Abstract
Thirteen patients who had 16 episodes of bacteremia were observed between 1993 and 1997 in a pediatric oncology ward with a high background isolation rate of cefotaxime- or aztreonam-resistant gram-negative bacteria. Four blood isolates were Escherichia coli and 12 were Klebsiella pneumoniae, and these isolates harbored extended-spectrum β-lactamases (ESBLs). All episodes of bacteremia were nosocomial, all except one of the episodes occurred in neutropenic patients, and all patients were treated with piperacillin or ceftazidime with amikacin and cefazolin prior to the onset of bacteremia. Nine of 13 patients were receiving extended-spectrum β-lactam treatment when the bacteremias caused by ESBL producers occurred. Molecular studies revealed that four K. pneumoniae SHV-2-producing isolates from 1994 were of the same clone. Other ESBL producers, including six that carried both TEM-1 and SHV-5, five that carried SHV-5, and one that carried SHV-2 alone, were unrelated. In conclusion, SHV-5 was present in 11 of the 16 isolates and coexisted with TEM-1 in 6 isolates. Acquisition of resistance genes probably occurred under antibiotic selection pressure. This study highlights the importance of routine checks for and detection of ESBL producers. Effective therapy against ESBL producers should be considered early for children who have malignancies and neutropenia and who are septic, despite treatment with a regimen that includes an extended-spectrum β-lactam, in a clinical setting of an increased incidence of ESBL-producing bacteria.
Neutropenic patients are at high risk for various infectious diseases even if cultures of clinical specimens are not positive. Extended-spectrum β-lactam monotherapy or extended-spectrum β-lactam therapy in combination with an aminoglycoside is generally accepted as the empirical regimen for febrile neutropenia following chemotherapy for a malignancy (6, 7, 17, 27, 40). However, the choice of empirical antimicrobial therapy should be evaluated periodically to prevent treatment failure due to antimicrobial resistance. Combination therapy with an antipseudomonal β-lactam antibiotic such as piperacillin or ceftazidime with an aminoglycoside (amikacin usually) had been used extensively to treat patients with malignancies and infectious diseases in the pediatric oncology ward at the National Taiwan University Hospital (NTUH) in the past 10 years. Such combination therapy was particularly used in febrile neutropenic patients. However, recent reports of treatment failure due to extended-spectrum β-lactamase (ESBL)-producing bacteria were of great concern to clinical practice (3, 21). High incidences of infections caused by ESBL-producing bacteria in intensive care units were reported in different areas (14, 25). Concern about such drug-resistant bacterial infections had extended to nursing homes (43), geriatric (8) and pediatric (10, 12, 13) populations, transplant recipients (12), and oncology patients (16). Those infected were often fragile and unable to tolerate infectious diseases. Failure to identify ESBL producers by routine susceptibility testing may lead to inappropriate antimicrobial treatment and may result in increased mortality.
Gram-negative aerobic bacteria accounted for the majority of cases of nosocomial infection at NTUH (5). The frequency of extended-spectrum β-lactam-resistant Klebsiella pneumoniae at NTUH increased from 3.4% in 1993 to 10.3% in 1997, as determined by the disk diffusion method (18). An increasing prevalence of extended-spectrum β-lactam-resistant Escherichia coli, from 2.5% in 1993 to 6.7% in 1997, was also observed (24). Compared with the average data from a hospital-wide surveillance, the frequencies of extended-spectrum β-lactam-resistant K. pneumoniae and E. coli isolates in a pediatric ward were found to be three to five times higher (see below). In the current study, a retrospective survey with detailed molecular analysis was conducted to investigate the clinical significance of ESBL-producing K. pneumoniae and E. coli bacteremia in children with malignancies.
MATERIALS AND METHODS
Patients and bacterial strains.
We searched the computerized database at NTUH, an 1,800-bed acute-care medical center, for E. coli and K. pneumoniae blood isolates resistant or intermediately susceptible to aztreonam or broad-spectrum cephalosporins in a pediatric oncology ward between 1993 and 1997. The pediatric oncology ward has 35 beds, and most patients were admitted for evaluation and management of malignant diseases. The medical records of patients harboring the studied microorganisms were reviewed. Neutropenia was defined as a polymorphonuclear cell count of ≤500/mm3. The bacterial strains were stored at −70°C before retrieval for testing.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility was determined by both the agar dilution and disk diffusion tests according to the National Committee for Clinical Laboratory Standards (NCCLS) (30, 31). For susceptibility testing by the agar dilution method, the following antimicrobial agents were obtained as standard reference powders of known potency for laboratory use: amoxicillin, ampicillin, and cephalothin (Sigma Chemical Co., St. Louis, Mo.); clavulanic acid (SmithKline Beecham, Brockhans Park, United Kingdom); piperacillin and tazobactam (Lederle Laboratories, Pearl River, N.Y.); cefmetazole (Sankyo Co., Hiratsuka, Japan); imipenem (Merck Sharp & Dohme, West Point, Pa.); cefotaxime (Hoechst Marion Roussel, Frankfurt, Germany); ceftazidime (Glaxo Group Research Limited, Greenford, United Kingdom); cefepime, amikacin, and aztreonam (Bristol-Myers Squibb Laboratories, Princeton, N.J.); meropenem (Sumitomo Pharmaceuticals Co., Osaka, Japan); and ciprofloxacin (Bayer Co., Leverkusen, Germany). All drugs were incorporated into Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, Md.) in serial twofold concentrations from 0.03 to 128 μg/ml. Two control strains, E. coli ATCC 35218 and ATCC 25922, were included in each test run. Inoculated plates were incubated in ambient air at 35°C for 16 to 18 h. The MIC of each antimicrobial agent was defined as the lowest concentration that inhibited visible growth of the organism.
Screening tests for ESBL-producing strains.
