Abstract
Morphological development of the fungal pathogen Candida albicans is profoundly affected by ambient pH. Acidic pH restricts growth to the yeast form, whereas neutral pH permits development of the filamentous form. Superimposed on the pH restriction is a temperature requirement of approximately 37°C for filamentation. The role of pH in development was investigated by selecting revertants of phr2Δ mutants that had gained the ability to grow at acid pH. The extragenic suppressors in two independent revertants were identified as nonsense mutations in the pH response regulator RIM101 (PRR2) that resulted in a carboxy-terminal truncation of the open reading frame. These dominant active alleles conferred the ability to filament at acidic pH, to express PHR1, an alkaline-expressed gene, at acidic pH, and to repress the acid-expressed gene PHR2. It was also observed that both the wild-type and mutant alleles could act as multicopy suppressors of the temperature restriction on filamentation, allowing extensive filamentation at 29°C. The ability of the activated alleles to promote filamentation was dependent upon the developmental regulator EFG1. The results suggest that RIM101 is responsible for the pH dependence of hyphal development.
Candida albicans is the predominant fungal pathogen of humans (13). It is commonly associated with the gut of humans as well as other warm-blooded animals and is considered an obligate saprophyte (8, 24). The most important reservoir of C. albicans in human disease is believed to be endogenous, and this pathogen can infect most tissues and organs, indicating that it is well adapted for survival within the diverse environmental niches of its host (24). C. albicans is polymorphic, able to change reversibly between round budding yeast and elongated hyphae or filamentous growth forms. This morphological flexibility appears to be a key contributor to virulence (21). The yeast form predominates under standard culture conditions, but hyphal development occurs in diverse host niches and can be promoted in vitro by diverse culture conditions.
Among the environmental variables that influence filamentation in vivo, ambient pH has a defining role (1, 4, 10). Optimal filamentation occurs near neutral pH and is much reduced at pH below 6.0. The yeast form is exclusively present at pH 4.0 (4). In conjunction with neutral pH, filamentation is favored by an elevated temperature of around 37°C and is largely absent below 34°C (4).
The molecular mechanisms that govern the relationship between environment and morphological development are not clear. CPH1 and EFG1 encode two transcription factors important in this process. CPH1 appears to lie at the end of a mitogen-activated protein kinase cascade analogous to STE12 of Saccharomyces cerevisiae (20). EFG1 lies downstream of TPK2, a cyclic AMP (cAMP)-dependent protein kinase required for filamentation (3, 35). The environmental signals that activate these two pathways are unknown.
The pH response depends upon a zinc finger transcription factor, RIM101/PRR2, hereafter referred to as RIM101, which is related to PacC of Aspergillus nidulans and RIM101 of S. cerevisiae (6, 19, 27, 39, 40). Mutants lacking RIM101 are defective in filamentation and pH-dependent gene regulation (6, 27, 40). C. albicans responds to changes in environmental pH by differential expression of several genes, including PHR1 and PHR2 (23, 29, 31). C. albicans can be confronted by the neutral pH of blood during the course of a generalized infection or the acidic pH of the vagina and skin (24). Differential, pH-dependent expression of PHR1 and PHR2 is not restricted to in vitro conditions but appears to be similarly controlled by the pH of the host niche (7). PHR2 is an acid-expressed gene that is not expressed at detectable levels above pH 6.5. Mutants lacking PHR2 are unable to grow at acidic pH and exhibit morphological defects (23). PHR1 is an alkaline-expressed gene with the inverse pattern of expression. PHR1 and PHR2 encode functionally homologous proteins involved in cell wall biosynthesis, which is pivotal in cell shape changes during dimorphism (11, 22, 23, 29). pH-regulated dimorphism and pH-dependent differential gene expression are also observed in the polymorphic fungal pathogen Candida dubliniensis, which is closely related to C. albicans (16, 30).
In this report, further insight into the roles of PHR1, PHR2, and the pH response in morphogenesis was sought by reversion analysis of a homozygous phr2Δ mutant. The revertants were selected by restoration of growth at acidic pH. The phenotype of revertants included pH-independent expression of PHR1 and the ability to filament at acidic pH. Detailed analysis of two revertants demonstrated that they had acquired dominant activating mutations in RIM101. Unexpectedly, RIM101 was found to act as a multicopy suppressor of the temperature requirement for filamentation. In addition, we show that pH-regulated dimorphism but not pH-dependent gene expression requires the transcription factor Efg1p.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. YPD and YNB media were prepared as described before (33). Medium 199 was prepared as previously described (23), and the medium of Lee et al. was prepared as described before (18). Media were supplemented with uridine (25 mg/ml) as needed. 5′-Fluoroorotic acid-containing medium was prepared as described by Boeke et al. (2), except that uridine was substituted for uracil. Media were solidified with 2% agar. To test the effect of acid culture conditions on filamentation in liquid medium, cells were cultured overnight to the stationary phase at 28 or 37°C in medium 199, YNB, or Lee's medium, each adjusted to pH 4.0. The stationary-phase cells were inoculated into fresh medium of the same composition at a density of 5 × 106 cells/ml and incubated at 29 or 37°C for 4 h on a rotary shaker. Filamentation on agar-solidified medium was assessed using medium 199 (pH 4.0). The plates were spotted with 106 cells in 5 μl of sterile water and incubated at 37°C for 3 to 6 days.
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| SC5314 | Clinical isolate | 15 | |
| CAF3-1 | ura3Δ::λimm434/ura3Δ::λimm434 | 12 | |
| CFM-2 | phr2Δ::hisG/phr2Δ::hisG-URA3-hisG ura3Δ::λimm434/ura3Δ::λimm434 | 23 | |
| CFM-4 | phr2Δ::hisG/phr2Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 | 23 | |
| CFM-5 | phr2Δ::hisG/phr2Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434-(pBSK-TEF1pr::PHR1-URA3) | 23 | |
| CEM-1 | CFM-4 | RIM101-1426/RIM101 phr2Δ::hisG/phr2Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 | This work |
| CEM-2 | CFM-2 | RIM101-1751/RIM101 phr2Δ::hisG/phr2Δ::hisG-URA3-hisG ura3Δ::λimm434/ura3Δ::λimm434 | This work |
| CEM-5 | CEM-1 | RIM101-1426/rim101Δ::hisG-URA3-hisG phr2Δ::hisG/phr2Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 | This work |
| CEM-5U | CEM-5 | RIM101-1426/rim101Δ::hisG phr2Δ::hisG/phr2Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 | This work |
| CEM-6 | CEM-1 | rim101-1426Δ::hisG-URA3-hisG/RIM101 phr2Δ::hisG/phr2Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 | This work |
| CEM-7 | CEM-1 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-1426-URA3) RIM101-1426/RIM101 phr2Δ::hisG/phr2Δ::hisG | This work |
| CEM-8 | CEM-1 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-1426-URA3)n≥2 RIM101-1426/RIM101 phr2Δ::hisG/phr2Δ::hisG/phr2Δ::hisG | This work |
| CEM-10 | CEM-5U | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-1426-URA3) RIM101-1426/rim101Δ::hisG phr2Δ::hisG/phr2Δ::hisG | This work |
| CEM-16-1 | CFM-4 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-1426-URA3) RIM101/RIM101 phr2Δ::hisG/phr2Δ::hisG | This work |
| CEM-31-1 | CFM-4 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-URA3)RIM101/RIM101 phr2Δ::hisG/phr2Δ::hisG | This work |
| CEM-16-3 | CFM-4 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-1751-URA3) RIM101/RIM101 phr2Δ::hisG/phr2Δ::hisG | This work |
| CAF3-16-1 | CAF3-1 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-1426-URA3) RIM101/RIM101 | This work |
| CAF3-31-1 | CAF3-1 | ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-URA3) RIM101/RIM101 | This work |
| JKC18 | cph1Δ::hisG/cph1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 | 20 | |
| JKC18-pSM2 | JKC18 | cph1Δ::hisG/cph1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -URA3) RIM101/RIM101 | This work |
| JKC18-16 | JKC18 | cph1Δ::hisG/cph1Δ::hisG/cph1Δ::hisG ura3Δ::λimm434 (-pBSK+ -RIM101-1426-URA3) RIM101/RIM101 | This work |
| JKC18-31 | JKC18 | cph1Δ::hisG/cph1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 -(pBSK+ -RIM101-URA3) RIM101/RIM101 | This work |
| HLC67 | efg1Δ::hisG/efg1Δ::hisG ura3Δ:: λimm434/ura3Δ::λimm434 RIM101/RIM101 | 21 | |
| HLC67-pSM2 | HLC67 | efg1Δ::hisG/efg1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 -(pBSK+ -URA3) RIM101/RIM101 | This work |
| HLC67-16 | HLC67 | efg1Δ::hisG/efg1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 −(pBSK+ -RIM101-1426-URA3) RIM101/RIM101 | This work |
| HLC67-31 | HLC67 | efg1Δ::hisG/efg1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 −(pBSK+ -RIM101-URAe) RIM101/RIM101 | This work |
| CDB1 | cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG ura3Δ::λimm434/ura3Δ::λimm434 RIM101/RIM101 | 21; Bockmühl, unpublished data | |
| CDB1-pSM2 | CDB1 | cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG | This work |
| ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -URA3) RIM101/RIM101 | |||
| CDB1-16 | CDB1 | cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG | This work |
| ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+-RIM101-1426-URA3) RIM101/RIM101 | |||
| CDB1-31 | CDB1 | cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG | This work |
| ura3Δ::λimm434/ura3Δ::λimm434-(pBSK+ -RIM101-URA3) RIM101/RIM101 |
Isolation of phr2Δ revertants.
