Abstract
(1) Background: Main Protease (Mpro) is an attractive therapeutic target that acts in the replication and transcription of the SARS-CoV-2 coronavirus. Mpro is rich in residues exposed to protonation/deprotonation changes which could affect its enzymatic function. This work aimed to explore the effect of the protonation/deprotonation states of Mpro at different pHs using computational techniques. (2) Methods: The different distribution charges were obtained in all the evaluated pHs by the Semi-Grand Canonical Monte Carlo (SGCMC) method. A set of Molecular Dynamics (MD) simulations was performed to consider the different protonation/deprotonation during 250 ns, verifying the structural stability of Mpro at different pHs. (3) Results: The present findings demonstrate that active site residues and residues that allow Mpro dimerisation was not affected by pH changes. However, Mpro substrate-binding residues were altered at low pHs, allowing the increased pocket volume. Additionally, the results of the solvent distribution around S, H, N1 and H1 atoms of the catalytic residues Cys145 and His41 showed a low and high-water affinity at acidic pH, respectively. It which could be crucial in the catalytic mechanism of SARS-CoV-2 Mpro at low pHs. Moreover, we analysed the docking interactions of PF-00835231 from Pfizer in the preclinical phase, which shows excellent affinity with the Mpro at different pHs. (4) Conclusion: Overall, these findings indicate that SARS-CoV-2 Mpro is highly stable at acidic pH conditions, and this inhibitor could have a desirable function at this condition.
Keywords: SARS-CoV-2 Mpro, pH, molecular dynamics, docking analysis, PF-00835231
1. Introduction
The COVID-19 pandemic is still an ongoing major health threat for the whole world, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,2,3]. The coronavirus family enters the host by fusion with the plasma or endosomal membranes [4,5]. This mechanism has been used by many positive-strand RNA viruses like the influenza A virus (IAV) [6,7], the human immunodeficiency virus (HIV) [8,9], coronaviruses and others. Among the therapeutical targets of these types of viruses are proteases, classified according to their catalytic site as cysteine proteases [10], serine proteases [11] aspartic proteases [12,13] and metalloproteases.
In the SARS-CoV-2 entry mechanism, the coronavirus spike protein binds to ACE2 receptors found on the surface of many human cells. The virus enters the cytoplasm by endocytosis, then its genetic material, a single-stranded RNA, is released into the cytoplasm. This material encoded two polyproteins (pp1a and pp1ab) which are necessary for viral replication and transcription [14,15,16]. The proteolytic process was carried out by the SARS-CoV-2 Main Protease (Mpro), a cysteine protease with a catalytic dyad that compromises two principal residues, Cys145 and His41 [17,18,19,20,21,22]. This protein is essential in the life cycle of the virus and is considered a relevant target against the replication of the coronavirus. The catalytic mechanism among coronavirus cysteine proteases involves either using a catalytic dyad to perform a nucleophilic attack that covalently links the protease to the substrate protein, releasing the first half of the product. This covalent acyl-enzyme intermediate is then deacylated by an active water molecule releasing the second half of the product [23,24].
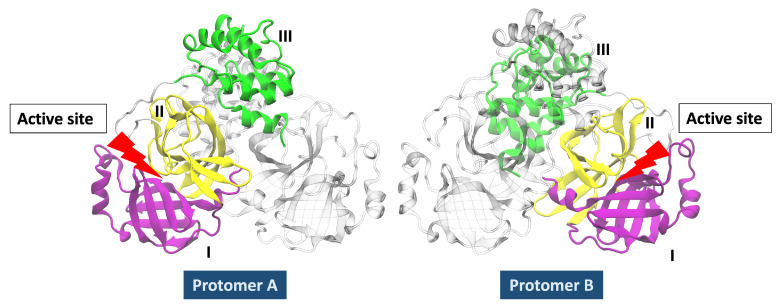
SARS-CoV-2 Mpro is active in its dimeric conformation, but the SARS-CoV-1 Mpro is found as a mixture of monomer and dimers in solution [25]. SARS-CoV-1 Mpro has been crystallised in different pH-dependent conformations suggesting flexibility, particularly around the active site [26]. The protease is highly conserved in all coronaviruses, including SARS-CoV-2, so targeting either dimerisation or enzymatic activity may give rise to drugs that can target multiple coronaviruses, known and yet unknown [27,28]. In Figure 1, the three-dimensional structure representation of SARS-CoV-2 Mpro (approx. 33kDa of total weight) shows two identical protomers (homodimer conformation). Each protomer has three domains, domain I (residue 10 to 99), domain II (residue 100 to 182), and domain III (residue 198 to 303); a cluster of five -helices characterises this last domain. Domain II is connected to domain III via a longer loop.
Figure 1.
SARS-CoV-2 Mpro, PDB ID: 5RE4. Domain I in purple colour, domain II in yellow colour and domain III in green colour.
Many macromolecular properties in the coronavirus family are pH-dependent [4,29]. As a starting point, it is important to mention that there are studies on the effect of pH in SARS-CoV-1 Mpro, showing the high proteolytic activity of SARS-CoV-1 Mpro around the physiological pH (7.0–7.6) [26,30,31,32,33]. The probability of the pH-activity profile of the SARS-CoV-1 Mpro was determined by protonation of His163 (inactivation at acidic pH) and deprotonation of His172 [26]. This behaviour is different for conformational stability; some viral proteases have shown their biological role at low pHs, as shown in the endogenous viral proteases from porcine coronavirus are active in a low-pH environment [34].