The double-disk synergy test and the Etest for ESBLs were used as screening tests to detect ESBL-producing strains. In the double-disk synergy test, the following antimicrobial disks were placed on Mueller-Hinton agar (BBL Microbiology Systems) adjacent to an amoxicillin-clavulanic acid disk (20 μg of amoxicillin plus 10 μg of clavulanate): cefotaxime (30 μg), ceftazidime (30 μg), aztreonam (30 μg), and cefepime (30 μg). All disks were purchased from Becton Dickinson Microbiology System (Sparks, Md.). The procedures and interpretation of the double-disk synergy test were as described previously (19).
The Etest ESBL screen (PDM Epsilometer; AB Biodisk, Solna, Sweden), based on the recognition of a reduction in the ceftazidime MIC in the presence of clavulanic acid, was performed according to the manufacturer's instructions.
Genomic fingerprinting by PFGE.
Total DNA was prepared and pulsed-field gel electrophoresis (PFGE) was performed as described previously (1, 41). The restriction enzyme XbaI (New England Biolabs, Beverly, Mass.) was used at the manufacturer's suggested temperature. Restriction fragments were separated by PFGE in a 1% agarose gel (Bio-Rad, Hercules, Calif.) in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1.0 mM EDTA [pH 8.0]) with the Bio-Rad CHEF-DRII apparatus (Bio-Rad Laboratories, Richmond, Calif.). The initial pulse time of 1 s was increased linearly to 35 s in 20 h at 200 V at 4°C. The gels were then stained with ethidium bromide and photographed under UV light.
The band patterns were visually compared and were classified as indistinguishable (clonal), closely related (clonal variants, three or fewer band differences), possibly related (four to six band differences), and unrelated according to previously described criteria (42).
Plasmid isolation and resistance transfer.
Plasmid profile analysis was performed by the alkaline extraction method (20). Resistance transfer was carried out by conjugation. A rifampin-resistant strain of E. coli (strain JP-995) was used as the recipient. Recipients and donors were separately inoculated into brain heart infusion broth (Oxoid, Basingstoke, England) and were incubated at 37°C for 4 h. They were then mixed at a volume ratio of 1:1 for overnight incubation at 37°C. A 0.01-ml volume of the overnight broth mixture was then spread onto a MacConkey agar plate containing rifampin (100 μg/ml) and either ceftazidime (1 μg/ml) or aztreonam (2 μg/ml).
Isoelectric focusing.
The isoelectric focusing procedure was generally performed as described previously (28). Bacteria were harvested from a 20-h brain heart infusion broth culture by centrifugation, and the pellet was resuspended in 1 ml of phosphate buffer (0.05 M; pH 7). The enzymes were released by two cycles of freezing (−70°C) and thawing (room temperature) and by sonication for 5 min in a sonicator in ice-cold water. Isoelectric focusing was performed in an ampholine gel (pH 3.0 to 10.0; Pharmacia, Uppsala, Sweden). Preparations from standard strains known to harbor TEM-1, SHV-1, and SHV-5 were used as standards. After isoelectric focusing, β-lactamases were detected by spreading nitrocefin (50 μg/ml) on the gel surface.
Plasmid profile by restriction enzyme digestion.
Plasmid DNA from the transconjugant was prepared as described previously (1). Restriction enzyme analysis of the plasmids from the transconjugants was performed according to the manufacturer's instructions. The restriction enzymes PvuII and PstI (Gibco BRL, Grand Island, N.Y.) were used. HindIII- and EcoRI-digested λ phage DNA was used as a molecular weight marker.
PCR amplification for blaTEM and blaSHV and direct DNA sequencing.
The oligonucleotide primers (Gibco BRL) used for the PCR assay were as follows: 5′-ATAAAATTCTTGAAGACGAAA (primer A), 5′-GACAGTTACCAATGCTTAATCA (primer B), 5′-GGGTAATTCTTATTTGTCGC (primer C), and 5′-TTAGCGTTGCCAGTGCTC (primer D). Primers A and B were known to be specific for blaTEM (26). Primers C and D were known to be specific for blaSHV (37). Reactions were performed in a DNA thermal cycler (Bio-Rad) in 50-μl mixtures containing 2.5 U of Taq polymerase (Promega, Madison, Wis.) and 1× buffer consisting of 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.01 μg of gelatin, each deoxynucleoside triphosphate at a concentration of 200 μM, and each oligonucleotide primer at a concentration of 2 μM. Thirty-five cycles were performed for each reaction, with the following temperature profile for each cycle: 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min.
For direct DNA sequencing, PCR products were purified with MicroSpin S-300 HR PCR purification columns (Pharmacia). Sequencing reactions were performed with corresponding primers specific for the blaTEM and blaSHV genes (26, 37) by the method of Sanger et al. (38). An automated sequencer (377; ABI Prism, Perkin-Elmer, Norwalk, Conn.) was used.
RESULTS
Bacterial strains and clinical features.
Forty-nine (56.3%) of 87 isolates of K. pneumoniae and 21 (18.6%) of 113 E. coli isolates obtained from all unselected clinical specimens from the pediatric oncology ward submitted to the clinical microbiology laboratory between 1993 and 1997 were resistant or intermediately susceptible to aztreonam or broad-spectrum cephalosporins by the disk diffusion method. Of the blood isolates, 46.2% (12 of 26) of the K. pneumoniae isolates and 23.1% (6 of 26) of the E. coli isolates were not susceptible to the extended-spectrum β-lactams. Of the 18 resistant blood isolates recovered from storage, one E. coli isolate was missing and another E. coli isolate was excluded because the MICs of aztreonam and the broad-spectrum cephalosporins for the strain were in the susceptible range (MICs, ≤2 μg/ml) when the MICs were determined by the agar dilution method. Twelve clinical isolates of K. pneumoniae (isolates kp1 to kp12) and four isolates of E. coli (isolates ec13 to ec16) were included in this study. Two isolates of E. coli (isolates ec13 and ec14) were obtained from the same patient on different occasions (2 days apart). Similarly, two pairs of K. pneumoniae isolates (isolates kp1 and kp2 and isolates kp10 and kp11) were from two individual patients and were recovered 2 and 4 days apart, respectively. A total of three episodes of E. coli bloodstream infection and 10 episodes of K. pneumoniae bloodstream infection in the pediatric ward from 1993 to 1997 were reviewed. Clinical data on 13 patients are summarized in Table 1. All patients had malignancies and received chemotherapy before their bacteremia occurred. Only one (patient 6) was not neutropenic when the multiple-drug-resistant strains were isolated. All these infection episodes were nosocomial. The infection focus could not be determined in most cases except for two episodes of catheter-related infection and two episodes of urinary tract infection (patients 1, 3, 4, and 12).
