Abstract
目的
探讨血管内皮生长因子(VEGF)对三阴性乳腺癌肿瘤干细胞的调控作用并探索其作用机制。
方法
通过Oncomine数据库、UALCAN数据库及Human Protein Atlas数据库分别验证VEGF在乳腺癌中的表达情况,分析VEGF表达与不同分子亚型的关系,利用在线数据库分析VEGF的表达情况与乳腺癌预后的关系。选取三阴性乳腺癌MDA-MB-231细胞,加入外源性hVEGF165通过体外微球形成实验观察体外成球能力,并利用Western blot、RT-qPCR等方法检测肿瘤干细胞相关标志物CD44、cMyc、Nanog、ALDH1的表达以及相关通路的激活情况。
结果
利用Oncomine、UALCAN、HPA数据库分析显示,VEGF在乳腺癌中表达较癌旁组织增高(P<0.0001),其表达与分子亚型相关,三阴性乳腺癌中VEGF表达显著升高。Kaplan-Meier Plotter数据库分析结果提示高表达VEGF的乳腺癌患者预后更差,总生存期OS缩短,差异具有统计学意义(P<0.0001)。与VEGF低表达患者相比,VEGF高表达的乳腺癌患者无进展生存期、无远处转移生存期以及复发后生存期均显著缩短,差异具有统计学意义(P<0.0001)。体外实验发现加入外源性的hVEGF165后三阴性乳腺癌MDA-MB-231细胞的成球能力显著增强,差异具有统计学意义(P=0.0029)。Western blotting、RT-qPCR结果表明乳腺癌MDA-MB-231微球体中VEGF/NRP-1及肿瘤干细胞相关标志物CD44、Nanog、c-Myc的表达量显著升高。加入外源性的hVEGF165促进了肿瘤干细胞相关标志物CD44、c-Myc、Nanog及ALDH1的表达,而CD24的表达量则显著下降,上述作用具有时间依赖性。Western blotting实验提示加入外源性的hVEGF165能够激活三阴性乳腺癌细胞ERK/MAPK通路。
结论
VEGF可能通过激活ERK/MAPK通路促进了三阴性乳腺癌肿瘤干细胞的形成。
Keywords: 三阴性乳腺癌, 血管内皮生长因子A, 肿瘤干细胞
Abstract
Objective
To investigate the role of vascular endothelial growth factor (VEGF) in regulating triple-negative breast cancer (TNBC) stem cells and the possible pathways involved in this regulatory mechanism.
Methods
The Oncomine database, UALCAN database and Human Protein Atlas (HPA) database were used to analyze the expression of VEGF in breast cancer and its association with the molecular subtypes and prognosis of breast cancer. Sphere formation assay was carried out to examine the effects of hVEGF165 on sphere formation ability of TNBC MDA-MB-231 cell line; Western blotting and RT-qPCR were performed to detect the expression of the tumor stem cell markers including CD44, c-Myc, Nanog, and ALDH1 and the activation of the related pathways.
Results
Data from the online databases all showed a significant increase of VEGF expression in breast cancer tissues than in the adjacent tissues (P < 0.0001), and its expression level was associated with the molecular subtypes of breast cancer. Specifically, the expression of VEGF was markedly higher in TNBC than in other subtypes of breast cancer. Survival analysis showed that breast cancer patients with a high VEGF expression had a significantly shortened overall survival (P < 0.0001). In the cell experiments, the sphere formation ability of MDA-MB-231 cells was significantly enhanced after treatment with hVEGF165 (P=0.0029). Compared with the monolayer cells, MDA-MB-231 spheres showed significantly increased expressions of VEGF, NRP-1, CD44, Nanog and c-Myc. Treatment with hVEGF165 resulted in significant time-dependent up-regulation of the expressions of CD44, c-Myc, Nanog and ALDH1 and down-regulation of CD24 expression in the cells. The results of Western blotting demonstrated that treatment with hVEGF165 caused significant activation of the ERK/MAPK pathway in MDA-MB-231 cells.
Conclusion
VEGF promotes cancer stemness of triple-negative breast cancer possibly through the ERK/MAPK pathway.