The SARS-CoV-2 Mpro activity is thought to exhibit a bell-shaped pH-rate profile as determined for the SARS-CoV-1 Mpro. For SARS-CoV-1, fitting kinetic data to model equations provides rate constants and two pKa values (6.2–6.4, 7.7–8.3) that presumably correspond to the catalytically competent His41 and Cys145 side chains [35,36]. Further understanding of pH effects can be gained from theoretical pKa estimations for all the titratable amino acids of Mpro, which have been also employed to determine their most likely protonation states [31]. Another recent work has demonstrated that the structure of SARS-CoV-2 Mpro shows pH dependence. At basic pH, the Mpro structure was less stable, and as pH becomes more acidic, the protein gets destabilised. Moreover, the SARS-CoV-2 Mpro showed maximum stability at neutral pH but in the other pHs analysed other than neutral pH, dimerisation and S1 binding pocket were found to be affected [37].
Moreover, due to the high homology between SARS-CoV-1 Mpro and SARS-CoV-2 Mpro, similar behaviour is expected. Indeed, the coronavirus family is characterised as an enveloped virus that may or may not be low pH dependent [26,38,39]. It has been reported that higher levels of ACE2 in tissues could be the consequence of some of the comorbidities associated with severe SARS-CoV-2 infection [40]. They showed that the acid pH associated with Barrett’s oesophagus patients might drive an increased expression of ACE2. In addition, they studied human monocytes cultured in acid pH, observing an increased ACE2 expression and higher viral load upon SARS-CoV-2 infection [40]. A recent review shows that acidosis is generally associated with the severity of SARS-CoV-2. Lactic acidosis produces an excess of the levels of lactate, which is associated with a reduction of pH [41]. The associated enzyme, lactate dehydrogenase (LDH), which transforms pyruvate to lactate, could be selected as one biomarker, as high LDH levels have been associated with worse outcomes in patients with the viral infection [42]. The excess of lactate, a stronger acid than pyruvate, leads to a decrease in the pH of the medium. Herein, we investigate the effect of acidic conditions on the pH-dependent conformational properties of SARS-CoV-2 Mpro, which may be involved in inhibition processes associated with severe SARS-CoV-2 infections. Currently, the literature shows us several studies of the effect of pH by docking calculations. These studies have shown that the pH dependence of the enzyme activity may reflect structural changes in the catalytic dyad [43,44,45,46,47,48,49]. For instance, Drazic et al. tested the influence of different pH values on substrate cleavage velocity and found no significant impact in the pH range from 7.0 to 8.0 [50]. Several publications described the influence of pH on the enzyme activity of the SARS-CoV-1 Mpro, which reported a peak of substrate cleavage at pH 7.0 [26,51], whereas other studies reported the highest processing at around pH 7.5 [31], or pH 8.0 [52,53].
The first analysis by molecular dynamic simulation at pH 7 of SARS-CoV-2 Mpro demonstrated the active site’s flexibility and effects induced by substrate binding [35]. On the other hand, SARS-CoV-2 Mpro showed high thermodynamic stability in a wide pH range and low stability in the presence of salts [45]. With respect to the catalytic site, the most likely hydrolysis mechanism assisted by the coronavirus Mpro, the reactive events would involve the proton transfer from Cys145(SH) to His41(N) and the nucleophilic attack of the S atom to the carbonyl group of a Gln. Thus, the general mechanism considers that Cys145 should be protonated and His41 acts as a base, as in the case of the neutral pH [35].
Verma et al. analysed the proton-coupled dynamics of SARS-CoV-2 Mpro that represented the first step toward the mechanistic understanding of the proton-coupled structure dynamics function at pH 5 to 9 [54]. Likewise, new advances in pH-dependence have shown the importance of appropriate histidine protonation states of SARS-CoV-2 Mpro in the coupling with inhibitors [55]. In the last two years, different SARS-CoV-2 inhibitors have been studied, such as synthetic and natural compounds [28,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71].
Our study provides evidence about the relation between flexibility and pH-dependent Mpro, which was observed at the low pH conditions. We applied Semi-Grand Canonical Monte Carlo (SGCMC) method [72,73,74,75] to assign different protein charge distributions, considering the protonation/deprotonation states for each pH. The homemade developed code within our group makes it possible to define different protonation/deprotonation microstates for a particular pH as shown in the work of Barazorda-Ccahuana et al. [76]. One step further was made by analysing the observed conformational changes due to protonation/deprotonation states by atomistic molecular dynamics simulations.
Along with the explored protonation/deprotonation impact presented in an atomistic level for the Mpro in the frame of molecular dynamics simulations, also we stressed the effect of interaction with PF-00835231 as a potent inhibitor of SARS-CoV-2 Mpro by docking calculations at different pHs. The finding of the therapeutic effect of the PF-00835231 has demonstrated promising data or outcomes in Pfizer preclinical studies [19,77,78,79,80,81,82]. Indeed, the SARS-CoV-2 Mpro and PF-00835231 coupling analysis could serve as a model to study other molecules of natural origin targeting SARS-CoV-2.
