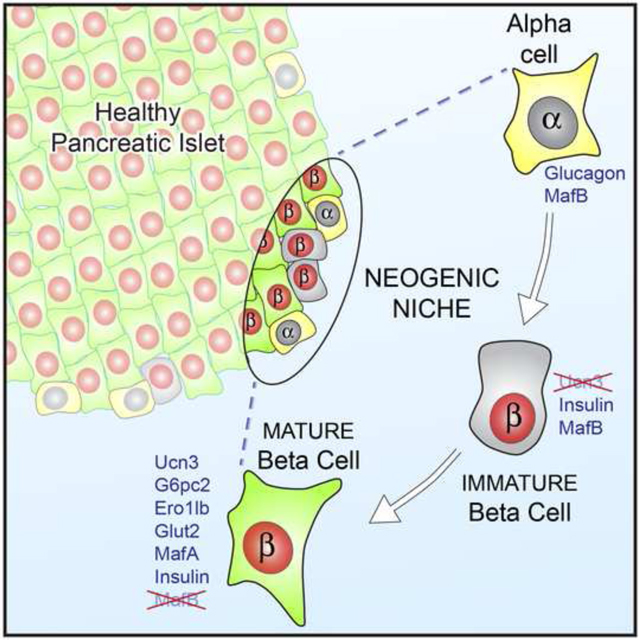
Summary
Postnatal maintenance or regeneration of pancreatic beta cells is considered to occur exclusively via the replication of existing beta cells, but clinically meaningful restoration of human beta cell mass by proliferation has never been achieved. We discovered a population of immature beta cells that is present throughout life and forms from non-beta precursors at a specialized micro-environment or ‘neogenic niche’ at the islet periphery. These cells express insulin, but lack other key beta cell markers, are transcriptionally immature, incapable of sensing glucose and unable to support calcium influx. They constitute an intermediate stage in the transdifferentiation of alpha cells to cells that are functionally indistinguishable from conventional beta cells. We thus identified a lifelong source of new beta cells at a specialized site within healthy islets. By comparing co-existing immature and mature beta cells within healthy islets we stand to learn how to mature insulin-expressing cells into functional beta cells.
Keywords: beta cell maturation, transdifferentiation, beta cell neogenesis, Urocortin3, islet architecture, stem cell, diabetes, GCaMP6, neogenic niche, alpha cell
Graphical Abstract
Introduction
The insulin deficiency that characterizes Type 1 Diabetes (T1D) and Type 2 Diabetes (T2D) has led to strong interest in processes that control beta cell mass. The current view is that beta cell mass is determined by the net effect of islet neogenesis, beta cell proliferation and hyperplasia, balanced by dedifferentiation and beta cell death through apoptosis (Bonner-Weir et al., 2010). Mice increase beta cell mass by self-replication to compensate for increased metabolic demand in the context of obesity or pregnancy (Cox et al., 2016; Parsons et al., 1992), although beta cell proliferation rates decline sharply with age (Brennand et al., 2007; Teta et al., 2005). In humans, increases in beta cell mass in response to similar conditions are modest at best. The mechanisms responsible for this increase are not fully understood. They have long been considered less reliant on beta cell replication (Butler et al., 2010), although standard replication markers may underestimate human beta cell proliferation rates during post-mortem conditions (Sullivan et al., 2015). Regardless, the existence of progenitors within the pancreas that support the regeneration of functional beta cells is a highly relevant - but controversial - topic in diabetes.
Several recent reports demonstrate that forced expression of lineage-inappropriate transcription factors causes islet cells to switch identity (Collombat et al., 2007; Collombat et al., 2009; Gao et al., 2014; Papizan et al., 2011; van der Meulen and Huising, 2015), which has been attributed to the similar chromatin states of alpha and beta cells (Bramswig et al., 2013). This establishes that beta cells can arise from non-beta endocrine cells in the islet via direct transdifferentiation. Indeed, near-complete ablation of pre-existing beta cells is eventually followed by the restoration of beta cell mass via transdifferentiation of non-beta endocrine cells in mice (Chera et al., 2014; Thorel et al., 2010). These studies provided important proof of principle that insulin independence can be regained by transdifferentiation, but have also led to the notion that it is triggered by severe beta cell ablation and the associated pancreas remodeling (Habener and Stanojevic, 2012).
Instead, we demonstrate here that the islet periphery contains a ‘neogenic niche’; a privileged microenvironment that supports lifelong conversion between alpha and beta cells. This is evident from the presence of a distinct population of immature beta cells at the islet periphery that does not yet express the late maturation marker Urocortin3 (Ucn3) (Blum et al., 2012; van der Meulen and Huising, 2014; van der Meulen et al., 2012). Similar Ucn3-negative beta cells are readily identifiable in human pancreas of different ages and in donors with T1D. Ucn3 negative beta cells are transcriptionally immature, lack cell surface Glut2, cannot sense glucose, and do not support calcium influx in response to depolarization. By lineage tracing we demonstrate that these Ucn3-negative beta cells are ‘virgin’ beta cells that represent an intermediate stage in the transdifferentiation of alpha cells into mature beta cells. On the basis of their transcriptional signature and functional responses within intact islets, we demonstrate that beta cells that arise from alpha cells at the islet edge are functionally indistinguishable from conventional beta cells. Conversely, we also observe mature beta cells that adopt an alpha cell fate and function at the islet edge. We propose that this ongoing plasticity within the neogenic niche can be targeted to regenerate beta cells.
Results
Ucn3-negative immature beta cells persist throughout life
Ucn3 is a late maturation marker for primary and stem cell-derived beta cells (Blum et al., 2012; van der Meulen et al., 2012) that continued to increase progressively weeks after beta cell expression of Nkx6–1 and Mafa reached steady state, and coincided with the gradual loss of Mafb from beta cells (Figure 1A) (Artner et al., 2010). However, Ucn3-negative beta cells persisted in islets of adult mice (Figure 1B). The fraction of beta cells that did not yet co-express Ucn3 comprised 15% of all beta cells at postnatal day 2 before stabilizing at approximately 1 – 2 % of all beta cells from 3 weeks to 14 months of age (Figure 1C). Ucn3-negative beta cells did not reflect proliferating beta cells that had transiently down regulated Ucn3, as Ki67-postive beta cells continued to stain for Ucn3 (Figure 1D).
Figure 1. The absence of Ucn3 marks beta cells in the neogenic niche.
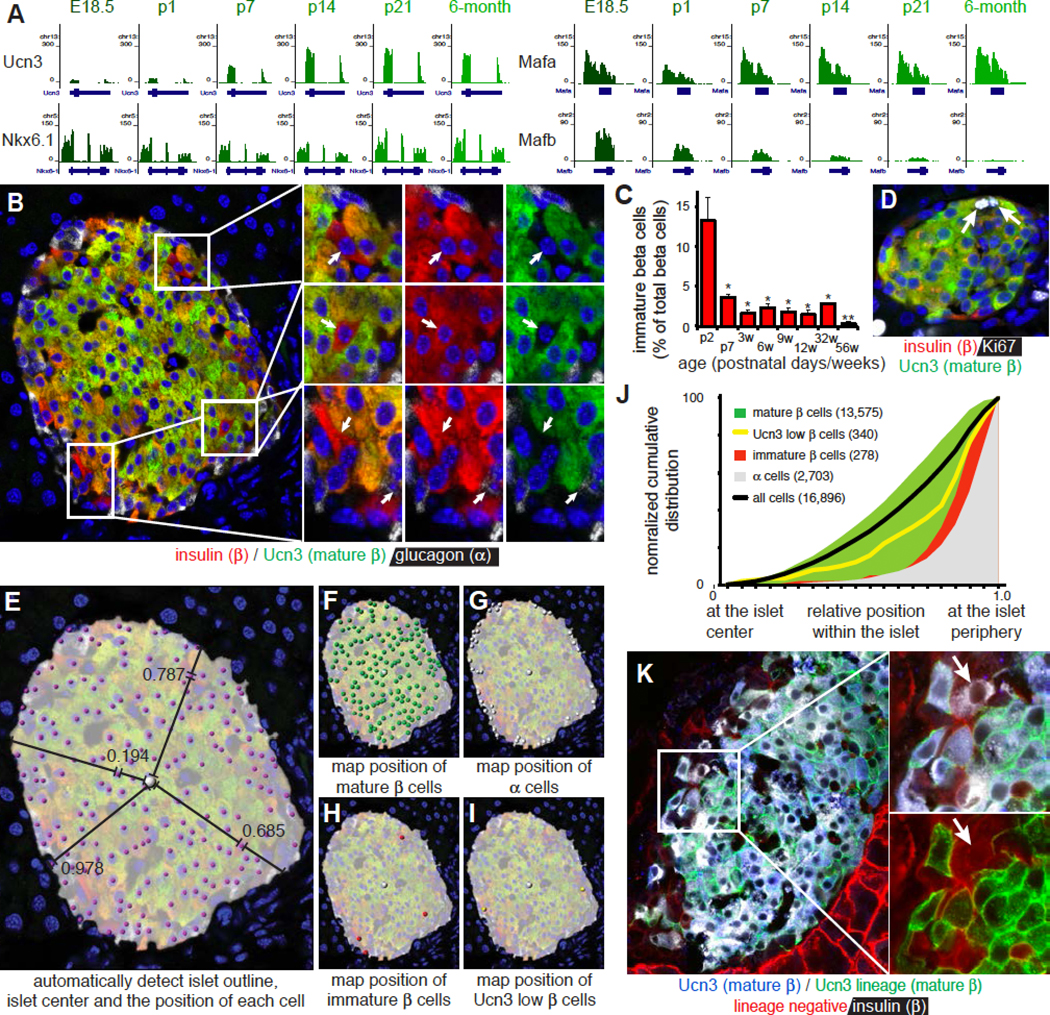
(A) Gene expression of Ucn3, Nkx6.1, Mafa and Mafb by RNAseq on FACS sorted beta cells during perinatal maturation. Gene structure and chromosome number are indicated for each panel.
(B) Detection of insulin, glucagon and Ucn3 in a 3-month old islet. Insets show Ucn3-negative beta cells.
(C) Fraction of Ucn3-negative beta cells at different ages (counted n=3 animals per time point, 10 islets each). Error bars reflect SEM, * P <0.05, ** P < 0.01.
(D) Ki67-positive beta cells maintain Ucn3 expression indicating that proliferating beta cells and Ucn3-negative beta cells are not the same.
(E-I) Image analysis to detect the islet outline and center of mass to compute the position of a cell relative to the center and nearest edge. Cells are manually classified as (F) mature beta cell (insulin and Ucn3 co-positive), (G) alpha cell (glucagon positive), (H) immature beta cell (insulin-positive, Ucn3-negative) or (I) Ucn3 low beta cell (insulin-positive, Ucn3-low).
(J) Normalized cumulative distribution of Ucn3-negative beta cells compared to mature beta cells of all ages combined (3, 6 and 9 weeks; 3, 8 and 14 months; 3 animals per age, 16,896 cells). See Figure S1 for distributions by age and Table S1A for the P values and D statistics for each pairwise comparison.
(K) Ucn3-negative, insulin positive beta cells (white, but not blue, indicated by the arrow) at the islet periphery of Ucn3-Cre x mT/mG mice are Ucn3-lineage negative (they express mTomato, instead of mGFP). These cells make up 0.75% ± 0.56% (n = 3) of all insulin-positive beta cells. See also Figure S2.