Keywords: triple-negative breast cancer, vascular endothelial growth factor, cancer stem cells
乳腺癌作为女性最常见的恶性肿瘤,其发病率及死亡率均呈现逐年上升的趋势,发病年龄也趋于年轻化[1, 2]。三阴性乳腺癌(TNBC)是一类雌、孕激素受体及人表皮生长因子受体HER-2均为阴性的乳腺癌亚型,约占所有乳腺癌患者的15%~20%[3]。与luminal型乳腺癌相比,三阴性乳腺癌表现出更多肿瘤干细胞特性,肿瘤干细胞被认为可能是维持TNBC高度侵袭性、远处转移及复发的重要原因[4]。由于缺乏有效的治疗靶点,三阴性乳腺癌的治疗仍是目前亟待解决的临床难点[5, 6]。
血管内皮生长因子(VEGF)是由肿瘤细胞和基质细胞分泌,能够促进肿瘤新生血管生成。越来越多的研究表明,VEGF可能参与了肿瘤细胞的迁移侵袭、上皮间质转化及肿瘤干细胞特性等的调控[7-10],多种肿瘤中发现VEGF的分泌及表达量增高与患者预后密切相关[11]。VEGF家族的受体主要有两大类:经典的酪氨酸激酶受体(包括VEGFR-1、VEGFR-2及VEGFR-3)以及协同受体Neuropilin-1(NRP-1)。多种肿瘤细胞及组织中有NRP-1的表达,且NRP-1高表达与肿瘤分期、侵袭性及不良预后显著相关,NRP-1可能广泛参与了不同肿瘤发生及发展的过程[12, 13]。
前期研究中发现,VEGF/NRP-1轴能够显著促进三阴性乳腺癌MDA-MB-231细胞的体外迁移、侵袭、上皮间质转化(EMT)及体内成瘤[14]。文献报道EMT转化能够促进肿瘤干细胞形成及“干性”维持[15]。因此,推测VEGF可能参与TNBC肿瘤干细胞生成,推动了肿瘤发生及复发转移。目前VEGF/NRP-1轴是否参与三阴性乳腺癌干细胞的调控有待进一步研究证实,其具体作用机制尚不明确。为了进一步探讨VEGF在三阴性乳腺癌干细胞中的作用及其机制,我们首先通过查询Oncomine数据库、UALCAN数据库及Human Protein Atlas数据库分析了VEGF在乳腺癌组织、癌旁组织及不同分子亚型乳腺癌组织中的表达情况,并进一步通过Kaplan-Meier Plotter数据库探讨了VEGF表达与乳腺癌患者预后的关系。体外实验中,选用具有高度致瘤性和侵袭性的人三阴性乳腺癌细胞系MDA-MB-231细胞作为研究模型,探讨了VEGF对三阴性乳腺癌细胞MDA-MB-231中肿瘤干细胞富集和发生的作用,本研究的开展为三阴性乳腺癌患者的治疗提供了新思路。
1. 材料和方法
1.1. 生物信息学分析
通过Oncomine数据库(<a href="https://www.oncomine.org/resource/login.html" target="_blank">https://www.oncomine.org/resource/login.html</a>)<sup>[<xref ref-type="bibr" rid="b16">16</xref>]</sup>, UALCAN数据库(<a href="http://ualcan.path.uab.edu" target="_blank">http://ualcan.path.uab.edu</a>)<sup>[<xref ref-type="bibr" rid="b17">17</xref>]</sup>及Human Protein Atlas(<a href="https://www.proteinatlas.org/" target="_blank">https://www.proteinatlas.org/</a>)<sup>[<xref ref-type="bibr" rid="b18">18</xref>]</sup>在线数据库对VEGF的表达及其与临床病理指标的相关性进行分析,并利用在线预后分析数据库K-M plotter(<a href="http://kmplot.com/analysis/" target="_blank">http://kmplot.com/analysis/</a>)<sup>[<xref ref-type="bibr" rid="b19">19</xref>]</sup>检索VEGF与乳腺癌预后的相关性并下载数据制图。
1.2. 细胞株
人乳腺癌MDA-MB-231细胞来自中国科学院典型培养物保藏委员会上海细胞库,由本实验室保存。
1.3. 主要试剂
胎牛血清购自以色列BI公司,L-15培养基购自江苏凯基公司,人NRP-l抗体、人VEGF抗体(Abcam,稀释比例1∶1000),人Nanog抗体(稀释比例1∶4000)、人c-Myc抗体(稀释比例1∶1000)(武汉三鹰生物),人GAPDH抗体(稀释比例1∶1000,SantaCruz),人ERK1/ 2抗体(稀释比例1∶1000,SantaCruz),人p-ERK1/2抗体(稀释比例1∶1000,SantaCruz),人hVEGF165因子(Cell Signaling Technology),EFG、bFGF、B27细胞因子(Sigma),反转录试剂盒、实时荧光定量PCR试剂盒(TaKaRa),GAPDH、Nanog、c-Myc、Sox2、Oct4、CD44、CD24、VEGF、NRP-1引物均由北京鼎国生物有限公司合成。