TABLE 1.
Clinical features and review of antimicrobial agent use by patients with respect to detection of ESBL-producing bacteremiaa
| Patient | Isolate | Age (yr) | Sex | Infection focus | Malignancy | Antibiotic used when bacteremia occurred | Antibiotic used after detection (timing after bacteremia)b | Outcome |
|---|---|---|---|---|---|---|---|---|
| P1 | kp1 | 14 | F | Catheter-related infection | Ewing's sarcoma | Cefazolin, amikacin, piperacillin, amphotericin B | No change | Died of Candida tropicalis fungemia on day 34 after K. pneumoniae kp2 bacteremia |
| kp2 (isolated 2 days after kp1 was isolated) | Cefmetazole, aztreonam, amphotericin B (within 1 day) | |||||||
| P2 | kp3 | 2 | M | FUO | AML | Piperacillin, cefazolin, amikacin | Imipenem, gentamicin, amphotericin B (within 1 day) | Recovered |
| P3 | kp4 | 1 | F | Urinary tract infection | Neuroblastoma | Nil | Cefazolin, amikacin, ceftazidime (2 days) | Recovered |
| P4 | kp5 | 9 | M | Catheter-related infection | ALL | Cefazolin, amikacin, piperacillin, ceftazidime | Vancomycin, amikacin, ceftazidime, amphotericin B (within 1 day) | Died of S. maltophilia infection on day 36 after K. pneumoniae kp5 bacteremia |
| P5 | kp6 | 6 | M | FUO | AML | Cefazolin, amikacin, ceftazidime, amphotericin B, vancomycin | Imipenem, cefmetazole, amphotericin B (1 day) | Died of S. maltophilia infection on day 74 after K. pneumoniae kp6 bacteremia |
| P6 | kp7 | 13 | M | FUO | Ewing's sarcoma | Amikin, piperacillin, amphotericin B, cefazolin | Imipenem, gentamicin (4 days) | Died of seizure and apnea on day 34 after K. pneumoniae kp7 bacteremia |
| P7 | kp8 | 16 | M | FUO | ALL | Cefazolin, ceftazidime, amikacin | Vancomycin, imipenem, ciprofloxacin (within 1 day) | Died on day 2 after K. pneumoniae bacteremia |
| P8 | kp9 | 13 | M | FUO | ALL | Cefazolin, netilmicin, piperacillin, amphotericin B | Imipenem, cefmetazole, amphotericin B (2 days) | Recovered |
| P9 | kp10 | 2 | F | FUO | AML | Cefazolin, ceftazidime, amikacin | Imipenem (3 days) | Died on day 2 after K. pneumoniae kp11 and Citrobacter freundii bacteremia (K. pneumoniae and C. freundii were isolated on 2 separate days) |
| kp11 (isolated 4 days after kp10 was isolated) | Imipenem | Imipenem, amphotericin B (within 1 day) | ||||||
| P10 | kp12 | 4 | M | FUO | AML | Cefazolin, amikacin, ceftazidime, amphotericin B | No change | Died on day of K. pneumoniae bacteremia |
| P11 | ec13 | 5 | F | FUO | ALL | Ceftazidime | Imipenem (1 day) | Recovered |
| ec14 (isolated 2 days after kp13 was isolated) | Imipenem | Imipenem, gentamicin (1 day) | ||||||
| P12 | ec15 | 3 | M | Urinary tract infection | ALL | Nil | Imipenem, amikacin (2 days) | Recovered |
| P13 | ec16 | 2 | M | FUO | AML | Nil | Cefazolin, piperacillin, and tobramycin (within 1 day) and then imipenem and vancomycin (3 days) | Died of sepsis caused by unidentified pathogen on day 116 after E. coli ec16 bacteremia |
Abbreviations: F, female; M, male; FUO, fever of unknown origin; AML, acute myeloid leukemia; ALL, acute lymphoid leukemia.
The antibiotics used after the detection of bacteremia, including the antibiotics used empirically and antibiotics used after the culture report became available. The timing of the change in antibiotic when antibiotics were adjusted on the day of bacteremia is presented as “within 1 day.”
Patients had previously received piperacillin and/or ceftazidime with amikacin and/or cefazolin. Nine of the 10 patients were receiving an extended-spectrum β-lactam when they developed bacteremia caused by an ESBL-producing member of the family Enterobacteriaceae. Thereafter, nine patients received imipenem-containing regimens. Of these nine patients, four patients recovered, three died of causes not attributable to infection with an ESBL producer, and two died of mixed bacteremia or K. pneumoniae infection within 1 day of blood culturing. The other 4 patients (of a total of 13 patients) were treated with an extended-spectrum β-lactam-containing regimen, and only one of these patients died as a result of the infection with an ESBL-producing K. pneumoniae strain. One patient survived, one died of fungemia, and one died of Stenotrophomonas maltophilia infection.
