Abstract
To determine if there is variability in virulence among strains of Porphyromonas gingivalis in human periodontitis, their distribution in a group of subjects with clear indicators of periodontitis and in a healthy, age-matched control group was examined. The presence of heteroduplex types of P. gingivalis in the two groups was determined with a PCR-based assay. This assay relied on detection of polymorphisms in the ribosomal internal spacer region (ISR). ISR fragments generated by PCR with P. gingivalis-specific primers were hybridized to fragments from reference strains, and the formation of heteroduplexes from the hybridization of nonidentical sequences was observed by polyacrylamide gel electrophoresis. Characteristic fingerprints from comparison with a panel of reference strains allowed the identification of heteroduplex types in clinical samples. One hundred thirty adults with periodontitis and 181 controls were sampled. With this approach, 11 heteroduplex types of P. gingivalis were detected in the population. Sufficient numbers were available for statistical analysis of six of these types. Heteroduplex type hW83 was found to be very strongly associated with periodontitis (P = 0.0000), and two additional types, h49417 and hHG1691, were also significantly associated with disease. The remaining types, h23A4, h381, and hA7A1, were detected more frequently in subjects with periodontitis than in healthy subjects, but the difference was not significant. These data indicate that virulence in human periodontitis varies among strains of P. gingivalis, and they identify an apparently highly virulent subgroup.
Of the several bacterial species suspected of playing an important role in the pathogenesis of adult-onset periodontitis, Porphyromonas gingivalis has been the most consistently and strongly associated with disease (2, 11, 13, 14, 23, 25, 28, 30). However, it has also been detected in periodontally healthy subjects. By using a sensitive, PCR-based assay, P. gingivalis was detected in 25% of a mature, periodontally healthy control group (11). The presence of P. gingivalis in this group without significant disease raises questions regarding the existence of strains of low virulence.
Several studies have examined the effect of subcutaneous injection of P. gingivalis into rodents (6, 7, 9, 17, 18, 27, 33). All of these studies have shown differences among strains in the ability to cause localized or systemic infections. Katz et al. also found differences among strains in the ability to cause bone loss in an acute oral infection in the rat (17). However, no correlation was observed between severity of infections in subcutaneous experiments and oral infections. These studies demonstrate a clear difference in phenotype among strains of P. gingivalis but do not necessarily identify strains most likely to be involved in chronic periodontitis in humans.
Various strain-typing approaches have been used to examine the phylogeny of P. gingivalis and to track P. gingivalis in small cohorts, including whole genomic restriction fragment length polymorphism (10, 34), ribotyping (15, 34), PCR with arbitrary primers (26, 34), serotyping (8, 16, 29), and multilocus enzyme electrophoresis (21). These techniques have not been used to study the distribution of strain types in health and disease. Because extensive genetic variability was observed within the species, and because a large variety of strain types were observed in subjects with disease, it has been assumed that all strains are equally virulent (21).
The purpose of this study was to determine if all strains of P. gingivalis exhibit similar strengths of association with human periodontitis or if indeed there are relatively more- and less-virulent strains. To do this we examined the distributions of strains of P. gingivalis in subjects with periodontitis and a periodontally healthy control group. Differences in the ribosomal intergenic spacer region (ISR) sequence provided a means for distinguishing among strains. This locus has been shown to be useful for distinguishing among strains of several species (1, 4, 32), including P. gingivalis (31). Heteroduplex analysis of DNA generated by PCR from this locus provides an efficient method for analysis of large numbers of samples. It also allows the resolution of multiple strains in a single sample and avoids the bias toward predominant and easily cultured organisms inherent with cultivation-based methods. Heteroduplex analysis of the ISR of P. gingivalis has yielded 22 distinct groups within the species (20). Eleven of these heteroduplex types were observed in the study population examined here. Differences in the strength of association with disease were observed among heteroduplex types, and one type in particular was very strongly associated with disease.
MATERIALS AND METHODS
Study population and sampling.
