Abstract
DNA-protein crosslinks (DPCs) are unusually bulky DNA lesions that form when cellular proteins become trapped on DNA following exposure to UV light, free radicals, aldehydes, and transition metals. DPCs can also form endogenously when naturally occurring epigenetic marks (5-formyl cytosine, 5fC) in DNA react with lysine and arginine residues of histones to form Schiff base conjugates. Our previous studies revealed that DPCs inhibit DNA replication and transcription, but can undergo proteolytic cleavage to produce smaller DNA-peptide conjugates. We have shown that 5fC conjugated DNA-peptide crosslinks (DpCs) placed within CXA sequence (X = DpC) can be bypassed by human translesion synthesis (TLS) polymerases η and κ in an error-prone manner. However, local nucleotide sequence context can have a large effect on replication bypass of bulky lesions by influencing the geometry of the ternary complex between DNA template, polymerase, and the incoming dNTP. In the present work, we investigated polymerase bypass of 5fC-DNA-11-mer peptide crosslinks placed in seven different sequence contexts (CXC, CXG, CXT, CXA, AXA, GXA, and TXA) in the presence of human TLS polymerase η. Primer extension products were analyzed by gel electrophoresis, and steady state kinetics of dAMP misinscorporation opposite the DpC lesion in different base sequence contexts was investigated. Our results revealed a strong impact of nearest neighbor base identity on polymerase η activity both in the absence and in the presence of a DpC lesion. Molecular dynamics simulations were used to structurally explain the experimental findings. Our results reveal a possible role of local DNA sequence in promoting TLS related mutational hotspots both in the presence and in the absence of DpC lesions.
For Table of Contents use only
INTRODUCTION
The epigenetic DNA mark 5-formyl cytosine (5fC) is an intermediate in the DNA demethylation pathway catalyzed by ten-eleven translocase (TET) dioxygenases.1 5fC is endogenously present in all mammalian tissues, albeit at relatively low levels (less than 0.002% of total cytosines).2 We and others reported that the aldehyde group of 5fC can react with lysine and arginine side chains of histone proteins in human cells to form DNA-protein crosslinks (DPCs), potentially influencing chromatin structure and gene expression levels.3–6 DPCs are unusually bulky DNA lesions that can block DNA replication, transcription, and repair, as well as interfere with chromatin architecture and induce mutations, genomic instability, and cell death.7–9 In addition to 5fC mediated DNA-histone crosslinking, DPCs can form with a variety of cellular proteins after exposure to UV radiation, heavy metals, free radicals, and anti-cancer agents.10–13
Endogenously formed 5fC-histone DPCs are hydrolytically labile due to their imine structure and are not practical to use in biochemical experiments.5, 6, 14 Reducing agents such as NaCNBH4 can be used to stabilize the Schiff base conjugates; however, such treatment produces a mixture of products and leads to protein denaturation. Therefore, we have developed an oxime ligation methodology to generate site specific, hydrolytically stable model DNA-peptide crosslinks (DpCs).15 In our approach, an unnatural oxy-lysine amino acid was inserted into a peptide derived from the N-terminus of histone H4. Oxy-lysine within the histone-derived peptide spontaneously reacted with the aldehyde group of 5fC in DNA, forming a hydrolytically stable DNA-peptide conjugate.15 This same methodology has been used to conjugate oxy-lysine containing histone H3 to 5fC-containing DNA (Pujari and Tretyakova, unpublished observations). The resulting conjugates are structurally analogous to DPCs found in human cells.6, 15
Previous studies in our laboratory have investigated DNA replication in the presence of 5fC mediated DNA-protein conjugates and found that they completely blocked translesion synthesis (TLS) DNA polymerases η and κ, which are specialized polymerases known to bypass other bulky DNA lesions.16 This raised a question of how human cells can cope with endogenous DPCs formed at epigenetic DNA marks. Several groups reported the ability of specialized proteases, such as the metalloprotease Spartan, to degrade DNA-conjugated proteins to DNA-peptide conjugates, allowing for continued replication via bypass of the resulting DNA-peptide crosslinks by translesion synthesis polymerases.14, 16–19 DPCs may also be a substrate for ubiquitinylation and proteasomal degradation.20–22
Although proteolytic cleavage of DPC removes the replication block, the resulting DNA-peptide lesions are potentially mutagenic. We have previously observed targeted C→T transitions and deletion mutations when human translesion synthesis polymerase η (hPol η) bypassed a 5fC-11-mer peptide conjugate in the CXA sequence context.16 TLS polymerases such as Y-family Pols η, ι, κ, and Rev 1, and B-family Pol ζ lack normal proofreading mechanisms and are known to induce mutations during DNA replication.23–25 Molecular dynamics simulations revealed the formation of a stable wobble base pair mismatch between the modified C within the 5fC-peptide DpC lesion and the incoming dATP nucleotide in the active site of hPol η.16
Local DNA sequence context can have a profound effect on polymerase bypass of nucleobase lesions. This has been previously demonstrated for (6–4) pyrimidine-pyrimidinone photoproducts and O6-benzylguanine lesions.26–29 However, the ability of TLS polymerases to bypass DpC lesions located in different sequence contexts has not been previously investigated. In the present work, we investigated hPol η-catalyzed in vitro replication in the presence of 5-formylC DpCs lesions placed in seven different local DNA sequence contexts (CXC, CXG, CXT, CXA, AXA, GXA, and TXA). Polymerase η (hPol η) was chosen based on our previous studies demonstrating its ability to bypass DNA-polypeptide crosslinks,16 while the peptide sequence was derived from the N-terminal region oh histone H4. Following primer extension in the presence of hPol η, steady state kinetics experiments were used to examine the effects of DNA sequence context on the ability of hPol η to add correct (dGMP) and incorrect nucleotides (dAMP) opposite the DNA-peptide crosslink. Molecular modeling and molecular dynamics simulations were employed to gain a new structural understanding of the effects of the local DNA sequence context on DpC lesion bypass by hPol η.
METHODS
Synthesis, purification, and characterization of 5fC-containing oligonucleotides.
17-mer oligonucleotides (ODNs) containing site specific 5fC (A-G in Table 1) were prepared using solid phase synthesis on a MerMade 8 DNA synthesizer (Bioautomation, Irving, TX, USA) using 5-Formyl-dC-III-CE phosphoramidite (Glen Research, Sterling, VA, USA) under standard coupling conditions. ODNs were deprotected in 30% ammonium hydroxide at room temperature for 17 h followed by 80% acetic acid at RT for 6h. ODNs were purified by semi-preparative HPLC, desalted by Illustra Nap-5 columns (GE Healthcare, Pittsburgh, PA, USA), and characterized by HPLC-ESI-MS (A-G in Table 1). All unmodified oligodeoxynucleotides were purchased from IDT (Coralville, IA, USA), purified and characterized as described above.