Strain CFM-4 (phr2Δ ura3Δ) (23) was grown to stationary phase in YPD (pH 7.0). After washing with sterile distilled H2O, 108 cells were spread on each of five YNB agar plates adjusted to pH 4.0 and containing 25 μg of uridine per ml. The plates were incubated for 3 days at 30°C. Colonies growing at this restrictive pH were recovered with a median frequency of approximately 1.5 × 10−7. One of the CFM-4 revertants, named CEM-1, was chosen for further analysis. A second revertant, designated CEM-2, was derived from strain CFM-2 (phr2Δ URA3) (23).
Disruption of RIM101 in CEM-1.
Plasmid pARA3 (27) was used to delete one or the other of the two RIM101 alleles in CEM-1. Plasmid DNA was digested with HindIII and SspBI to release a 5-kb RIM101 disruption construct (27), and 8 μg of the gel-purified fragment was used to transform CEM-1 to Uri+ using lithium acetate (14). Transformants were selected on YNB buffered to pH 7.0 with 150 mM HEPES. The resulting colonies were replica plated in parallel to YNB adjusted to either pH 4.0 or 7.0 and assessed for growth following incubation for 2 days at 30°C. Transformants that retained the CEM-1 phenotype, growth at both pH 4 and pH 7, were designated type-1 transformants. CEM-5 is a representative type-1 transformant. Transformants that had lost the ability to grow at pH 4.0 were classified as type-2 transformants, as represented by strain CEM-6. Integration of the transforming DNA at the RIM101 locus was confirmed by Southern blotting.
Cloning and sequencing of RIM101 alleles from the revertants.
RIM101 was amplified by PCR using oligonucleotide primers PRR2-3 (5′-ACGACCTTATATGCGTAATCC-3′) and PRR2-4 (5′-GAACCATGTAAATAGAGAACGG-3′). The primers are located 753 nucleotides 5′ and 181 nucleotides 3′, respectively, of the 1,986-bp RIM101 coding region. Amplification was performed with an initial denaturation step of 95°C for 3 min, followed by 15 cycles, each cycle consisting of 40 s at 95°C, 1 min at 58°C, and 2 min at 72°C and a final step of 10 min at 72°C. When 1 μg of genomic DNA from type-1 and type-2 transformants was used as the template, two amplification products of 2.9 and 5 kb were obtained. These corresponded to the functional and disrupted alleles, respectively. The 2.9-kb product from two independent type-1 transformants was cloned into vector pCR2.1 (Invitrogen) to generate plasmids pCR-16-1 and pCR-16-2. Analogous cloning from two independent type-2 transformants yielded plasmids pCR-31-1 and pCR-31-2. The inserts from all four plasmids were sequenced on both strands. Four additional independent clones were generated and partially sequenced to verify sequence differences.
In analyzing the RIM101 locus of revertant CEM-2, both alleles were amplified as a mixed PCR product using primers PRR2-3 and PRR2-4, 1 μg of genomic DNA as the template, and 15 cycles of amplification. The resulting 2.9-kb PCR product was gel purified and cloned into vector pCR2.1. The PCR product was sequenced directly using primer PRR2-11 (5′-CCTCAACAGCAACACCCAC-3′) or following cloning into pCR2.1. Sequence analysis identified two cloned alleles, the wild type, represented by plasmid pCR-31-3, and a mutant allele, represented by plasmid pCR-16-3.
Introduction of recovered RIM101 alleles by transformation.
The RIM101 alleles recovered from the revertants were introduced into various C. albicans strains by transformation. The inserts from plasmids pCR-16-1, pCR-16-3, pCR-31-1, and pCR-31-3 were released by XbaI-SpeI digestion and cloned into the XbaI site of plasmid pSM-2. Plasmid pSM-2 consists of a 3.85-kb XbaI fragment containing URA3 blunt-end ligated into the SmaI site of pBSK(+) (Stratagene). The resulting plasmids, pEM-16-1, pEM-16-3, pEM-31-1, and pEM-31-3, respectively, were made linear by digestion at the unique HpaI site adjacent to URA3 prior to transformation. Uri+ transformants were selected, and proper targeting of the plasmid was confirmed by Southern blot analysis. The transformed strains included CFM-4 (phr2Δ ura3Δ), CAF3-1 (ura3Δ), CEM-1 (ura3Δ RIM101/RIM101-1426), CEM-5U (ura3Δ rim101/RIM101-1426), JKC18 (ura3Δ cph1Δ), HLC67 (ura3Δ efg1Δ), and CDB1 (ura3Δ cph1Δ efg1Δ). CDB1 (21; D. P. Bockmühl, unpublished data), and CEM-5U are Uri− derivatives of HLC54 and CEM-5A (rim101Δ/RIM101-1426), respectively, obtained by selection on medium containing 5-fluoroorotic acid. All transformation events were confirmed by Southern blot analysis.
Southern and Northern blot analysis.
Hybridization of Southern and Northern blots was conducted as previously described (23; Bockmühl, unpublished). Hybridization probes used to detect PHR1, PHR2, and RIM101 have been described previously (23, 27, 29). For hybridization with HWP1, a PCR product amplified with nucleotide primers HWP1-1 (5′-ATGAGATTATCAACTGCTCAA-3′), HWP1-2 (5′-TTAGATCAAGAATGCAGCAAT-3′), and genomic DNA from strain SC5314 as the template, was used (36). Hybridization with ACT1 was conducted with a PCR amplification product generated with primers OK1 (5′-TGTTTTCCCATCCCTCGT-3′) and OK2 (5′-TTCGTCGTATTCTTGTTT-3′) and genomic DNA from SC5314 as the template. Both probes generated by PCR were partially sequenced prior to use to verify the presence of the desired amplification product.