1.4. 细胞培养
MDA-MB-231细胞培养于L-15细胞培养液,培养液均添加100mL/L胎牛血清和1%青霉素-链霉素,细胞均在37 ℃、50 mL/L CO2的细胞培养箱中培养。
1.5. 体外细胞成球培养
实验分为两组(hVEGF165处理的实验组及对照组),将人乳腺癌MDA-MB-231细胞消化后轻柔地反复吹打,得到单细胞悬液,进行细胞计数,将2000个细胞接种至含EGF(20 ng/mL)、bFGF(20 ng/mL)和B27(50×)的DMEM/F12的无血清培养液中,置于37 ℃、5%CO2体积分数的饱和湿度条件下培养。10 d后对两组悬浮成球情况进行观察,同时收集悬浮的细胞微球体,在光学显微镜下分别计数和拍照记录实验组和对照组形成的微球体的数量和直径。
1.6. Western blotting检测蛋白表达量
冷PBS冲洗后,RIPA冰上裂解乳腺癌细胞10 min并收集细胞碎片以12 000 r/min、4 ℃离心15 min。小心吸出上清并与5×蛋白缓冲液混合,金属浴锅煮沸10 min,-20 ℃保存备用。将蛋白质进行SDS-PAGE胶电泳,后转移至PVDF膜上。5% 脱脂奶室温封闭1 h,一抗4 ℃孵育过夜。隔日TBST洗涤3次后二抗室温孵育1 h,再次清洗3次,显影检测。
1.7. 荧光定量聚合酶链式反应(RT-qPCR)检测mRNA水平的表达
使用FASTA200提取乳腺癌细胞中总RNA,应用Nano-500超微量核酸分析仪检测其浓度,参照逆转录及SYBR试剂说明进行操作,得到cDNA,将相应试剂、目的基因引物和cDNA加入八联管中,于定量PCR仪进行扩增,重复40个循环。相对表达量使用2-ΔΔCT法进行计算,每个样本按照相同条件进行3次重复实验,各基因引物见表 1。
1.
RT-qPCR引物
Primers for RT-qPCR
| Gene name | Primer sequences(5'to 3') | |
| Forward primer | Reverse primer | |
| GAPDH | ACCACAGTCCATGCCATCA | TCCACCACCCTGTTGCTGT |
| VEGF | AACTTTCTGCTGTCTTGG | ACTTCGTGATGATTCTGC |
| Nanog | GGGATTGGGAGGCTTTGCTT | GCACAACCAACAAATTAGGGGA |
| OCT-4 | GTAGTCCCTTCGCAAGCCC | AGGTCCGAGGATCAACCCAG |
| SOX-2 | CATGAAGGAGCACCCGGATT | ATGTGCGCGTAACTGTCCAT |
| c-Myc | GTCAAGAGGCGAACACACAAC | TTGGACGGACAGGATGTATGC |
| CD44 | CTGCCGCTTTGCAGGTGTA | CATTGTGGGCAAGGTGCTATT |
| CD24 | CACGCAGATTTATTCCAGTGAAAC | GACCACGAAGAGACTGGCTGTT |
| NRP-1 | AGGACAGAG ACTGCAAGTATGAC | AACATTCAGGACCTCTCTTGA |
1.8. 统计学方法
采用SPSS23.0、Graphpad Prism 7.0及EXCEL软件对实验结果进行统计分析及绘图。各项实验均独立重复3次,计量资料以均数±标准差表示,两个独立样本均数间的比较采用t检验;ANOVA方差分析比较多组间均数差异,P<0.05认为差异具有统计学意义。
2. 结果
2.1. VEGF在乳腺癌中的表达情况及其与肿瘤预后的关系
利用Oncomine和UALCAN在线数据库进行再次验证,Oncomine数据库分析结果提示,有4个研究(共2753例乳腺癌标本)显示VEGF基因在乳腺肿瘤组织中呈高表达,1个研究(共675例乳腺癌标本)分析结果为低表达(表 2,图 1A)。与此相对应,UALCAN数据库结果同样支持上述差异,在1211例乳腺癌标本中,114例为癌旁组织,1097例为乳腺癌组织,与癌旁组织相比,VEGF在乳腺癌组织中呈现高表达状态,且差异具有统计学意义(P<0.0001)。VEGF在不同分子分型的乳腺癌中表达情况不同,在luminal型中表达最低,而在HER-2阳性型及三阴性乳腺癌中表达量较高,差异具有统计学意义(luminal型 vs HER- 2阳性型,P<0.01;luminal型vs三阴性型P<0.001,图 1B、C)。HPA数据库通过免疫组化验证了乳腺癌中VEGF在蛋白水平上表达量也显著升高(图 1D)。通过在线数据库KaplanMeier Plotter分析了VEGF表达量与肿瘤预后的关系,结果显示与VEGF低表达组相比,VEGF高表达患者预后更差,总生存期(OS)、无进展生存期(PFS)、无远处转移生存期(DMFS)、复发后生存期(PPS)均显著缩短(图 1E)。
2.