Immature beta cells are located at the islet edge
A striking aspect unique to Ucn3-negative immature beta cells was their frequent proximity to the islet edge. We therefore quantified the relative position of approximately 17,000 individual islet cells across hundreds of islets from 23 individual animals across a range of ages, normalized for islet size (Figure 1E). Identity of each cell was assigned manually. Most cells were either alpha or mature beta cells (Figure 1F, G). We reserved the designation of immaturity for only those insulin+ cells where no Ucn3 was detected (Figure 1H) and separately quantified the distribution of insulin+ beta cells with weak, but detectable, Ucn3 (Ucn3-low beta cells). We reasoned that the latter cells may reflect maturing beta cells, or may have resulted from the inherent limitations in quantifying gradual increases in signal intensities by antibody-based methods and concluded that the prudent approach was to account for these cells separately (Figure 1I). We then compared the normalized cumulative distribution of each population (Figure 1J). Mature beta and alpha cells distributed preferentially at the islet core and periphery, respectively, which recapitulated the characteristic mouse islet architecture and validated our approach. Both Ucn3 low and Ucn3 negative beta cells localized peripherally, just internally from alpha cells in the mouse islet (Figure 1J). When broken down by age, immature and mature beta cell populations in the first postnatal week distributed in proximity, but segregated as adult architecture is established (Shih et al., 2013) (Figure S1).
To confirm these findings independent of staining we crossed mIns1-H2b-mCherry (Benner et al., 2014) and Ucn3-eGFP reporter mice (Figure S2A, B). Islets from bitransgenic offspring revealed a small population of mCherry single positive beta cells at the islet periphery, while beta cells in the rest of the islet were consistently co-positive for eGFP and mCherry (Figure 2A, B; Movie S1).
Figure 2. Comparison of mature and immature beta cells from the same islets.
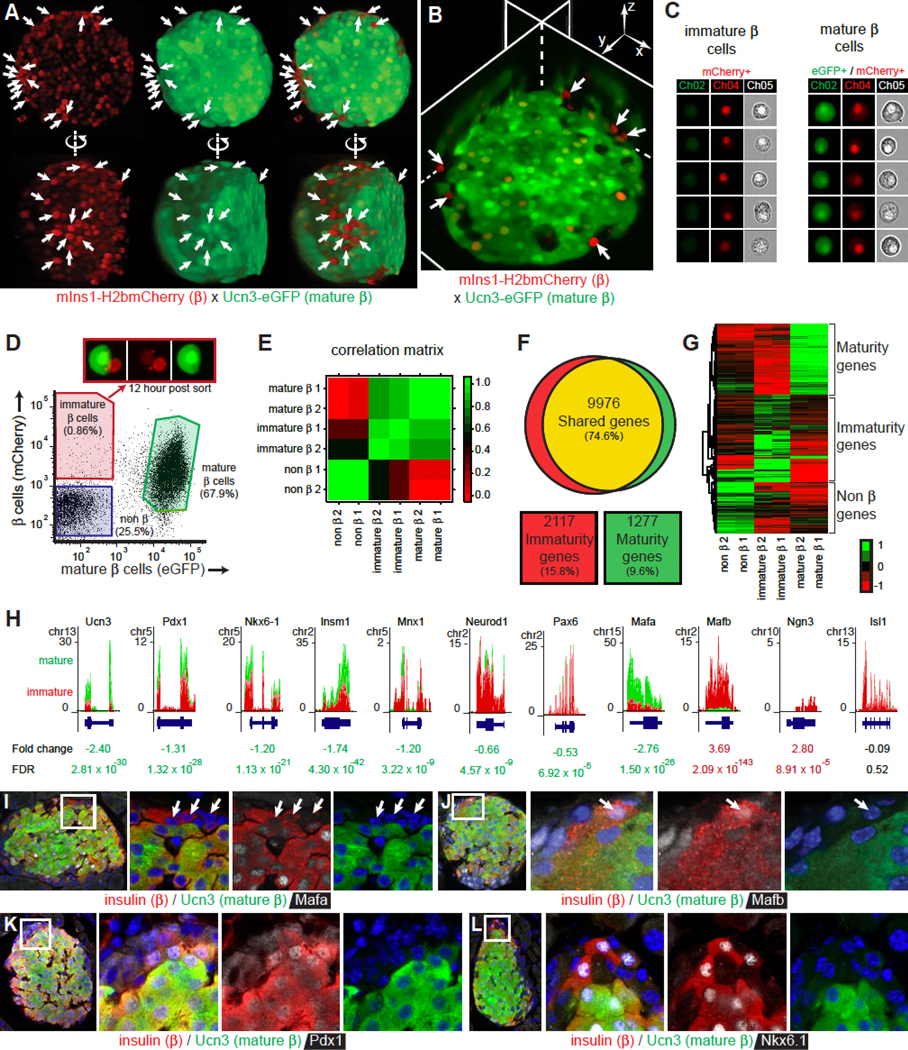
(A) View from two angles of a 3D reconstructed islet expressing mIns1-H2b-mCherry (all beta cells) and Ucn3-EGFP (mature beta cells only). Arrows indicate immature beta cells. See also Figure S2.
(B) Immature beta cells (arrows) in the neogenic niche revealed by virtual slicing of a 3D reconstructed islet. See also Movie S1.
(C) Imaging cytometer analysis of individual immature and mature beta cells.
(D) FACS strategy to obtain Ucn3-eGFP mIns1-H2b-mCherry co-positive mature and mCherry single positive immature beta cells from the same islets. Immature beta cells start expressing Ucn3-eGFP in culture 12 hours after sorting (inset).
(E) Correlation matrix of the 200 top differentially expressed (100 enriched, 100 depleted) genes between mature and immature cells.
(F) Venn diagram comparing gene expression (RPKM>1 in either population) between mature and immature beta cells. Expression was considered different when the absolute log2FC > 1 and FDR < 0.001.
(G) Heat map of the most differentially expressed genes between mature and immature beta cells.
(H) Expression of key genes in mature and immature beta cells by RNAseq. See also Figure S3.
(I-L) Colocalization of insulin, Ucn3 and Mafa (I), Mafb (J), Pdx1 (K), or Nkx6–1 (L) in an adult mouse islet. Arrows indicate immature beta cells.
Ucn3-negative beta cells at the islet edge are ‘virgin’ beta cells
While Ucn3 is an excellent beta cell maturation marker, dedifferentiating beta cells also lose Ucn3 (Blum et al., 2014; van der Meulen et al., 2015). Therefore, Ucn3 negative beta cells at the islet edge could either be new beta cells or have lost Ucn3 expression secondary to dedifferentiation. To distinguish between both scenarios, we lineage-traced mature beta cells by crossing Ucn3-Cre (Figure S2C–F) to the mT/mG reporter mouse that switches from membrane-tdTomato (mT) to membrane-eGFP (mG) upon Cre expression (Muzumdar et al., 2007). Ucn3-negative beta cells at the islet periphery had not acquired a green mature beta lineage-label and instead retained mT (Figure 1K). This ruled out that these were once mature beta cells that dedifferentiated, but is entirely consistent with the scenario that peripheral Ucn3-negative cells are ‘virgin’ beta cells that arose from a peripheral non-beta precursor within the islet.
Expression profile of the immature beta cells at the neogenic niche
We then purified mCherry positive immature beta cells and mCherry/eGFP co-positive mature beta cells (Figure 2C) from the same islets by FACS (Figure 2D) for transcriptome analysis. Overall, immature and mature beta cells were more similar to each other than to non-beta cells from the same islets (Figure 2E) and revealed no differential expression for 75% of detectable genes (Figure 2F). The remaining genes contained several clusters that were selectively enriched in either immature or mature beta cells (Figure 2G). Ucn3 and several transcription factors associated with beta cell identity, such as Pdx1, Nkx6–1, Insm1, Mnx1, Neurod1, Pax6, and Mafa were enriched in mature beta cells. (Figure 2H). In contrast, Mafb and Neurog3 were enriched in immature beta cells (Figure 2H). This was confirmed by immunofluorescence, where a majority of Ucn3-negative immature beta cells did not yet stain for Mafa and retained Mafb (Figure 2I, J). A minority of Ucn3-negative immature beta cells lacked nuclear Pdx1, but most immature cells already showed nuclear staining for Nkx6–1 (Figure 2K, L). On the basis of their transcriptome, immature beta cells resembled peri- and early post-natal time points, while mature beta cells resembled the later post-natal stages (Figure S3).
Ucn3 negative beta cells are transcriptionally immature
We next addressed if Ucn3 negative beta cells were functional. We observed significant reductions in immature cells of essential beta cell genes, including many genes required for glucose-stimulated insulin secretion, the TCA cycle and oxidative phosphorylation (Figure 3A–D). In contrast, key metabolic genes that are either considered ‘disallowed’ or contribute to their regulation (Dhawan et al., 2015; Martinez-Sanchez et al., 2016; Piccand et al., 2014; Pullen and Rutter, 2013) were enriched in immature beta cells (Figure 3E). Islets were also co-stained for insulin, Ucn3, and G6pc2, which hydrolyzes glucose-6-phosphate (Pound et al., 2013) or the oxidoreductase Ero1lb, which is required for disulfide bond formation in pro-insulin (Zito et al., 2010). Both markers co-labeled Ucn3-positive mature beta cells, and the majority of Ucn3 negative beta cells did not express G6pc2 or Ero1lb (Figure 3F–I). We did not observe G6pc2-negative beta cells with Ucn3, but detected a minority of Ucn3-positive beta cells without Ero1lb. This indicates that G6pc2 expression generally preceded Ero1lb and Ucn3.
Figure 3. Ucn3 negative beta cells are transcriptionally immature.
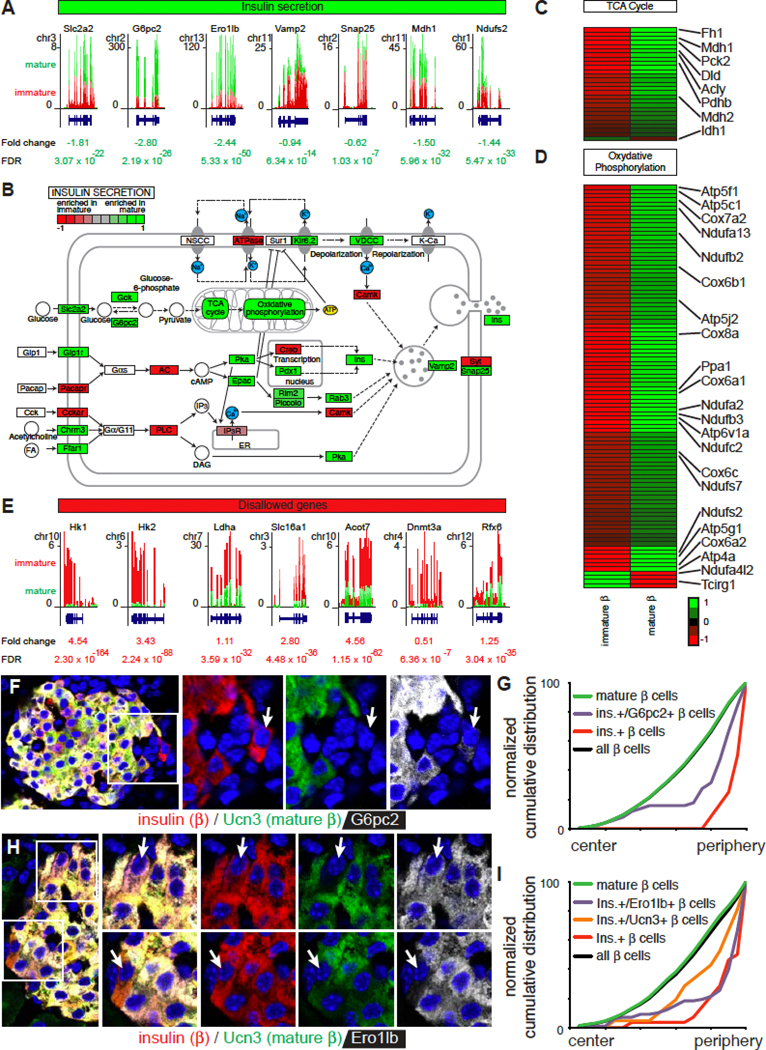
(A) Gene expression of select genes involved in insulin secretion in mature and immature beta cells by RNAseq.