2. Computational Methods
2.1. Protonation/Deprotonation States by SGCMC
We have used the SARS-COV-2 Mpro crystal structure reported in the Protein Data Bank (https://www.rcsb.org, accessed on 10 June 2020) by the access code PDB ID: 5RE4. It was determined by the X-ray diffraction method with a resolution of 1.88 Å. The first step was to determine the pKa values of each ionisable residue with the PROPKA v. 3 program [83,84]. Indeed, the summary of pKa values were used for the calculation of protonation states of Asp, Glu, Arg, Lys and His residues, and the C-terminal and N-terminal ends.
In this work, we use the homemade code (see Data Availability Statement) that calculates the protonation and deprotonation states by Semi-Grand Canonical Monte Carlo (SGCMC) procedure based on the free energy associated with the pKa of each i titratable residue, this is calculated using the Equation (1) or Equation (2) [72,73,74,75]
| (1) |
| (2) |
In this work, we applied 200,000 steps for protonation/deprotonation of SARS-CoV-2 Mpro residues generating ten independent microstates for five pHs (3 to 7).
2.2. Molecular Dynamics Simulation Details
Molecular Dynamics (MD) simulations have been performed for four microstates for each condition of pHs with the Gromacs (Groningen Machine for Chemical Simulations) v. 2019.1 software [85]. AMBER99SB-ILDN [86] force fields have been used in the simulations of SARS-CoV-2 Mpro. The periodic boundary conditions (PBC) [87] were considered to minimise edge effects in a finite system. For each system, the protein was in the centre of a cubic box with a minimum distance of 1.5 nm to the faces of the simulation box. The box system was solvated with TIP5P [88] water model. As well as, the total charge of each microstate was neutralised with Cl or Na to attain equilibration. Energy minimisation was carried out using the steepest-descent algorithm with 200,000 steps of simulation. WE performed 10 ns MD simulations of equilibrium in the canonical ensemble NVT with position restraint, and the temperature was regulated with the V-rescale thermostat at 309.65 K. Furthermore, we performed the production of simulation in the isothermal-isobaric ensemble with the Parrinello–Rahman barostat with a reference pressure of 1 bar, V-rescale thermostat with 309.65 K and 250 ns time of the simulation. To further corroborate our findings, the conformational and angles of residues in Mpro were analysed. This analysis of the quality of the structure was solved with the Ramachandran plot in the Molprobity server [89].
2.3. Molecular Docking
Molecular Docking calculations were performed using the AutoDock Vina software [90] integrated into the SAMSON molecular design platform as a SAMSON extension available at http://samson-connect.net, accessed on 10 June 2020. The extension provides additional functionality to prepare receptors and ligands easily, dock ligand libraries, analyse and export docking results. We used SAMSON and Vina extensions to configure calculations, export input files, and run docking calculations.
The number of modes was set to 100 with an energy range = 3 kcal/mol (default value). The energy range (kcal/mol) is a maximum energy difference between the best binding mode and the unfavourable one displayed. The energy (affinity) that differs more than 3 kcal/mol from the best mode are not saved among results. In the configuration file, the parameter called “exhaustiveness” was set to 8. This parameter controls how comprehensive will be the search space.
2.4. Simulation Data Analysis
The RMSD (Root-Mean Squared Deviation), RMSF (Root-Mean Squared Fluctuation), RG (Radius of Gyration), HB (Number of Hydrogen Bonds), SASA (Solvent Accessible Surface Area) and RDF (Radial Distribution Function) analysis were calculated by Gromacs tools.
Additionally, the Salt Bridges (SB) were analysed, considering the last frame of MD for each microstate with the “Salt Bridges” utility of the Visual Molecular Dynamic (VMD [91]) program. Finally, the plots were made in Gnuplot v. 5.4 [92] and the represented in the manuscript images were created with VMD.
3. Results and Discussion
3.1. Protonation/Deprotonation States of SARS-CoV-2 Mpro
The molecular structure of SARS-CoV-2 Mpro is a homodimeric structure with approximately 36% of acid/base residues (Asp, Glu, Arg, Lys, His), and almost 8% corresponds to Cys residue distributed throughout the protein structure. For this reason, the analysis of protonation and deprotonation of titratable residues could affect the total charge of Mpro. In this work, different charge distributions are used to assign the protonation state of Mpro acid/basic residues for each pH. For instance, in the case of acidic residues (Glu and Asp) of each microstate, these can be found in the protonated or deprotonated modes for pH studied; as well as for basic residues (His, Lys, and Arg). In the case of the residues of the catalytic dyad (His41 and Cys145), we considered Cys145 protonated and His41 in the neutral state.
Thus, as conventional MD simulations are used, the atomic charges and the associated protonation states are constant throughout the simulation. The effect of charge fluctuation on a pH is approximated by averaging different simulations with different charge distributions. This approach should be a good approximation of the pH effect, as no overall structural changes of the protein are observed that could significantly change the intrinsic acidity constants.