Three patients (patients 1, 3, and 4) who did not die as a result of ESBL-producing K. pneumoniae bacteremia and who received extended-spectrum β-lactams had identified sources of infection, i.e., urinary tract infection or catheter-related infection. Their infected catheters were removed, and their antimicrobial regimens contained effective antibiotics. For two patients (patients 1 and 4) with catheter-related ESBL-producing K. pneumoniae infection, cefmetazole (MIC = 1 μg/ml for kp1) or amikacin (MIC = 0.5 μg/ml for kp5) was used. For patient 3, who had a urinary tract infection and bacteremia due to ESBL-producing K. pneumoniae, the isolate was susceptible to amikacin (MIC = 16 μg/ml), and he was successfully treated with amikacin with cefazolin and ceftazidime.
Three patients (patients 1, 9, and 11) had bacteremias 2 to 4 days after a prior bacteremic episode. When the second episode of bacteremia occurred, one patient (patient 1) was not receiving therapy effective against an ESBL producer and two patients (patients 9 and 11) had just begun (within 1 day) to receive imipenem.
Plasmid profile and transfer of resistance determinants.
Comparison of plasmids isolated from clinical strains revealed that each contained one or two plasmids with molecular sizes of >90 kb. The gene encoding the ESBL could be transferred with either ceftazidime or aztreonam selection in most donors except for four K. pneumoniae isolates (isolates kp1 to kp4). Only one plasmid was transferred for each strain, and the resistance gene in each transconjugant was found to be located in a plasmid of >90 kb (data not shown).
PFGE analysis of donors and plasmid profiles of transconjugants.
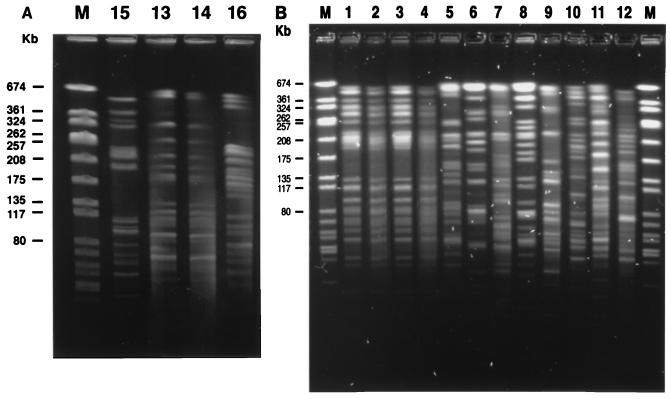
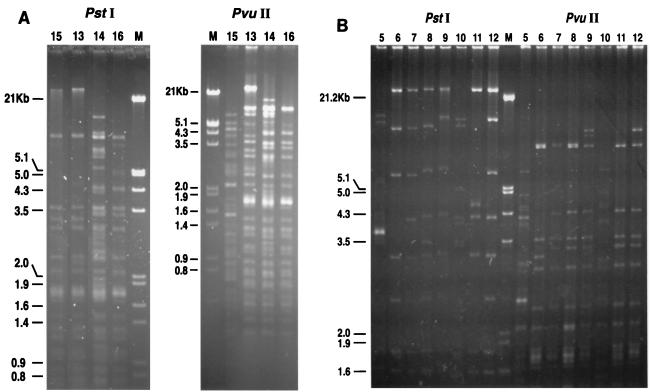
Two E. coli isolates (isolates ec13 and ec14) from the same patient had exactly the same total DNA profile by PFGE after XbaI digestion (Fig. 1A, lanes 13 and 14). Among the three patients with E. coli bacteremia, three different DNA profiles were identified by PFGE (Fig. 1A, lanes 15, 13 and 14, and 16). Regarding the K. pneumoniae isolates, four isolates (isolates kp1 to kp4) had indistinguishable PFGE patterns. The other eight K. pneumoniae isolates (Fig. 1B, lanes 5 to 12, respectively), including isolates kp10 and kp11 from the same patient, had unrelated PFGE patterns (Fig. 1B). Restriction enzyme digestion of plasmids showed that all four E. coli transconjugants (Fig. 2A) and eight K. pneumoniae transconjugants (isolates kp5 to kp12) (Fig. 2B) had different PstI or PuvII digestion profiles, indicating that their plasmids were all different.
FIG. 1.
Profiles produced by PFGE of macrorestriction fragments of ESBL-producing E. coli (A) and K. pneumoniae (B) isolates after digestion with XbaI. Lanes M, SmaI-digested Staphylococcus aureus NCTC 8325 DNA ladder as a molecular size marker. See Table 1 for the origins of the isolates. Lanes 1 to 12, isolates from patients 1 to 10, respectively; lane 15, isolate from patient 12; lanes 13 and 14, isolates from patient 11; lane 16, isolate from patient 13.
FIG. 2.
PstI and PvuII digestion profiles of plasmids from transconjugants of E. coli (A) and K. pneumoniae (B). Lanes M, EcoRI-HindIII-digested λ phage DNA ladder; lanes 5 to 12 and 13 to 16, transconjugants of clinical isolates kp5 to 12 and ec13 to 16, respectively. See Table 1 for the origins of the clinical isolates.
Antimicrobial susceptibility of clinical isolates and transconjugants.
Results of the in vitro antimicrobial susceptibility tests are shown in Table 2. All 4 E. coli isolates and all 12 K. pneumoniae isolates were resistant to ampicillin and cefazolin and susceptible to cefepime, cefmetazole, imipenem, meropenem, and ciprofloxacin. The proportions of isolates susceptible to amoxicillin-clavulanic acid and piperacillin-tazobactam were 66.6 and 93.8%, respectively. The MICs of extended-spectrum β-lactams were not above the resistance breakpoint according to NCCLS criteria (31) for all clinical isolates. A total of 20, 26.6, 46.6, and 100% of the clinical isolates were susceptible to aztreonam, ceftazidime, cefotaxime, and cefepime, respectively. However, when clavulanic acid at a fixed concentration of 4 μg/ml was combined with ceftazidime, cefotaxime, aztreonam, or cefepime individually for susceptibility testing, the MICs decreased by more than 16 times for all isolates and all isolates became susceptible. The MICs of various antimicrobial agents for the 12 transconjugants were similar to those for the clinical isolates from which they were derived.