(B) Visualization of differential expression of the Kegg pathway analysis for insulin secretion (FDR < 0.001).
(C) Heat map of the differential expression of tricarboxylic acid (TCA) cycle genes between immature and mature beta cells.
(D) Heat map of the differential expression of genes involved in oxidative phosphorylation between immature and mature beta cells.
(E) Gene expression of ‘disallowed’ genes in mature and immature beta cells by RNAseq.
(F) Immunofluorescence detection of G6pc2 (white), insulin (red) and Ucn3 (green) in a mouse islet. Arrow indicates an immature beta cell.
(G) Distribution of 2329 G6pc2-positive and negative beta cells within mouse islets (2329 cells). See Table S1B for the P values and D statistics for each pairwise comparison.
(H) Immunofluorescence detection of Ero1lb (white), insulin (red) and Ucn3 (green) in a mouse islet. Arrows indicate examples of beta cells with Ero1lb but not Ucn3 (top) and with Ucn3 but not Ero1lb (bottom).
(I) Distribution of 2043 Ero1lb-positive and negative beta cells within mouse islets (2073 cells).
Ucn3 negative beta cells are functionally immature
The reduced expression of genes encoding critical insulin secretion components, including Slc2a2, established that Ucn3 negative beta cells are transcriptionally immature, but did not directly demonstrate their functional status. We determined that Ucn3-negative peripheral beta cells lacked cell-surface expression of the essential Glut2 glucose transporter that is encoded by Slc2a2 (Figure 4A, B, S4), which suggested they were incapable of sensing glucose. Indeed, glucose uptake experiments in intact mIns1-H2b-mCherry islets using the fluorescent non-hydrolysable glucose analog 6-NBDG revealed rapid glucose uptake across the entire extracellular and intracellular space of the islet after 50 minutes, with the exception of a small number of mIns-H2b-mCherry positive beta cells at the islet periphery (Figure 4C, D; Movie S2).
Figure 4. Ucn3 negative beta cells are functionally immature.
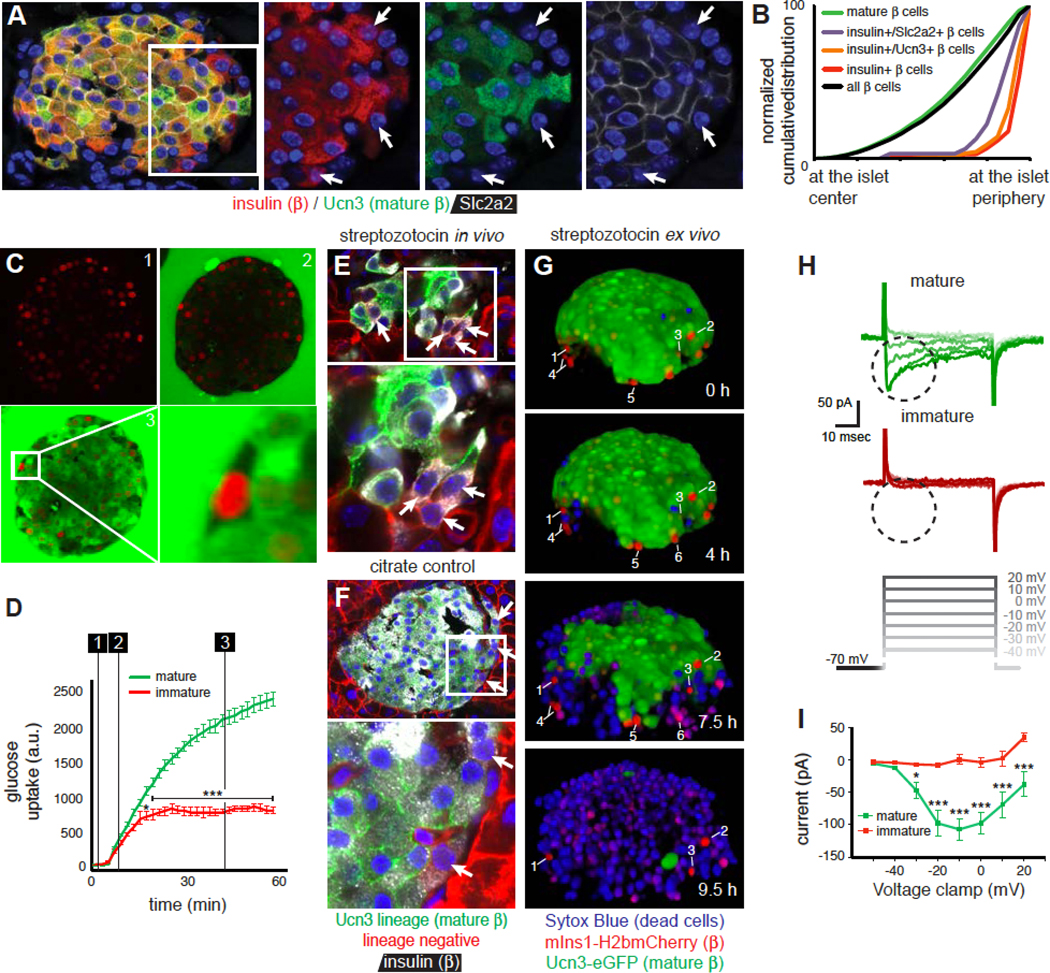
(A) Ucn3-negative beta cells at the periphery (arrows) do not express cell-surface Glut2.
(B) Distribution of Glut2-negative beta cells at the periphery of the islet (6783 cells total). See Table S1B for the P values and D statistics for each pairwise comparison and Figure S4 for a comparison of immature beta cells with other heterogeneous populations of beta cells.
(C) Uptake of the glucose analog 6-NBDG over time by all beta cells except for immature beta cells at the periphery of intact islets.
(D) Quantification of 6-NBDG uptake. Numbered lines corresponds to panel C. Data represent mean ± SEM for 3 immature and 7 mature beta cells. * P < 0.05, *** P < 0.001. See also Movie S2.
(E) Treatment with a high dose of the Glut2-dependent beta cell toxin streptozotocin ablates a majority of beta cells. Remaining Ucn3 lineage-positive beta cells are often insulin negative, while immature beta cells (arrows) survive owing to the lack of cell surface Glut2.
(F) Ucn3 lineage-negative beta cells at the islet edge (arrows) in citrate controls.
(G) STZ-induced death of Ucn3-eGFP x mIns1-H2b-mCherry mature beta cells in intact islets in real time. The death of beta cells is marked by the acute loss of eGFP protein and the nuclear uptake of the dead cell marker Sytox Blue. In contrast, mCherry single positive immature beta cells (numbered) do not take up Sytox Blue. Individual immature beta cells are labeled for clarity. Note that immature beta cells #4–6 disappear from the Z-stack as the islet volume expands due to the extensive cell death. See also Movie S3.
(H) Immature beta cells cannot support calcium influx following depolarization. Representative traces of depolarization-induced inward calcium currents (circled) from mIns1-mCherry+ immature beta cells and Ucn3-eGFP positive mature beta cells from the same preparations.
(I) Full Current-Voltage (I-V) plot contrasting the voltage-dependent inward calcium current in Ucn3-eGFP positive mature beta cells with the lack thereof in mIns1-mCherry+ immature beta cells from the same preparations. Data represent mean ± SEM for 4 immature and 8 mature beta cells from four individual animals. * P < 0.05, ** P < 0.01, *** P < 0.001.
We next administered the beta cell toxin Streptozotocin (STZ; 120 mg/kg on 2 consecutive days), which is taken up specifically by beta cells in a Glut2-dependent manner, to acutely ablate beta cells. While STZ treatment kills a large fraction of beta cells, beta cells that survive STZ treatment dedifferentiate, downregulate Ucn3, and are therefore difficult to distinguish from immature beta cells. We therefore used Ucn3-Cre x mT/mG bitransgenic mice where immature beta cells would remain Ucn3 lineage-negative (red), while dedifferentiated beta cells remain Ucn3 lineage-positive (green) despite the down regulation of Ucn3 expression. Indeed, STZ treatment killed most mature beta cells, leading to a collapse in islet architecture with remaining Ucn3 lineage-positive beta cells down regulating insulin at 48 hours after the second STZ injection. Ucn3 lineage-negative beta cells on the other hand persisted and continued to show normal insulin expression (Figure 4E). Citrate controls once again revealed clusters of Ucn3 lineage-negative immature beta cells at the periphery of otherwise uniformly Ucn3 lineage-positive islets (Figure 4F). As the purpose of this experiment was to test the ability of immature beta cells to resist acute STZ-mediated cell death, we did not track their fate long-term.
As STZ administration in vivo provides only a snapshot of the islet at the conclusion of the experiment, we applied STZ (5 mM) ex vivo to Ucn3-eGFP x mIns1-H2b-mCherry bitransgenic islets, while imaging continuously in the presence of the nuclear dead cell marker Sytox Blue. STZ proceeded to kill all Ucn3-eGFP positive beta cells over the course of the 10-hour treatment (Figure 4G). In contrast, mCherry single positive immature beta cells are among the last beta cells to take up Sytox Blue (Movie S3). Collectively, these three experiments demonstrate that immature beta cells have impaired glucose uptake and are protected from STZ-mediated beta cell death, in line with their lack of cell-surface Glut2.
As ATP generated by glycolytic flux triggers beta cell depolarization downstream of Glut2, this experiment did not address if immature beta cells were capable of supporting voltage-triggered inward calcium currents. We therefore directly recorded these cells in voltage-clamp mode and observed that immature beta cells were unable to respond to stepwise depolarization with the opening of L-type voltage-gated calcium channels (Figure 4H, I).
Immature beta cells are present in human islets
In human neonatal islets, Ucn3-negative beta cells preferentially localize to the interface between clusters of alpha and beta cells or to the periphery. (Figure 5A–B). This suggests that a spatially distinct neogenic niche exists at an early age in human islets before alpha and beta cells progressively intermingle into the adult human islet architecture. Moreover, we observed residual insulin-positive, Ucn3-negative beta cells in human donors with diagnosed Type 1 Diabetes (Figure 5D, E), although it is impossible to know without lineage tracing whether these cells reflect regeneration of new beta cells or dedifferentiation of existing ones.
Figure 5. Ucn3-negative beta cells are present in human islets of young and adult donors and donors with T1D.