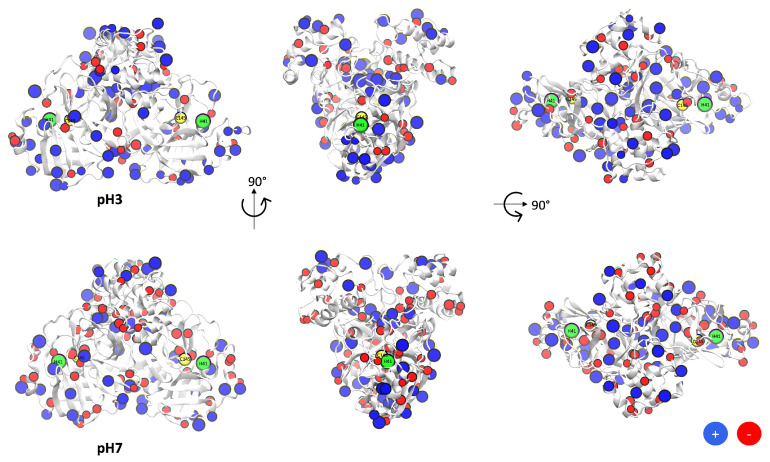
For instance, Figure 2 shows the Mpro at pH 3 and pH 7, where the beads of blue colour represent the protonated residues and the red colour the deprotonated residues. We notice that at pH 3 there is a low concentration of red beads, the microstates are more protonated at this condition. In contrast, at pH7 a more equilibrated distribution of red and blue beads are seen in the structure of the Mpro. Whereas, in both states (pH 3 and pH 7) the yellow (Cys145) and green (His41) beads represented the residues from the active site with a neutral charge.
Figure 2.
SARS-CoV-2 Main Protease (Mpro) with titratable group in beads representation. Yellow beads (Cys145), green beads (His41), blue beads (basic amino acids) and red beads (acidic amino acids).
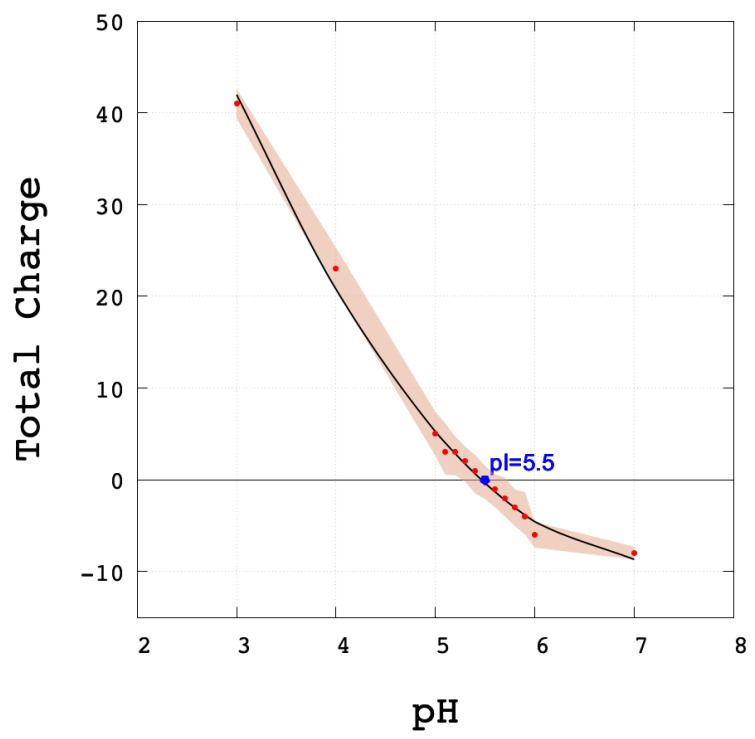
Indeed, in Table 1 we evaluate the total charge of 10 microstates generated by the SGCMC method, where the average total charge was +(41 ± 2) (pH 3) and −(8.4 ± 0.7) (pH 7), and zero value around pH 5, as shown in Figure 3. These results allowed us to find the estimated computationally isoelectric point (pI) of SARS-CoV-2 Mpro (pI = 5.5) similar to other studies [93].
Table 1.
Total charge of Mpro as a function of pH for the different microstates (MicroS).
| MicroS-1 | MicroS-2 | MicroS-3 | MicroS-4 | MicroS-5 | MicroS-6 | MicroS-7 | MicroS-8 | MicroS-9 | MicroS-10 | Average Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| pH3 | 38 | 40 | 41 | 42 | 43 | 43 | 42 | 41 | 39 | 41 | 41 ± 2 |
| pH4 | 24 | 27 | 20 | 22 | 23 | 20 | 26 | 22 | 22 | 22 | 23 ± 2 |
| pH5 | 7 | 4 | 8 | 6 | 2 | 5 | 1 | 5 | 2 | 7 | 5 ± 2 |
| pH6 | −6 | −5 | −4 | −4 | −5 | −6 | −8 | −7 | −4 | −6 | −(5.5 ± 1.4) |
| pH7 | −8 | −9 | −9 | −8 | −8 | −7 | −8 | −9 | −9 | −9 | −(8.4 ± 0.7) |
Figure 3.
Total charge of Mpro at different pHs. The representation of smooth (colour) indicates the average and the error for each microstate, the estimated computationally pI = 5.5 is in blue colour, and the points were fitted to a polynomial regression line (black line).