Table 1:
Nucleobase sequence and mass spectrometry characterization of synthetic DNA/DpC oligodeoxynucleotides.
| Sequence | Expected mass (Da) | Observed mass (Da) | |
|---|---|---|---|
| A | 5′-d(TTC CFC GTC ACG ACG TT)-3′ | 5125.4 | 5124.6 |
| B | 5′-d(TTC CFG GTC ACG ACG TT)-3′ | 5165.4 | 5164.5 |
| C | 5′-d(TTC CFT GTC ACG ACG TT)-3′ | 5140.4 | 5139.6 |
| D | 5′-d(TTC AFA GTC ACG ACG TT)-3′ | 5173.4 | 5172.6 |
| E | 5′-d(TTC GFA GTC ACG ACG TT)-3′ | 5189.4 | 5188.6 |
| F | 5′-d(TTC TFA GTC ACG ACG TT)-3′ | 5164.4 | 5163.6 |
| G | 5′-d(TTC CFA GTC ACG ACG TT)-3′ | 5149.3 | 5149.4 |
| H | 5’-d(TTC CXC GTC ACG ACG TT)-3’ | 5967.2 | 5966.2 |
| I | 5’-d(TTC CXG GTC ACG ACG TT)-3’ | 6007.3 | 6006.2 |
| J | 5’-d(TTC CXT GTC ACG ACG TT)-3’ | 5982.2 | 5981.6 |
| K | 5’-d(TTC AXA GTC ACG ACG TT)-3’ | 6015.3 | 6014.5 |
| L | 5’-d(TTC GXA GTC ACG ACG TT)-3’ | 6031.3 | 6030.0 |
| M | 5’-d(TTC TXA GTC ACG ACG TT)-3’ | 6006.3 | 6005.0 |
| N | 5’-d(TTC CXA GTC ACG ACG TT)-3’ | 5989.2 | 5988.2 |
=5-formyl-dC
=5-formyl-dC crosslinked to NH2-GGG KGL GK*G GA, where K*=oxy-lysine
Synthesis, purification, and characterization of DNA-peptide crosslinks.
DNA-peptide crosslinks were synthesized as previously described.15 Briefly, synthetic lysine analogue containing an oxygen at the 𝜀-CH2 position (oxy-lysine) was site-specifically incorporated into a 11-mer peptide (NH2-GGG KGL GK*G GA). This oxy-lysine -containing peptide was then conjugated to the 5fC-containing DNA by incubating with DNA oligonucleotides (H-N in Table 1) in 100 mM NH4OAc buffer (pH 4.5) containing 100 mM aniline, at 37 °C for 17 h. The resulting DNA-peptide crosslinks (DpCs) were purified by HPLC, desalted by C-18 Sep Pak columns (Waters corporation, Milford, MA, USA) and characterized by HPLC-ESI-MS (H-N in Table 1).
Primer extension assays using human DNA polymerase η.
32P-labeled primer-template complexes, containing unmodified dC or 5fC-11-mer peptide crosslinks (NH2-GGGKGLGK*GGA, K*=oxy-lysine), were prepared as described previously.16 These primer-template complexes were incubated at 37 °C with all four dNTPs (500 μM final concentration) in a buffer containing 50 mM Tris/HCl (pH 7.5), 5 mM MgCl2, 50 mM NaCl, 5 mM DTT, 100 μg/mL BSA, and 10% glycerol (v/v), (total reaction volume 30 μL). The polymerization reaction was initiated by the addition of human DNA polymerase η (0.17 pmol). hPol η was expressed in bacteria and purified as described previously; full details are provided in Supplemental Materials.30, 31 Aliquots (4.5 μL) of the reaction mixtures were quenched with a gel loading buffer (20 mM EDTA in 95 % formamide including 0.05 % bromophenol blue and xylene cyanol) at pre-selected time points (0, 5, 15, 30 min). The extension products were then loaded onto 20% (w/v) denaturing PAGE containing 7 M urea, denatured at 80 W for 2 h in 1 x TBE buffer, and visualized using the Typhoon FLA 7000 phosphorimager (GH Healthcare, Pittsburgh, PA, USA).
Single-nucleotide incorporation assays.
32P-labeled primer–template complexes containing 5fC cross-links to 11-mer peptide (NH2-GGGKGLGK*G GA, K*=oxy-Lysine) (1 pmol) or unmodified template containing native C were incubated with individual dNTPs (50 μM final concentration) in the same buffer system described above at 37 °C. Human DNA polymerase η (0.17 pmol) was added to initiate the polymerization reaction in a final volume of 30 μL. Aliquots (4 μL) were quenched with gel loading buffer after 0, 5, 15 or 30 min, and the extension products were denatured and visualized as described above. Extension products at 30 min were quantified by volume analysis using ImageQuant TL 8.0 software (GE Healthcare).
Steady-state kinetics analysis.
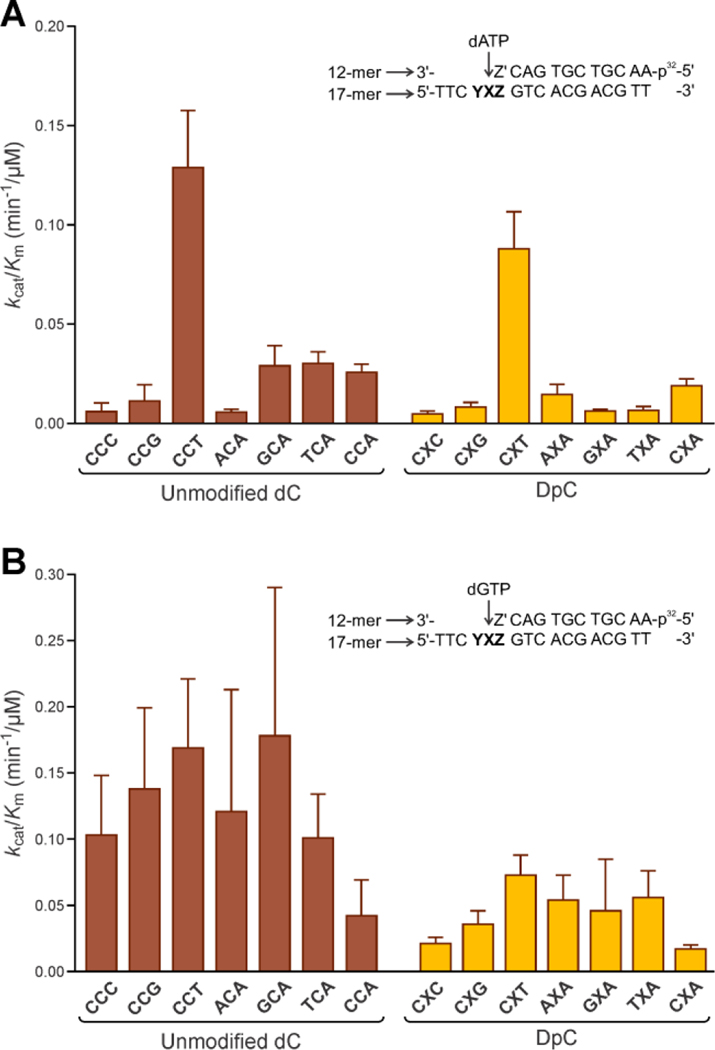
Steady-state kinetics for incorporation of individual dNTPs opposite unmodified dC, 5fC, or 5fC-conjugated 11mer-peptide (NH2-GGGKGLGK*GGA, K*=oxy-Lysine) was examined by performing single nucleotide insertion assays in the presence of human DNA polymerase η and increasing concentrations of specific dNTPs. Additional control experiments were performed for 5fC placed in the CfCT sequence context. Primer–template duplexes (30 nM) were incubated with hPol η (0.3–1.0 nM) in the presence of individual dNTPs (10, 25, 50, 100, 150, 250, 500, 800 μM) for specified time periods (0–30 min). Primer extension products were visualized with a Typhoon FLA 7000 system and quantified by volume analysis using the ImageQuant TL 8.0 software (GE Healthcare). Steady-state kinetic parameters were calculated by nonlinear regression analysis using one-site hyperbolic fits in Prism 4.0 (GraphPad Software, La Jolla, CA, USA). Error bars in Figure 1 were calculated by standard deviation of the mean of catalytic efficiency values. Error values for Vmax, Km, and kcat are standard deviations, and were calculated in Prism 4.0.