TABLE 2.
In vitro susceptibilities of the 4 E. coli isolates, 12 K. pneumoniae isolates, and their transconjugants determined by the agar dilution method
| Antibiotica | MIC (μg/ml)b
|
% Susceptible strains | ||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| ATM | 2–64 | 16 | 64 | 25 |
| CAZ | 4–≥256 | 128 | ≥256 | 25 |
| CAZ+CLA | 0.06–4 | 0.5 | 4 | |
| CTX | 2–32 | 16 | 32 | 43.8 |
| CTX+CLA | ≤0.03–2 | 0.5 | 2 | |
| FEP | 0.12–2 | 1 | 2 | 100 |
| AMP | 16–≥256 | ≥256 | ≥256 | 0 |
| AMX+CLA | 2–32 | 8 | 32 | 62.5 |
| CFZ | 16–≥256 | 64 | 128 | 0 |
| CMZ | 0.5–16 | 2 | 16 | 100 |
| AMK | 0.5–16 | 4 | 16 | 100 |
| IPM | 0.06–0.5 | 0.12 | 0.25 | 100 |
| CIP | ≤0.03–1 | ≤0.03 | 0.5 | 100 |
| PIP+TZB | 1–128 | 2 | 16 | 93.8 |
ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; AMP, ampicillin; AMX, amoxicillin; CFZ, cefazolin; CMZ, cefmetazole; AMK, amikacin; IPM, imipenem; CIP, ciprofloxacin; PIP, piperacillin; CLA, clavulanic acid (which was used at a fixed concentration of 4 μg/ml when tested with ceftazidime, cefotaxime, aztreonam, and cefepime and at a ratio of 2:1 when it was tested with amoxicillin); TZB, tazobactam (fixed concentration of 4 μg/ml).
50% and 90%, MICs at which 50 and 90%, respectively, are inhibited.
All 16 isolates from 13 patients yielded positive results by the double-disk synergy test and the Etest ESBL screening test.
Isoelectric focusing.
Isoelectric focusing of sonic extracts of the strains followed by nitrocefin screening revealed two major bands with pIs of 5.4 and 8.2 for both the clinical isolates and the transconjugants of two K. pneumoniae isolates (isolates kp9 and kp12) and all four E. coli isolates. Five isolates (isolates kp1 to kp4 and kp10) had only one band with a pI of 7.6 and five isolates (isolates kp5 to kp8 and kp11) had a band with a pI of 8.2 (Table 3).
TABLE 3.
Molecular characterization of ESBL-producing E. coli and K. pneumoniae isolates
| Patient | Isolate | SHV | TEM | Macrorestriction genotypea | Plasmid digestion profileb | β-Lactamase pIc |
|---|---|---|---|---|---|---|
| P1 | kp1 | SHV-2 | B | ND | 7.6 | |
| P1 | kp2 | SHV-2 | B | ND | 7.6 | |
| P2 | kp3 | SHV-2 | B | ND | 7.6 | |
| P3 | kp4 | SHV-2 | B | ND | 7.6 | |
| P4 | kp5 | SHV-5 | E | e | 8.2 | |
| P5 | kp6 | SHV-5 | F | f | 8.2 | |
| P6 | kp7 | SHV-5 | G | g | 8.2 | |
| P7 | kp8 | SHV-5 | H | h | 8.2 | |
| P8 | kp9 | SHV-5 | TEM-1 | I | i | 5.4, 8.2 |
| P9 | kp10 | SHV-2 | J | j | 7.6 | |
| P9 | kp11 | SHV-5 | K | k | 8.2 | |
| P10 | kp12 | SHV-5 | TEM-1 | L | l | 5.4, 8.2 |
| P11 | ec13 | SHV-5 | TEM-1 | A | a | 5.4, 8.2 |
| P11 | ec14 | SHV-5 | TEM-1 | A | c | 5.4, 8.2 |
| P12 | ec15 | SHV-5 | TEM-1 | C | b | 5.4, 8.2 |
| P13 | ec16 | SHV-5 | TEM-1 | D | d | 5.4, 8.2 |
Macrorestriction genotypes were determined by PFGE after digestion with XbaI.
Plasmid digestion profile for tranconjugants digested with PvuII and PstI. ND, not determined.
β-Lactamase pIs were determined by isoelectric focusing.
PCR amplification and sequencing of PCR products for blaTEM and blaSHV.
All four E. coli transconjugants were positive for both blaTEM and blaSHV by PCR amplification. The entire blaTEM sequence including the promoter region from the four E. coli isolates was found to be identical to the blaTEM-1-encoding Tn2 sequence (11). Comparison of the sequences at the blaSHV region of the four E. coli strains to the published SHV-1 gene sequence (29) revealed three nucleotide substitutions that caused two amino acid changes (position 238, Gly [GGC]→Ser [AGC]; position 240, Glu [GAG]→Lys [AAG]), with the remaining one being silent (position 268, Thr [ACG]→Thr [ACC]). The amino acid sequences at the blaSHV region of all four E. coli isolates were found to be identical to the published SHV-5 sequence (2). Two isolates of K. pneumoniae (isolates kp9 and kp12) also harbored both TEM-1 and SHV-5 genes, as did the four E. coli isolates. The other 10 K. pneumoniae isolates had only one SHV-type gene (Table 3). Five isolates had the SHV-5 gene. Five isolates had an SHV-2 gene with the characteristic of one nucleotide substitution that caused one amino acid change (position 238, Gly [GGC]→Ser [AGC]). Four SHV-2-producing K. pneumoniae isolates (isolates kp1 to kp4) did not have the silent mutation (position 268, Thr [ACG]→Thr [ACC]), whereas another SHV-2 producer (isolate kp10) and all the SHV-5-containing isolates had the same silent mutation that the E. coli isolates had.