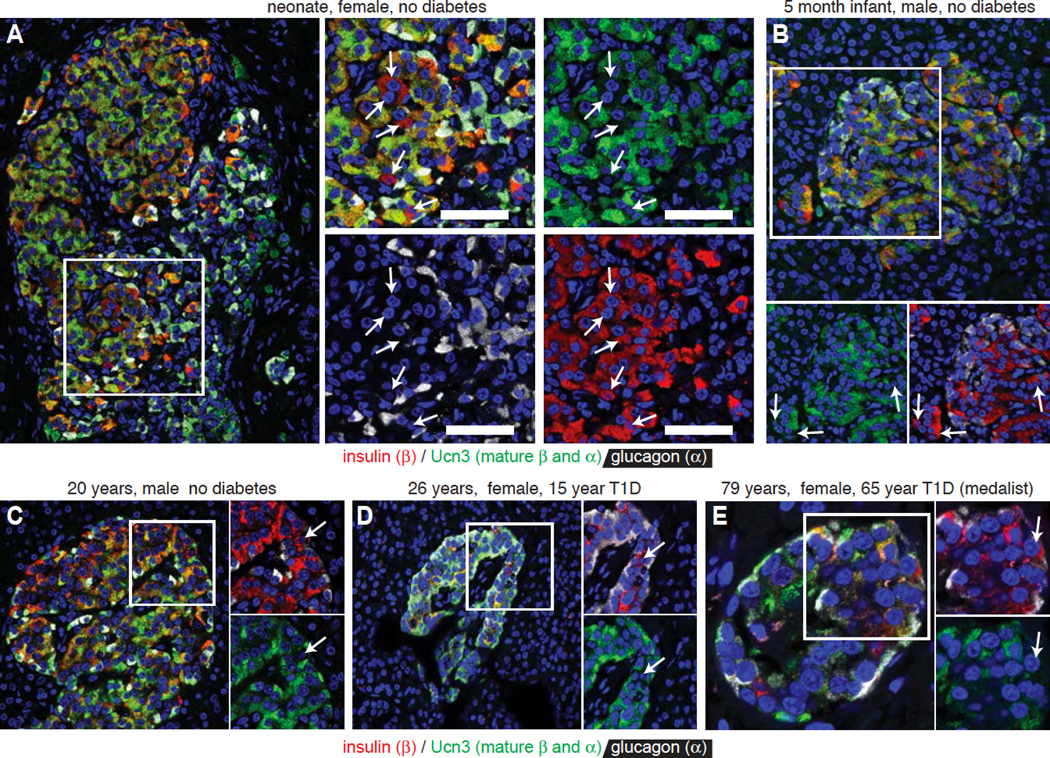
Expression of insulin, glucagon and Ucn3. Arrows indicate Ucn3-negative beta cells.
(A) Neonate, female, no diabetes, nPOD #6200.
(B) Infant, 5 months old, male, no diabetes, nPOD #6115.
(C) Adult, 20 years old, male, no diabetes, nPOD #6238.
(D) Adult, 26 years old, female, 15 years with diagnosed T1D, nPOD #6196.
(E) Adult, 79 years old, female, 56 years with diagnosed T1D (medalist), nPOD #6065.
Immature beta cells are an intermediate stage in alpha to beta transdifferentiation
The location of immature beta cells at the neogenic niche suggested the presence of a local precursor distinct from existing mature beta cells. This may be a progenitor sensu strictu that is destined to become a beta cell, or it could be another differentiated cell type that, under the right circumstances, becomes a beta cell. This led us to hypothesize that the persistent population of immature beta cells we discovered reflects an intermediate stage in the transdifferentiation of alpha to mature beta cells. We therefore lineage-labeled alpha cells using bitransgenic offspring of a cross between Gcg-Cre (Herrera, 2000) and mT/mG reporter mice and readily observed cells with an alpha cell lineage-label (green) that expressed Ucn3 instead of glucagon (Figure 6A). The acquisition of an alpha cell lineage-label by beta cells could reflect an artifact of transgenesis. Under such a stochastic scenario, one would expect lineage-labeled beta cells to occur randomly across the cross-sectional area of the islet (Figure 6B). Alternatively, beta cells may acquire an alpha cell lineage-label if they go through an insulin/glucagon bi-hormonal progenitor stage during embryonic development. As beta cell mass increases rapidly in early postnatal life by clonal expansion, this would lead to clusters of clonally related beta cells with an alpha cell lineage-label. Indeed, such a distribution was observed in adult islets of a Gcg-Cre line that labeled bi-hormonal cells during pancreas development (Shiota et al., 2013), but is never observed in the line we used (Herrera, 2000). However, if beta cells that carry an alpha cell lineage-label reflect transdifferentiation of alpha to beta cells at the periphery of adult islets, one would expect that beta cells with an alpha cell lineage-label preferentially co-distribute with alpha cells (Figure 6B). Indeed, we observed these cells to be peripherally distributed just internally from the alpha cell population (Figure 6C). Moreover, we occasionally observed immature beta cells with an alpha cell lineage-label that did not yet co-express Ucn3 at the islet edge (Figure 6D). Nevertheless, to more firmly establish that alpha to beta transdifferentiation takes place in adult islets at the neogenic niche, we used Gcg-CreER x lsl-eYFP bitransgenic reporter mice (Ackermann et al., 2017) in a pulse-chase approach to lineage-label all alpha cells at 2 months of age. Two days after the lineage-labeling, the fraction of beta cells with an alpha cell lineage-label was < 0.2%, but this fraction had increased significantly 4 months after tamoxifen (Figure 6E, F). This is consistent with a scenario where alpha cells transdifferentiate via a transient, immature stage that expresses insulin, but not Ucn3. Interestingly, the total fraction of glucagon-positive cells that carries an alpha cell lineage-label also dropped from nearly 100% to slightly less than 97% over the same period (Figure 6G), suggesting that new alpha cells arose from a non-alpha source during the same window.
Figure 6. Immature beta cells reflect a transient stage in the transdifferentiation between alpha and beta cells at the islet edge.
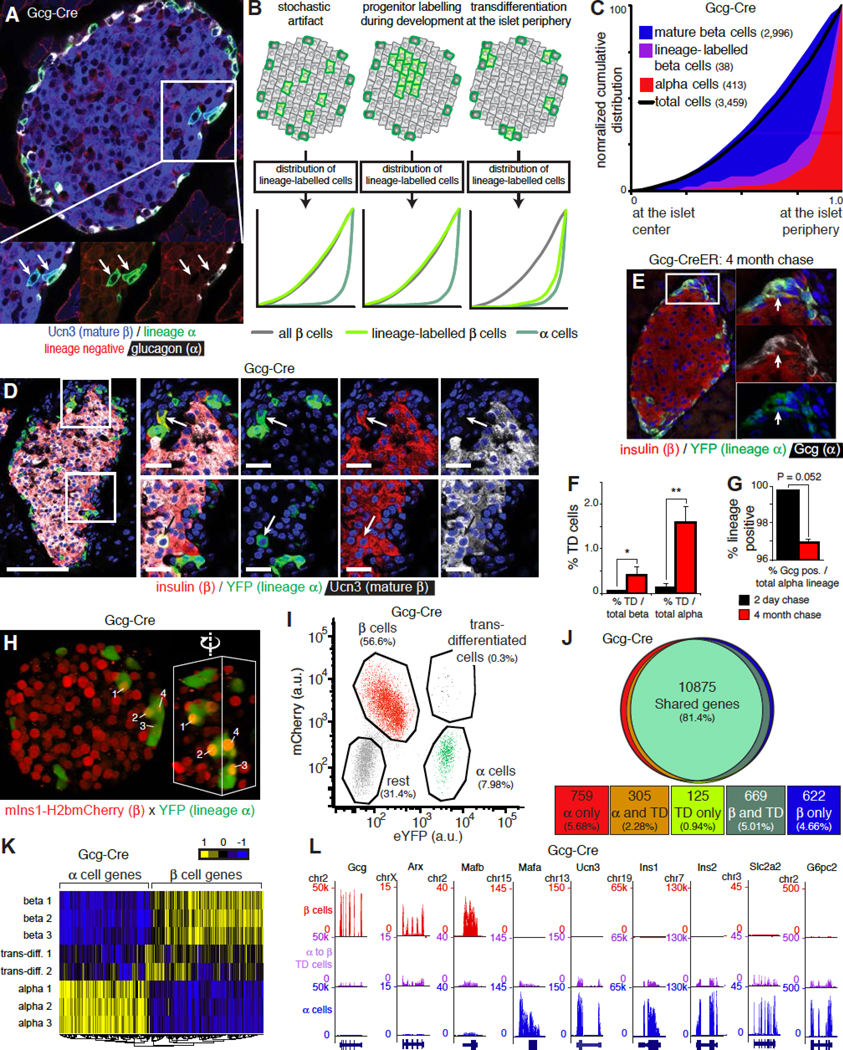
(A) A pair of cells at the islet periphery with an alpha lineage mark (arrows) that now express Ucn3 instead of glucagon.
(B) Three potential scenarios that could account for the detection of beta cells with an alpha cell lineage-label and their predicted distribution across the cross-sectional islet area: 1) random labeling of beta cells by the ‘leaky’ expression of Cre recombinase in beta cells, 2) labeling of bi-hormonal progenitors during development is predicted to lead to randomly localized clusters of lineage-labeled beta cells that expanded from a single bi-hormonal progenitor, 3) transdifferentiation of alpha cells into beta cells at the periphery of adult islets. See also Figure S5.
(C) Observed distribution of alpha lineage-labeled beta cells (3459 cells). See Table S1C for P values and D statistics for each pairwise comparison.
(D) Alpha lineage-labeled islet, visualized by lsl-YFP, that features two transdifferentiated cells. One of these co-expresses Ucn3 and is mature (bottom, arrow), the other cell is an immature beta cell of alpha cell-descent that expresses insulin but not yet Ucn3 (top, arrow).
(E) Lineage-labeling all alpha cells in 2-month old mice via Gcg-CreER followed by a 4-month chase demonstrates that alpha cells continue to transdifferentiate into beta cells. (F) The fraction of beta cells of alpha cell descent increased significantly four months after lineage-labeling all alpha cells, as measured relative to either total beta cell or alpha cell number. * P < 0.05, ** P < 0.01.
(G) Conversely, the alpha cell fraction, which is nearly completely lineage-labeled immediately after tamoxifen administration, is notably diluted 4 months later by alpha cells without a lineage-label.
(H) Islets from triple transgenic offspring of a cross between mIns1-mCherry x Gcg-Cre x lsl-YFP reveal the presence of mCherry positive beta cells that carry the YFP alpha cell lineage-label at the edge of the intact islet. Inset shows detail of the same cluster from different angle to emphasize the nuclear mCherry in transdifferentiated cells. See also Movie S4.
(I) FACS strategy to isolate transdifferentiated cells along with alpha and beta cells from dissociated islets of mIns1-mCherry x Gcg-Cre x lsl-YFP triple transgenic islets. See also Figure S6.
(J) Venn diagram of genes that are detectably expressed (RPKM > 1) among alpha, beta and transdifferentiated cells. Expression was considered different when the absolute log2FC > 1 and FDR < 0.001.
(K) Heat map of alpha, beta and transdifferentiated cells based on the most differentially expressed genes from panel 6J.
(L) Expression of key genes in alpha, beta and transdifferentiated cells by RNAseq. Gene structure and chromosome number are indicated for each panel.
Based on the absence of mT/mG co-positive ‘yellow’ cells in the pancreas of bitransgenic Ins-Cre x LSL-mT/mG reporter mice it had previously been concluded that there was no postnatal contribution of non-beta cells to the beta cell mass (Xiao et al., 2013); a conclusion that appears to contradict our current observations. However, beta cells are only (mT/mG co-positive) for a short, 4-day window after activation of Ins1-CreER with a single tamoxifen pulse (Figure S5). Moreover, this study attempted to detect this transient ‘yellow’ intermediate stage in dissociated whole pancreas (which consists of >98% non-islet cells) and was by design underpowered to detect a relatively small but ongoing contribution of non-beta cells to the beta cell mass. By using isolated islets and a constitutive Cre to accumulate transdifferentiation events, we achieved a markedly better sensitivity.