Figure 1:
Catalytic efficiencies (kcat/Km) for single nucleotide insertion of mismatch dAMP (A) and correct insertion of dGMP (B) opposite unmodified dC or 11-mer peptide (NH2-GGGKGLGK*GGA) conjugated to the C5 position of cytosine by hPol η. Catalytic efficiency values for nucleotide insertion opposite unmodified dC are shown in red, while the corresponding parameters for DpC templates are shown in yellow. YXZ in the sequence schematic corresponds to the sequence context used, as listed below the X-axis. Z’ corresponds to the complementary nucleotide to Z in each sequence context. Michaelis-Menten curves were calculated by nonlinear regression analysis using one-site hyperbolic fits in Prism 4.0 (Graphpad Software, La Jolla, CA, USA).
Molecular modeling and molecular dynamics simulations.
Full details concerning initial models, force fields and the DNA-peptide crosslink parametrization, and molecular dynamics protocols are given in the Supplementary Materials.32–45
RESULTS AND DISCUSSION
Preparation of DpC containing templates.
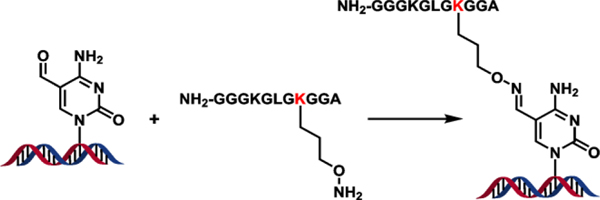
Model DpCs were generated via the previously published oxime ligation methodology.15 In brief, 5fC-containing DNA 17-mer oligodeoxynucleotides (A-G in Table 1) were site-specifically conjugated to an oxy-lysine containing 11-mer peptide to form hydrolytically stable DNA-peptide crosslinks (Scheme 1). These conjugates are structurally analogous to Schiff base conjugates between 5fC and histone proteins endogenously formed in human cells.6 Peptide sequence (NH2-GGGKGLGK*GGA) was derived from the N-terminal region of histone H4, where Lys-8 was replaced with oxy-Lys (K*). The original DNA sequence 5’-TTCCXAGTCACGACGTT-3’, where X is 5fC, is derived from the M13mp2 vector and was used in our earlier publication.16 A series of related DNA sequences were engineered by altering nucleobases at the +1 and the −1 positions from the 5fC-peptide lesion (Table 1). To study the impact of the neighboring bases on polymerase activity, we grouped them as follows: 5’C (+1 position) with variable 3’-base (−1 position = C, G, T, A) to study the effect of the 3’-bases; 3’-A (−1 position) with variable 5’ base (+1 position = C, G, T, A) to study the effect of the 5’ base. This yields a total of 7 sequences: CXC, CXG, CXT, CXA and AXA, GXA, and TXA (A-G in Table 1). Synthetic DpCs were purified by HPLC and characterized by HPLC-ESI–-MS as reported previously (H-N in Table 1, see Supplementary Figure S1 for representative data).16
Scheme 1:
Site specific DNA-peptide crosslinking via oxime ligation.
In vitro replication of DNA containing 5fC DpC lesions by human polymerase η.
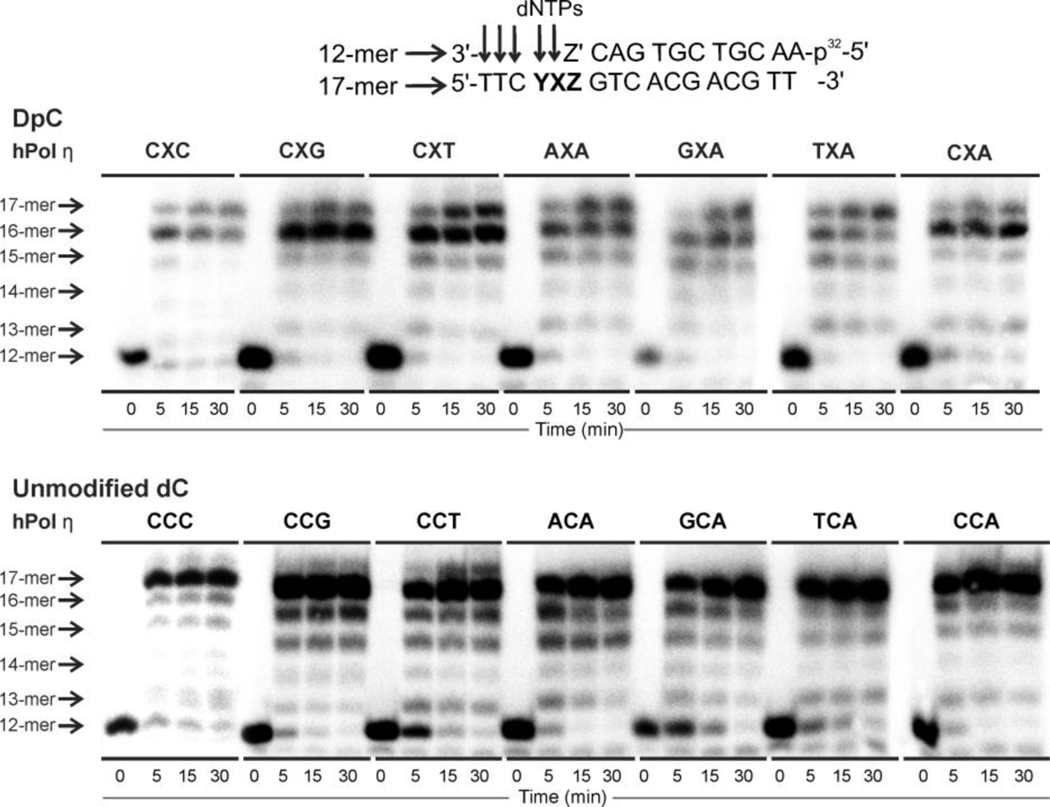
To investigate the effects of local DNA sequence on polymerase bypass of 5fC-peptide lesions (DpC), primer-template complexes containing either DpC or unmodified dC (negative control) in defined sequence context were subjected to in vitro replication in the presence of human polymerase η (6:1 enzyme:DNA molar ratio) and all four dNTPs. Primer extension products were analyzed by denaturing PAGE (Figure 2). For all sequences examined, hPol η was able to extend the primer past the DpC lesion, but to varying degrees of completion (Figure 2, top panel). For CXC, CXG, CXT, AXA, and TXA sequences, full primer extension products (17-mers) were observed as early as at the 5 min timepoint, while templates containing GXA and CXA were replicated less efficiently, requiring longer incubation times (Figure 2). By comparison, sequences containing unmodified dC were all replicated very efficiently by hPol η, reaching full extension at 5 minutes (Figure 2, bottom panel). Overall, these primer extension experiments provided initial evidence that hPol η activity in the presence of DpC is influenced by the local sequence context.
Figure 2:
Primer extension assays for replication bypass of covalent DNA–peptide cross-links by hPol η. 32P-labeled 12-mer primer was annealed with 17-mer DNA containing 5-formyl dC conjugated to an 11-mer peptide (X=DpC) (top panel) or unmodified dC (X=dC) (bottom panel) via oxime ligation. YXZ corresponds to the sequence context used, as listed above the X-axis. Z’ corresponds to the nucleotide complementary to Z in each sequence context. Primer extension reactions were initiated by the addition of hPol η and a mixture of dNTPs. Reactions were quenched at specific time points (0, 5, 15, 30 min), and loaded onto a 20% PAGE gel containing 7M urea.