DISCUSSION
In the hospital studied, routine susceptibility testing was performed by the Kirby-Bauer disk diffusion test, and cefotaxime was the antibiotic representative of broad-spectrum cephalosporins, as recommended by NCCLS (30), unless a special request was made to test for susceptibility to another antimicrobial agent or the isolate was resistant to cefotaxime. The double-disk synergy test was not routinely used in our clinical microbiology laboratory. Since some ESBL producers may not be detectable by the routine use of only one extended-spectrum β-lactam, the prevalence of ESBL-producing members of the family Enterobacteriaceae might have been even higher than what was reported.
In this study, clonal spread was detected by PFGE in four isolates of K. pneumoniae from 3 patients (patients 1 to 3) that produced SHV-2 in 1994. The four isolates also shared the characteristic of not transferring their resistance by conjugation and lacked a silent mutation (position 268, Thr [ACG]→Thr [ACC]). The other K. pneumoniae isolates from different patients proved to be unrelated by PFGE. The differences in their plasmid digestion profiles also ruled out the possibility of plasmid dissemination in the ward studied.
Two plasmid-mediated β-lactamases, TEM-1 and SHV-5, were simultaneously found in two K. pneumoniae isolates (isolates kp9 and kp12) and in all four E. coli isolates. These resistance determinants were conjugatively transferable, and a plasmid of >90 kb was identified in all six transconjugants. However, the plasmid digestion profiles of these isolates revealed six different patterns, even for the two isolates from the same patient (isolates ec13 and ec14). The circulation of one resistant plasmid harboring two β-lactamase genes was thus excluded.
It is estimated that worldwide about 50% of clinical E. coli isolates produce a TEM-1 β-lactamase (23, 36), and TEM-1 accounts for about 80% of all plasmid-encoded β-lactamases in clinical members of the family Enterobacteriaceae (9). Although the prevalence of TEM-1 in Taiwan was not studied, considering the worldwide distribution of TEM-1 and the finding that, in 1998, 82% of the E. coli isolates in our hospital were ampicillin resistant, as determined by the routine disk diffusion method, the existence of TEM-1 in our E. coli isolates seems reasonable. The gene for SHV-5 has been reported to be the most common ESBL gene in K. pneumoniae isolates in another Taiwan hospital (22) and was the predominant ESBL gene in our K. pneumoniae isolates as well. That the spread of ESBL is mainly by patient-to-patient transfer rather than by direct selection of point mutation derivatives has been postulated on the basis of a clinical intervention study (34). The coexistence of TEM-1 and SHV-5 instead of the occurrence of TEM-1 mutant ESBLs in our E. coli isolates suggests a greater ease of acquisition of another ESBL gene compared with the development of a TEM-1 ESBL gene with a mutation.
There were three pairs of sequential isolates from three patients (Table 3). Two E. coli isolates from patient 11 recovered 2 days apart were of the same clone but had different plasmids. Although the existence of completely different plasmids in the two isolates was not impossible, this finding suggests the possible existence of a unit smaller than a plasmid, such as integrons or transposons (15), or the existence of an unstable plasmid that may change easily. The two K. pneumoniae isolates from patient 1 (isolates kp1 and kp2) and another two strains (isolates kp3 and kp4) had the same macrorestriction pattern by PFGE, suggesting a clonal origin. As for isolates kp10 and kp11, which were isolated from patient 9 4 days apart, the totally different macrorestriction patterns, ESBL genes, and plasmid digestion patterns suggest that these two strains were acquired independently of each other. There were thus three different bacteriologic patterns in patients with bacteremia caused by consecutive ESBL producers.
In the current study, most of the ESBL producers had initially been reported to be intermediately susceptible or resistant to cefotaxime. With even further determination of MICs by the agar dilution method, some isolates were susceptible to cefotaxime, ceftazidime, or aztreonam. According to the new recommendations by NCCLS in 1999 (32) for susceptibility testing of E. coli and K. pneumoniae, strains that are inhibited to a lesser degree than the normal susceptible population, even though the MIC is lower than the standard breakpoint for resistance, should be screened for a potential ESBL. Resistance should be reported if the strain shows a positive inhibition test result (32). Since the SHV-5 β-lactamase is generally classified as a ceftazidimase and hydrolyzes cefotaxime less efficiently (4), if organisms harboring this enzyme are not tested according to the guidelines stated above but are tested only by a cefotaxime disk test, an inability to detect resistance and treatment failure might ensue (21).
Schiappa et al. (39) found that patients with bacteremia caused by ESBL-producing E. coli strains were more likely to survive if they received appropriate treatment within 3 days of the onset of the infection. A pediatric patient with leukemia who developed bacteremia caused by a cefotaxime-susceptible but ceftazidime-resistant E. coli strain died after receiving cefotaxime for less than 1 day (39). In addition, another study suggested that mortality was significantly lower when a carbapenem regimen instead of a noncarbapenem regimen was used in the first 5 days of bacteremia (35). Delays in the use of antibiotics effective against ESBL producers may result in delayed clearance of bacteremia, as demonstrated in the three patients (patients 1, 9, and 11) with two episodes of bacteremia. For patient 9 the outcome of the second bacteremia episode was fatal because of mixed ESBL-producing K. pneumoniae and Citrobacter freundii infections. Thus, timely identification of an ESBL producer is important, and policies for laboratory performance and antimicrobial therapy may need to be reevaluated to take this into account, especially when the prevalence of an ESBL producer is increasing in a specific setting.