RESULTS
Spontaneous revertants of phr2Δ mutants are altered in growth and filamentation.
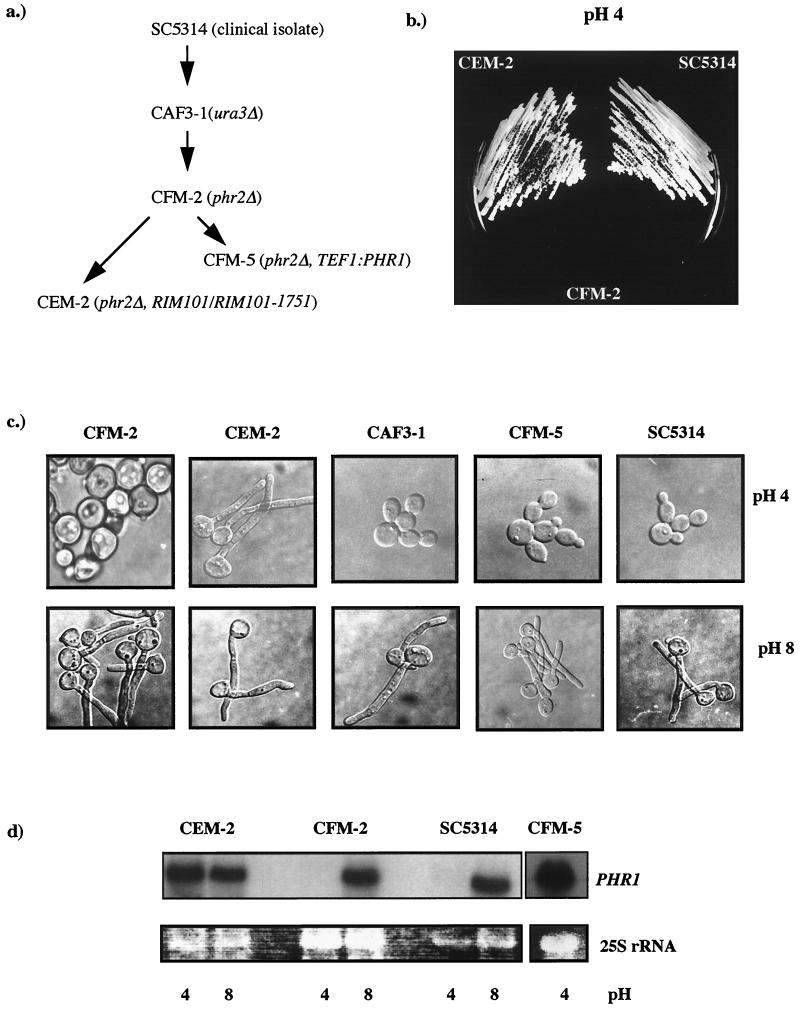
Following a shift from a permissive pH of 7.0 to a restrictive pH of 4.0, phr2Δ mutants undergo one mass doubling and then cease growth (23). This phenotype allowed easy selection of revertants in which growth was restored at the restrictive pH (Fig. 1b). Spontaneous revertants arose at a median frequency of 1.5 × 10−7. In addition to the growth defect, phr2Δ mutants incubated at the restrictive pH form enlarged, morphologically abnormal cells due to defective cell wall assembly (11, 23). This aberrant morphology was no longer evident upon microscopic examination of cells of the revertant, suggesting that, in addition to the growth defect, the defect in cell wall biosynthesis was also suppressed. An unexpected phenotype associated with the revertants was the ability to filament at acidic pH. It was noted that the colonies of the revertants on either neutral or acidic medium exhibited an atypical rough colony morphology, which has been associated with the presence of hyphae and pseudohyphae (20). Microscopic examination of cells from these colonies verified that they were composed of a mixture of cell forms, including yeasts, hyphae, and pseudohyphae. This observation was surprising because acidic pH suppresses filamentation of wild-type C. albicans. Therefore, the effect of pH on the ability of the revertant strains to initiate hyphal development in the form of germ tubes was examined. When tested in medium 199 (pH 7.5) at 37°C, conditions conducive to germ tube formation, there was no notable difference between the wild-type control strain, SC5314, and the revertant, CEM-1. The percent germination in three independent experiments averaged 96% ± 1% for SC5314 and 88% ± 6% for CEM-1. However, a dramatic difference was observed when the medium was adjusted to pH 4.0. Less than 1% of the wild-type cells formed germ tubes, but germination of CEM-1 or CEM-2 was comparable to that at neutral pH, 88% ± 1% (Fig. 1c; see Fig. 4b; and data not shown). Similar frequencies of germ tube formation at pH 4 were observed with the revertant CEM-2 and when alternative media were used, including the medium of Lee et al. and YNB. Additional strain controls included the phr2Δ parents of the revertants, CFM-2 and CFM-4, and strain CAF3-1, from which CFM-2 and CFM-4 were derived. At pH 4.0 CFM-2 and CFM-4 exhibited the previously reported abnormalities (23) and strain CAF3-1 failed to form germ tubes, indicating that the phenotype was not due to extraneous mutations introduced in the construction of CFM-2 and CFM-4 (Fig. 1c). The ability of the revertants to filament at acidic pH was not restricted to liquid suspension culture but was also observed on agar-solidified medium 199 and YNB (data not shown).
FIG. 1.
Phenotypes of revertant CEM-2. (a) Genealogy of CEM-2. The relevant genotypes are shown. (b) Growth of wild-type strain SC5314, the phr2Δ mutant CFM-2, and the revertant CEM-2 after 48 h of incubation on YNB at pH 4.0 and 30°C. (c) Morphology of the indicated strains 3 h postinoculation into medium 199 at pH 4.0 or pH 8 and 37°C. (d) PHR1 expression as a function of ambient pH. RNA was isolated from the indicated strains cultured in medium 199 at the pH indicated below the panels. The upper panel shows the Northern blot hybridized with PHR1. The lower panel shows the corresponding ethidium bromide-stained agarose gel prior to blotting.
FIG. 4.
Multicopy suppression of temperature restriction. (a) Equal amounts of genomic DNA from strain CEM-8 [RIM101/(RIM101-1426)n≥3], CEM-7 (RIM101/RIM101-1426/RIM101-1426), and the parental strain CEM-1 (RIM101/RIM101-1426) were digested with either PstI (left panel) or EcoRI (right panel). The Southern blots were hybridized with RIM101 or ACT1, as indicated. (b) Morphology of CEM-1, CEM-7, and CEM-8 following 3 h of incubation in medium 199 at pH 4.0 or pH 7.0 and 29 or 37°C.
PHR1 expression is no longer pH dependent in the revertants.
Previous studies demonstrated that Phr1p and Phr2p are functionally analogous (23). Since PHR1 is normally expressed only at a pH of ≥5.5, its ability to complement a phr2Δ mutation was demonstrated by using the constitutive promoter from TEF1 to drive PHR1 expression at acidic pH (23). Expression of PHR1 in this manner complemented both the growth and morphological defects of the phr2Δ mutant. Based on these observations, we asked whether the pH dependence of PHR1 expression was altered in the spontaneous revertants. The phr2Δ mutant exhibited the expected pH-dependent pattern of PHR1 expression, as assessed by Northern blot analysis (Fig. 1d). In contrast, expression of PHR1 in the revertant strains CEM-1 and CEM-2, as well as three other independent revertants, was unaffected by the pH of the culture medium; comparable levels of mRNA were present in cells cultured both at neutral and at acidic pH (Fig. 1d and data not shown).