VEGF在各个数据集中的表达情况
Expression of VEGF in different studies
| Type of breast cancer versus normal breast tissue | Sample | Fold Change | P | t | Source/Reference |
| Lobular breast carcinoma | 415 | 2.144 | <0.0001 | 7.524 | Zhao Breast[20] |
| Invasive ductal breast carcinoma | 609 | 2.598 | <0.0001 | 9.986 | Zhao Breast[20] |
| Intraductal cribriform breast adenocarcinoma | 623 | 2.849 | <0.0001 | 8.493 | TCGA Breast |
| Ductal breast carcinoma | 1106 | 2.300 | <0.0001 | <0.0001 | Richardson Breast 2[21] |
| Invasive breast carcinoma stroma | 675 | -7.071 | <0.0001 | -17.841 | Finak Breast[22] |
1.

VEGF在乳腺癌中的表达情况及其预后
Expression of VEGF in breast cancer and its association with the patients' prognosis. A: Expression of VEGF in 20 different tumors (ONCOMINE database); B: Expression of VEGF expression in breast cancer (UALCAN database); C: Expression of VEGF in breast cancer of different molecular subtypes (UALCAN database); D: The VEGF protein expression in breast cancer(Human Protein Atlas database); E: Kaplan-Meier survival analysis of the association of VEGF expression with the prognosis of breast cancer patients. **P < 0.01, ***P < 0.001, ****P < 0.0001.
2.2. VEGF对三阴性乳腺癌MDA-MB-231成球能力影响
微球体形成实验结果显示对照组成球数量为(66± 11.53)/1000细胞,而加入hVEGF165组成球数量为(142.3±16.86)/1000细胞,差异具有统计学意义,P= 0.0029,且加入hVEGF165组所形成的微球直径显著增大(图 2)。加入外源性hVEGF165后三阴性乳腺癌MDAMB-231细胞体外成球数量显著增多,微球体直径显著增大。
2.
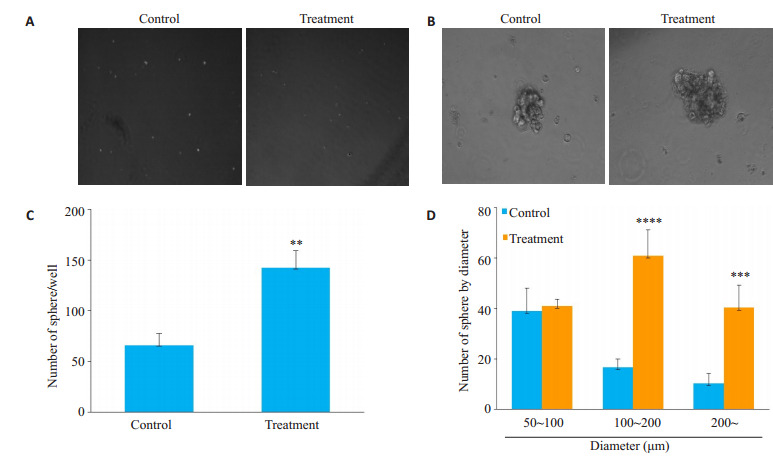
VEGF对三阴性乳腺癌细胞系MDA-MB-231体外成球能力影响
Effects of VEGF on sphere formation ability of TNBC MDA-MB-231 cells
2.3. 三阴性乳腺癌MDA- MB- 231细胞微球体中VEGF、NRP-1及肿瘤干细胞标志物的表达情况
Western Blotting结果显示与对照组普通细胞相比,MDA-MB-231微球体细胞中VEGF及其受体NRP-1的表达量显著升高(图 3A、B,P<0.0001及P<0.001),与此同时,干性相关标志物CD44、Nanog、c-Myc表达增加,而CD24表达降低(图 3A、B,P<0.0001)。采用RTqPCR实验验证mRNA水平上干性标志物的变化,结果与Western blotting一致。与对照组相比,MDA-MB-231微球体细胞中SOX-2、Nanog、c-Myc mRNA水平表达均显著升高(图 3C,P<0.0001)。
3.