Beta cells of alpha cell descent are transcriptionally similar to conventional beta cells
We compared the transcriptomes of beta cells of alpha cell descent with conventional beta cells from the same islets, using mIns1-H2b-mCherry mice crossed to Gcg-Cre and lsl-YFP mice (DiGruccio et al., 2016). In this strategy, alpha cells that transdifferentiated into beta cells acquired mCherry, but maintained the YFP lineage-label and can be FACS-sorted accordingly (Figure 6H, I, Movie S4). We applied a stringent doublet exclusion strategy to ensure that the 0.3% mCherry/YFP co-positive events did not reflect doublets. We validated this by imaging cytometry and confirmed that transdifferentiated cells expressed mRNA for both mCherry and YFP (Figure S6). Approximately 81% of all detectable genes were shared by all three cell populations. The remaining 19% of detectable genes were split between alpha and beta cells. Less than 1% of all genes were enriched in trans-differentiated cells (Figure 6J). This is relevant, as these genes may contain markers to identify beta cells from alpha cell descent in human islets, where lineage-tracing is not possible. Transdifferentiated cells more closely resembled beta cells, but clustered intermediate from the alpha cells that they once were and the beta cells that they became (Figure 6K, L).
Beta cells that arise via transdifferentiation from alpha cells are functionally mature
To determine whether transdifferentiation from alpha cells at the neogenic niche generates functionally mature beta cells, we assessed their ability to respond to glucose stimulation using a floxed allele of the genetically encoded calcium indicator GCaMP6. This strategy served the dual purpose of a traditional Cre-dependent lineage-labeling approach with the ability to dynamically record the behavior of the lineage-labeled cells within intact islets in real time. We conducted these experiments on the mIns1-H2b-mCherry background to distinguish beta from non-beta cells. We first established the prototypical responses of alpha and beta cells to known alpha and beta cell-selective cues. To selectively stimulate alpha cells, we applied the peptide hormone AVP, which activates V1b receptors that are selectively expressed by alpha cells (DiGruccio et al., 2016) (Figure 7A) and dose-dependently stimulated glucagon release (Dunning et al., 1984) (Figure 7B). Alpha cells within intact islets responded simultaneously with a calcium response to the initial application of each escalating dose of AVP, which was followed by a period of uncoordinated, alpha cell-autonomous calcium activity (Figure 7C; Movie S5). To selectively stimulate beta cells, we continuously perfused with 16.8 mM glucose, which activated a pulsatile, synchronized calcium response uniquely characteristic of healthy beta cells (Figure 7D; Movie S5). To establish the functional responses of transdifferentiated cells at the neogenic niche, we used Gcg-Cre to lineage-label alpha cells with GCaMP6. As before, AVP stimulated an alpha cell-autonomous response. However, upon switching to 16.8 mM glucose, a new population of cells at the islet surface that had not responded to AVP now started firing in the synchronized, pulsatile fashion that is characteristic of functionally mature beta cells (Figure 7E; Movie S6). While their GCaMP6 expression indicated alpha cell lineage history, their functional behavior and Ins1-dependent expression of nuclear mCherry each confirmed current beta cell identity; AVP-responsive alpha cells maintained a dark nuclear shade (Figure 7E, F). A brief depolarization (30 mM KCl) activated all GCaMP6-expressing cells, regardless of current identity to indicate cell viability throughout the experiment.
Figure 7. Transdifferentiated cells are functionally mature.
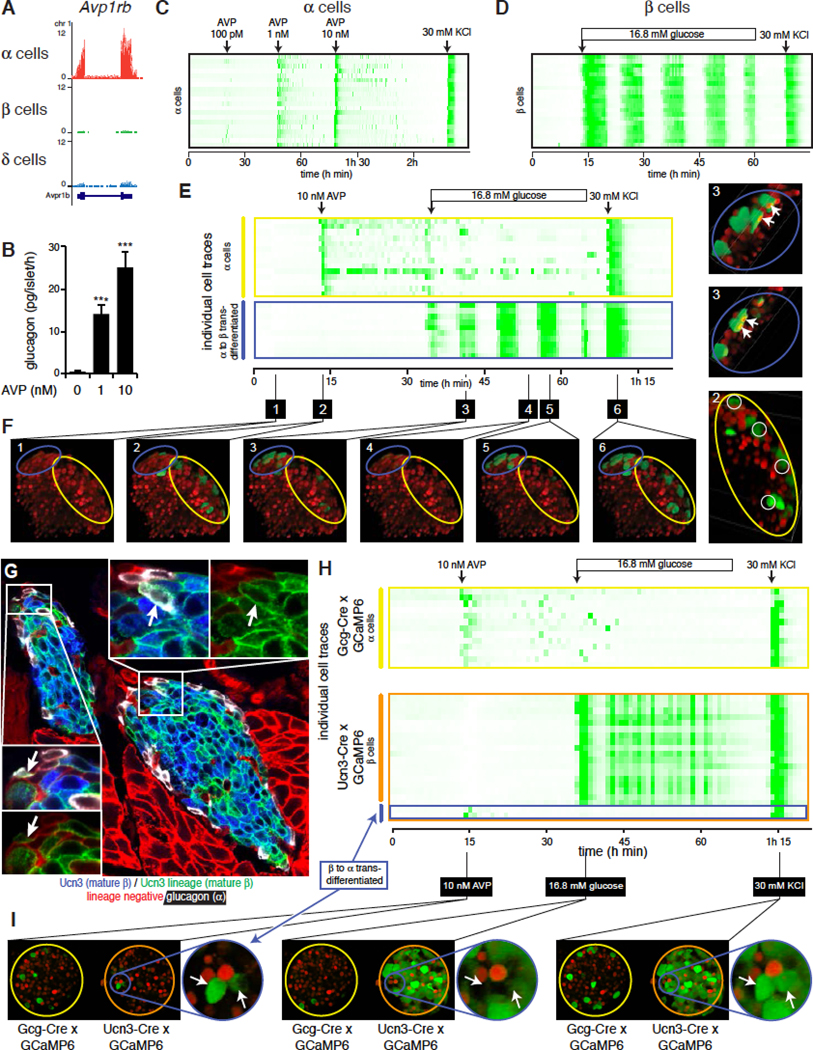
(A) Expression of arginine vasopressin (AVP) receptor Avp1rb in alpha, beta and delta cells.
(B) Glucagon secretion in response to AVP. Data represent mean ± SEM; n = 4. *** P < 0.001.
(C) Alpha cell calcium activity in response to brief stimulation with increasing doses of AVP, indicated by arrows. Depolarization (30 mM KCl) serves as positive control. See also Movie S5.
(D) Coordinated beta cell calcium activity in response to continuous stimulation with 16.8 mM glucose. Arrow indicates the start of continuous stimulation. See also Movie S5.
(E) Consecutive stimulation with a brief pulse of AVP (arrow) followed by continuous stimulation with 16.8 mM glucose in an islet where GCaMP6 expression is restricted to the alpha cell lineage and nuclear mCherry marks current beta cells. See also Movie S6.
(F) Still images of key frames in at the indicated times in (E). Yellow ellipsoid contains conventional alpha cells; blue ellipsoid contains alpha-to-beta transdifferentiated cells.
(G) Beta-to-alpha transdifferentiated cells revealed by the absence of Ucn3 and expression of glucagon in Ucn3-Cre lineage cells at the islet periphery.
(H) Responses of two islets where expression of GCaMP6 is restricted to the alpha cell lineage (Gcg-Cre, top) or mature beta cell lineage (Ucn3-Cre; bottom). Stimulation protocol as in Figure 6E. Both islets were imaged simultaneously in a single recording. See also Movie S7.
(I) Still images of key frames in (H).
Mature beta cells transdifferentiate into alpha cells at the islet edge
Our findings thus far raised the question whether the privileged micro-environment of the neogenic niche specifically supports the conversion of alpha cells into beta cells, or supports lineage plasticity in general. The observation that new lineage-negative alpha cells appeared during the 4 months following the tamoxifen-induced labeling of the entire alpha cells (Figure 6G) suggests that new alpha cells are continuously generated from a non-alpha progenitor. Indeed, when we used Ucn3-Cre to lineage-label mature beta cells, we observed Ucn3 lineage-labeled cells at the islet edge that expressed glucagon instead of Ucn3 (Figure 7G). To determine if such alpha cells of mature beta descent fully completed their transdifferentiation, islets from mIns1-H2b-mCherry mice that expressed GCaMP6 in the alpha (Gcg-Cre) or mature beta lineage (Ucn3-Cre) were simultaneously subjected to the same set of alpha and beta cell-specific stimuli as before (Movie S7). We readily identified Ucn3 lineage-labeled GCaMP6-positive cells that nevertheless responded in typical alpha cell fashion to AVP under basal glucose conditions and were indistinguishable from conventional Gcg-Cre-expressing alpha cells in the adjacent islet (Figure 7H). These cells no longer participated in the synchronized pulsatile calcium response following subsequent stimulation with 16.8 mM glucose, which implies that they were no longer coupled to the beta cell mass via gap junctions. They also no longer had an mCherry+ nucleus, but responded robustly to depolarization by KCl (Figure 7H, I). This established that alpha cells can transdifferentiate from mature beta cells at the edge of healthy islets.
Discussion
Here we describe a novel population of Ucn3-negative beta cells that occurs throughout life at a spatially distinct ‘neogenic niche’ at the islet periphery. These cells express insulin, but are transcriptionally immature and lack key beta cell markers such as G6pc2, Ero1lb and cell-surface Glut2. Indeed, these beta cells do not sense glucose and cannot support calcium influx following depolarization, indicating that they are functionally immature. They represent virgin beta cells that constitute a transitional stage in the transdifferentiation from alpha into mature, functional beta cells. We also observe the converse transition from mature beta into functional alpha cells. Based on the totality of these observations, we propose that the islet periphery contains a specialized ‘neogenic niche’; a specific micro-environment where the necessary cues and conditions to facilitate islet cell plasticity are met. The differentiation of pancreatic beta cells during embryonic development is driven by spatio-temporal gradients that determine the formation of endocrine progenitors at the trunk of the budding ducts followed by their delamination from the ductal epithelia to eventually cluster into islets (Shih et al., 2013). Our discovery suggests the continued importance of the 3-dimensional environment in determining the plasticity and maintenance of islet cell fate.
The continuous formation of new beta cells from a progenitor pool in adult islets is a distinct departure from the long-standing paradigm that maintenance of beta cell mass occurs exclusively through beta cell self-replication (Brennand et al., 2007; Dor et al., 2004). However, since the latter study was conducted using a low-efficiency (30%) lineage-labeling strategy dependent on the rat insulin promoter, it was underpowered to rule out a relatively small contribution from non-beta cells. Moreover, this strategy could not have differentiated between the immature and mature beta cells we report here, as both express insulin. Our observations are easily reconciled with the common view that beta cell self-renewal is the major mechanism to regenerate or expand beta cell mass in mice, provided sufficient beta cells remain (Cox et al., 2016). Our discovery of the neogenic niche extends this model to establish that beta cells can and do arise via alternative paths within islets of healthy, non-diabetic individuals. This is particularly relevant to established T1D, where proliferation of any residual beta cells by itself is unlikely to support meaningful regeneration of functional beta cell mass, given that human beta cells rarely proliferate (Meier et al., 2008). This emphasizes the need to pursue alternative beta cell sources to restore functional beta cell mass.