The constitutive expression of PHR1 in the revertants could account for the restoration of growth and cell morphology, but it was not known if this could account for the ability of the revertants to filament at acidic pH. To address this issue, strain CFM-5, in which PHR1 is constitutively expressed from the TEF1 promoter (23), was examined for its ability to filament at pH 4.0. No filamentation was observed (Fig. 1c) even though the steady-state levels of PHR1 mRNA in CFM-5 were similar to that in the revertants (Fig. 1d). Thus, the acidic filamentation phenotype of the revertants was not directly related to the pH-independent expression of PHR1.
Evidence of heterozygosity at the RIM101 locus of the revertants.
The foregoing suggested either that the revertants contained a single mutation that simultaneously affected PHR1 expression and filamentation or that the revertants had acquired a minimum of two mutations, one specific to each of these phenotypes. Two pieces of evidence suggested that the revertants had acquired a single dominant mutation. The frequency of revertants was much higher than expected for a spontaneous homozygous recessive mutation but possibly lower than expected for other events that might uncover an existing recessive mutation. Furthermore, low-dose UV treatment resulted in a high frequency of back-mutation to the parental phenotype, consistent with a heterozygous dominant mutation (F. A. Mühlschlegel and W. A. Fonzi, unpublished data). These observations were coupled with recent data showing that the pH response regulator RIM101 controls pH-dependent expression of PHR1 and PHR2 and is required for filamentous growth (6, 27). Furthermore, dominant active mutations of the RIM101 orthologs of Aspergillus nidulans and Yarrowia lipolytica result in pH-independent expression of alkaline-induced genes (17, 39), analogous to the expression pattern of PHR1 in the revertants. Thus, we tested the hypothesis that a heterozygous dominant mutation in RIM101 gave rise to the revertants. This was tested by targeted deletion of one allele of RIM101. If in the revertants this locus harbored a heterozygous dominant allele, then disruption of the dominant allele would restore the parental phenotypes, whereas disruption of the wild-type allele would not. These alternate outcomes were expected to occur with roughly similar frequency.
The revertant CEM-1 was transformed with a hisG-URA3-hisG cassette replacing 1,270 bp of the RIM101 coding region, and 40 Uri+ transformants were recovered at pH 7.0 from two independent transformations. Twelve representative isolates were examined by Southern analysis, and all were disrupted at the desired locus, indicating that the majority of transformants resulted from homologous recombination at the RIM101 locus.
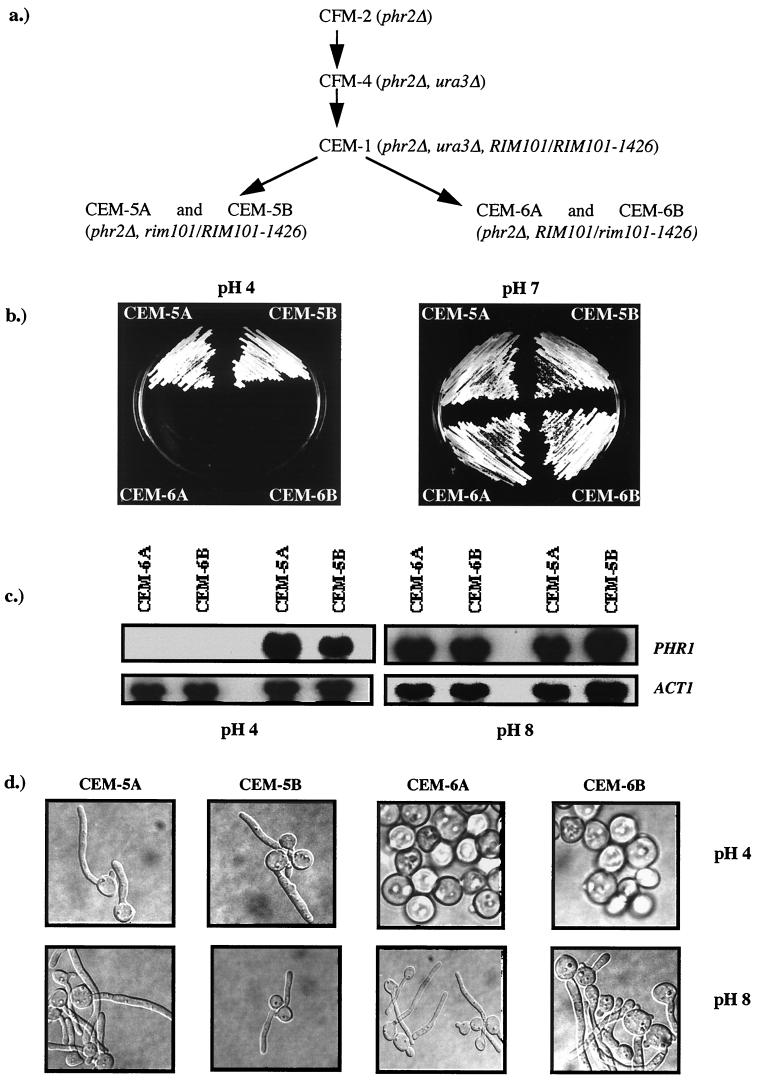
The growth phenotype of the transformants was consistent with the hypothesized heterozygosity. Twenty-five of the 40 transformants were the same as CEM-1, that is, they retained the ability to grow at both acidic and alkaline pH. These were designated type-1 transformants and are represented by strains CEM-5A and CEM-5B (Fig. 2b). The other 15 transformants, however, were unable to grow at pH 4.0, indicating a loss of the revertant phenotype and restoration of the parental growth pattern. This is the phenotype expected for disruption of the dominant allele. These were called type-2 transformants and are represented by strains CEM-6A and CEM-6B (Fig. 2b).
FIG. 2.
Effect of RIM101 disruption in CEM-1. (a) Genealogy of revertant CEM-1 and transformants CEM-5A, CEM-5B, CEM-6A, and CEM-6B. The relevant genotypes are indicated. (b) Growth of strains on YNB at pH 4.0 or 7.0 following 48 h at 30°C. (c) PHR1 expression as a function of ambient pH. RNA was isolated from the indicated strains cultured in medium 199 at the pH indicated below the panels. The upper panel shows the Northern blot hybridized with PHR1. The lower panel was hybridized with ACT1. (d) Morphology of the indicated strains 3 h postinoculation into medium 199 at pH 4.0 or pH 8 and 37°C.
Restoration of the parental growth phenotype was accompanied by the simultaneous restoration of the pH dependence of PHR1 expression. This was demonstrated with Northern blots of the type-2 strains CEM-6A and CEM-6B, in which PHR1 mRNA was abundant in alkaline-grown cells and absent in acid-grown cells (Fig. 2c). In contrast, the type-1 transformants CEM-5A and CEM-5B maintained the pH-independent expression pattern characteristic of the revertants. In addition, all 25 type-1 transformants maintained the capacity to filament at pH 4.0, while all 15 type-2 transformants lost this ability (Fig. 2d). Thus, all three phenotypes of the revertants could be simultaneously restored to parental types by disruption of one allele of RIM101. These findings were consistent with the hypothesis that the revertants were heterozygous for a dominant active mutation at the RIM101 locus.