Two of nine patients receiving imipenem-containing regimens died within 1 day of septicemia, and three of four patients receiving noncarbapenem therapy did not die as a result of bacteremia caused by an ESBL producer. No increased mortality among patients treated with antibiotics to which ESBL producers are susceptible was found. However, we cannot provide a definite suggestion about the appropriate treatment regimen for bacteremia caused by ESBL producers from the results of this study due to the small number of cases and the lack of a case-control design. Among the four patients receiving antibiotics to which ESBL producers are susceptible, two had catheter-related infections and one had a urinary tract infection. For the patients with catheter-related bacteremia caused by ESBL producers, immediate removal of the infected catheter and the use of an active antimicrobial agent other than imipenem may still produce a good outcome. Naumovski et al. (33) reported that seven children with nonbacteremic urinary tract infections were successfully treated with ceftazidime therapy, and they proposed that a high level of ceftazidime in urine was the reason for successful therapy. Ceftazidime and amikacin therapy for bacteremic urinary tract infection was shown to be effective for one patient in the study.
Prior administration of any antibiotic and prior ceftazidime or aztreonam administration were reported to be risk factors for the acquisition of ESBL-producing organisms (39). Every patient included in current study had received the standardized regimen containing an antipseudomonal β-lactam, amikacin, and cefazolin 3 weeks prior to bacteremia. However, the retrospective nature of this study could not prove definitely that prior antimicrobial administration was the risk factor. Nevertheless, the finding that 9 of 13 patients developed bacteremia caused by an ESBL-producing strain while under extended-spectrum β-lactam therapy argues for the occurrence of selection pressure.
In conclusion, we report on the clonal spread of ESBL-producing K. pneumoniae and the identification of E. coli and K. pneumoniae isolates from blood with transferable resistance plasmids carrying both extended-spectrum (SHV-5) and restricted-spectrum (TEM-1) β-lactamase genes in a pediatric oncology ward. SHV-5 was the predominant ESBL detected in this study. The possibility that the ESBL gene may disseminate to other members of the family Enterobacteriaceae necessitates close monitoring of the ESBL producer to prevent such an occurrence. In a ward with high percentages of ESBL-producing E. coli and K. pneumoniae isolates, if symptomatic improvement is not seen during empirical treatment with a combination of an antipseudomonal β-lactam and an aminoglycoside, a change in the antibiotic treatment regimen to include agents that are active against ESBL producers, such as imipenem, should be promptly considered. The new recommendation by NCCLS to screen for ESBLs in E. coli and K. pneumoniae isolates should also be strictly enforced to avoid a false designation of susceptibility to broad-spectrum cephalosporins and a resulting clinical failure.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Morre D D, Seidman J G, Smith J A. Current protocols in molecular biology, unit 2.4.2. Boston, Mass: Massachusetts General Hospital and Harvard Medical School; 1995. Miniprep of bacterial genomic DNA. [Google Scholar]
- 2.Billot-Klein D, Gutmann L, Collatz E. Nucleotide sequence of the SHV-5 β-lactamase gene of a Klebsiella pneumoniae plasmid. Antimicrob Agents Chemother. 1990;34:2439–2441. doi: 10.1128/aac.34.12.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;ii:302–306. doi: 10.1016/s0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M L, Chen Y C, Pan H J, Chang S C, Yang L S, Ho S W, Luh K T, Hsieh W C, Chuang C Y. Secular trends in the etiology of nosocomial infection at a teaching hospital in Taiwan, 1981–1994. Chin J Microbiol Immunol. 1995;28:203–217. [PubMed] [Google Scholar]
- 6.Chong C Y, Tan A M, Lou J. Infections in acute lymphoblastic leukaemia. Ann Acad Med Singapore. 1998;27:491–495. [PubMed] [Google Scholar]
- 7.Cordonnier C, Herbrecht R, Pico J L, Gardembas M, Delmer A, Delain M, Moreau P, Ladeb S, Nalet V, Rollin C, Gres J J. Cefepine/amikacin versus ceftazidime/amikacin as empirical therapy for febrile episodes in neutropenic patients: a comparative study. Clin Infect Dis. 1997;24:41–51. doi: 10.1093/clinids/24.1.41. [DOI] [PubMed] [Google Scholar]
- 8.Cukier L, Lutzler P, Bizien A, Avril J L. Investigation of an epidemic of an extended spectrum beta-lactamase producing Escherichia coli in a geriatrics department. Pathol Biol. 1999;47:440–444. [PubMed] [Google Scholar]
- 9.Du Bois S K, Marriott M S, Amyes S G B. TEM- and SHV-derived extended-spectrum β-lactamases: relationship between selection, structure and function. J Antimicrob Chemother. 1995;35:7–22. doi: 10.1093/jac/35.1.7. [DOI] [PubMed] [Google Scholar]
- 10.Gniadkowski M, Palucha A, Grzesiowski P, Hryniewicz W. Outbreak of ceftazidime-resistant Klebsiella pneumoniae in a pediatric hospital in Warsaw, Poland: clonal spread of the TEM-47 extended-spectrum beta-lactamase (ESBL)-producing strain and transfer of a plasmid carrying the SHV-5-like ESBL-encoding gene. Antimicrob Agents Chemother. 1998;42:3079–3085. doi: 10.1128/aac.42.12.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goussard S, Courvalin P. Sequence of the genes blaT-1B and blaT-2. Gene. 1991;15:71–73. doi: 10.1016/0378-1119(91)90540-r. [DOI] [PubMed] [Google Scholar]
- 12.Green M, Barbadora K. Recovery of ceftazidime-resistant Klebsiella pneumoniae from pediatric liver and intestinal transplant recipients. Pediatr Transplant. 1998;2:224–230. [PubMed] [Google Scholar]
- 13.Grogan J, Murphy H, Butler K. Extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a Dublin paediatric hospital. Br J Biomed Sci. 1998;55:111–117. [PubMed] [Google Scholar]