Transdifferentiation from alpha to beta cells without genetic perturbation of transcription factor expression was first described following near-complete beta cell ablation in mice (Thorel et al., 2010). The process is commonly presumed to be induced by insulin deficiency or pancreatic injury and remodeling (Chera et al., 2014; Habener and Stanojevic, 2012). Instead, our observations demonstrate that transdifferentiation between alpha and beta cells takes place within a specialized niche at the periphery of healthy islets. It follows that the near-complete removal of existing beta cells as the predominant source of beta cell regeneration may have merely unmasked transdifferentiation as an alternate contributor to beta cell regeneration. This represents an important conceptual shift: it suggests that the signal(s) that control plasticity at the neogenic niche are associated with the constellation of cell types and autonomic nerves that converge at the periphery of healthy islets instead of being induced in response to major pancreas remodeling. Of course, this does not rule out a scenario where transdifferentiation is further stimulated under certain (patho)physiological circumstances, as was recently demonstrated when blockade of glucagon-dependent amino acid clearance by the liver promoted alpha cell proliferation, accompanied by increased alpha to beta transdifferentiation (Solloway et al., 2015).
Islet architecture differs markedly between adult human and mouse islets (Brissova et al., 2005; Cabrera et al., 2006; Dolensek et al., 2015; Kim et al., 2009). However, in younger human donor islets, alpha and beta cells segregate into distinct areas or adopt a mantle/core topology similar to mouse islets (Figure 5). We readily observe Ucn3-negative beta cells at the alpha/beta interface in islets of neonatal and infant pancreas donors and can also identify such cells in adult human pancreas. This suggests that a neogenic niche similar to the one we discovered in mice may exist in human islets, at least in younger ages. Moreover, we continue to observe Ucn3 negative beta cells within islets of patients with decades-long history of Type 1 Diabetes, possibly reflecting subclinical beta cell regeneration supported by the islet plasticity we describe here. However, human islet architecture becomes less defined with age and Ucn3 expression is lost from beta cells in diabetes (Blum et al., 2014; van der Meulen et al., 2015). The fact that a lack of Ucn3 marks both immature and de-differentiated beta cells complicates its utility as a marker to establish whether immature beta cells continue to congregate in a spatially distinct niche. Nevertheless, our observations could indicate human alpha cell-autonomous potential to adopt a beta cell identity that may be re-engaged by recreating the appropriate conditions that converge at the neogenic niche.
Our discovery that a population of immature beta cells persists in the islets of healthy adult mice and, possibly, humans, is also a strong testament to the significant heterogeneity that has long been known amongst beta cells (Giordano et al., 1991; Kiekens et al., 1992). Insulin-expressing Pancreatic Multipotent Progenitor cells (PMPs) were reported that are capable of extensive self-replication, self-renewal and contribute to multiple pancreatic and neural cell types (Smukler et al., 2011). These and similar cells found in small clusters outside of normal islets (Beamish et al., 2016) share a lack of cell-surface Glut2 with the immature beta cells we discovered at the niche. However, the genes enriched in the PMP transcriptome (Razavi et al., 2015) demonstrate virtually no overlap with those enriched in immature beta cells (Figure S4). Moreover, while we consider Ucn3-negative immature beta cells to be a reservoir of beta cell progenitors, we do not have evidence to suggest that they possess the multipotent progenitor properties described for PMPs. Recently, a distinct population of approximately 15% of all beta cells was described to resist immune attack in non-obese diabetic (NOD) mice. These beta cells are characterized by reduced expression of beta cell identity genes including Glut2 and likely reflect the de-differentiation of existing beta cells (Rui et al., 2017). While these cells are distinct from the virgin beta cells that we describe, these observations nevertheless suggest that immature beta cells by virtue of their reduced expression of beta cell identity genes – including the major auto-antigens insulin and G6pc2 – may escape auto-immune diabetes like they resist STZ-mediated beta cell death.
Other recent papers describe a distinct population of beta ‘hub’ cells that control the response of islets to stimulation (Johnston et al., 2016) or report heterogeneity based on a transgenic reporter strain for Flattop (Bader et al., 2016). However, based on several lines of evidence, it is clear that the Ucn3-negative immature beta cells we discovered are completely distinct from both. Flattop negative beta cells constitute approximately 20% of the total beta cell mass and hub cells make up 5 – 10% of the beta cells, with both beta cell types found throughout the islet (Figure S4). This contrasts with the 1.5% of beta cell number and the peripheral location of Ucn3-negative immature beta cells. Moreover, Flattop positive and negative beta cells both robustly express Ucn3 protein (Bader et al., 2016); vice versa Ucn3-negative immature and Ucn3-expressing mature beta cells express similar levels of Flattop mRNA (Figure S4), indicating that Flattop and Ucn3 do not mark the same beta cell population. In human islets four sub-populations of beta cells were recently described based on the expression of ST8SIA1 and CD9 (Dorrell et al., 2016). ST8SIA1 positive cells share traits with the immature beta cells we describe here (Figure S4), although others (including ST8SIA1 itself) are distinct between these beta cell sub-populations.
In summary, the plasticity of beta cell identity is a double-edged sword that provides an opportunity to regenerate beta cells from endogenous progenitors, while contributing to their dedifferentiation in diabetes (Jonas et al., 1999; Talchai et al., 2012). The lessons we learn from beta cell maturation at the neogenic niche will be widely applicable under different scenarios where beta cell maturity is either desired or compromised. This includes the generation of functionally mature human beta cells from stem cells to alleviate the pressing need for an unlimited source of human beta cells to cure T1D - a feat that despite impressive recent progress in the field (Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015) - has not been achieved. We envision that the signaling responses and epigenetic mechanisms that underlie beta cell maturation at the niche overlap with those that are impaired when beta cells dedifferentiate in diabetes. It follows that the discovery of the specific spatio-temporal cues that govern islet cell fate at the neogenic niche can be wielded to take advantage of the significant plasticity in beta cell regeneration. These insights could apply to generate functionally mature beta cells from endogenous sources or stem cell-derived progenitors and to block the adverse consequences of the beta cell identity crisis in diabetes.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mark Huising (mhuising@ucdavis.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Commercial male C57BL6/NHsd mice were obtained from Harlan (Indianapolis, IN; now Envigo) at 3, 6 and 9 weeks and 3, 8 and 14 months of age and used immediately for pancreas collection. A number of transgenic mouse lines were employed. The dual color lsl-mT/mG reporter mouse (Gt(ROSA)26Sortm4(ACTB-tdTomato,-eGFP)Luo/J, JAX strain 007576) (Muzumdar et al., 2007). The lsl-eYFP reporter mouse (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J) (Srinivas et al., 2001). Calcium levels in the cell were visualized using the lsl-GCaMP6 mouse line (B6;129S6-Gt(ROSA)26Sortm96(CAG-GCaMP6s)Hze/J) (Madisen et al., 2015). Two Ucn3 BAC transgenic reporter mice were used that are based on BAC clone RP23–332L13, which contains the Ucn3 gene flanked by more than 197 kb of genomic context: the Ucn3-Cre line (B6.FVB(Cg)-Tg(Ucn3-cre)KF43Gsat/Mmucd and the Ucn3-eGFP line (Tg(Ucn3-eGFP)HY36Gsat/Mmucd). For alpha cell labeling we employed a Gcg-Cre mouse line (Herrera, 2000) or a Gcg-CreER line (Ackermann et al., 2017). To label beta cells we used the B6.Cg-Tg(Ins1-HIST1H2BB/mCherry)5091Mhsg/J mouse line (Benner et al., 2014). To lineage label beta cells in Figure S6, we used the Ins1-CreER mouse line (B6.Cg-Tg(Ins1-cre/ERT)1Lphi/J) (Wicksteed et al., 2010). All transgenic lines are maintained by back crossing to commercially obtained C57BL6/NHsd (Envigo). Unless indicated otherwise, adult mice of both sexes were used between 3 and 9 months of age. Animals were maintained in group-housing on a 12-h light/12-h dark cycle with free access to water and standard rodent chow. All animal procedures were approved by the Salk Institute for Biological Studies and UC Davis Institutional Animal Care and Use Committees and performed in compliance with the Animal Welfare Act and the ILAR Guide to the Care and Use of Laboratory Animals.
Validation of novel transgenic mouse models
We observed close overlap between the expression of Ucn3 peptide and the expression of GFP in the Ucn3-eGFP mouse line Tg(Ucn3-eGFP)HY36Gsat/Mmucd (Figure S2). We evaluated two distinct Ucn3-Cre lines and validated the Ucn3-Cre(KF43) line to selectively and specifically label the Ucn3 expression domain in within the islets (Figure S2). In this line, over 98% of all insulin-positive beta cells in bitransgenic offspring from the Ucn3-Cre x mT/mG cross carries the mG lineage-label, validating this reporter line. We also tested the B6.FVB(Cg)-Tg(Ucn3-cre)KF31Gsat/Mmucd) line, but found the efficiency and specificity of this line to be undesirable (Figure S2).
Primary cell cultures
Primary islets were cultured in RPMI (5.5 mM glucose, 10% FBS, pen/strep) under 5% CO2 at 37° C in 10 cm petri dishes (i.e. not tissue culture treated). Islets for microscopy or dissociated islet cells for electrophysiology experiments were cultured overnight on uncoated #1.5 glass-bottom 35 mM culture dishes (MatTek Corporation, Ashland, MA). No cell lines were used in this study.
Human Subjects
Human donor pancreata were obtained via the Network of Pancreatic Islet Donors with Diabetes (nPOD). Gender and age are indicated in the figure legend. The UC Davis Institutional Review Board declared the human pancreas histological specimens used in this study exempt from IRB review under 45 CFR 46.101 (b) Category (4) on December 11, 2014.
METHOD DETAILS
Islet isolation
Islets were isolated by injecting collagenaseP (0.8 mg/mL in HBSS; Roche Diagnostics) (Invitrogen) via the common bile duct while the ampulla of Vater was clamped (Huising et al., 2010). The entire pancreas was collected following the injection of 2mL collagenase solution and, after addition of 2 more ml of collagenase solution, was incubated at 37°C for 13 min. Pancreata were dissociated by gentle manual shaking followed by three washes with cold HBSS containing 5% NCS. The digested suspension was passed through a nylon mesh (pore size 425 μm; Small Parts Inc.), and islets were isolated by density gradient centrifugation on a Histopaque gradient (1.077 g/mL density; Sigma) for 20 min at 1400 × g without brake. Islets were collected from the interface, washed once with cold HBSS containing 5% NCS, and hand-picked several times under a dissecting microscope prior to culture in RPMI (5.5 mM glucose, 10% FBS, pen/strep).
Flow and imaging cytometry
Islets were dissociated by incubation in 0.25% trypsin/EDTA for 2 minutes complemented by gentle trituration with a p200 pipette, washed once in RPMI (5.5 mM glucose, 10% FBS, pen/strep) and immediately processed on the cytometer. Conventional FACS separation was conducted based on red and green fluorescence, using 1 μg/ml Dapi for live/dead exclusion, as previously described (DiGruccio et al., 2016; van der Meulen et al., 2015) and samples were collected directly in Trizol reagent for library preparation. Samples for imaging cytometry were run on an Amnis Imagestream MarkII imaging cytometer (EMD Millipore, Billerica, MA) at the Gladstone Institutes in San Francisco.
Next generation sequencing
FACS-sorted samples were collected directly into Trizol reagent. RNA isolation and library construction was done by Illumina’s TruSeq RNA sample Prep Kit v2 (Illumina Inc. San Diego, CA), sequenced at 50 cycles, and single read on an Illumina HiSeq 2000 platform as previously described (DiGruccio et al., 2016).