One allele of RIM101 is mutated in the revertants.
Direct evidence for a heterozygous mutation at the RIM101 locus was obtained by cloning and sequencing both alleles. Because one allele was deleted and genetically tagged in the type-1 and type-2 transformants, the functional allele in these strains could be specifically identified and cloned. The intact allele was amplified by PCR from the type-1 transformants CEM-5A and CEM-5B and the type-2 transformants CEM-6A and CEM-6B. The resulting 2.9-kb fragment encompassed the 1,986-bp open reading frame plus 753 bp 5′ and 184 bp 3′. The allele recovered from CEM-5A and CEM-5B contained a single base substitution, a C-to-T transition at position +1426 of the open reading frame. This mutation lies in the first position of glutamine codon 476 and converts it to a nonsense codon. As a result, the 661-amino-acid protein predicted for the wild-type gene is truncated by 186 amino acids. Thus, the revertant phenotype exhibited by these type-1 transformants was associated with this mutant allele. In contrast, the product recovered from CEM-6A and CEM-6B was identical in sequence to that previously reported for the wild-type allele (27). Thus, the loss of the revertant phenotype in these type-2 transformants was associated with disruption of the mutant allele and retention of the wild-type allele.
The sequence of RIM101 was examined in a second independent revertant, CEM-2. PCR amplification using genomic DNA as the template produced a mixed product derived from both alleles. Direct sequencing of this product demonstrated that the chromatographic peak corresponding to position +1751 of the open reading frame consistently exhibited overlapping nucleotides of C and A. Sequence analysis of multiple subclones of the PCR product demonstrated two types of clones, one having a C at position 1751, the other having an A. The cytosine corresponds to the wild-type sequence. The adenine transversion converts serine codon 584 to a nonsense codon. This would truncate the protein by 78 residues. Thus, two independent revertants, CEM-1 and CEM-2, had both acquired a nonsense mutation in one allele of RIM101 and both mutations resulted in a carboxy-terminal truncation of the predicted protein.
The mutant alleles of RIM101 confer the revertant phenotypes.
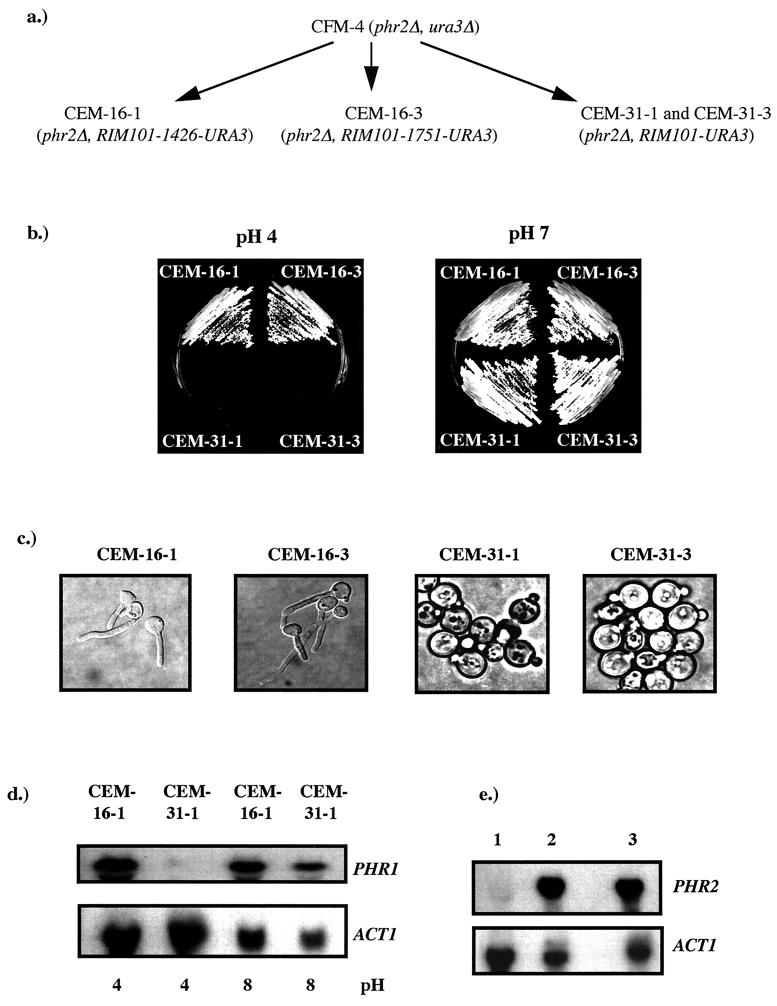
To test whether the mutant alleles of RIM101 were causative of the revertant phenotype, the mutant allele recovered from CEM-1, designated RIM101-1426, or the one from CEM-2, designated RIM101-1751, were transformed into the phr2Δ mutant CFM-4 and integrated at the URA3 locus. The Uri+ transformants were selected at pH 7.0, and 10 RIM101-1426 and 10 RIM101-1751 transformants were characterized. Representative transformants CEM-16-1, containing RIM101-1426, and CEM-16-3, containing RIM101-1751, are shown in Fig. 3. Like the revertants CEM-1 and CEM-2, the transformants acquired the ability to grow at pH 4.0 (Fig. 3b). Similarly, the transformants had gained the ability to filament at pH 4.0, and the frequency of germ tube formation was similar to that in the revertants, 89% ± 4% (Fig. 3c). Northern blot analysis demonstrated that, as in the revertants, expression of PHR1 was pH independent (Fig. 3d). Identical phenotypes were observed in all 10 transformants, and no differences were observed between cells receiving RIM101-1426 or RIM101-1751. In contrast, introduction of the wild-type allele recovered from revertant CEM-1 or CEM-2 had no obvious phenotypic consequences in the 10 transformants examined. Representative transformants CEM-31-1 and CEM-31-3 are shown in Fig. 3. Thus, a single copy of the mutant allele was sufficient to impart the pleiotropic phenotypes observed in the revertants. Also, since these strains contained, in addition to the mutant allele, both wild-type alleles at the RIM101 locus, RIM101-1426 and RIM101-1751 were at least partially dominant to the wild type.
FIG. 3.
Effect of mutant alleles of RIM101. (a) Genealogy of strains. Transformation of the mutant alleles RIM101-1426 and RIM101-1751 into CFM-4 produced strains CEM-16-1 and CEM-16-3, respectively. Introduction of the wild-type allele produced control strains CEM-31-1 and CEM-31-3. (b) Growth of strains on YNB at pH 4.0 or 7.0 after 48 h at 30°C. (c) Morphology of the indicated strains 3 h postinoculation into medium 199 at pH 4.0 and 37°C. (d) PHR1 expression as a function of ambient pH. RNA was isolated from the indicated strains cultured in medium 199 at the pH indicated below the panels. The upper panel shows the Northern blot hybridized with PHR1. The lower panel was hybridized with ACT1. (e) Effect of RIM101-1426 on PHR2 expression. RNA was isolated from CAF3-1-16 (lane 1), CAF3-1-31 (lane 2), and SC5314 (lane 3) grown in medium 199 at pH 4. The Northern blots were hybridized with PHR2 or ACT1, as indicated. CAF3-16-1 contains RIM101-1426. CAF3-31-1 contains the wild-type allele. SC5314 is a wild-type control.