- 14.Gunseren F, Mamikoglu L, Ozturk S, Yucesoy M, Biberoglu K, Yulug N, Doganay M, Sumerkan B, Kocagoz S, Unal S, Cetin S, Calangu S, Koksal I, Leblebicioglu H, Gunaydin M. A surveillance study of antimicrobial resistance of gram-negative bacteria isolated from intensive care units in eight hospitals in Turkey. J Antimicrob Chemother. 1999;43:373–378. doi: 10.1093/jac/43.3.373. [DOI] [PubMed] [Google Scholar]
- 15.Heritage J, Hawkey P M, Todd N, Lewis I J. Transposition of the gene encoding a TEM-12 extended-spectrum beta-lactamase. Antimicrob Agents Chemother. 1992;36:1981–1986. doi: 10.1128/aac.36.9.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibbert-Rogers L C, Heritage J, Gascoyne-Binzi D M, Hawkey P M, Todd N, Lewis I J, Bailey C. Molecular epidemiology of ceftazidime resistant Enterobacteriaceae from patients on a paediatric oncology ward. J Antimicrob Chemother. 1995;36:65–82. doi: 10.1093/jac/36.1.65. [DOI] [PubMed] [Google Scholar]
- 17.Hughes W T, Armstrong D, Bodey G P, Brown A E, Edwards J E, Feld R, Pizzo P, Rolston K V, Shenep J L, Young L S. 1997 guidelines for the use of antimicrobial agents in neutropenic patients with unexplained fever. Clin Infect Dis. 1997;25:551–573. doi: 10.1086/513764. [DOI] [PubMed] [Google Scholar]
- 18.Jan I S, Hsueh P R, Teng T J, Ho S W, Luh K T. Antimicrobial susceptibility testing for Klebsiella pneumoniae isolates resistant to extended spectrum beta-lactam antibiotics. J Formos Med Assoc. 1998;97:661–666. [PubMed] [Google Scholar]
- 19.Jarlier V, Nicolas M H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1998;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 20.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karas J A, Pillay D G, Muckart D, Sturm A W. Treatment failure due to extended spectrum beta-lactamase. J Antimicrob Chemother. 1996;37:203–204. doi: 10.1093/jac/37.1.203. [DOI] [PubMed] [Google Scholar]
- 22.Liu P Y, Tung J C, Ke S C, Chen S L. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a district hospital in Taiwan. J Clin Microbiol. 1998;36:2759–2762. doi: 10.1128/jcm.36.9.2759-2762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu P L, Hsueh P R, Hung C C, Chang S C, Luh K T. Abstracts of the 1998 Annual Meeting of the Infectious Disease Society of Republic of China. 1998. Clinical feature of extended spectrum beta-lactam antibiotics resistant Escherichia coli bacteremia; p. 47. [Google Scholar]
- 25.Lucet J C, Chevret S, Decre D, Vanjak D, Macrez A, Bedos J P, Wolff M, Regnier B. Outbreak of multiply resistant Enterobacteriaceae in an intensive care unit: epidemiology and risk factors for acquisition. Clin Infect Dis. 1996;22:430–436. doi: 10.1093/clinids/22.3.430. [DOI] [PubMed] [Google Scholar]
- 26.Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 553–563. [Google Scholar]
- 27.Maschmeyer G, Hiddemann W, Link H, Cornely O A, Buchheidt D, Glass B, Adam D. Management of infections during intensive treatment of hematologic malignancies. Ann Hematol. 1997;75:9–16. doi: 10.1007/s002770050306. [DOI] [PubMed] [Google Scholar]
- 28.Matthew M, Harris A M. Identification of beta-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J Gen Microbiol. 1976;94:55–67. doi: 10.1099/00221287-94-1-55. [DOI] [PubMed] [Google Scholar]
- 29.Mercier J, Levesque R C. Cloning of SHV-2, OHIO-1, and OXA-6 beta-lactamases and cloning and sequencing of SHV-1 beta-lactamase. Antimicrob Agents Chemother. 1990;34:1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. Performance standards for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: ninth information supplement, (M100-S9) vol. 19, no. 36. M2-A6 and M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 33.Naumovski L, Quinn J P, Miyashiro D, Patel M, Bush K, Singer S B, Graves D, Palzkill T, Arvin A M. Outbreak of ceftazidime resistance caused by extended-spectrum beta-lactamase in isolates from cancer patients. Antimicrob Agents Chemother. 1992;36:1991–1996. doi: 10.1128/aac.36.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nordmann P. Trends in beta-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27(Suppl. 1):S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 35.Paterson D L, Ko W, Gottberg A V, Mohapatra S, Casellas J M, Mulazimoglu L, Goossens H, Trenholme F, Klugman K, Rice L B, Bonomo R A, Yu V L. Abstracts of the 36th Annual Meeting of the Infectious Diseases Society of America. 1998. In vitro susceptibility and clinical outcome of bacteremia due to extended spectrum beta-lactamase (ESBL) producing K. pneumoniae, abstr. 188. [Google Scholar]
- 36.Philippon A, Arlet G, Lagrange P H. Origin and impact of plasmid-mediated extended-spectrum beta-lactamases. Eur J Clin Microbiol Infect Dis. 1994;13:S17–S29. doi: 10.1007/BF02390681. [DOI] [PubMed] [Google Scholar]
- 37.Rasheed J K, Jay C, Metchock B, Berkowitz F, Weigel L, Crellin J, Steward C, Hill B, Medeiros A A, Tenover F C. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41:647–653. doi: 10.1128/aac.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiappa D A, Hayden M K, Matushek M G, Hashemi F N, Sullivan J, Smith K Y, Miyashiro D, Quinn J P, Weinstein R A, Trenholme G M. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli bloodstream infection: a case-control and molecular epidemiologic investigation. J Infect Dis. 1996;174:529–536. doi: 10.1093/infdis/174.3.529. [DOI] [PubMed] [Google Scholar]
- 40.Schimpff S C. Empiric antibiotic therapy for granulocytopenic cancer patients. Am J Med. 1986;80:13–20. [PubMed] [Google Scholar]
- 41.Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992;30:1491–1498. doi: 10.1128/jcm.30.6.1491-1498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiener J, Quinn J P, Bradford P A, Goering R V, Nathan C, Bush K, Weinstein R A. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA. 1999;281:517–523. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]