To identify the nucleotides being inserted opposite the DpC lesion by Pol η, single nucleotide insertion experiments were conducted in the presence of individual dNTPs. The products were analyzed by denaturing PAGE (See Supplementary Figure S2 for representative gel images) and all extension products of single nucleotide insertion at 30 minutes were quantified by volume analysis (Supplementary Figure S2). For control templates, polymerase ƞ was able to incorporate any one of the four dNMPs opposite unmodified dC: dGMP was preferentially added in most cases; while in ACA and GCA, the wrong nucleotide dAMP was slightly preferred over the correct nucleotide dGMP. In the presence of the DpC, the correct nucleotide dGMP was inserted most efficiently opposite the lesion, and relatively smaller amounts of the other three nucleotides were also added (Supplementary Figure S2).
Among all mutagenic events, misincorporation of dAMP opposite the 5fC-polypeptide adduct was the most efficient (Supplementary Figure S2). This was true in all sequence contexts examined except for CXC. dAMP misincorporation opposite to the 5fC-conjugated peptide or unmodified C is anticipated to induce C→T transitions, which are among the most common mutations in the human genome.46 Therefore, our further experiments have focused on the kinetics of Pol η-catalyzed dAMP insertion opposite the DpC lesion placed in different local sequence contexts (see below).
Steady state kinetics experiments for incorporation of dA and dG opposite unmodified dC or the DpC lesion.
To determine to what extent local DNA sequence influences the kinetics of Pol η-mediated dAMP misincorporation opposite the DpC lesion and unmodified dC, steady state kinetics experiments were conducted. For comparison, the kinetics of incorporation of the correct nucleotide (dGMP) was also investigated. To ensure steady state conditions, 30–100-fold molar excess of DNA over hPol η was employed (see Supplementary Figure S3 for representative gel images). Primer extension reactions were repeated in the presence of increasing amounts of dATP (10–800 μM). The physiological concentration of dATP in dividing cells is 24 ± 22 μM.47 Local DNA sequence surrounding the lesion was systematically varied (H-N in Table 1). Catalytic efficiency (kcat/Km) was calculated by plotting reaction velocities against the concentration of dATP (Table 2) or dGTP (Supplementary Table S1) by using the Michaelis–Menten equation. Additionally, one sequence (CXT) was selected to detect any kinetic differences between 5fC and dC at the X position for dAMP misincorporation (CfCT in Table 2).
Table 2:
Steady-state kinetics parameters for single nucleotide insertion of mismatch dAMP opposite unmodified dC or 11-mer peptide (NH2-GGGKGLGKGGA) conjugated to C5 position of cytosine (X) by hPol η. Catalytic efficiency values for unmodified dC and DpC sequences are shown in the fifth column (kcat/Km). Average effect is the percentage difference between the catalytic efficiency values of each DpC and its corresponding control. Michaelis Menten curves were calculated by nonlinear regression analysis using one-site hyperbolic fits in Prism 4.0 (Graphpad Software, La Jolla, CA, USA). Error was calculated by standard deviation of the mean in Prism 4.0.
| Template | Vmax (μM/min) | Km (μM) | kcat (min−1) | kcat/Km (min−1/μM) | Average Effect (%) |
|---|---|---|---|---|---|
|
| |||||
| CCC | 4.99 ± 1.47 | 251 ± 171 | 1.50 ± 0.44 | 0.006 ± 0.004 | ---- |
| CXC | 1.79 ± 0.18 | 113 ± 35 | 0.54 ± 0.05 | 0.005 ± 0.002 | −16.7% |
|
| |||||
| CCG | 2.46 ± 0.62 | 147 ± 103 | 1.65 ± 0.41 | 0.011 ± 0.008 | ---- |
| CXG | 2.20 ± 0.22 | 178 ± 47 | 1.47 ± 0.15 | 0.008 ± 0.002 | −27.3% |
|
| |||||
| CCT | 5.01 ± 0.20 | 18 ± 3.8 | 2.26 ± 0.09 | 0.129 ± 0.029 | ---- |
| CfCT | 2.78 ± 0.24 | 24 ± 9.9 | 2.23 ± 0.19 | 0.094 ± 0.004 | −27.1% |
| CXT | 5.29 ± 0.23 | 27 ± 5.6 | 2.40 ± 0.10 | 0.088 ± 0.020 | −31.8% |
|
| |||||
| ACA | 2.74 ± 0.23 | 107 ± 26 | 0.60 ± 0.07 | 0.006 ± 0.001 | ---- |
| AXA | 6.28 ± 0.73 | 130 ± 44 | 1.88 ± 0.22 | 0.015 ± 0.005 | +150% |
|
| |||||
| GCA | 5.47 ± 0.45 | 41 ± 14 | 1.20 ± 0.09 | 0.025 ± 0.010 | ---- |
| GXA | 1.92 ± 0.07 | 67 ± 8.6 | 0.42 ± 0.01 | 0.006 ± 0.001 | −76.0% |
|
| |||||
| TCA | 1.33 ± 0.06 | 30 ± 5.7 | 0.89 ± 0.02 | 0.030 ± 0.006 | ---- |
| TXA | 2.43 ± 0.27 | 246 ± 65 | 1.63 ± 0.08 | 0.007 ± 0.001 | −76.7% |
|
| |||||
| CCA | 4.63 ± 0.21 | 81 ± 12 | 2.08 ± 0.09 | 0.026 ± 0.004 | ---- |
| CXA | 4.86 ± 0.29 | 115 ± 21 | 2.19 ± 0.12 | 0.019 ± 0.003 | −26.9% |
Our results revealed that in most sequence contexts, the efficiency of dAMP misincorporation opposite the DpC lesion was slightly lower than that for unmodified dC (Table 2). Unlike our previous data for rigid DNA-substance P peptide crosslinks,16 the presence of a flexible histone H4 derived peptide at the C5 position of C had a relatively modest effect on the efficiency of dAMP incorporation by hPol ƞ (Table 2). An opposite trend was observed for AXA sequence, where the presence of the peptide crosslink increased the catalytic efficiency for dAMP misincorporation (Table 2).
Local sequence context influenced the kinetics of hPol ƞ-catalyzed dAMP incorporation opposite DpC and unmodified dC (Figure 1A, Table 2). Notably, in control sequences, the highest catalytic efficiency of dAMP misincorporation opposite unmodified dC was revealed in the CCT context, with the value of kcat/Km being 0.129 μM−1min−1; in other sequences, the values of kcat/Km for the dA misincorporation were less than 0.03 μM−1min−1 (Figure 1A, Table 2). A similar trend was also observed in the DpC sequences: the highest catalytic efficiency of dA incorporation opposite the DpC lesion was seen in the CXT context, with the value of kcat/Km being 0.088 μM−1min-1. For the CfCT control, we also observed relatively high catalytic efficiency with the value of kcat/Km of 0.094 μM−1min−1, which was comparable to that of unmodified CCT (see CfCT in Table 2).
Steady state kinetics results for incorporation of the correct nucleotide (dGMP) opposite DpC lesion and unmodified dC are shown in Supplementary Table S1 and Figure 1B. Depending on local sequence context, the presence of the peptide lesion reduced catalytic efficiency for nucleotide addition by 44–80 %. Furthermore, as compared to our result for dAMP, the effect of sequence context on dGMP addition was less pronounced (Figure 1B, Supplementary Table S1); the kcat/Km values for incorporation of correct nucleotide (dGMP) opposite unmodified dC were in the similar range of 0.1~0.18 in all cases except CCA, which showed lower efficiency.