The study population and sampling methods have been described previously (11). Briefly, subjects for this institutionally approved study were recruited from the clinics of the Ohio State University College of Dentistry. Potential subjects were screened and were selected for participation if they met certain criteria for periodontal health or disease. These criteria were established to include approximately the healthiest and the least healthy one-fourth of the population based on periodontal probing depths and attachment levels. The healthy group was age matched to the disease group. Subgingival plaque samples were collected on sterile endodontic paper points. All teeth were sampled to maximize the possibility of detection of P. gingivalis if it was present at any site. Samples from each individual were pooled in a sterile 2-ml microcentrifuge tube and frozen for later analysis.
Detection of P. gingivalis and determination of heteroduplex type.
DNA was isolated, and samples were analyzed for the presence of P. gingivalis, as previously described (11, 24), with a PCR assay that did not require that the samples be cultured. The target sequence was the ribosomal DNA (rDNA) ISR between the 16S and 23S ribosomal genes. The samples were analyzed for the presence of P. gingivalis by using a nested, two-step PCR procedure with a P. gingivalis-specific primer for the second amplification. DNA fragments were separated by agarose gel electrophoresis. Positive samples were prepared and heteroduplex analysis was performed as previously described (20). Briefly, heteroduplexes were formed by mixing amplified ISR DNA from two samples. The mixtures were incubated at 95°C for 5 min to melt double strands and then cooled to 25°C at the rate of 1°C per min in a thermal cycler to reanneal. Heteroduplexes were formed between amplicons generated from similar but nonidentical ISR DNA fragments amplified from different strains of P. gingivalis. They were resolved by polyacrylamide gel electrophoresis, stained with ethidium bromide, and visualized with UV light.
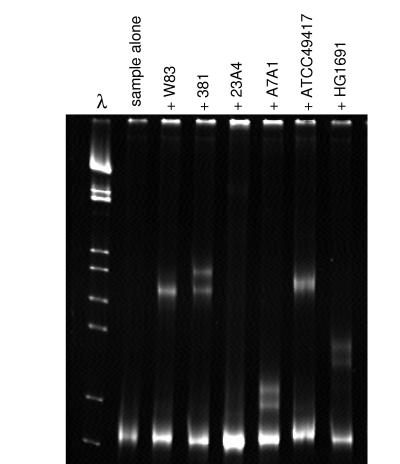
To identify the heteroduplex types present in a sample, heteroduplexes were formed between ISR amplicons from samples and a panel of ISR DNA fragments generated from laboratory strains. Characteristic migration patterns were observed as shown in Fig. 1. Twenty-two heteroduplex types have been identified (19, 20); several of these include characterized laboratory strains and are named accordingly.
FIG. 1.
Heteroduplex gel of a sample containing P. gingivalis heteroduplex type h23A4. ISR DNA amplified from a sample was hybridized to ISR DNA from a series of reference strains of P. gingivalis. Heteroduplex bands were seen for duplexes formed with all strains except 23A4. The migration pattern of heteroduplex bands generated with the sample was identical to that observed for 23A4 duplexed to the same strains (data not shown).
Multiple strains present in a single sample were detected by first examining each sample without the addition of reference DNA. When multiple types were present in a single sample, they were identified from the mixed sample by using the same regimen of comparison to a panel of ISR DNA fragments from laboratory strains.
Statistical analysis.
The prevalences of heteroduplex types of P. gingivalis among healthy and diseased subjects were compared by both univariate chi-square modeling and multivariate modeling with a nominal logistic regression. Odds ratios with 95% confidence intervals and likelihood ratio chi-square values were computed. Chi-square analysis was used to test the frequency with which various strains occurred together, and Bonferroni's correction was applied to correct for multiple tests. t tests were used to examine the relationship of the presence or absence of each strain to parameters of periodontal health within the healthy group and the group with periodontitis. The binomial probability function was used to calculate the expected distribution of multiple strains based on the number of strains detected within each group. Expected values were calculated for each group separately for this analysis because the overall prevalences of P. gingivalis in the two groups differed.