Table S1 from Supporting Information shows the distribution of the total charge of the protein among the different titratable residues of the protein. It could be seen that the charge state of acidic residues (Asp and Glu) was the most affected by pH in the studied range, where a large part of these residues at pH 3 was in their protonated state, generating an average of −(9 ± 1) (Asp) and −(4 ± 1) (Glu) partial charge. For pH 4, there are values of −(23 ± 2) (Asp) and −(7 ± 1) (Glu). For the case of pH 5, pH 6 and pH 7, they have had a similar number of Asp and Glu deprotonated. The basic residues (Lys and Arg) were always charged independently of the studied pH. On the contrary, the average total charge of His was +10, +(9 ± 1), +(6 ± 1), +(2 ± 1) and +(1 ± 1) for pH 3, pH 4, pH 5, pH 6 and pH 7, respectively (as shown in Table S1 from Supporting Information). Additionally, we focused on the charge state of two important histidines of the active site, His41 and His172. In general, these two histidines were protonated at pH 3 and pH 4 and deprotonated at pH 5, pH 6 and pH 7 (Table S2 from Supporting Information).
3.2. Analysis of Molecular Dynamic Simulation
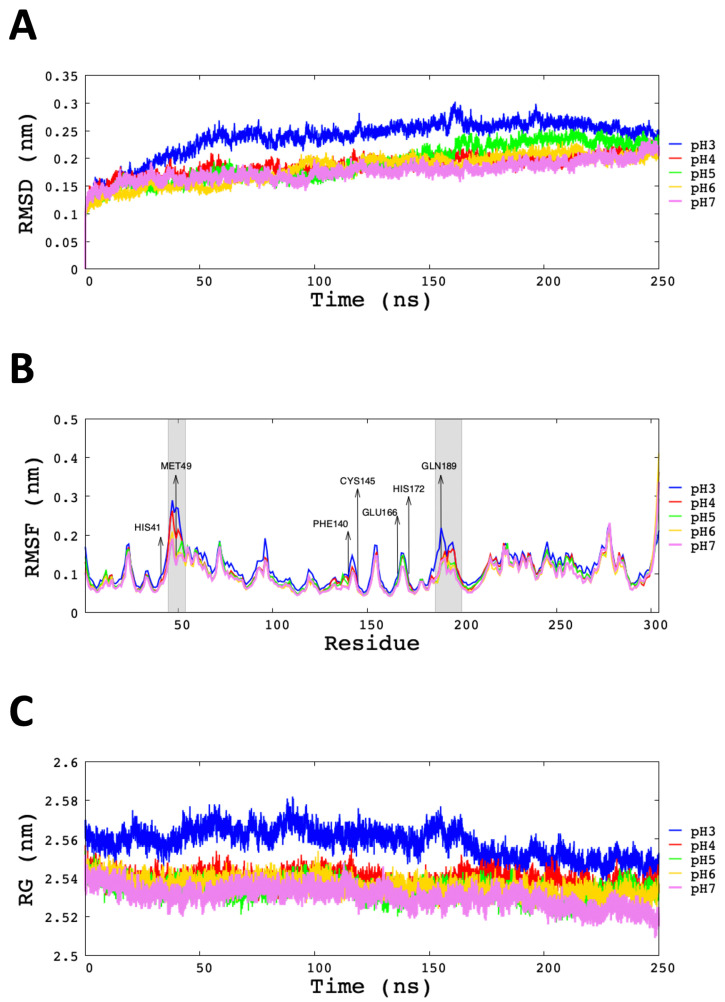
To envisage the role and the importance of conformational flexibility of the explored cases 20 MD simulations were conducted (4 independent microstates for each of 5 pHs considered). In Figure 4A the RMSD diagram indicates that the Mpro in explicit solvent TIP5P water model starts to be relatively stable around 50 ns at the different conditions of pHs.
Figure 4.
RMSD, RMSF and RG graph of backbone for each pH modelled. (A) RMSD presented as a function of time. (B) RMSF per residue of C-, highlighting the most important residues of Mpro, during last 100 ns of MD simulations. (C) RG analysis of structural compactness.
It can be seen in Table 2 that the average value of the RMSD at pH 3 was (0.26 ± 0.06) nm; this was the highest value among all simulated pHs. In the range of pHs 4 to 7, the average RMSD value was very similar. In order to analyse the dispersion of RMSD values among different microstates, individual RMSD values are given in Table S3 from Supporting Information.
Table 2.
Values of the MD were fitted against the average structure of the last 100 ns of the dynamics was used.
