation defects after a-SMN and FL-SMN silencing in primary hippocampal cultures. PLoS One 2018;13:e0199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins LF, Costa RO, Pedro JR, Aguiar P, Serra SC, Teixeira FG, et al. Mesenchymal stem cells secretome-induced axonal outgrowth is mediated by BDNF. Sci Rep 2017;7:4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xin H, Katakowski M, Wang F, Qian JY, Liu XS, Ali MM, et al. MicroRNA cluster miR-17-92 cluster in exosomes enhance neuroplasticity and functional recovery after stroke in rats. Stroke 2017;48:747-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Chopp M, Liu XS, Katakowski M, Wang X, Tian X, et al. Exosomes derived from mesenchymal stromal cells promote axonal growth of cortical neurons. Mol Neurobiol 2017;54:2659-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettger WJ, McKeehan WL. Mechanisms of cellular nutrition. Physiol Rev 1986;66:1-35. [DOI] [PubMed] [Google Scholar]
- 38.Thomson AC, Schuhmann T, de Graaf TA, Sack AT, Rutten BPF, Kenis G. The effects of serum removal on gene expression and morphological plasticity markers in differentiated SH-SY5Y cells. Cell Mol Neurobiol 2021. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]