Immunohistochemistry
Immunohistochemistry was conducted as follows: slides were washed 3 times for 5 minutes each in KPBS, antibodies were diluted in KPBS supplemented with 2% donkey serum and 0.4% Triton-X100 and applied overnight at 4 degrees Celsius. After 3 more washes in KPBS, slides were incubated with secondary antibodies (obtained from Jackson ImmunoResearch (Westgrove, PA) and used at 1:600 final dilution), also in donkey block, now for 45 minutes at room temperature. Three more washes completed the procedure. Where applicable, nuclei were counterstained by Dapi at 1 μg/ml final concentration and slides were embedded in Prolong Gold Antifade (Thermo Fisher Scientific, Waltham, MA) and imaged on either a Zeiss LSM780 confocal microscope or a Nikon A1R+ confocal microscope. The antiserum for Ero1lb (Zito et al., 2010) was generously provided by Dr. David Ron, the antiserum for G6pc2 (Hutton and Eisenbarth, 2003) was originally generated by the late Dr. John Hutton and generously provided by Drs. Jay Walters and Howard Davidson.
Glucose uptake
To measure glucose uptake, we incubated intact islets from freshly isolated mIns1-H2b-mCherry reporter mice overnight on uncoated #1.5 glass-bottom 35 mM culture dishes (MatTek Corporation, Ashland, MA) in RPMI (10% FBS, 5.5 mM glucose, pen/strep). The next day, Z-stacks of islets were continuously acquired as the non-hydrolysable glucose analog 6-NBDG (6-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-6-Deoxyglucose; Thermo Fisher Scientific, Waltham, MA) was added at a final concentration of 0.3 mM, using a Nikon A1R+ confocal microscope in resonant scanning mode. The relative rate of glucose uptake was determined by drawing ROIs of individual mIns1-H2b-mCherry+ beta cells that either had or had not taken up glucose.
Streptozotocin treatment
Ucn3-Cre x mTmG mice were treated with two doses of 120 mg/kg of streptozotocin (EMD Millipore, Billerica MA) dissolved fresh in 100 mM sodium citrate (pH 4.5) on consecutive days. Citrate controls were included and animals were monitored closely around the clock to prevent hypoglycemia from STZ-induced insulin release. Mice were euthanized 48 hours after the second STZ injection when hyperglycemia indicated the destruction of most beta cells and processed for immunohistochemistry. To study streptozotocin-induced beta cell death ex vivo, we cultured mIns1-H2b-mCherry x Ucn3-eGFP bitransgenic islets as described above and followed them for 15 hours in the presence of 5 mM STZ and the nuclear dead cell marker Sytox Blue (500 nM, Thermo Fisher Scientific, Waltham, MA) while acquiring a Z-stack every 30 minutes on a Nikon A1R+ confocal microscope in resonant scanning mode.
Pulse-chase of Gcg-CreER mice
Gcg-CreER x lsl-eYFP bitransgenic mice were treated at 2 months of age with tamoxifen (100 μg/g BW) 3 times over the course of 5 days and collected 2 days (pulse) or 4 months (chase) after the last injection. These samples were embedded in paraffin and a GFP antiserum was used to detect eYFP protein following antigen retrieval by microwave in 10 mM sodium citrate pH 6.0.
Electrophysiology
Dispersed immature and mature beta cells from mIns1-H2b-mCherry x Ucn3-eGFP bitransgenic islets were patched in the whole cell configuration and held at −70 mV. All cells were subjected to the voltage clamp protocol depicted in Figure 4H. Recordings were performed using the following internal solution: (mM) Cs-aspartate 87, CsCl 20, MgCl2 1, MgATP 5, HEPES 10 pH 7.2, EGTA 10 and external solution: (mM) NaCl 138, KCl 5.4, MgCl2 1, CaCl210, HEPES 10 pH 7.4, glucose 2.8. Glass capillary electrodes (Suttter Instruments) were pulled to have a resistance of 7–12 MΩ. All electrical signals were acquired using a HEKA EPC 10 USB dual head stage amplifier. Each head stage was mounted on a single Sutter micromanipulator driven by the MPC 225 controller. All cells were visualized using a Zeiss Axiovert inverted microscope with 10x, 40x and 63x objectives with filters for blue, GFP, YFP, and red fluorophores. The scope was mounted upon a Siskiyou x y translator bolted to a TMC anti-vibration table. All islet cells were continuously perfused with external solution maintained at 33°C via a stage mounted incubation system. Temperature control was achieved with a SF-20 in line heater (Harvard Apparatus). Data was acquired using HEKA Patchmaster software version 2×90.2. Signal data was analyzed using Clampfit 10 (Molecular devices).
Glucagon secretion
Static glucagon secretion experiments were carried out on 30 wild type mouse islets per well in Krebs Ringer Buffer (KRB). Islets were isolated the day prior to the secretion assay, cultured overnight in RPMI (5.5 mM glucose, 10% FBS, pen/strep) and transferred to KRB containing 5.5 mM glucose an hour before the start of the assay. Islets were picked to the wells plate for final secretion in 10% of the final assay volume. The remaining 90% of volume consisted of KRB without glucose, with the indicated concentration of AVP added, to effectively reduce the final glucose concentration to 0.5 mM at the start of the secretion assay. AVP peptide was synthesized in-house and generously provided by Dr. Jean Rivier (Salk Institute).
Calcium responses in intact islets
We used islets from a triple transgenic offspring of a cross between mIns1-H2b-mCherry, lsl-GCaMP6 and either Gcg-Cre or Ucn3-Cre to label the alpha or mature beta cell lineages, respectively. Live islets were cultured overnight after the islet prep, placed on 35mm dishes with glass bottom (#1.5; MatTek Corporation), allowed to attach overnight and imaged in x, y, z and t on a Nikon A1R+ confocal microscope using a 40x lens with a long working distance. Treatments were continuously perfused over the islets using a Masterflex peristaltic pump at 2.5 ml per minute. Each protocol concluded with a 30 mM potassium chloride pulse to demonstrate viability and responsiveness throughout the treatment. Individual cells in individual z-planes were defined as regions of interest (ROI) and the green fluorescence intensity within the ROIs was plotted over time as a measure of calcium activity.
QUANTIFICATION AND STATISTICAL ANALYSIS
Bio-informatics analysis
Read refinement for quality and adapter contamination was performed using FASTQC, Scythe, and Sickle. After quality control and filtering, the libraries comparing immature to mature beta cells had an average library size of 21.4 million reads. The libraries comparing alpha and beta cells to beta cells of alpha cell-descent had an average library size of 19.2 million reads. The libraries of the postnatal maturation time series had an average library size of 12.8 million reads. Libraries were then aligned to mouse genome version GenCode M8 (GRCm38.p4 M8; mm10) using STAR (Dobin et al., 2013) with the two-pass method with default parameters, with the exception of a tighter mismatch restriction of no greater than 3% per read. Average unique read alignment was 87.5%, 86.6% and 86.9% for the three sets of libraries, respectively. Bigwigs were generated using the wiggle output option in STAR and Genome Utilities in the UCSC Genome Browser (Kent et al., 2002). Gene-level quantification was performed on sorted BAM files using featureCounts (Liao et al., 2014) with default parameters, counted by Gencode defined exons, and aggregated to the gene level. Differential expression analyses were performed using the edgeR generalized linear model approach and maximum likelihood method testing (Robinson et al., 2010). Results were filtered for statistical significance using the thresholds: FDR < 0.001 and an absolute log2FC > 1 across all experiments. In comparing immature and mature beta cells from adult islets, we observed 3394 differentially expressed genes (2117 Immature enriched, 1277 Mature enriched) that met these thresholds. Using the same criteria, we observed 2480 genes to be differentially expressed between any of the pair-wise comparisons in comparing alpha and beta cells with trans-differentiated cells. Heat maps were generated based on the n most up- and n most down-regulated genes between each pairwise comparison, with duplicate genes removed, where n = 750 for Figure 2G and n = 200 for Figure 6K. To enable the most direct comparison, the same analysis pipeline—from raw sequencing reads, to quantification and differential analyses— was also applied to compare our data against mouse PMPs (Razavi et al., 2015) and human beta cell subtypes (Dorrell et al., 2016). Distance matrices for the latter comparison were generated using the R package lattice, with RPKM values from each experiment used to correlate relatedness.
Bio-informatics visualization
All clustering and heat map visuals were generated using Gene Cluster 3.0 (de Hoon et al., 2004) on log transformed RPKM values normalized by the median and hierarchically clustered by the city block distance method. The R-package ComplexHeatmap (Gu et al., 2016) was used on the clustering output using default parameters, with the exception of the maturation time series experiment where the Pearson instead of the Euclidean distance method was used. All Venn diagrams were generated using the R library package VennDiagram (Chen and Boutros, 2011), using the edgeR output and our FDR and log2FC filtering criteria for gene selection. We included a lower RPKM threshold of 1 when comparing our immature beta cell transcriptomes with those of the PMPs (Razavi et al., 2015). KEGG gene set enrichment testing was performed on the immature beta data set using the R Bioconductor package GAGE (Luo et al., 2009) on the edgeR output that met the statistical threshold. Pathway visuals were created using the R-package Pathview (Luo and Brouwer, 2013).
Distribution algorithm
The distribution algorithm we developed to quantify the distribution of cell types across the cross-sectional surface of the islet is build up into three main parts: segmentation, classification, and distribution measurement. All classification was done manually for each cell by a person. The segmentation and the distribution measurements are handled using two separate Matlab functions that interface with Bitplane’s Imaris software. We utilized Imaris in this process to provide an accurate visual representation of the segmentations for verification and classification. After loading the data set into Imaris and converting the data to 32-bit float, the segmentation function is called in to segment the islet volume and nuclear volumes separately using a simple intensity based threshold for binary segmentation. The user is prompted to input which channel(s) should be used for each of these segmentation steps. The segmentation function then creates two surface objects in Imaris, one for the islet sub volume and the second for the nuclei. Both the islet and nuclei surface objects are visually verified for accuracy, followed by the manual classification of each individual cell into distinct bins by cell type. Once the classification is complete, the distribution measurement function is called to find the distributions of each classified group created by the researcher. The distribution measurement function creates a new channel that is the distance transformation with respect to the interior edge of the islet. The distance transformation channel intensity is then recorded at the center of each classified nucleus. Recorded measurements are written to a csv file containing all measurements along with the sample information.
Statistical Analysis
Data were analyzed by t-test, corrected for multiple comparisons using the Holm-Sidak method where appropriate. The distribution of cells across the islet was tested using a non-parametric Kruskal-Wallis test using Dunn’s multiple comparisons test. A post-hoc Kolmogorov-Smirnov test was conducted to compute the D statistic on the basis of the cumulative frequency distributions computed from raw data. Data are represented as mean ± SEM across, with n defined in the corresponding figure legend. Differences were considered significant when p<0.05. Statistics were computed using Prism (GraphPad Software, La Jolla, CA).
DATA AND SOFTWARE AVAILABILITY
Sequencing data sets
Sequencing data associated with this publication have been deposited in GEO under GSE88778, GSE88779 and GSE90766.
Distribution algorithm
The Matlab routine for the distribution algorithm described in this paper can be found here: http://huisinglab.com/cell_metabolism_2017/index.html.
ADDITIONAL RESOURCES
A resource website featuring UCSC browser plots searchable by plain text query for the RNAseq data presented in this paper can be found at: http://huisinglab.com/cell_metabolism_2017/index.html.
Supplementary Material
Acknowledgements
We gratefully acknowledge Dr. David Ron for sharing the Ero1lb antiserum and Drs. Jay Walters and Howard Davidson for sharing the G6pc2 antiserum generated by the late Dr. John Hutton. We thank Ms. Bridget McLaughlin from the UC Davis Flow Cytometry Shared Resource Laboratory (funded by NCI P30 CA093373 and NIH NCRR C06-RR12088, S10 RR12964 and S10 RR 026825) for help with flow cytometry and Dr. Marielle Cavrois for help with the Amnis Imagestream imaging cytometer (DOD grant W81XWH-11-1-0562). We thank Ms. Giselle Blanco for technical assistance. The research described in this paper was funded by a Career Development Award from the Juvenile Diabetes Research Foundation (2-2013-54) and an Individual Biomedical Research Award from the Hartwell Foundation to MOH.