| pH | RMSD | RG | SASA | Hbond | Hbond |
|---|---|---|---|---|---|
| (nm) | (nm) | (nm) | (Protein–Protein) | (Protein–Waters) | |
| pH3 | 0.26 ± 0.06 | 2.553 ± 0.010 | 268 ± 2 | 401 ± 1 | (107 ± 4) × 10 |
| pH4 | 0.20 ± 0.02 | 2.538 ± 0.002 | 261 ± 2 | 437 ± 10 | (110 ± 2) × 10 |
| pH5 | 0.21 ± 0.04 | 2.537 ± 0.005 | 259 ± 3 | 448 ± 3 | (114 ± 8) × 10 |
| pH6 | 0.20 ± 0.02 | 2.532 ± 0.005 | 260 ± 2 | 456 ± 6 | (114 ± 12) × 10 |
| pH7 | 0.19 ± 0.02 | 2.526 ± 0.007 | 259 ± 1 | 461 ± 1 | (114 ± 12) × 10 |
The RMSF for C- atoms per residue of Mpro were computed at different pHs (Figure 4B). In all cases, the behaviour seems to be similar for all pHs; for example, the catalytic dyad’s residues (His41 and Cys145) presented low fluctuation independently of the studied pH. On the other hand, the average RMSF of the regions that compromise residues 45 to 50 (Thr45, Ser46, Glu47, Asp48, Met49, Leu50) and 186 to 190 (Val186, Asp187, Arg188, Gln189, Thr190) showed us high fluctuation at pH 3 and pH 4 (Figure 4B). Regarding the Glu166 residue, an important residue that participates in the protomers dimerisation, its degree of fluctuation is not affected by the studied pHs. Therefore, the relative stability of global RMSD in conjunction with the relatively low values of RMSF of the main part of the amino acids implies that the global structure of Mpro does not disaggregate at low pH conditions, dimeric conformation is stable.
The RG values determined the compactness of the Mpro and were plotted as a function of time. It can be seen in Table 2 that at pH 3 and pH 4 are obtained the highest RG value 2.55 ± 0.01 and 2.54 ± 0.01, respectively. In contrast, at pH 5, pH 6 and pH 7 the stability and compactness of the protein were similar (approx. 2.53 nm). The SASA analysis of each pH studied is given in Table 2. The average SASA value at pH 3 exhibited increased compared to the other systems ( = (268 ± 2) nm). Therefore, the SASA value increase in average as pH decreases. The temporal evolution of the SASA values at different pHs are shown in Figure S1a from Supporting Information.
However, significant differences could be observed in the number of protein–protein and protein–waters hydrogen bonds. In particular, the average number of hydrogen bonds, protein–protein and protein–waters increase in average as pH increases (Figure S1b,c from Supporting Information show additional information of each pH studied). In summary, these results show that Mpro at pH 3 presented a lower quantity of hydrogen bonds (protein–protein) than other pHs analysed. The fact that Mpro has many aspartic residues led to constraints for hydrogen bonds formation when the residues were protonated at low pH.
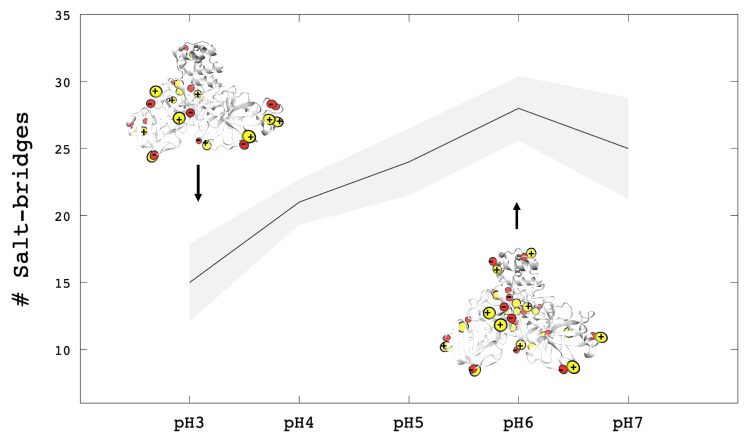
Another parameter that determines the stability of Mpro, was the number of salt bridges that depend on the binding of positively and negatively group charges [94]. In this work, the change in the protonated and deprotonated states of Mpro determined that at low pHs exist a loss of salt-bridges. Usually, since the salt bridges depend on the pH, we saw that at pH 3, there is a lower number of salt bridges than the other pHs (Figure 5). Since these systems are further from the isoelectric point (pI = 5.5), the positive and negative charge is unbalanced, so a lower amount of salt bridges is expected.
Figure 5.
Salt bridges of whole Mpro structure at different conditions of pHs. At low pHs, Mpro is more protonated than pH 6 and pH 7, this graphic show less salt bridges at pH 3. The line-tendency (black) represent the average value and the smooth tendency (gray) represent the statistical error.