Collectively, our steady state kinetics results reveal a relatively modest effect of the flexible peptide lesion on polymerase ƞ activity. The efficiency of dAMP incorporation cytosine-conjugated peptide lesion and unmodified dC was strongly affected by the local sequence context, with CCT showing the highest catalytic efficiency in both cases (Table 2, Figure 1). In contrast, nearest neighbor identities had little influence on the kinetics of dGMP addition (Figure 1B, Supplementary Table S1).
Molecular modeling and molecular dynamics simulations for misincorporation of incorrect base A opposite the unmodified dC or the DpC lesion.
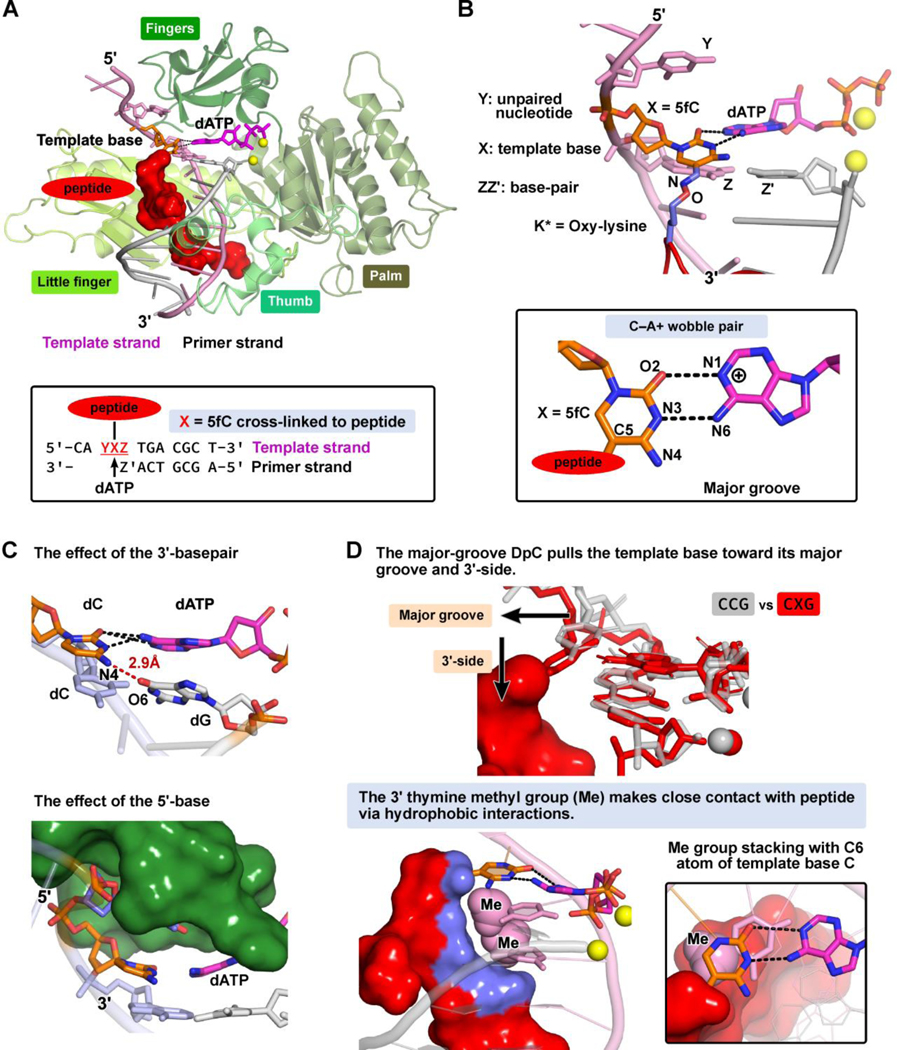
The goal of our MD simulations was to explain the molecular basis of the experimentally obtained steady-state kinetic parameters for misincorporation of dAMP opposite the unmodified dC or the DpC lesion by human Pol ƞ (Figure 1A, Table 2). We wished to understand why misincorporation of dA opposite DpC (and unmodified dC) was strikingly more efficient in the presence of a 3’ thymine (CCT and CXT results in Figure 1A and Table 2). We modeled the hPol ƞ ternary complex with the incoming dATP opposite unmodified dC or the 5fC conjugated to the 11-mer peptide that is housed in the major groove (Figure 3A). At the active site, the template base and the incorrect base A of incoming nucleotide form a C─A mismatch with wobble pairing scheme, as in our prior work (Figure 3B).16
Figure 3: (A).
Overall structure of the human Pol ƞ-DNA ternary complex in the presence of the 5fC-conjugated to the 11-mer peptide with incorrect incoming dATP at the insertion site. Inset box: the DNA sequences are indicated. See Movie S1.
(B) In the local sequence YXZ, Y is the unpaired nucleotide on the 5’-side to the template base; Z, on the 3’-side to the template base, forms a Watson-Crick base pair with Z’ at the primer terminus; X is the modified template 5fC with its major-groove C5 atom conjugated to the oxy-lysine (K*) via a −C=N− linker. In oxy-lysine, the ɛ-CH2 group was replaced with an oxygen atom. The incoming dATP forms a C─A mismatched base pair with template 5fC-conjugated to the 11mer-peptide. The C─A mismatch is modeled with a two-hydrogen bond scheme as a C─A wobble pair (inset box).
(C) The effects of the neighbor sequences to the template base on the alignments of the C─A mismatch. The effect of the base-pair on the 3’-side of the template base: a slipped H-bond between the template base and the primer terminus can form to distort the alignment of the C-A mismatch. The effect of the unpaired nucleotide on the 5’-side of the template base: the thymine with its small size and methyl group fits well and stably into a pocket in the finger domain via hydrophobic interactions; it therefore stacks well with the template base which stabilizes the alignment of the C-A mismatch.
(D) The effect of the major-groove-positioned DpC: Superimposed are the structures of CCG (grey) and CXG (red) sequences, showing that the peptide, which is housed in the major groove, pulls the template base toward the major groove and its 3’-side. The ability of the peptide to pull the template base is most restrained in the CXT sequence. This is because the 3’-thymine restrains the DpC (the close contact highlighted as blue surface) via hydrophobic interactions with its methyl group. Furthermore, this 3’-thymine methyl group stacks well with the C6 atom of the template base, and thus inhibits the template base from being pulled away by the DpC.
Polymerase bypass efficiency is directly related to the alignment quality of the C─A mismatch at the active site. Our MD results show that the incoming dATP is well-positioned for phosphodiester bond formation in all sequences investigated (Supplementary Figure S4). The distance between the O3’ of the primer terminus and the Pα of the dNTP is close to the near reaction-ready state of ~ 3.4 Å, as observed in a well-organized crystal structure of a ternary complex.48 These results indicate that the neighboring sequences surrounding the template base, and the presence of the DpCs does not affect the positioning of dATP. Therefore, the alignment of the C─A mismatch at the active site depends predominantly on the positioning and orientation of the template base, which, by contrast, is greatly impacted by the nature of the neighboring bases and the presence of the DpCs.