乳腺癌MDA-MB-231细胞微球体中VEGF、NRP-1及肿瘤干细胞标志物表达情况
Expression of VEGF, NRP-1 and tumor stem cell markers in MDA-MB-231 spheres. A: Western blot analysis of VEGF, NRP-1 and tumor stem cell markers in MDA-MB-231 monolayer and sphere cells with GAPDH as the loading control. B: Quantification of Western blot analysis. C: RT-qPCR analysis of the mRNA expressions of VEGF, NRP-1 and tumor stem cell markers in MDA-MB-231 monolayer and sphere cells. ***P < 0.001, ****P < 0.0001 vs monolayer cell.
2.4. 外源性hVEGF165对三阴性乳腺癌MDA-MB-231细胞“干性”的影响
Western blot结果显示,加入hVEGF165后肿瘤干细胞相关标志物Nanog、CD44、ALDH1、c-Myc表达增加,而CD24表达下降,在加入hVEGF165后60min表达水平达到高峰(图 4A)。
4.

外源性hVEGF165对三阴性乳腺癌MDA-MB-231细胞干性指标的影响
Effects of exogenous hVEGF165 on expressions of tumor stem cell markers in TNBC MDA-MB-231 cells. A: Western blot analysis for detecting expressions of tumor stem cell markers in TNBC MDA-MB-231 cells. B: RT-qPCR analysis for detecting mRNA expressions of tumor stem cell markers in TNBC MDA-MB-231 cells. MDA-MB-231 cells treated with hVEGF165 for 60 min vs 0 min, *P < 0.05, ***P < 0.001, ****P < 0.0001 vs 0 min.
采用RT-qPCR实验验证mRNA水平上干性标志物的变化,结果与Western blotting基本一致(图 4B,P<0.05)。
2.5. 外源性hVEGF165激活三阴性乳腺癌细胞ERK/ MAPK通路
外源性hVEGF165能够促进乳腺癌MDA-MB-231细胞p-ERK的表达,并且该激活作用具有时间依赖性,在加入外源性hVEGF165后30~60 min p-ERK的表达水平达到高峰,随后逐渐下降(图 5,P<0.01)。
5.

外源性hVEGF165对三阴性乳腺癌细胞ERK/MAPK通路的影响
Effect of exogenous hVEGF165 on ERK/MAPK pathway in MDA-MB-231cells. A: Western blot analysis of the expressions of p-ERK and ERK1/2 in MDA-MB-231 cells. B: Quantitative analysis of the blots. **P<0.01, ****P<0.0001 vs 0 min.
3. 讨论
三阴性乳腺癌5年生存率仅有70%,是目前临床治疗的难点[23, 24]。血管内皮生长因子VEGF是促进肿瘤血管生成的主要调节因子[31]。研究表明VEGF可以调控肿瘤细胞的迁移侵袭、上皮间质转化及肿瘤干细胞[7-9]。大量文献表明,VEGF在乳腺癌中表达增高且与多个临床病理特征相关,高表达VEGF的患者预后普遍较差[14, 32, 33]。本研究利用多个在线生物信息数据库对VEGF在乳腺癌中的表达、其表达与临床病理特征的关系及预后进行了梳理,结果显示与癌旁组织相比,VEGF在乳腺癌组织中表达增高,进一步分析发现,VEGF在不同分子分型的乳腺癌中表达情况不同,在luminal型中表达最低,而在HER-2阳性型及三阴性乳腺癌中表达量较高,差异具有统计学意义。通过在线数据库Kaplan-Meier Plotter分析了VEGF表达量与肿瘤预后的关系,结果显示VEGF高表达组患者预后更差,OS、PFS、DMFS及PPS均显著缩短。上述结果提示VEGF可能在乳腺癌的发生发展及复发转移中发挥了重要作用。
文献报道TNBC分子分型多属于basal-like型,表现出一系列间质细胞的特性,与EMT过程存在密切联系。研究表明EMT与肿瘤干细胞的形成和干细胞特性的维持有密切联系。肿瘤干细胞是肿瘤组织中一群具有自我更新、无限增殖、多潜能分化特征的细胞[25]。研究表明[26-30],Lin-ESA+/CD44+/CD24-/low、ALDH1、SOX-2、OCT-4、Nanog、c-Myc是乳腺癌干细胞的重要标志。Zhang等[8]研究显示乳腺癌细胞微球体中VEGF表达增高且VEGF/NRP-1轴通过Wnt通路发挥对乳腺癌干细胞的调节作用。我们前期研究显示三阴性乳腺癌细胞系MDA-MB-231细胞中存在VEGF及其受体NRP-1的高表达,VEGF/NRP-1轴的激活能够促进TNBC细胞系MDA-MB-231的EMT转化及裸鼠体内乳腺癌原位移植瘤的生长和转移[14]。目前,VEGF及其受体是否参与了TNBC中肿瘤干细胞功能的调控,上述调控作用机制尚不明确。为了探索三阴性乳腺癌中VEGF与肿瘤干细胞的关系,我们分别进行了体外成球培养、Western blotting、RT-qPCR等实验。结果显示与对照组相比,加入外源性hVEGF165后三阴性乳腺癌MDA-MB-231细胞体外成球数量显著增多,微球体直径显著增大,差异具有统计学意义,表明外源性hVEGF165加入促进了三阴性乳腺癌MDA-MB-231细胞的体外成球能力,提示外源性VEGF可能促进了三阴性乳腺癌干细胞的形成。我们通过Western blotting、RT-qPCR进一步检测了MDA-MB-231微球体与普通MDA-MB-231细胞中VEGF、NRP-1及CD44、CD24、Nanog、SOX-2、c-Myc等肿瘤干细胞相关标志物的表达情况,结果显示微球体细胞中肿瘤干细胞相关标志物及VEGF/NRP-1的表达同步升高,提示VEGF可能参与了三阴性乳腺癌细胞“干性”的调控。通过体外加入外源性hVEGF165,发现hVEGF165的加入促进了三阴性乳腺癌MDA-MB-231细胞Nanog、CD44、ALDH1、c-Myc等的表达,且上述作用具有时间依赖性,在加入hVEGF165后60 min表达水平达到高峰。
既往研究表明VEGF能够通过受体酪氨酸激酶(RTKs)激活细胞内ERK/MAPK及PI3K/Akt等信号通路,从而发挥生物学功能[34]。我们研究发现,加入外源性hVEGF165 30~60 min后MDA-MB-231细胞中p-ERK的表达水平达到高峰,初步证实VEGF能够激活三阴性乳腺癌细胞系MDA-MB-231细胞内ERK/MAPK信号通路,且该作用存在时间依赖性。
综上所述,我们通过生物信息学分析及体外相关实验发现,乳腺癌中VEGF表达显著升高,高表达VEGF的乳腺癌患者预后不良。加入外源性的hVEGF165后三阴性乳腺癌MDA-MB-231细胞的体外成球能力显著增强,乳腺癌MDA-MB-231微球体中VEGF、NRP-1及肿瘤干细胞相关标志物CD44、Nanog、c-Myc的表达量显著升高。外源性的hVEGF165促进了三阴性乳腺癌细胞MDA-MB-231肿瘤干细胞相关标志物CD44、c-Myc、Nanog及ALDH1的表达,并能够激活MDA-MB-231细胞ERK/MAPK通路,上述作用具有时间依赖性。提示VEGF/NRP-1轴可能参与了三阴性乳腺癌肿瘤干细胞的形成,上述作用的发挥可能与ERK/MAPK信号通路激活相关。但VEGF如何激活ERK/MAPK信号通路进而调控肿瘤干细胞形成,其具体作用机制仍有待进一步研究。