The phenotype conferred by the mutant alleles was independent of the status of the PHR2 locus. When RIM101-1426 was transformed into CAF3-1, which is wild type for PHR2, filamentation and PHR1 expression became pH independent (data not shown). Since this transformant, strain CAF3-1-16, was wild type for PHR2, this allowed examination of the effect of the mutation on PHR2 expression. In wild-type cells PHR2 is repressed under alkaline growth conditions and expressed at acidic pH. In rim101Δ mutants, PHR2 is constitutively expressed (6, 27). In the presence of RIM101-1426, expression of PHR2 was not detected at either acidic or alkaline pH (Fig. 3e). As a control, the wild-type allele of RIM101 allele was introduced into CAF3-1, producing strain CAF3-1-31. This had no effect on filamentation or expression of PHR2 (Fig. 3e and data not shown).
Multiple copies of RIM101 partially suppress the temperature requirement for filamentation.
Although the mutant alleles bypassed the pH requirement for filamentation, the strains were not constitutively hyphal and still required a temperature of approximately 37°C for filamentation to occur. Fortuitously, we observed that the mutant allele could act as a multicopy suppressor of the temperature requirement. The revertant CEM-1 was transformed with plasmid pEM-16-1 containing the mutant allele RIM101-1426. Southern blot analysis of the transformants demonstrated that some had integrated a single copy of the plasmid at the URA3 locus and others had integrated multiple tandem copies. Integration of a single copy was indicated by a 1-kb band in addition to the 8.2-kb PstI hybridization band present in the parental strain CEM-1 (Fig. 4a). The intensity of the 1-kb band was enhanced in some of the transformants (Fig. 4a), in comparison to control hybridizations with the ACT1 probe, indicating integration of multiple tandem copies. Two transformants containing a single copy of the plasmid, represented by strain CEM-7, and four containing multiple tandem copies, represented by strain CEM-8, were examined. All of these transformants maintained the ability to filament at pH 4.0 or pH 7.5 at 37°C (Fig. 4b). As with wild-type strains, no filamentation occurred at 25°C regardless of the pH (data not shown). However, at 29°C, germ tube formation was >70% for those transformants containing multiple copies of the plasmid but <1% for transformants with a single integrated copy. Wild-type cells and the parental strain CEM-1 also failed to filament at this temperature. Low-temperature filamentation was independent of pH and was observed at both pH 4.0 and pH 7.5. The multicopy integrants also exhibited extensive filamentation on agar-solidified medium at 29°C (data not shown). Low-temperature filamentation was also observed when multiple copies of RIM101-1426 were introduced into CAF3-1, indicating that the phenotype was independent of the presence or absence of PHR2 (data not shown).
The requirement for multiple copies of RIM101-1426 was not due to incomplete dominance of the mutant allele. This possibility was raised by the fact that the CEM-1 transformants contain one wild-type allele of RIM101 and CAF3-1 transformants contain two. The wild-type allele could conceivably interfere with complete expression of the mutant allele. However, germ tube formation by strain CEM-5A was restricted to 37°C. CEM-5A was constructed by disruption of the wild-type allele of RIM101 in the revertant CEM-1 and thus contains a single mutant allele, RIM101-1426. Similarly, germ tube formation by strain CEM-10 was also restricted to 37°C. CEM-10 was derived from strain CEM-5A by integration of a single copy of plasmid pEM-16-1. Thus, CEM-10 lacked the wild-type allele and contained two copies of RIM101-1426. Analysis of 11 additional strains analogous to CEM-10 gave the same result. These data indicated that more than two copies of RIM101-1426 are required to suppress the temperature requirement for filamentation.
As a control, strains containing multiple copies of the wild-type allele were constructed by transformation of CAF3-1 (PHR2/PHR2) and CFM-4 (phr2Δ/phr2Δ) with plasmid pEM-31-1. Unexpectedly, in both backgrounds, strains containing multiple tandem copies of the plasmid exhibited a high frequency of germ tube formation at 29°C, comparable to strains with multiple copies of the mutant allele. Unlike the transformants harboring RIM101-1426, however, filamentation was restricted to neutral pH (data not shown). To ensure that suppression was not due to multiple copies of the vector sequences or the marker gene, strains CEM-1 and CAF3-1 were transformed with plasmid pSM-2, which lacks the RIM101 sequences present in pEM-16-1 and pEM-31-1. Germ tube formation by strains containing multiple copies of pSM-2 was restricted to 37°C. Thus, multiple copies of either the wild-type or mutant allele of RIM101 can partially suppress the temperature requirement for filamentation.
RIM101-1426 does not bypass EFG1.
EFG1 encodes a transcription factor that lies downstream of a cAMP-dependent protein kinase signal pathway (21, 35, 37). Deletion of EFG1 prevents germ tube formation and restricts morphological development largely to pseudohyphae under a wide variety of inducing conditions (21, 35). The developmental defect imparted by an efg1Δ mutation is potentiated by deletion of CPH1, which encodes a transcription factor at the terminus of a mitogen-activated protein kinase cascade (20, 21). An efg1Δ cph1Δ mutant is restricted entirely to the yeast morphology under most, but not all, conditions (21, 28, 34), even though the cph1Δ mutation alone does not prevent germ tube formation in liquid medium and affects filamentation on only a few solid induction media (20). The dominant active alleles of RIM101 provided the opportunity to access the interaction of the pH response pathway with these other developmental pathways.
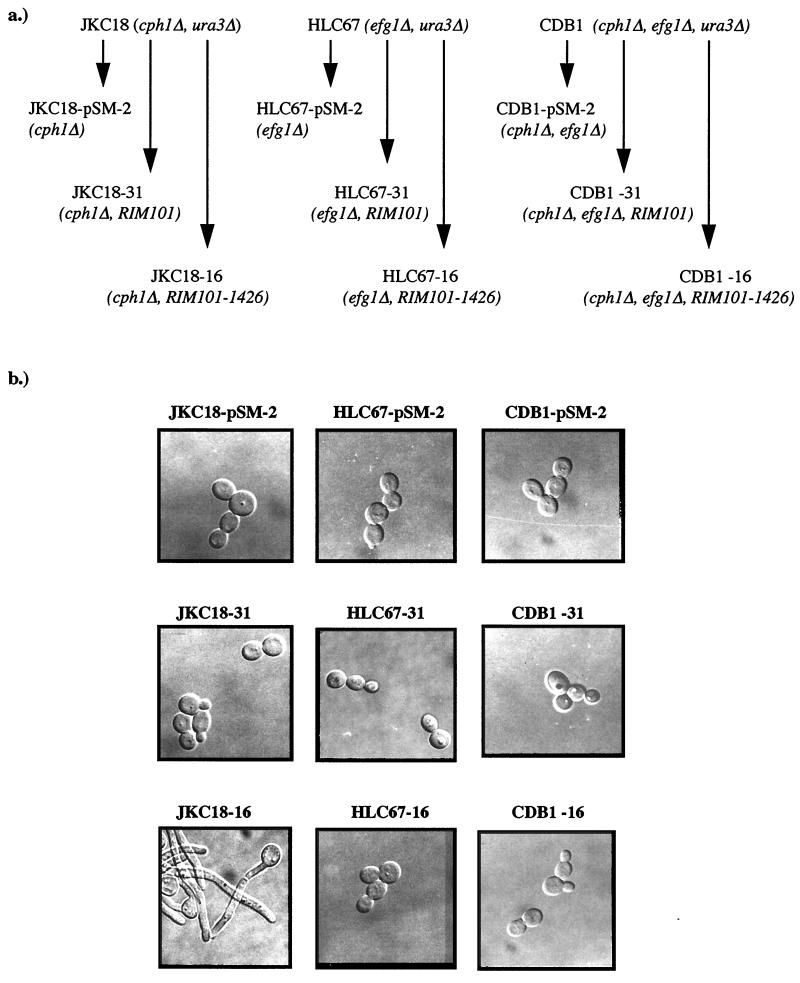
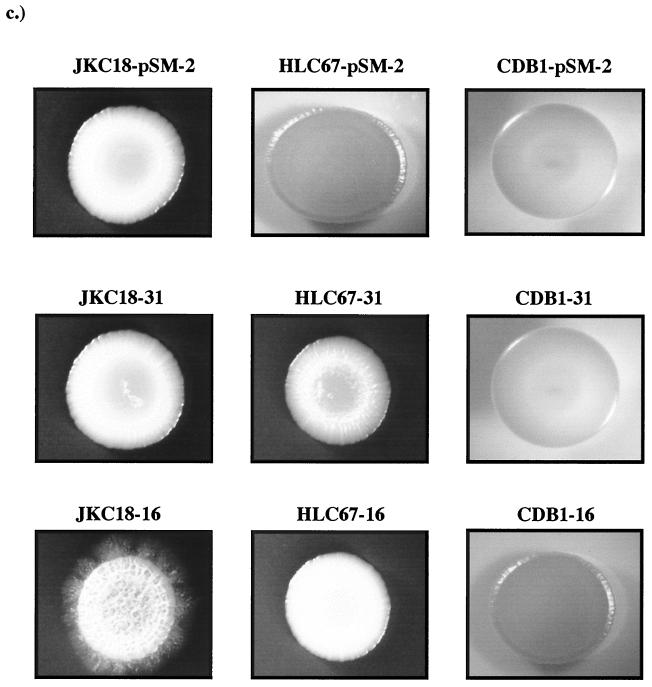