To elucidate the intrinsic effects of the neighboring bases on the positioning of the template base, we initially focused on polymerase complexes with templates containing unmodified dC. Our results showed that the alignment quality and the geometry of the hydrogen bonds in the C─A mismatch do depend notably on the nature of the neighboring bases, which differ on both the 3’- (Supplementary Figure S5) and the 5’- (Supplementary Figure S6) side to the template base C. We first investigated the role of the 3’-neighbor to the template base on the quality of the C─A mismatch. MD simulations revealed that among the four sequences examined (CCT, CCG, CCA, and CCC), the best alignment of the C-A mismatched wobble pair was observed in the CCT sequence (Supplementary Table S2, Figure S5): the 3’-thymine methyl (Me) group interacts favorably with the template base C via Me/π stacking interactions; accordingly, this 3’-T has the closest contact to the template among all the 3’-bases (Supplementary S7A). Thus, the template base (C) is particularly favored to align well with dATP when the 3’-base is T (Supplementary Figure S7B). This explains why dAMP misincorporation by hPol ƞ is most efficient in the CCT sequence context (Figure 1A, Table 2). By contrast, the worst alignment of the C─A mismatch was found in the CCC sequence, where we observed a slipped H-bond between the template C (N4) and the primer terminus base G (O6), present in about 52% of the population (Figure 3C, Supplementary Figure S5). Such slippage was very modest or absent in other sequences. The slipped hydrogen bond causes the C─A wobble pair at the active site to be very distorted (Supplementary Table S2), explaining the lowest bypass efficiency observed in the CCC sequence (Table 2, Figure 1A). Such slipped hydrogen bonds and their key role in mutagenesis during replication were seminally delineated by Streisinger49 and elaborated by Goodman.50
To uncover the influence of the 5’-neighboring nucleotide on dAMP incorporation opposite DpC lesion or unmodified C, the geometry of polymerase complexes with CCA, TCA, GCA, and ACA sequences was examined. As shown in Table 2, the lowest catalytic efficiency for dAMP incorporation opposite C was observed for the ACA sequence, while the others (GCA, TCA, and CCA) were comparable. Our MD simulations provide an insight into these findings. We observed that the alignment quality of the C−A mismatch varies depending on the identity of the 5’ base (Figure 3C, Supplementary Table S2, Figure S6). This in turn impacts the positioning of the template base and its alignment with dATP. MD results revealed that the unpaired 5’-neighboring nucleotide is naturally flexible and dynamic. As a result of the large hPol ƞ active site, the 5’-base can either stack with the template base or flip out into the major groove, depending on the nature of this 5’ base. Interestingly, in the polymerase active site, a flexible, induced pocket can form, which consists of several hydrophobic residues in the β-sheet of the finger domain (Supplemental Figure S8). When the 5’-neighboring base is a pyrimidine base (T or C), it fits well into this hydrophobic pocket (Supplementary Figure S8A) to stabilize the template base’s position via stacking interactions; the 5’-T interacts particularly well with the pocket hydrophobic residues via its methyl group. However, when the 5’- base is a purine, it either becomes more dynamic while still in the catalytic pocket (as in the case of GCA), or it is flipped out into the major groove (as in the ACA sequence, see Supplementary Figure S8B). When the 5’-A flips out into the major groove and thus does not stack with the template base, the C−A mismatch may rupture, as observed in one of our simulations (Supplementary Figure S9). This explains the low efficiency of dAMP addition opposite C in the ACA sequence context (Table 2). On the other hand, the 5’-base in TCA, CCA, and GCA does occupy the catalytic pocket and stacks with the template base to stabilize the C−A mismatch. This helps explain the comparable range of the bypass efficiencies revealed in these three cases (Table 2).
The highest bypass efficiency was observed for CXT, where the thymine is on the 3’-side of the template base (Table 2). We wished to understand how the presence of a major groove DpC lesion affects the sequence-dependent mutagenic bypass efficiency of hPol ƞ. In particular, we wanted to elucidate why the insertion of dAMP opposite to the modified 5fC-conjugated peptide is most preferred in the CXT context (Figure 1A, Table 2). Therefore, we examined MD simulations of dATP misincorporation in the hPol ƞ ternary complexes with the DpCs in the CXT, CXG, CXC, and CXA sequences and compared them to their unmodified partners discussed above.
Our MD simulations revealed that the presence of the DpC distorts the alignment quality of the C−A mismatch compared to the unmodified cases (Figure 3D, Supplementary Table S2, Figure S10). We found that there are notable disturbances to shear, buckle, and propeller twist (Supplementary Table S2). Because the peptide is accommodated in the major groove of DNA, it pulls the template base toward the major groove side, which enlarges the shear of the C-A mismatch. The peptide also pulls the template base toward its 3’-side, which increases the propeller twist and inverts the buckle of the C─A mismatch (Figure 3D, Supplementary Table S2). These DpC-induced distortions to the C−A mismatch explain the overall lower catalytic efficiency upon replication bypass of DpC (Table 2).
The conformations and dynamics of the DpC peptide determine to what extent it pulls the template base away from its ideal position, so that the alignment of the C─A mismatch is distorted. The backbone of the unstructured peptide is flexible and not tightly bound to the major groove of DNA due to the presence of several glycine residues which lack the Cβ atom (Scheme 1). Because the peptide adopts different conformations with varied flexibilities in different DNA sequences (Supplementary Figure S11), the disturbances induced by the presence of the peptide are sequence-dependent (Supplementary Table S2), leading to differences in bypass efficiency (Table 2). Notably, the DpC-induced disturbances are the least pronounced in the CXT sequence context where the peptide is least flexible, being restrained by interaction with the 3’-T base (Supplementary Figures S11–12). The 3’-neighboring T has the closest contact with the conjugated peptide due to hydrophobic interactions via its 5-methyl group (Figure 3D). Furthermore, this 3’-T also stacks more favorably with the template base than in the other sequences (Supplementary Figure S12) (also seen in the unmodified CCT sequence, Supplementary Figure S7). Thus, the 3’-T whose methyl group stacks favorably with the template base while also manifesting hydrophobic interactions with the peptide, largely prevents the template base from being pulled away from its undistorted position. In contrast, for the other 3’-bases, the peptide is less restrained; it has significantly less contact with the 3’-base and consequently pulls the template base away from its normal position, as seen in the unmodified cases (Figure 3D, Supplementary Figure S12). Hence, the geometry of the C−A mispair is more distorted (Supplementary Table S2), which explains why the bypass efficiency for misincorporation of dATP is much lower in these three sequences as compared to CXT (Table 2).
In our earlier paper,16 we examined the fidelity of hPol ƞ in CXA context, where X is dC or a DpC lesion containing 11-mer peptide RPKPQQFFGLM. We investigated the efficiency of hPol ƞ catalyzed incorporation of all four dNTPs opposite DpC or unmodified dC in a single sequence context (CXA). We found that dAMP incorporation opposite DpC was preferred over correct nucleotide (dGMP), with misinsertion frequency (f) of 1.92.16 Our MD simulations revealed that the C-A mismatched base-pair was distorted in the case of unmodified dC. However, the rigid 11-mer peptide interacted with the DNA major groove and anchored the templating cytosine base, pulling it toward the 3’-side. As a result, the C-A mismatched base-pair was more ideally positioned in the presence of the 11-mer peptide than for unmodified dC. In contract, in the tertiary complex with incoming dGTP (correct base), the presence of the 11-mer peptide weakened the hydrogen bonds for the modified C:G base pair, because the rigid major-groove-positioned 11mer-peptide caused significant tilting of the conjugated templating base C, pulling the base C toward the major groove and its 3’-side. Without the DpC lesion, the C-G Watson-Crick pairing was normal.
In the present study, we focused on nucleotide incorporation opposite a different type of DpC lesion constructed using the N-terminal sequence of histone H4 (GGG KGLGK*G GA). Unlike structurally rigid peptide investigated previously (RPKPQQFFGLM),16 histone H4 peptide contains multiple glycine residues. Our MD simulations revealed that the flexible 11-mer peptide adopts different conformations depending on DNA sequence contexts (Supplementary Figure S11), so that the DpC-induced disturbances are sequence-dependent, which accounts for their different bypass efficiencies. Overall, the variety in bypass efficiencies in hPol ƞ stems from the different chemical structures of the investigated peptide lesions and their local DNA sequence contexts, as they are differently accommodated in the spacious active site of the polymerase.