RESULTS
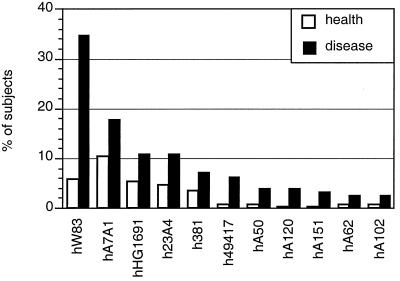
Samples from 311 subjects were analyzed for this study, 130 from the periodontitis group and 181 from the healthy group. The demographics of the samples have been described previously (11). As reported, P. gingivalis was detected in 79% of subjects with periodontitis and in only 25% of periodontally healthy subjects (P < 0.0001) (11). In the present investigation the strains present in these samples were identified by heteroduplex analysis. Eleven of 22 previously described heteroduplex types (20) were detected in the study population, as shown in Fig. 2.
FIG. 2.
Prevalence of heteroduplex types of P. gingivalis in the healthy group and the group with periodontitis. Since multiple strains were present in many subjects, the total percentage is more than 100% for each group.
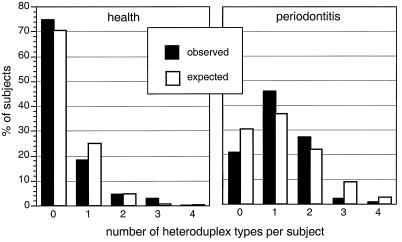
Many subjects harbored multiple strains of P. gingivalis, as shown in Fig. 3. The expected distribution of numbers of strains per individual was compared to the observed distribution (Fig. 3) by chi-square analysis.
FIG. 3.
Presence of multiple heteroduplex types of P. gingivalis in periodontally healthy subjects and subjects with periodontitis. The expected distribution of number of heteroduplex types is also shown. This was calculated for each group based on the binomial probability function and the number of heteroduplex types observed within the group. Both the healthy group and the group with periodontitis showed significant differences between the observed distribution of numbers of strains per individual and the expected distribution. In the healthy group fewer subjects harbored a single strain than expected (P = 0.0003). In the periodontitis group more subjects harbored a single strain than expected (P = 0.026).
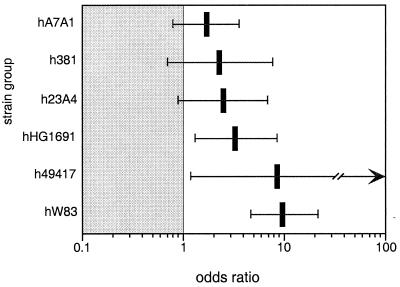
In order to determine if some strains are more virulent than others, the distributions of the various heteroduplex types of P. gingivalis in periodontitis patients and healthy subjects were determined. Sufficient numbers of subjects were available for statistical analysis for 6 of the 11 heteroduplex types observed in the study population. The remaining five heteroduplex types were each found in six or fewer subjects and were excluded from analysis. The univariate model for the relationship of heteroduplex type to disease status is shown in Table 1, and the multivariate model is shown in Table 2. Due to the presence of multiple strains in many subjects, the multivariate model better reflects the relationship of strains to disease. Odds ratios determined by the multivariate model are shown in Fig. 4. Three heteroduplex types, hW83, h49417, and hHG1691, were clearly associated with periodontitis. The three remaining types, h23A4, h381, and hA7A1, were not statistically significantly associated with disease.
TABLE 1.
Univariate model for association of heteroduplex types of P. gingivalis with periodontitis
| Heteroduplex type | Odds ratio (95% confidence interval) | Likelihood ratio chi-square | Likelihood ratio P value |
|---|---|---|---|
| hW83 | 8.9 (4.4–19.5) | 44.3 | 0.0000 |
| h49417 | 11.6 (2.1–216) | 8.9 | 0.0029 |
| hHG1691 | 2.3 (0.96–5.6) | 3.5 | 0.0614 |
| h23A4 | 2.6 (1.1–6.6) | 4.4 | 0.0359 |
| h381 | 2.1 (0.75–6.5) | 2.0 | 0.1557 |
| hA7A1 | 1.9 (0.99–3.8) | 3.7 | 0.0547 |
TABLE 2.
Multivariate model for association of heteroduplex types of P. gingivalis with periodontitis
| Heteroduplex type | Odds ratio (95% confidence interval) | Likelihood ratio chi-square | Likelihood ratio P value |
|---|---|---|---|
| hW83 | 9.6 (4.7–21.5) | 44.1 | 0.0000 |
| h49417 | 8.6 (1.3–170) | 4.9 | 0.0261 |
| hHG1691 | 3.3 (1.3–8.5) | 6.5 | 0.0106 |
| h23A4 | 2.5 (0.91–6.9) | 3.1 | 0.0759 |
| h381 | 2.3 (0.71–7.7) | 2.0 | 0.1593 |
| hA7A1 | 1.7 (0.80–3.6) | 1.9 | 0.1700 |
FIG. 4.