Strains JKC18 (cph1Δ), HLC67 (efg1Δ), and CDB1 (efg1Δ cph1Δ) were transformed with plasmid pEM-16-1 containing RIM101-1426, plasmid pEM-31-1 containing the wild-type allele, or pSM-2, the base vector. Single-copy integration events were verified by Southern analysis, and five such isolates from each transformation were assessed for their ability to form germ tubes and to form hyphae on solid medium at pH 4.0 and 37°C. As shown in Fig. 5b and c, germ tube formation and filamentation in liquid medium and on agar were both blocked by the efg1Δ mutation either alone or in combination with the cph1Δ mutation. The cph1Δ mutation alone did not interfere with the ability of RIM101-1426 to promote filamentation at acidic pH (Fig. 5b and c). Control strains transformed with the wild-type allele or vector alone were identical to the parental mutants. These results demonstrate that the filamentation phenotype conferred by RIM101-1426 is EFG1 dependent.
FIG. 5.
Effect of mutations in CPH1 and EFG1 on expression of RIM101-1426. (a) Genealogy of strains. (b) Morphology of strains 3 h postinoculation into medium 199 at p/H 4.0 and 37°C. (c) Colony morphology following 6 days of incubation on medium 199 at pH 4.0 and 37°C.
The inability of RIM101-1426 to bypass the efg1Δ mutation was also evident in its lack of effect on developmental gene expression. HWP1 encodes a cell wall protein whose expression is induced in filamentous cells and requires EFG1 for expression (32, 36). HWP1 was not expressed in the efg1Δ mutant in the presence or absence of RIM101-1426 (Fig. 6).
FIG. 6.
Effect of EFG1 on expression of RIM101-1426. RNA was isolated from HLC67-16-1 (efg1Δ CPH1 RIM101-1426), CDB1-16-1 (efg1Δ cph1Δ RIM101-1426), and CEM-16-1 (EFG1 CPH1 RIM101-1426) 3 h postinoculation into medium 199 at pH 4 and 37°C. The Northern blot was hybridized with PHR1, HWP1, or ACT1, as indicated.
pH-dependent gene expression does not require EFG1.
Since EFG1 was required for expression of the filamentation phenotype, the effect of EFG1 on the gene expression phenotype of the RIM101-1426 mutation was examined. The presence of the activated allele of RIM101 in HLC67-16 (efg1Δ RIM101-1426) resulted in expression of PHR1 at pH 4.0, as was observed in an EFG1 wild-type background (Fig. 3 and 6). As previously observed (29), PHR1 was not detected in the wild-type strain SC5314 at pH 4.0 (data not shown). Thus, the interaction between RIM101 and EFG1 that is required for filamentation is not required for pH-dependent gene regulation.
DISCUSSION
In examining spontaneous extragenic suppressors of the pH-conditional growth defect of phr2Δ mutants, we identified mutations in the C. albicans pH response regulator RIM101 that bypassed the pH requirement for hyphal development. These results suggest that RIM101 is the key element controlling the pH dependence of dimorphism in vitro. The mutations resulted in a gain of function at acidic pH, as evidence by the expression of PHR1 at pH 4.0. Normally PHR1 is not expressed below pH 5.5, and its expression is dependent upon PRR1/RIM101 (26, 27). The aberrant expression of PHR1 in the revertants is likely to account for the restoration of growth, since forced expression of PHR1 was previously shown to complement the loss of PHR2 (23).
The dominant gain of function associated with these mutations is clearly consistent with the current model of pH-dependent gene expression developed in A. nidulans and with related data from studies of other fungi (17, 19, 39). Both mutant alleles contained a nonsense mutation resulting in premature termination of the open reading frame. In RIM101-1426, a C-to-T transition at nucleotide 1426 introduced an ochre codon at position 476 of the coding region, truncating the 661-residue native protein by 186 amino acids. Similarly, a C-to-A transversion at position 1751 of RIM101-1751 converted the codon for Ser-584 to an ochre stop codon, truncating the protein by 78 residues. Rim101p is homologous to the zinc finger-containing transcription factor PacC of A. nidulans, Rim101p of S. cerevisiae, and the Y. lipolytica homolog YlRim101p (17, 19, 27, 39). PacC is synthesized as a 687-residue inactive precursor, which is activated when cells are cultured at an alkaline pH. Activation occurs by proteolysis around residues 252 to 254, approximately 90 amino acids carboxy-terminal to the zinc finger domain (9, 25). The carboxy terminus is essential to the pH dependence of activation and appears to control accessibility of the proteolytic site (9, 25). Thus, mutations that cause premature termination and loss of the carboxy terminus result in proteolytic activation irrespective of ambient pH (9, 25). Since PacC is required for transcriptional activation of alkaline-expressed genes and repression of acid-expressed genes, these truncating mutations can result in constitutive expression of alkaline-expressed genes and constitutive repression of acid-expressed genes (39). Similar effects are observed for truncated alleles of yeast RIM101 and YlRIM101 (17, 19). The observation that RIM101-1426 and RIM101-1751 allow expression of the alkaline-induced gene PHR1 at acidic pH and cause constitutive repression of PHR2 is entirely analogous to PacC.
The ability of RIM101-1426 and RIM101-1751 to relieve the pH dependence of hyphal development suggests that activation of Rim101p is the limiting factor for filamentation at acidic ambient pH. This result complements previous studies demonstrating that RIM101 is required for filamentation (27). However, the phenotype of the RIM101-1426 and RIM101-1751 mutations is in contrast to a previous report in which site-specific mutagenesis was used to introduce a premature stop codon in RIM101 (6). This mutation, located at codon 405 of the reported sequence (6), corresponding to codon 462 of the putative full-length open reading frame (27), was shown to suppress the loss of upstream components of the pH response pathway but did not overcome the inhibition of filamentation at acidic pH (6). These phenotypic differences may reflect intrinsic differences in the mutant alleles that affect, for instance, the proteolytic processing or stability of the truncated protein. Allele-specific variations in phenotype have been observed for PacC mutations (39).