Biography
王璐,在读博士研究生,E-mail: 346111310@qq.com
Funding Statement
国家自然科学基金青年科学基金(8180111330);陕西省自然科学基础研究计划(2018JQ8046)
Supported by Natural Science Foundation for the Youth (NSFY) of China (8180111330)
Contributor Information
王 璐 (Lu WANG), Email: 346111310@qq.com.
罗 敏娜 (Minna LUO), Email: maria1204@163.com.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012[J]. Int J Cancer, 2015, 136(5): E359-86.] [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012 [J]. CA Cancer J Clin, 2015, 65(2): 87-108.] [DOI] [PubMed] [Google Scholar]
- 3.Eroles P, Bosch A, Pérez-Fidalgo JA, et al. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38(6):698–707. doi: 10.1016/j.ctrv.2011.11.005. [Eroles P, Bosch A, Pérez-Fidalgo JA, et al. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways[J]. Cancer Treat Rev, 2012, 38(6): 698-707.] [DOI] [PubMed] [Google Scholar]
- 4.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. PNAS. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells[J]. PNAS, 2003, 100(7): 3983-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee KL, Kuo YC, Ho YS, et al. Triple-negative breast cancer: current understanding and future therapeutic breakthrough targeting cancer stemness. Cancers. 2019;11(9):1334. doi: 10.3390/cancers11091334. [Lee KL, Kuo YC, Ho YS, et al. Triple-negative breast cancer: current understanding and future therapeutic breakthrough targeting cancer stemness[J]. Cancers, 2019, 11(9): 1334.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–90. doi: 10.1038/nrclinonc.2016.66. [Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease[J]. Nat Rev Clin Oncol, 2016, 13(11): 674-90.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanami LS, Chen HY, Peiró S, et al. Vascular endothelial growth factor-A stimulates Snail expression in breast tumor cells: implications for tumor progression. Exp Cell Res. 2008;314(13):2448–53. doi: 10.1016/j.yexcr.2008.05.004. [Wanami LS, Chen HY, Peiró S, et al. Vascular endothelial growth factor-A stimulates Snail expression in breast tumor cells: implications for tumor progression[J]. Exp Cell Res, 2008, 314 (13): 2448-53.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Wang H, Li C, et al. VEGF-A/Neuropilin 1 Pathway Confers Cancer Stemness via Activating Wnt/beta-Catenin Axis in Breast Cancer Cells. Cell Physiol Biochem. 2017;44(3):1251–62. doi: 10.1159/000485455. [Zhang L, Wang H, Li C, et al. VEGF-A/Neuropilin 1 Pathway Confers Cancer Stemness via Activating Wnt/beta-Catenin Axis in Breast Cancer Cells[J]. Cell Physiol Biochem, 2017, 44(3): 1251-62.] [DOI] [PubMed] [Google Scholar]
- 9.Ciccone V, Terzuoli E, Donnini S, et al. Stemness marker ALDH1A1 promotes tumor angiogenesis via retinoic acid/HIF-1α/ VEGF signalling in MCF-7 breast cancer cells. J Exp Clin Cancer Res. 2018;37(1):311. doi: 10.1186/s13046-018-0975-0. [Ciccone V, Terzuoli E, Donnini S, et al. Stemness marker ALDH1A1 promotes tumor angiogenesis via retinoic acid/HIF-1α/ VEGF signalling in MCF-7 breast cancer cells[J]. J Exp Clin Cancer Res, 2018, 37(1): 311.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao D, Pan C, Sun J, et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene. 2015;34(24):3107–19. doi: 10.1038/onc.2014.257. [Zhao D, Pan C, Sun J, et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2 [J]. Oncogene, 2015, 34(24): 3107-19.] [DOI] [PubMed] [Google Scholar]