Odds (log scale) of finding heteroduplex types of P. gingivalis in subjects with periodontitis compared to healthy subjects as determined by a nominal logistic regression. The 95% confidence intervals are shown as horizontal bars. The odds are greater than 1 for hW83, h49417, and HG1691 at an α level of 0.05.
The frequency with which various heteroduplex types were found together in the same individual was examined in a matrix shown in Table 3. A significant association was seen for hW83 and hA7A1, hW83 and hHG1691, and hHG1691 and h23A4. The interaction was negative: these pairs of heteroduplex types were significantly less likely to be found together than would be expected based on their prevalences in the population. To apply Bonferroni's correction for multiple chi-square tests, significance levels are adjusted by dividing the α level by the number of tests. In this case, with 15 tests, P must equal 0.003 for significance at the 0.05 level. When this very conservative correction for multiple tests was applied, hHG1691 and hW83 still showed a significant negative interaction, as did hHG1691 and h23A4.
TABLE 3.
Matrix of simultaneous colonization by different heteroduplex types
| Heteroduplex type | Observed frequencya (expected frequencyb) of simultaneous colonization by the indicated pairs of heteroduplex types
|
||||
|---|---|---|---|---|---|
| hA7A1 | h381 | h23A4 | hHG1691 | h49417 | |
| hW83 | 9 (15.1) | 3 (5.5) | 6 (8.1) | 2 (8.5) | 4 (3.3) |
| P = 0.02 | P = 0.14 | P = 0.30 | P = 0.001 | P = 0.63 | |
| h49417 | 4 (2.5) | 1 (0.9) | 2 (1.3) | 1 (1.4) | |
| P = 0.26 | P = 0.92 | P = 0.54 | P = 0.70 | ||
| hHG1691 | 3 (6.3) | 1 (2.3) | 0 (3.4) | ||
| P = 0.07 | P = 0.28 | P = 0.0005 | |||
| h23A4 | 7 (6.1) | 2 (2.2) | |||
| P = 0.63 | P = 0.87 | ||||
| h381 | 2 (4.1) | ||||
| P = 0.16 | |||||
In 149 colonized subjects.
Based on the likelihood ratio chi-square test.
t tests were performed for indicators of periodontal health and the presence or absence of heteroduplex types within the healthy group and the group with periodontitis. The results are shown in Table 4. Only one comparison showed a statistically significant difference: the average probing depth was greater among healthy subjects who harbored heteroduplex type hHG1691 than among those who did not.
TABLE 4.
Comparison of mean attachment loss, probing depth, and P values for t tests for healthy subjects and subjects with periodontitis with and without various heteroduplex types of P. gingivalis
| Heteroduplex type | Greatest probing depth
|
Greatest attachment loss
|
||||||
|---|---|---|---|---|---|---|---|---|
| Healthy group
|
Periodontitis group
|
Healthy group
|
Periodontitis group
|
|||||
| Mean (SD)a | P | Mean (SD)a | P | Mean (SD)a | P | Mean (SD)a | P | |
| W83 | ||||||||
| + | 3.7 (0.8) | 7.7 (1.3) | 4.2 (0.9) | 8.3 (1.9) | ||||
| − | 3.8 (0.8) | 0.697 | 7.5 (1.3) | 0.454 | 4.3 (0.8) | 0.705 | 8.1 (1.6) | 0.343 |
| h49417 | ||||||||
| + | 4.0 (0) | 7.4 (1.5) | 4.0 (0) | 9.1 (1.8) | ||||
| − | 3.8 (0.8) | 0.797 | 7.6 (1.3) | 0.630 | 4.3 (0.8) | 0.718 | 8.1 (1.7) | 0.103 |
| hHG1691 | ||||||||
| + | 4.7 (0.5) | 7.9 (1.6) | 4.8 (0.4) | 8.1 (1.6) | ||||
| − | 3.7 (0.8) | 0.0006 | 7.5 (1.3) | 0.311 | 4.3 (0.8) | 0.069 | 8.2 (1.7) | 0.791 |
| h23A4 | ||||||||
| + | 4.1 (0.8) | 7.4 (1.1) | 4.1 (0.8) | 8.7 (2.7) | ||||
| − | 3.8 (0.8) | 0.231 | 7.6 (1.3) | 0.479 | 4.3 (0.8) | 0.548 | 8.1 (1.5) | 0.214 |
| h381 | ||||||||
| + | 4.3 (0.8) | 7.7 (2.1) | 4.3 (0.8) | 8.3 (2.2) | ||||
| − | 3.8 (0.8) | 0.092 | 7.6 (1.3) | 0.861 | 4.3 (0.8) | 0.908 | 8.2 (1.7) | 0.785 |
| hA7A1 | ||||||||
| + | 3.6 (0.8) | 7.6 (1.2) | 3.9 (0.9) | 8.3 (2.0) | ||||
| − | 3.8 (0.8) | 0.470 | 7.6 (1.3) | 0.914 | 4.3 (0.8) | 0.054 | 8.1 (1.6) | 0.610 |
Measured in millimeters.