An unexpected observation was that RIM101 could act as a multicopy suppressor of the temperature requirement for filamentation. In typical pH-regulated dimorphism, a shift from acid to neutral induces filamentation only if the ambient temperature is near 37°C. Work by Soll and colleagues (4) showed that lowering the temperature below a critical threshold of 34°C prevents filamentation irrespective of the ambient pH. The presence of four or more copies of the wild-type RIM101 permitted extensive filamentation to occur at temperatures below 30°C. Despite the diminished temperature restriction, morphological development was still pH dependent. If, however, multiple copies of the activated alleles were introduced, both the temperature and pH restrictions were removed. Suppression of the temperature requirement may be a direct effect if the stability of Rim101p or its proteolytic activation is normally limiting at temperatures below 34°C. Alternatively, elevated Rim101p may bypass temperature-dependent limitations on expression of a downstream function. In either case, this observation suggests that two environmental signals, pH and temperature, converge on common molecular targets. Integration of these signals may be important in establishing infection of external versus internal niches or survival outside the host or may facilitate transmission between hosts.
Despite the prominent role of RIM101 in controlling hyphal development, it does not act independently. An efg1Δ mutation was epistatic to RIM101-1426 and prevented filamentation. This result does not distinguish whether RIM101 and EFG1 act in parallel or within a single pathway. If RIM101 and EFG1 function within the same regulatory pathway, then this result suggests that EFG1 lies downstream of RIM101. The observation that the efg1 mutation did not prevent RIM101-1426 activation of PHR1 expression demonstrated that EFG1 is not interposed within the pathway controlling pH-dependent gene expression. However, this does not rule out the possibility of their functioning within the same pathway to control filamentation. In this regard it might be noted that EFG1 lies downstream of TPK2, which encodes a cAMP-dependent protein kinase (35). Rim101p of S. cerevisiae contains a functionally significant recognition site for cAMP-dependent protein kinases (38). Although this site is not conserved in the C. albicans homolog, two potential phosphorylation sites are present, and this could provide a regulatory connection between TPK2, RIM101, and EFG1.
Although RIM101 appears to impose a pH dependence on filamentation in vivo, its role in controlling morphological development during infection remains unclear, since extensive filamentation occurs during infection of acidic host niches (7). This suggests that in the acid niche, the requirement for RIM101 is bypassed or an alternative signal promotes Rim101p activation.
ACKNOWLEDGMENTS
We especially acknowledge the expert technical assistance of Stefanie Mücksch. We thank G. R. Fink for strains JKC18 and HLC67 and D. P. Bockmühl and J. F. Ernst for strain CDB1. C. Schmidt is acknowledged for assistance with the artwork. We are grateful to S. Suerbaum and H. Karch for critical reading of the manuscript. A.P. thanks B. Maresca for continuous support.
This work was supported by grant MU1212/2-1 from the Deutsche Forschungsgemeinschaft (to F.A.M.). A.E. held a predoctoral fellowship from the Deutsche Forschungsgemeinschaft, and O.K. is supported by a student fellowship from the Studienstiftung des Deutschen Volkes. A.P., A.R., and W.A.F. were supported by Public Health Service grant GM47727 from the National Institutes of Health and the Burroughs Wellcome Fund Scholar Award in Molecular Pathogenic Mycology.
REFERENCES
- 1.Auger P, Joly J. Factors influencing germ tube production in Candida albicans. Mycopathologia. 1977;61:183–186. doi: 10.1007/BF00468014. [DOI] [PubMed] [Google Scholar]
- 2.Boecke J P, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5′-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 3.Brown A J P, Gow N A R. Regulatory network controlling Candida albicans morphogenesis. Trends Microbiol. 1999;7:333–338. doi: 10.1016/s0966-842x(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 4.Buffo J, Herman M A, Soll D R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- 5.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 6.Davis D, Wilson R B, Mitchell A P. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Bernardis F, Mühlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do Carmo-Sousa L. Distribution of yeasts in nature. In: Alt R, Harrison J S, editors. The yeasts. Vol. 1. London, U.K: Academic Press; 1969. pp. 79–105. [Google Scholar]
- 9.Espeso E A, Roncal T, Díez E, Rainbow L, Bignell E, Álvaro J, Suárez T, Denison S H, Tilburn J, Arst H N, Jr, Peñalva M A. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 2000;19:719–728. doi: 10.1093/emboj/19.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans E G, Odds F C, Richardson M D, Holland K T. Optimum conditions for initiation of filamentation in Candida albicans. Can J Microbiol. 1974;21:338–342. doi: 10.1139/m75-048. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi W A. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans. J Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridkin S K, Jarvis W R. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz D, Jean A S, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillum A M, Tsay E Y H, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of Saccharomyces cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 16.Heinz, W. J., O. Kurzai, A. A. Brackhage, W. A. Fonzi, H. C. Korting, M. Frosch, and F. A. Mühlschlegel. Molecular responses to changes in the environmental pH are conserved between the fungal pathogens Candida dubliniensis and Candida albicans. Int. J. Med. Microbiol., in press. [DOI] [PubMed]
- 17.Lambert M, Blanchin-Roland S, Le Louedec F, Lépingle A, Gaillardin C. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol Cell Biol. 1997;17:3966–3976. doi: 10.1128/mcb.17.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K L, Buckley H R, Campell C C. An amino acid liquid synthetic medium for development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Mitchell A P. Proteolytic activation of Rimp1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 21.Lo H J, Köhler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous Candida albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 22.Mouyna, I., M. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J. P. Latgé. 2000. GPI-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem., in press. [DOI] [PubMed]
- 23.Mühlschlegel F A, Fonzi W A. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odds F C. Candida and candidosis. London, United Kingdom: Baillière Tindall; 1988. [Google Scholar]
- 25.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 26.Porta A, Ramon A M, Fonzi W A. PRR1, the homolog of the Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol. 1999;181:7516–7523. doi: 10.1128/jb.181.24.7516-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramon A M, Porta A, Fonzi W A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–7530. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riggle P J, Andrutis K A, Chen X, Tzipori S R, Kumamoto C A. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect Immun. 1999;67:3649–3652. doi: 10.1128/iai.67.7.3649-3652.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schorling, S. R., H. C. Korting, M. Frosch, and F. A. Mühlschlegel. The role of Candida dubliniensis in oral candidiasis in human immunodeficiency virus-infected individuals. Crit. Rev. Microbiol., in press. [DOI] [PubMed]
- 31.Sentandreu M, Elorza M V, Sentandreu R, Fonzi W A. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J Bacteriol. 1998;180:282–289. doi: 10.1128/jb.180.2.282-289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharkey L L, McNemar M D, Saporito-Irwin S M, Sypherd P S, Fonzi W A. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999;181:5273–5279. doi: 10.1128/jb.181.17.5273-5279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1986. [Google Scholar]
- 34.Sonneborn A, Bockmühl D P, Ernst J F. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect Immun. 1999;67:5514–5517. doi: 10.1128/iai.67.10.5514-5517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonneborn A, Bockmühl D P, Gerads M, Kurpanek K, Sanglard D, Ernst J. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- 36.Staab J F, Bradway S D, Fidel P L, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 37.Stoldt V R, Sonneborn A, Leuker C, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su S S Y, Mitchell A P. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 1993;21:3789–3797. doi: 10.1093/nar/21.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]