- 11.Senger DR. Vascular endothelial growth factor: much more than an angiogenesis factor. Mol Biol Cell. 2010;21(3):377–9. doi: 10.1091/mbc.e09-07-0591. [Senger DR. Vascular endothelial growth factor: much more than an angiogenesis factor[J]. Mol Biol Cell, 2010, 21(3): 377-9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding M, Liu L, Hu C, et al. Expression of VEGFR2 and NRP-1 in non-small cell lung cancer and their clinical significance. https://d.wanfangdata.com.cn/periodical/zgazyj201406007. Chin J Cancer Res. 2014;26(6):669–77. doi: 10.3978/j.issn.1000-9604.2014.12.04. [Ding M, Liu L, Hu C, et al. Expression of VEGFR2 and NRP-1 in non-small cell lung cancer and their clinical significance[J]. Chin J Cancer Res, 2014, 26(6): 669-77.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grun D, Adhikary G, Eckert RL. NRP-1 interacts with GIPC1 and α 6/β4-integrins to increase YAP1/? Np63α-dependent epidermal cancer stem cell survival. Oncogene. 2018;37(34):4711–22. doi: 10.1038/s41388-018-0290-4. [Grun D, Adhikary G, Eckert RL. NRP-1 interacts with GIPC1 and α 6/β4-integrins to increase YAP1/∆ Np63α-dependent epidermal cancer stem cell survival[J]. Oncogene, 2018, 37(34): 4711-22.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M, Hou L, Li J, et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-κB and β-catenin. Cancer Lett. 2016;373(1):1–11. doi: 10.1016/j.canlet.2016.01.010. [Luo M, Hou L, Li J, et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-κB and β-catenin[J]. Cancer Lett, 2016, 373 (1): 1-11.] [DOI] [PubMed] [Google Scholar]
- 15.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–29. doi: 10.1038/nrclinonc.2017.44. [Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications[J]. Nat Rev Clin Oncol, 2017, 14(10): 611-29.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18, 000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18, 000 cancer gene expression profiles[J]. Neoplasia, 2007, 9(2): 166-80.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–58. doi: 10.1016/j.neo.2017.05.002. [Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses[J]. Neoplasia, 2017, 19(8): 649-58.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asplund A, Edqvist PH, Schwenk JM, et al. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12(13):2067–77. doi: 10.1002/pmic.201100504. [Asplund A, Edqvist PH, Schwenk JM, et al. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research[J]. Proteomics, 2012, 12(13): 2067-77.] [DOI] [PubMed] [Google Scholar]
- 19.Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y. [Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets[J]. Sci Rep, 2018, 8(1): 9227.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Langerød A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15(6):2523–36. doi: 10.1091/mbc.e03-11-0786. [Zhao H, Langerød A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast[J]. Mol Biol Cell, 2004, 15(6): 2523-36.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal- like human breast cancer. Cancer Cell. 2006;9(2):121–32. doi: 10.1016/j.ccr.2006.01.013. [Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal- like human breast cancer[J]. Cancer Cell, 2006, 9(2): 121-32.] [DOI] [PubMed] [Google Scholar]
- 22.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–27. doi: 10.1038/nm1764. [Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer[J]. Nat Med, 2008, 14(5): 518-27.] [DOI] [PubMed] [Google Scholar]