DISCUSSION
To determine if there are virulent and avirulent strains of P. gingivalis, a group of subjects with clear indicators of periodontitis and a relatively healthier age-matched control group were identified. We previously reported a very strong association between periodontal disease and the presence of P. gingivalis in this study population. However, P. gingivalis was detected in 25% of the healthy group, raising the question of whether these individuals are carrying different, possibly less-virulent strains of P. gingivalis. For this study, the presence of strains of P. gingivalis in the two groups was determined by heteroduplex analysis. This assay relied on detection of polymorphisms in the ribosomal ISR. ISR fragments generated from samples with P. gingivalis-specific primers were hybridized to fragments from reference strains, and the formation of heteroduplexes from the hybridization of similar but nonidentical sequences was observed by polyacrylamide gel electrophoresis. Characteristic fingerprints from comparison with a panel of reference strains allowed the identification of heteroduplex types in samples, including those containing multiple strains.
The ribosomal operon has become the standard target for phylogenetic reconstruction, and the small-subunit gene has become the standard for identification of both prokaryotic and eukaryotic species. However, the small-subunit gene does not provide sufficient variability to distinguish among strains of P. gingivalis. The ISR, flanked by the 16S and 23S ribosomal genes, has recently been shown to be sufficiently variable to distinguish among strains of several species of bacteria, including P. gingivalis (1, 4, 20, 32). Most polymorphisms in the P. gingivalis ISR are resolved by heteroduplex analysis, and sequence analysis has shown that each heteroduplex type consists of a group of closely related samples that are very similar but do not always have completely identical ISR sequences (31). Heteroduplex analysis of the ISR has provided a method for identifying and tracking strains of P. gingivalis suitable for large-scale studies of human populations.
Eleven heteroduplex types of P. gingivalis were detected in the population examined for this study. Sufficient numbers were available for statistical analysis of six of these types. The use of univariate modeling to analyze the association of each strain with disease independently (Table 1) does not take into account cocolonization with other, possibly pathogenic strains which may account for the disease. Since many subjects harbored multiple strains, multivariate modeling with a nominal logistic regression provided a more informative analysis (Table 2). With multivariate analysis, three heteroduplex types were found to be clearly associated with disease. Type hW83 was by far the most strongly associated with periodontitis, as can be seen by its contribution to the overall chi-square value shown in Table 2. The odds of finding hW83 and h49417 were each almost 10 times higher in periodontitis patients than in healthy subjects, but the 95% confidence interval is much tighter for W83, as can be seen in Fig. 4. The wider confidence interval for h49417 may be attributed to the relatively smaller sample size for this type, as can be seen in Fig. 1. Heteroduplex type hHG1691 was also statistically significantly associated with periodontitis, but with a lower odds ratio. The remaining three heteroduplex types were found more often in the group of subjects with periodontitis, but the association with disease was not statistically significant. This was evidenced by the fact that the lower bounds of confidence intervals for the odds ratios were less than 1, as seen in Table 2. The sample sizes, particularly for hA7A1, were large enough relative to those for the other types that they probably do not account for the lack of a significant association. These data indicate that there are relatively more- and less-virulent strains of the periodontal pathogen P. gingivalis.