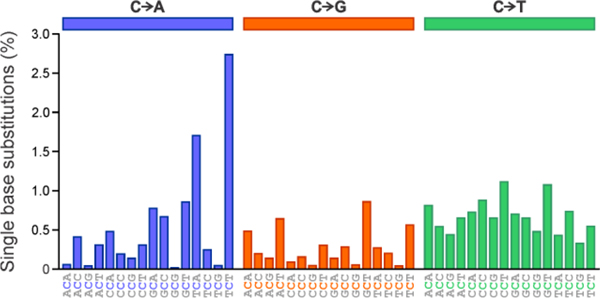
To our knowledge, this study is the first to systematically examine the effects of local DNA sequence on the kinetics of DNA replication catalyzed by hPol ƞ. Our findings may help provide a mechanistic explanation of the mutational signatures observed in cancer. Mutational signature profile of hPol ƞ from the catalogue of somatic mutations of cancer (COSMIC) reveals that C to T transitions preferentially occur in the CCT sequence context (Figure 4).51, 52 The reference human genome version GRCh37 displays a single base substitution from C to T for CCT at 1.10% of total CCT trinucleotides.52 For other sequences examined by molecular modeling and steady-state kinetics, the mutation frequency was below 0.9%. In human genome version GRCh38, the frequency of C to T mutation in the CCT context is slightly greater, at 1.11%, but other trinucleotide sequences align with similar increases.51 This data correlates with our experimental and molecular dynamics studies described above, identifying CCT and CXT as the most efficient sequences in allowing for dAMP incorporation opposite the central nucleotide. However, in some sequences (e.g. TCA), the rates of C to A transversion mutations within the signature plot are higher than the levels of C to T transition mutations for this same trinucleotide sequence (Figure 4). Future investigation of C to A transversion mutations through further molecular modeling and kinetics studies may be essential to understand the underlying role of these mutation signatures.
Figure 4:
Mutational signature profile of SBS9 (hPol η) in reference human genome version GRCh37, using the conventional 96-mutation type classification. Data obtained through the Catalogue of Somatic Mutations in Cancer (COSMIC).51 Substitution mutation frequencies are presented as respective proportions of mutations of each trinucleotide signature based on the actual frequency of each trinucleotide sequence found in the genome version GRCh37. C to A transversions appear in blue (left panel), C to G transversions appear in orange (middle panel), and C to T transitions appear in green (right panel).
CONCLUSIONS
Overall, our results indicate that human polymerase ƞ catalyzes replication bypass of 5fC-mediated DNA-peptide crosslinks derived from N-terminal sequence of histone H4, but the outcomes vary considerably depending on the local DNA sequence on either side of the DpC lesion. The neighboring bases significantly affect the efficiency of dAMP incorporation opposite the adduct, while the addition of the correct nucleotide (dGMP) is uniform across the seven sequences examined. Molecular dynamics simulations provided structural explanations for the sequence-governed bypass efficiency in the misincorporation of incoming dATP both with and without the DpC lesion. The observed differences in the outcomes of in vitro bypass of a DNA-peptide crosslink placed in different sequence contexts provides a new understanding of the biological outcomes of this type of DNA damage and their contributions to DPC induced mutagenesis in human cells. Furthermore, our observation of sequence dependent replication of unmodified DNA by hPol ƞ sheds new light on the origins of mutational hot spots which play important roles in carcinogenesis. Future studies are needed to investigate the effects of peptide identities and DNA sequence context on replication bypass by other TLS polymerases, as well as in vivo studies to determine whether similar trends are observed in living cells.
Supplementary Material
ACKNOWLEDGEMENT:
We thank Dr. Shaofei Ji, Xiaotong Lu, Emina Dzafic, and Nicholas Weirath for their assistance with kinetics analysis. We thank Maram Essawy (University of Minnesota) for her help in expression and purification of hPol η, Dr. F. Peter Guengerich (Vanderbilt University) for the gift of the hPol η plasmid vector and Dr. Jiri Zavadil (IACR) for his advice regarding mutational signatures in cancer. We thank Robert Carlson (University of Minnesota) for his help with figure preparation.
FUNDING:
National Institute of Environmental Health Sciences [R01 ES-023350 to N.T., R01 ES-025987 to S.B.]
Footnotes
All authors have given approval to the final version of the manuscript. J.T. and I.F. contributed equally to this work.
ACCESSION CODES: POLH-HUMAN: Q9Y253
Supporting Information Available: Methods for Human Polymerase η Expression and Purification, Molecular modeling and molecular dynamics simulations (Initial models, MD parameters for DNA-peptide crosslinks, Molecular dynamics simulations, Molecular dynamics simulation protocols, Structural analyses), Supplementary Tables, Supplementary Figures, Supplementary Movie. This material is available free of charge via the Internet.
REFERENCES
- (1).Lu X, Zhao BS, and He C. (2015) TET family proteins: oxidation activity, interacting molecules, and functions in diseases, Chem Rev 115, 2225–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gackowski D, Zarakowska E, Starczak M, Modrzejewska M, and Olinski R (2015) Tissue-specific differences in DNA modifications (5-hydroxymethylcytosine, 5-formylcytosine, 5-carboxylcytosine and 5-hydroxymethyluracil) and their interrelationships, PLoS One 10, e0144859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jones PA, and Takai D. (2001) The role of DNA methylation in mammalian epigenetics, Science 293, 1068–1070. [DOI] [PubMed] [Google Scholar]
- (4).Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, and Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine, Science 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Li F, Zhang Y, Bai J, Greenberg MM, Xi Z, and Zhou C. (2017) 5-Formylcytosine yields DNA-protein cross-links in nucleosome core particles, J Am Chem Soc 139, 10617–10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ji S, Shao H, Han Q, Seiler CL, and Tretyakova NY (2017) Reversible DNA-protein cross-linking at epigenetic DNA marks, Angew Chem Int Ed Engl 56, 14130–14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Barker S, Weinfeld M, and Murray D. (2005) DNA-protein crosslinks: their induction, repair, and biological consequences, Mutat Res 589, 111–135. [DOI] [PubMed] [Google Scholar]
- (8).Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, and Salem AM (2011) Repair and biochemical effects of DNA-protein crosslinks, Mutat Res 711, 113–122. [DOI] [PubMed] [Google Scholar]
- (9).Tretyakova NY, Michaelson-Richie ED, Gherezghiher TB, Kurtz J, Ming X, Wickramaratne S, Campion M, Kanugula S, Pegg AE, and Campbell C. (2013) DNA-reactive protein monoepoxides induce cell death and mutagenesis in mammalian cells, Biochemistry 52, 3171–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Nakamura J, and Nakamura M. (2020) DNA-protein crosslink formation by endogenous aldehydes and AP sites, DNA Repair (Amst) 88, 102806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shang M, Ren M, and Zhou C. (2019) Nitrogen mustard induces formation of DNA-histone cross-links in nucleosome core particles, Chem Res Toxicol 32, 2517–2525. [DOI] [PubMed] [Google Scholar]
- (12).Nakano T, Xu X, Salem AMH, Shoulkamy MI, and Ide H. (2017) Radiation-induced DNA-protein cross-links: Mechanisms and biological significance, Free Radic Biol Med 107, 136–145. [DOI] [PubMed] [Google Scholar]
- (13).Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, and Swenberg JA (2010) Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers, J Am Chem Soc 132, 3388–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Naldiga S, Ji S, Thomforde J, Nicolae CM, Lee M, Zhang Z, Moldovan GL, Tretyakova NY, and Basu AK (2019) Error-prone replication of a 5-formylcytosine-mediated DNA-peptide cross-link in human cells, J Biol Chem 294, 10619–10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pujari SS, Zhang Y, Ji S, Distefano MD, and Tretyakova NY (2018) Site-specific cross-linking of proteins to DNA via a new bioorthogonal approach employing oxime ligation, Chem Commun (Camb) 54, 6296–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ji S, Fu I, Naldiga S, Shao H, Basu AK, Broyde S, and Tretyakova NY (2018) 5-Formylcytosine mediated DNA-protein cross-links block DNA replication and induce mutations in human cells, Nucleic Acids Res 46, 6455–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Vaz B, Popovic M, and Ramadan K. (2017) DNA-protein crosslink proteolysis repair, Trends Biochem Sci 42, 483–495. [DOI] [PubMed] [Google Scholar]