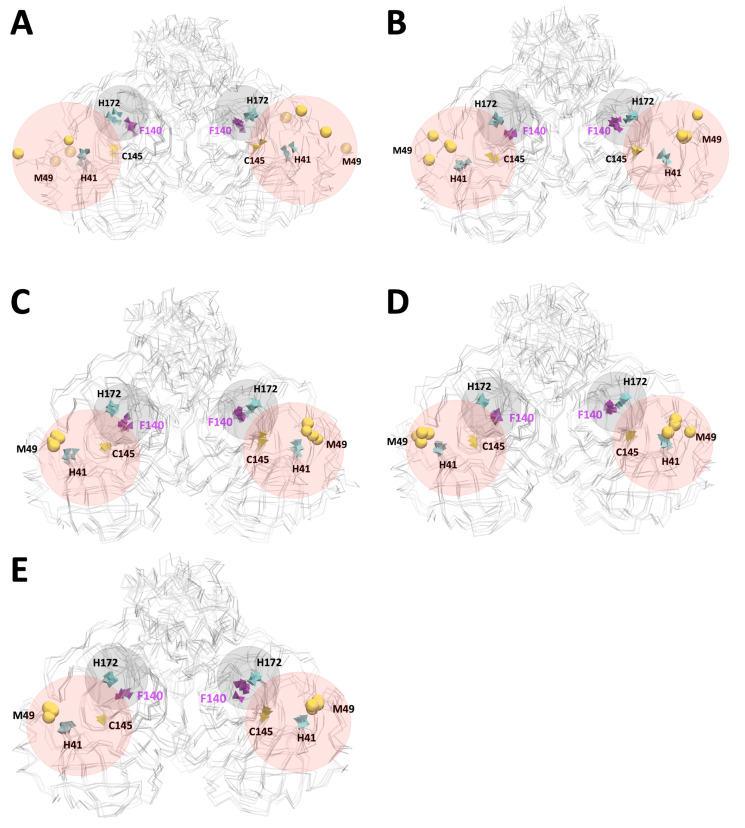
The flexibility of residues close to the active site was the object of interest and for other studies dedicated to the same stated problem. The S1 pocket was crucial in the protein dimerisation and residues involved in the substrate-binding sites of SARS-CoV-2 Mpro. For these reasons, we focused on the residues of the catalytic dyad (Cys145 and His41), two residues of the S1 pocket (His172 and Phe140) proposed by Verma et al. [54] and additionally the most fluctuating residues of RMSF analysis (Met49 and Gln189). In Figure 6, the relative distance of His41 and Met49 to Cys145 was analysed for the last frame of each microstate. The most remarkable difference is observed at pH 3 and pH 4, where Met49 is located with the most significant distance to Cys145 (in both protomers). Despite this behaviour in Met49, the other residues (His41, Cys145, Phe140, and His172) were not altered by changing the pH.
Figure 6.
Snapshots of the last frame of all microstates in Mpro, highlighting the active site (pink area) and pocket S1 (gray area). (A,B) pH 3 and pH 4 did not show a constant Met49–Cys145 distance, which was also analysed in the RMSF, where the Met49 residue showed high fluctuations at these pHs. (C–E) pH 5, pH 6 and pH 7 with a constant distance between Met49-Cys145, His41-Cys145 and His172-Phe140. The His41, His172, Cys145 and Phe49 residues (polyhedral graphic representation) and Met49 (bead graphic representation) are shown.
The calculation of the average distance of the last 100ns between C atoms showed us a constant distance value between Cys145-His41 and His172-Phe140; therefore, the active centre and pocket S1 of Mpro is not significantly altered by pH effect. However, the average distance between Met49 and Gln189 shows significant differences at pH 3 (see Figure S2 in the Supporting Information for more details). This final result indicates that the pocket increases its volume as there is a greater distance between these residues at pH 3 and pH 4. Therefore, the size of the pocket can compromise the selectivity of the substrate.
Two main steps are identified in the catalytic dyad: the acylation (acyl-enzyme complex) and deacylation (acyl-enzyme hydrolysed). Świderek and Moliner analysed the deacylation step mediated by a water molecule activated by His41 [95]. Therefore we have evaluated how the pH affects the solvation of the reactive atoms of this catalytic residue.
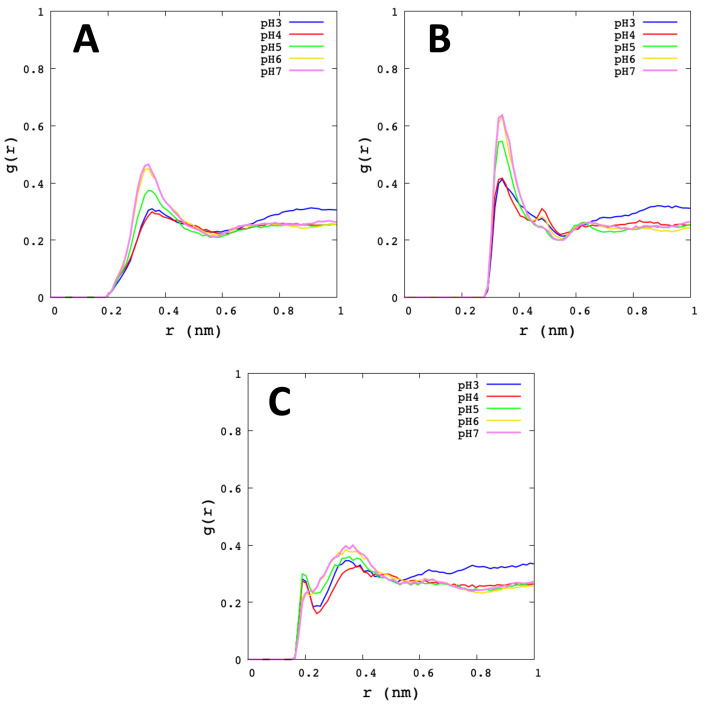
The radial distribution function (RDF) is the probability distribution to find a particle at a distance r away from a given reference particle. In this work, the RDF of specific atoms of Cys145 (S, H), His41 (N, H), Met49 (S), water sites (H, O), and ions, were analysed. In Figure 7, the high of the first peak-height of S–H and S–O appear at r distance of 0.36 nm and 0.34 nm, respectively. The most striking variation is seen at pH 7 and pH 6, followed by pH 5 and lower peaks at pH 4 and pH 3. In general, the RDF of the solvent distribution on S and H shows a low water affinity at low pHs, which could be crucial to understand the role of environmental conditions in the catalytic mechanism of SARS-CoV-2 Mpro. Additionally, the RDF analysis of ions around S from Cys145, shows the peak height at r distance of 0.5 nm for pH 3 and pH 4. It is worth noting that there is a significatively presence of mobile ions close to the active site (see Figure S3a from Supporting Information).