It is interesting that none of the disease-associated types were found in nearly half of subjects with disease. Some of these subjects were colonized by rare heteroduplex types that may be pathogens, but no P. gingivalis at all was detected in more than 20% of subjects with periodontitis. It seems unlikely that these represent false-negative results. The assay has been shown to be sensitive to as few as 10 cells (12), a threshold that should be exceeded by an organism capable of causing disease. The sampling strategy included all the teeth, so it is unlikely that P. gingivalis was missed in sample collection. Finally, the organism appears to be difficult to eradicate, even with treatment (5, 35), so it is unlikely that the absence of P. gingivalis could be accounted for by loss of the organism in subjects subsequent to disease activity. It appears that P. gingivalis is not responsible for all cases of periodontitis. This is consistent with data from other studies on adult-onset periodontitis in which additional species have been implicated (2, 13, 14, 23, 28, 30).
The effects of different strains of P. gingivalis have been compared in several animal studies (6, 7, 9, 17, 18, 27). Although a number of different approaches were used, all of the studies showed differences among strains in their effects on the host. In most animal studies, strain W50 or W83 is highly virulent. This correlates well with the findings of our in vivo study, where hW83 (W50 is indistinguishable by heteroduplex analysis and is included in this type) is the type most strongly associated with disease. Many of the animal studies have identified A7A1-28 as a highly virulent strain; however, we found that heteroduplex type hA7A1 (which includes A7A1-28) is the least often associated with human disease of all strains tested. Strain 381 has a relatively small effect in animal studies, and it also showed a weak association with human periodontal disease. A recent study of the distribution of the P. gingivalis fimA gene in a group of Japanese subjects with periodontitis showed one type to be more commonly found in deeper pockets (3). Strains ATCC 49417, 381, and W50 were all reported not to have the disease-associated fimA genotype. These findings do not correlate well with the disease association of strains in our Columbus, Ohio, cohort. Possible explanations include geographic variation and the larger sample size and multivariate analysis used in our study.
Criteria for selecting a group with periodontitis and a periodontally healthy control group were adopted based on previously reported age-based population norms for probing depth and attachment levels (22). Criteria were selected to allow the inclusion of no more than the healthiest and most-diseased quartiles of the population. Particular care was taken to match the age of the control group to that of the periodontitis group in order to avoid the inclusion of younger individuals who might harbor pathogenic oral flora that had not yet produced measurable periodontal destruction. For this reason the “healthy” group included individuals who might not meet conservative criteria for periodontal health but nonetheless were among the healthiest members of their age cohort. Based on the number of potential subjects rejected in screening exams (11), the criteria for health and disease were very selective. But in order to determine if differences in flora could be seen within the healthy group, the mean greatest probing depth and the site of greatest attachment loss in the presence of each heteroduplex type were compared with those in its absence (Table 4). Only one significant difference was seen, and that was between the greatest probing depths in the presence and absence of hHG1691 in the healthy group. This finding is consistent with the association of hHG1691 with disease, but it should also be regarded with some skepticism, since 24 t tests were performed with no correction in the α level. Overall the data were consistent within but not between the healthy group and the group with periodontitis, suggesting that they did adequately represent periodontal health and disease.
The presence of multiple strains was commonly observed in both the healthy group and the group with periodontitis (Fig. 3), but the distributions were different in the two groups. Healthy subjects were less likely to harbor a single strain than would be expected, and subjects with periodontitis were more likely to do so. It is possible that virulent strains present in subjects with disease are more likely to dominate the ecologic niche and inhibit the colonization or survival of less-virulent strains. When individual heteroduplex types were examined for simultaneous colonization of a single subject (Table 3), this appeared to be the case. Types hW83 and hHG1691, both strongly associated with disease, were highly unlikely to be found together, and both of these types were observed to exclude at least one of the less-virulent groups.