- (18).Stingele J, Bellelli R, and Boulton SJ (2017) Mechanisms of DNA-protein crosslink repair, Nat Rev Mol Cell Biol 18, 563–573. [DOI] [PubMed] [Google Scholar]
- (19).Morocz M, Zsigmond E, Toth R, Enyedi MZ, Pinter L, and Haracska L. (2017) DNA-dependent protease activity of human Spartan facilitates replication of DNA-protein crosslink-containing DNA, Nucleic Acids Res 45, 3172–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sun Y, Saha LK, Saha S, Jo U, and Pommier Y. (2020) Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways, DNA Repair (Amst) 94, 102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Gao R, Schellenberg MJ, Huang SY, Abdelmalak M, Marchand C, Nitiss KC, Nitiss JL, Williams RS, and Pommier Y. (2014) Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2·DNA and Top2·RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2), J Biol Chem 289, 17960–17969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Quinones JL, Thapar U, Wilson SH, Ramsden DA, and Demple B. (2020) Oxidative DNA-protein crosslinks formed in mammalian cells by abasic site lyases involved in DNA repair, DNA Repair (Amst) 87, 102773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yoon JH, McArthur MJ, Park J, Basu D, Wakamiya M, Prakash L, and Prakash S. (2019) Error-prone replication through UV lesions by DNA polymerase theta protects against skin cancers, Cell 176, 1295–1309.e1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Vaisman A, and Woodgate R. (2017) Translesion DNA polymerases in eukaryotes: what makes them tick?, Crit Rev Biochem Mol Biol 52, 274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jung H, Rayala NK, and Lee S. (2020) Translesion synthesis of the major nitrogen mustard-induced DNA lesion by human DNA polymerase eta, Biochem J 477, 4543–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Akagi J-I, Hashimoto K, Suzuki K, Yokoi M, de Wind N, Iwai S, Ohmori H, Moriya M, and Hanaoka F. (2019) Effect of sequence context on Polζ-dependent error-prone extension past (6–4) photoproducts, DNA repair 87, 102771–102771. [DOI] [PubMed] [Google Scholar]
- (27).Shriber P, Leitner-Dagan Y, Geacintov N, Paz-Elizur T, and Livneh Z. (2015) DNA sequence context greatly affects the accuracy of bypass across an ultraviolet light 6–4 photoproduct in mammalian cells, Mutat Res 780, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gahlon HL, Schweizer WB, and Sturla SJ (2013) Tolerance of base pair size and shape in postlesion DNA synthesis, J Am Chem Soc 135, 6384–6387. [DOI] [PubMed] [Google Scholar]
- (29).Laverty DJ, and Greenberg MM (2017) In vitro bypass of thymidine glycol by DNA polymerase theta forms sequence-dependent frameshift mutations, Biochemistry 56, 6726–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Su Y, Patra A, Harp JM, Egli M, and Guengerich FP (2015) Roles of residues Arg-61 and Gln-38 of human DNA polymerase η in bypass of deoxyguanosine and 7,8-dihydro-8-oxo-2’-deoxyguanosine, J Biol Chem 290, 15921–15933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, and Yang W. (2010) Structure and mechanism of human DNA polymerase eta, Nature 465, 1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Case DA, Babin V, Berryman J, Betz RM, Cai Q, Cerutti DS, Cheatham T, Darden T, Duke R, Gohlke H, Götz A, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee T-S, and Kollman PA (2014) AMBER 14, University of California, San Francisco. [Google Scholar]
- (33).Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C. (2015) ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB, J Chem Theory Comput 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ivani I, Dans PD, Noy A, Pérez A, Faustino I, Hospital A, Walther J, Andrio P, Goñi R, Balaceanu A, Portella G, Battistini F, Gelpí JL, González C, Vendruscolo M, Laughton CA, Harris SA, Case DA, and Orozco M. (2016) Parmbsc1: a refined force field for DNA simulations, Nat Methods 13, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wang J, Wolf RM, Caldwell JW, Kollman PA, and Case DA (2004) Development and testing of a general amber force field, J Comput Chem 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- (36).Bayly CI, Cieplak P, Cornell W, and Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model, J Phys Chem 97, 10269–10280. [Google Scholar]
- (37).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams Ding, F., Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, and Fox DJ (2016) Gaussian 16 Rev. C.01, Wallingford, CT. [Google Scholar]
- (38).Jorgensen W, Chandrasekhar J, Madura J, Impey R, and Klein M. (1983) Comparison of simple potential functions for simulating liquid water, J. Chem. Phys. 79, 926–935. [Google Scholar]
- (39).Joung IS, and Cheatham TE 3rd. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations, J Phys Chem B 112, 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, and Walker RC (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald, J Chem Theory Comput 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- (41).Gotz AW, Williamson MJ, Xu D, Poole D, Le Grand S, and Walker RC (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born, J Chem Theory Comput 8, 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Darden T, York D, and Pedersen L. (1993) Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems, J Chem Phys 98, 10089–10092. [Google Scholar]
- (43).Roe DR, and Cheatham TE 3rd. (2013) PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data, J Chem Theory Comput 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- (44).Lavery R, Moakher M, Maddocks JH, Petkeviciute D, and Zakrzewska K. (2009) Conformational analysis of nucleic acids revisited: Curves+, Nucleic Acids Res 37, 5917–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Case D, Betz R, Cerutti DS, Cheatham T, Darden T, Duke R, Giese TJ, Gohlke H, Götz A, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee T-S, LeGrand S, Li P, Lin C, Luchko T, and Kollman P. (2016) Amber 16, University of California, San Francisco. [Google Scholar]
- (46).Jiang C, and Zhao Z. (2006) Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms, Genomics 88, 527–534. [DOI] [PubMed] [Google Scholar]
- (47).Traut TW (1994) Physiological concentrations of purines and pyrimidines, Mol Cell Biochem 140, 1–22. [DOI] [PubMed] [Google Scholar]
- (48).Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, and Wilson SH (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex, Structure 14, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, and Inouye M. (1966) Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday, Cold Spring Harb Symp Quant Biol 31, 77–84. [DOI] [PubMed] [Google Scholar]
- (50).Efrati E, Tocco G, Eritja R, Wilson SH, and Goodman MF (1997) Abasic translesion synthesis by DNA polymerase beta violates the “A-rule”. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts, J Biol Chem 272, 2559–2569. [DOI] [PubMed] [Google Scholar]
- (51).Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, and Forbes SA (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer, Nucleic Acids Res 47, D941–d947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, and Wooster R. (2004) The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website, Br J Cancer 91, 355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.