Footnotes
References
- Ackermann AM, Zhang J, Heller A, Briker A, and Kaestner KH (2017). High-fidelity Glucagon-CreER mouse line generated by CRISPR-Cas9 assisted gene targeting. Mol Metab in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Hang Y, Mazur M, Yamamoto T, Guo M, Lindner J, Magnuson MA, and Stein R. (2010). MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes 59, 2530–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader E, Migliorini A, Gegg M, Moruzzi N, Gerdes J, Roscioni SS, Bakhti M, Brandl E, Irmler M, Beckers J, et al. (2016). Identification of proliferative and mature beta-cells in the islets of Langerhans. Nature 535, 430–434. [DOI] [PubMed] [Google Scholar]
- Beamish CA, Strutt BJ, Arany EJ, and Hill DJ (2016). Insulin-positive, Glut2-low cells present within mouse pancreas exhibit lineage plasticity and are enriched within extra-islet endocrine cell clusters. Islets 8, 65–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner C, van der Meulen T, Caceres E, Tigyi K, Donaldson CJ, and Huising MO (2014). The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics 15, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Hrvatin SS, Schuetz C, Bonal C, Rezania A, and Melton DA (2012). Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol 30, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Roose AN, Barrandon O, Maehr R, Arvanites AC, Davidow LS, Davis JC, Peterson QP, Rubin LL, and Melton DA (2014). Reversal of beta cell de-differentiation by a small molecule inhibitor of the TGFbeta pathway. eLife 3, e02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner-Weir S, Li WC, Ouziel-Yahalom L, Guo L, Weir GC, and Sharma A. (2010). Beta-cell growth and regeneration: replication is only part of the story. Diabetes 59, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, and Kaestner KH (2013). Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. J Clin Invest 123, 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Huangfu D, and Melton D. (2007). All beta Cells Contribute Equally to Islet Growth and Maintenance. PLoS Biol 5, e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M, Fowler MJ, Nicholson WE, Chu A, Hirshberg B, Harlan DM, and Powers AC (2005). Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem 53, 1087–1097. [DOI] [PubMed] [Google Scholar]
- Butler AE, Cao-Minh L, Galasso R, Rizza RA, Corradin A, Cobelli C, and Butler PC (2010). Adaptive changes in pancreatic beta cell fractional area and beta cell turnover in human pregnancy. Diabetologia 53, 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, and Caicedo A. (2006). The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci U S A 103, 2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, and Boutros PC (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, Baronnier D, Ghila L, Cigliola V, Jensen JN, Gu G, Furuyama K, Thorel F, Gribble FM, Reimann F, et al. (2014). Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, and Mansouri A. (2007). Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest 117, 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, and Mansouri A. (2009). The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell 138, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AR, Lam CJ, Rankin MM, King KA, Chen P, Martinez R, Li C, and Kushner JA (2016). Extreme obesity induces massive beta cell expansion in mice through self-renewal and does not alter the beta cell lineage. Diabetologia 59, 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, and Miyano S. (2004). Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, and Bhushan A. (2015). DNA methylation directs functional maturation of pancreatic beta cells. J Clin Invest 125, 2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGruccio MR, Mawla AM, Donaldson CJ, Noguchi GM, Vaughan J, Cowing-Zitron C, van der Meulen T, and Huising MO (2016). Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol Metab 5, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolensek J, Rupnik MS, and Stozer A. (2015). Structural similarities and differences between the human and the mouse pancreas. Islets 7, e1024405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, and Melton DA (2004). Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429, 41–46. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Schug J, Canaday PS, Russ HA, Tarlow BD, Grompe MT, Horton T, Hebrok M, Streeter PR, Kaestner KH, et al. (2016). Human islets contain four distinct subtypes of beta cells. Nat Commun 7, 11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning BE, Moltz JH, and Fawcett CP (1984). Actions of neurohypophysial peptides on pancreatic hormone release. Am J Physiol 246, E108–114. [DOI] [PubMed] [Google Scholar]
- Gao T, McKenna B, Li C, Reichert M, Nguyen J, Singh T, Yang C, Pannikar A, Doliba N, Zhang T, et al. (2014). Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metab 19, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano E, Bosco D, Cirulli V, and Meda P. (1991). Repeated glucose stimulation reveals distinct and lasting secretion patterns of individual rat pancreatic B cells. J Clin Invest 87, 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Habener JF, and Stanojevic V. (2012). alpha-cell role in beta-cell generation and regeneration. Islets 4, 188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera PL (2000). Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127, 2317–2322. [DOI] [PubMed] [Google Scholar]
- Huising MO, van der Meulen T, Vaughan JM, Matsumoto M, Donaldson CJ, Park H, Billestrup N, and Vale WW (2010). CRFR1 is expressed on pancreatic beta cells, promotes beta cell proliferation, and potentiates insulin secretion in a glucose-dependent manner. Proc. Natl. Acad. Sci. USA 107, 912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton JC, and Eisenbarth GS (2003). A pancreatic beta-cell-specific homolog of glucose-6-phosphatase emerges as a major target of cell-mediated autoimmunity in diabetes. Proc Natl Acad Sci U S A 100, 8626–8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston NR, Mitchell RK, Haythorne E, Pessoa MP, Semplici F, Ferrer J, Piemonti L, Marchetti P, Bugliani M, Bosco D, et al. (2016). Beta Cell Hubs Dictate Pancreatic Islet Responses to Glucose. Cell Metab 24, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas JC, Sharma A, Hasenkamp W, Ilkova H, Patane G, Laybutt R, Bonner-Weir S, and Weir GC (1999). Chronic hyperglycemia triggers loss of pancreatic beta cell differentiation in an animal model of diabetes. J Biol Chem 274, 14112–14121. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D. (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekens R, In ‘t Veld P, Mahler T, Schuit F, Van De Winkel M, and Pipeleers D. (1992). Differences in glucose recognition by individual rat pancreatic B cells are associated with intercellular differences in glucose-induced biosynthetic activity. J Clin Invest 89, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Miller K, Jo J, Kilimnik G, Wojcik P, and Hara M. (2009). Islet architecture: A comparative study. Islets 1, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Luo W, and Brouwer C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, and Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Pullen TJ, Chabosseau P, Zhang Q, Haythorne E, Cane MC, Nguyen-Tu MS, Sayers SR, and Rutter GA (2016). Disallowance of Acot7 in beta-Cells Is Required for Normal Glucose Tolerance and Insulin Secretion. Diabetes 65, 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JJ, Butler AE, Saisho Y, Monchamp T, Galasso R, Bhushan A, Rizza RA, and Butler PC (2008). Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes 57, 1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, and Melton DA (2014). Generation of functional human pancreatic beta cells in vitro. Cell 159, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, and Sussel L. (2011). Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev 25, 2291–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JA, Brelje TC, and Sorenson RL (1992). Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology 130, 1459–1466. [DOI] [PubMed] [Google Scholar]
- Piccand J, Strasser P, Hodson DJ, Meunier A, Ye T, Keime C, Birling MC, Rutter GA, and Gradwohl G. (2014). Rfx6 maintains the functional identity of adult pancreatic beta cells. Cell reports 9, 2219–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound LD, Oeser JK, O’Brien TP, Wang Y, Faulman CJ, Dadi PK, Jacobson DA, Hutton JC, McGuinness OP, Shiota M, et al. (2013). G6PC2: a negative regulator of basal glucose-stimulated insulin secretion. Diabetes 62, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen TJ, and Rutter GA (2013). When less is more: the forbidden fruits of gene repression in the adult beta-cell. Diabetes Obes Metab 15, 503–512. [DOI] [PubMed] [Google Scholar]
- Razavi R, Najafabadi HS, Abdullah S, Smukler S, Arntfield M, and van der Kooy D. (2015). Diabetes enhances the proliferation of adult pancreatic multipotent progenitor cells and biases their differentiation to more beta-cell production. Diabetes 64, 1311–1323. [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui J, Deng S, Arazi A, Perdigoto AL, Liu Z, and Herold KC (2017). Beta Cells that Resist Immunological Attach Develop during Progression of Autoimmune Diabetes in NOD Mice. Cell Metab in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. (2015). Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J 34, 1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Wang A, and Sander M. (2013). Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol 29, 81–105. [DOI] [PubMed] [Google Scholar]
- Shiota C, Prasadan K, Guo P, El-Gohary Y, Wiersch J, Xiao X, Esni F, and Gittes GK (2013). alpha-Cells are dispensable in postnatal morphogenesis and maturation of mouse pancreatic islets. Am J Physiol Endocrinol Metab 305, E1030–1040. [DOI] [PubMed] [Google Scholar]
- Smukler SR, Arntfield ME, Razavi R, Bikopoulos G, Karpowicz P, Seaberg R, Dai F, Lee S, Ahrens R, Fraser PE, et al. (2011). The adult mouse and human pancreas contain rare multipotent stem cells that express insulin. Cell Stem Cell 8, 281–293. [DOI] [PubMed] [Google Scholar]
- Solloway MJ, Madjidi A, Gu C, Eastham-Anderson J, Clarke HJ, Kljavin N, Zavala-Solorio J, Kates L, Friedman B, Brauer M, et al. (2015). Glucagon Couples Hepatic Amino Acid Catabolism to mTOR-Dependent Regulation of alpha-Cell Mass. Cell reports 12, 495–510. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Hollister-Lock J, Bonner-Weir S, and Weir GC (2015). Reduced Ki67 Staining in the Postmortem State Calls Into Question Past Conclusions About the Lack of Turnover of Adult Human beta-Cells. Diabetes 64, 1698–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talchai C, Xuan S, Lin HV, Sussel L, and Accili D. (2012). Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150, 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta M, Long SY, Wartschow LM, Rankin MM, and Kushner JA (2005). Very slow turnover of beta-cells in aged adult mice. Diabetes 54, 2557–2567. [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, and Herrera PL (2010). Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen T, Donaldson CJ, Caceres E, Hunter AE, Cowing-Zitron C, Pound LD, Adams MW, Zembrzycki A, Grove KL, and Huising MO (2015). Urocortin3 mediates somatostatin-dependent negative feedback control of insulin secretion. Nat Med 21, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen T, and Huising MO (2014). Maturation of Stem Cell-Derived Beta cells Guided by the Expression of Urocortin 3. Rev. Diabet. Stud 11, 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen T, and Huising MO (2015). Role of transcription factors in the transdifferentiation of pancreatic islet cells. J Mol Endocrinol 54, R103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen T, Xie R, Kelly OG, Vale WW, Sander M, and Huising MO (2012). Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PLoS One 7, e52181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicksteed B, Brissova M, Yan W, Opland DM, Plank JL, Reinert RB, Dickson LM, Tamarina NA, Philipson LH, Shostak A, et al. (2010). Conditional gene targeting in mouse pancreatic ss-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes 59, 3090–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen Z, Shiota C, Prasadan K, Guo P, El-Gohary Y, Paredes J, Welsh C, Wiersch J, and Gittes GK (2013). No evidence for beta cell neogenesis in murine adult pancreas. J Clin Invest 123, 2207–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E, Chin KT, Blais J, Harding HP, and Ron D. (2010). ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol 188, 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data sets
Sequencing data associated with this publication have been deposited in GEO under GSE88778, GSE88779 and GSE90766.
Distribution algorithm
The Matlab routine for the distribution algorithm described in this paper can be found here: http://huisinglab.com/cell_metabolism_2017/index.html.