In summary, when the association of individual heteroduplex types of P. gingivalis with periodontitis was examined, one type, hW83, was found to be highly statistically significantly associated with disease. Two additional types, h49417 and hHG1691, were also significantly associated with disease. The remaining types, h23A4, h381, and hA7A1, were not significantly associated with periodontitis. These data indicate that virulence in human periodontitis varies among strains of P. gingivalis, and they identify an apparently highly virulent subgroup. Further investigation into differences among these groups may offer insight into mechanisms of pathogenesis.
ACKNOWLEDGMENTS
We gratefully acknowledge Phillip Marucha and Steven Bradway for helpful conversations on the diagnosis and pathogenesis of periodontitis.
This work was supported by NIH grant DE10467.
REFERENCES
- 1.Aakra A, Utaker J B, Nes I F. RFLP of rRNA genes and sequencing of the 16S–23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int J Syst Bacteriol. 1999;49:123–130. doi: 10.1099/00207713-49-1-123. [DOI] [PubMed] [Google Scholar]
- 2.Alpagot T, Wolff L F, Smith Q T, Tran S D. Risk indicators for periodontal disease in a racially diverse urban population. J Clin Periodontol. 1996;23:982–988. doi: 10.1111/j.1600-051x.1996.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 3.Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–1430. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun J, Huq A, Colwell R R. Analysis of 16S–23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl Environ Microbiol. 1999;65:2202–2208. doi: 10.1128/aem.65.5.2202-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danser M M, Timmerman M F, van Winkelhoff A J, van der Velden U. The effect of periodontal treatment on periodontal bacteria on the oral mucous membranes. J Periodontol. 1996;67:478–485. doi: 10.1902/jop.1996.67.5.478. [DOI] [PubMed] [Google Scholar]
- 6.Ebersole J L, Kesavalu L, Schneider S L, Machen R L, Holt S C. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1995;1:115–128. doi: 10.1111/j.1601-0825.1995.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 7.Evans R T, Klausen B, Ramamurthy N S, Golub L M, Sfintescu C, Genco R J. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch Oral Biol. 1992;37:813–819. doi: 10.1016/0003-9969(92)90115-o. [DOI] [PubMed] [Google Scholar]
- 8.Fisher J G, Zambon J J, Genco R J. Identification of serogroup-specific antigens among Bacteroides gingivalis. J Dent Res. 1987;66:222. [Google Scholar]
- 9.Genco C A, Cutler C W, Kapczynski D, Maloney K, Arnold R R. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect Immun. 1991;59:1255–1263. doi: 10.1128/iai.59.4.1255-1263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genco R J, Loos B G. The use of genomic DNA fingerprinting in studies of the epidemiology of bacteria in periodontitis. J Clin Periodontol. 1991;18:396–405. doi: 10.1111/j.1600-051x.1991.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 11.Griffen A L, Becker M R, Lyons S R, Moeschberger M L, Leys E J. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffen A L, Leys E J, Fuerst P A. Strain identification of Actinobacillus actinomycetemcomitans using the polymerase chain reaction. Oral Microbiol Immunol. 1992;7:240–243. doi: 10.1111/j.1399-302x.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 13.Grossi S G, Zambon J J, Ho A W, Koch G, Dunford R G, Machtei E E, Norderyd O M, Genco R J. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 14.Haffajee A D, Cugini M A, Tanner A, Pollack R P, Smith C, Kent R L, Jr, Socransky S S. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 15.Hillman J D, Maiden M F, Pfaller S P, Martin L, Duncan M J, Socransky S S. Characterization of hemolytic bacteria in subgingival plaque. J Periodontal Res. 1993;28:173–179. doi: 10.1111/j.1600-0765.1993.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa E, Kumada H, Umemoto T. Serological classification of Bacteroides gingivalis and purification of group specific antigens. Shika Kiso Igakkai Zasshi. 1989;31:647–655. doi: 10.2330/joralbiosci1965.31.647. . (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 17.Katz J, Ward D C, Michalek S M. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11:309–318. doi: 10.1111/j.1399-302x.1996.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 18.Laine M L, van Winkelhoff A J. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13:322–325. doi: 10.1111/j.1399-302x.1998.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 19.Leys, E. J., and A. L. Griffen. 9 June 1999, revision date. Heteroduplex data. [Online.] http://www.dent.ohio-state.edu/griffen_leys. [9 June 1999, last date accessed.]