- 23.Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9(2):176–98. doi: 10.1158/2159-8290.CD-18-1177. [Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment[J]. Cancer Discov, 2019, 9(2): 176-98.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao D, Hou H, Zhang X. Progress in the treatment of solid tumors with apatinib: a systematic review. Onco Targets Ther. 2018;11:4137–47. doi: 10.2147/OTT.S172305. [Zhao D, Hou H, Zhang X. Progress in the treatment of solid tumors with apatinib: a systematic review[J]. Onco Targets Ther, 2018, 11: 4137-47.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? https://www.sciencedirect.com/science/article/pii/S1934590915000715. Cell Stem Cell. 2015;16(225):38. doi: 10.1016/j.stem.2015.02.015. [Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells[J]? Cell Stem Cell, 2015, 16(3): 225-38.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–67. doi: 10.1016/j.stem.2007.08.014. [Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome[J]. Cell Stem Cell, 2007, 1(5): 555-67.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufhold S, Garbán H, Bonavida B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J Exp Clin Cancer Res. 2016;35:84. doi: 10.1186/s13046-016-0359-2. [Kaufhold S, Garbán H, Bonavida B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication[J]. J Exp Clin Cancer Res, 2016, 35: 84.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tafani M, Perrone GA, Pucci B, et al. Reprogramming cancer cells in endocrine-related tumors: open issues. https://pubmed.ncbi.nlm.nih.gov/24304280/ Curr Med Chem. 2014;21(9):1146–51. doi: 10.2174/0929867321666131129125624. [Tafani M, Perrone GA, Pucci B, et al. Reprogramming cancer cells in endocrine-related tumors: open issues[J]. Curr Med Chem, 2014, 21(9): 1146-51.] [DOI] [PubMed] [Google Scholar]
- 29.Iv Santaliz-Ruiz LE, Xie X, Old M, et al. Emerging role of nanog in tumorigenesis and cancer stem cells. Int J Cancer. 2014;135(12):2741–8. doi: 10.1002/ijc.28690. [Iv Santaliz-Ruiz LE, Xie X, Old M, et al. Emerging role of nanog in tumorigenesis and cancer stem cells[J]. Int J Cancer, 2014, 135 (12): 2741-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dittmer J. Breast cancer stem cells: Features, key drivers and treatment options. Semin Cancer Biol. 2018;53:59–74. doi: 10.1016/j.semcancer.2018.07.007. [Dittmer J. Breast cancer stem cells: Features, key drivers and treatment options[J]. Semin Cancer Biol, 2018, 53: 59-74.] [DOI] [PubMed] [Google Scholar]
- 31.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114–27. doi: 10.1111/joim.12019. [Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis[J]. J Intern Med, 2013, 273(2): 114-27.] [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Liu Y, Wang Y, et al. Quantification of STAT3 and VEGF expression for molecular diagnosis of lymph node metastasis in breast cancer. Medicine: Baltimore. 2017;96(45):e8488. doi: 10.1097/MD.0000000000008488. [Chen Y, Liu Y, Wang Y, et al. Quantification of STAT3 and VEGF expression for molecular diagnosis of lymph node metastasis in breast cancer[J]. Medicine: Baltimore, 2017, 96(45): e8488.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.黄 清南, 黄 耀, 李 明, et al. 血管内皮生长因子(VEGF)高表达和血小板应答蛋白1(TSP-1)低表达与乳腺癌患者预后相关分析. https://www.cnki.com.cn/Article/CJFDTOTAL-XBFM201909012.htm. 细胞与分子免疫学杂志. 2019;35(9):828–31. [黄清南, 黄耀, 李明, 等. 血管内皮生长因子(VEGF)高表达和血小板应答蛋白1(TSP-1)低表达与乳腺癌患者预后相关分析[J]. 细胞与分子免疫学杂志, 2019, 35(9): 828-31.] [PubMed] [Google Scholar]
- 34.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–7. doi: 10.1016/S1470-2045(05)70168-6. [Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer[J]. Lancet Oncol, 2005, 6(5): 322-7.] [DOI] [PubMed] [Google Scholar]