- 20.Leys E J, Smith J H, Lyons S R, Griffen A L. Strain identification and detection of multiple strains of Porphyromonas gingivalis by heteroduplex analysis. J Clin Microbiol. 1999;37:3906–3911. doi: 10.1128/jcm.37.12.3906-3911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loos B G, Dyer D W, Whittam T S, Selander R K. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect Immun. 1993;61:204–212. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machtei E E, Christersson L A, Grossi S G, Dunford R, Zambon J J, Genco R J. Clinical criteria for the definition of “established periodontitis.”. J Periodontol. 1992;63:206–214. doi: 10.1902/jop.1992.63.3.206. [DOI] [PubMed] [Google Scholar]
- 23.Machtei E E, Dunford R, Hausmann E, Grossi S G, Powell J, Cummins D, Zambon J J, Genco R J. Longitudinal study of prognostic factors in established periodontitis patients. J Clin Periodontol. 1997;24:102–109. doi: 10.1111/j.1600-051x.1997.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 24.McClellan D L, Griffen A L, Leys E J. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1996;34:2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melvin W L, Assad D A, Miller G A, Gher M E, Simonson L, York A K. Comparison of DNA probe and ELISA microbial analysis methods and their association with adult periodontitis. J Periodontol. 1994;65:576–582. doi: 10.1902/jop.1994.65.6.576. [DOI] [PubMed] [Google Scholar]
- 26.Menard C, Brousseau R, Mouton C. Application of polymerase chain reaction with arbitrary primer (AP-PCR) to strain identification of Porphyromonas (Bacteroides) gingivalis. FEMS Microbiol Lett. 1992;74:163–168. doi: 10.1111/j.1574-6968.1992.tb05360.x. [DOI] [PubMed] [Google Scholar]
- 27.Neiders M E, Chen P B, Suido H, Reynolds H S, Zambon J J, Shlossman M, Genco R J. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 28.Papapanou P N, Baelum V, Luan W M, Madianos P N, Chen X, Fejerskov O, Dahlen G. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. J Periodontol. 1997;68:651–666. doi: 10.1902/jop.1997.68.7.651. [DOI] [PubMed] [Google Scholar]
- 29.Parent R, Mouton C, Lamonde L, Bouchard D. Human and animal serotypes of Bacteroides gingivalis defined by crossed immunoelectrophoresis. Infect Immun. 1986;51:909–918. doi: 10.1128/iai.51.3.909-918.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preus H R, Anerud A, Boysen H, Dunford R G, Zambon J J, Loe H. The natural history of periodontal disease. The correlation of selected microbiological parameters with disease severity in Sri Lankan tea workers. J Clin Periodontol. 1995;22:674–678. doi: 10.1111/j.1600-051x.1995.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 31.Rumpf R W, Griffen A L, Wen B G, Leys E J. Sequencing of the ribosomal intergenic spacer region for strain identification of Porphyromonas gingivalis. J Clin Microbiol. 1999;37:2723–2725. doi: 10.1128/jcm.37.8.2723-2725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stubbs S L, Brazier J S, O'Neill G L, Duerden B I. PCR targeted to the 16S–23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37:461–463. doi: 10.1128/jcm.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Steenbergen T J, Delemarre F G, Namavar F, De Graaff J. Differences in virulence within the species Bacteroides gingivalis. Antonie Leeuwenhoek. 1987;53:233–244. doi: 10.1007/BF00393930. [DOI] [PubMed] [Google Scholar]
- 34.Van Steenbergen T J, Menard C, Tijhof C J, Mouton C, De Graaff J. Comparison of three molecular typing methods in studies of transmission of Porphyromonas gingivalis. J Med Microbiol. 1993;39:416–421. doi: 10.1099/00222615-39-6-416. [DOI] [PubMed] [Google Scholar]
- 35.von Troil-Linden B, Saarela M, Matto J, Alaluusua S, Jousimies-Somer H, Asikainen S. Source of suspected periodontal pathogens re-emerging after periodontal treatment. J Clin Periodontol. 1996;23:601–607. doi: 10.1111/j.1600-051x.1996.tb01831.x. [DOI] [PubMed] [Google Scholar]