Figure 7.
Radial distribution function of Cys145. The graphs show the average value of the Mpro protomers and the different microstates for each pH. (A) S–H. (B) S–O. (C) H–O.
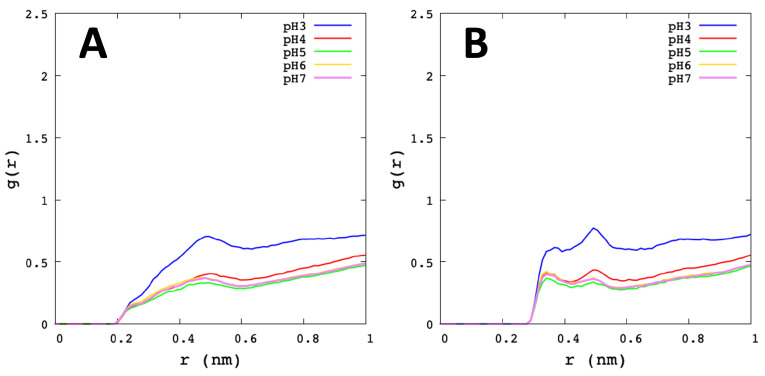
His41 reported more affinity for water at pH 3 than pH 7 (Figure 8). Indeed, N–H and N–O show the high first of the peak-height at pH 3 with r = 0.33 nm and r = 0.28 nm, respectively. Likewise, the H–O showed a large peak at r distance of 0.18 nm, where the highest was pH 3. Therefore, with this result, we can observe that the N and H atoms from His41 are more related to water at low pHs. Furthermore, His41 with ions showed a similar behaviour to Cys145 (see Figure S3b from Supporting Information).
Figure 8.
Radial distribution function of His41. The graphs show the average value of the Mpro protomers and the different microstates for each pH. (A) N1–H. (B) N1–O. (C) H1–O.
The RDF analysis of Met49 is seen in Figure 9. The S–H obtained the peak-height at r distance of 0.49 nm, similar to S–O at pH 3. Although Met49 is highly fluctuating at pH 3, this does not influence the early interaction with water molecules.
Figure 9.
Radial distribution function analysis of Met49. The graphs show the average value of the Mpro protomers and the different microstates for each pH. (A) S–H. (B) S–O.
In summary, all findings of MD simulations allowed us to understand the high stability of Mpro at low conditions of pHs. The conformational stability of Mpro during the simulation time remains stable at the explored pHs scales. The significant number of residues at low pHs led to increased SASA. The impact of this effect was translated to hydrogen bond formation (intramolecular and intermolecular), which was confined. Although these results are not unexpected, they show that the ensemble of the Mpro dimer remains stable and that the most acidic pH conditions seem to be unable to disaggregate the enzyme. Furthermore, we have shown that the distance between the residues of the active site and the S1 pocket of the Mpro remained constant during the simulation time, which indicated that the structure of Mpro could not disaggregate at low pH conditions. On the contrary, the residues of the zone of binding-substrate supported conformational changes at pH 3, which increased the pocket volume. This result has related to the high fluctuations are presented in the RMSF diagram. Taking into account the importance of the solvent in the reaction mechanism of SARS-CoV-2 Mpro. In the RDF analysis, we have found that the solvent showed a high and low affinity towards the atoms of His41 and Cys145, respectively. Finally, the last frame of each trajectory was utilised to analyse the accessible values of the torsion angles and to create a Ramachandran plot. This analysis allowed us to validate the protein structures of each pH. Mpro at different pHs conditions (3 to 7) had a similar percentage of residues in allowed (∼99%) and favoured (∼92%) regions (more details are in Table S5 from Supporting Information).
3.3. Molecular Docking Analysis
To analyse the effect of environmental pH in the interaction with protease inhibitors, a recent ligand of Mpro protease has been selected in the docking study. Molecular docking calculations with PF-00835231 ligand [19] and the 40 receptors (last frame of five pHs x five microstates x two monomers) in different conformations according to the protonation states have been conducted. (See Figure S4 from Supporting Information, where PF-00835231 presents an estimated pKa equal to 12.76 ± 0.10 predicted using ACDlabs software v11.02, which allows it to be neutral and does not suffer alteration with the change in pH.) Appropriated protonation states were preserved to both receptors and ligand. Both the number of flexible side chains and the size of the search domain were different for all the cases because of the receptor’s conformation (i.e., chain orientation, the position of residues). On average, there were about 30 flexible side chains (from about 27 to about 35 flexible side chains). Table S6 from Supporting Information represents the data for the SARS-CoV-2 Mpro with different pH: sizes of the search domain, the volume of the search domain, and the number of the flexible side chains.
The search space was defined by a docking box that wraps the space around the receiver for each system with 40 different docking boxes. Active pocket amino acid residues were used to centre the docking box. Therefore, the pocket was placed around Cys145.
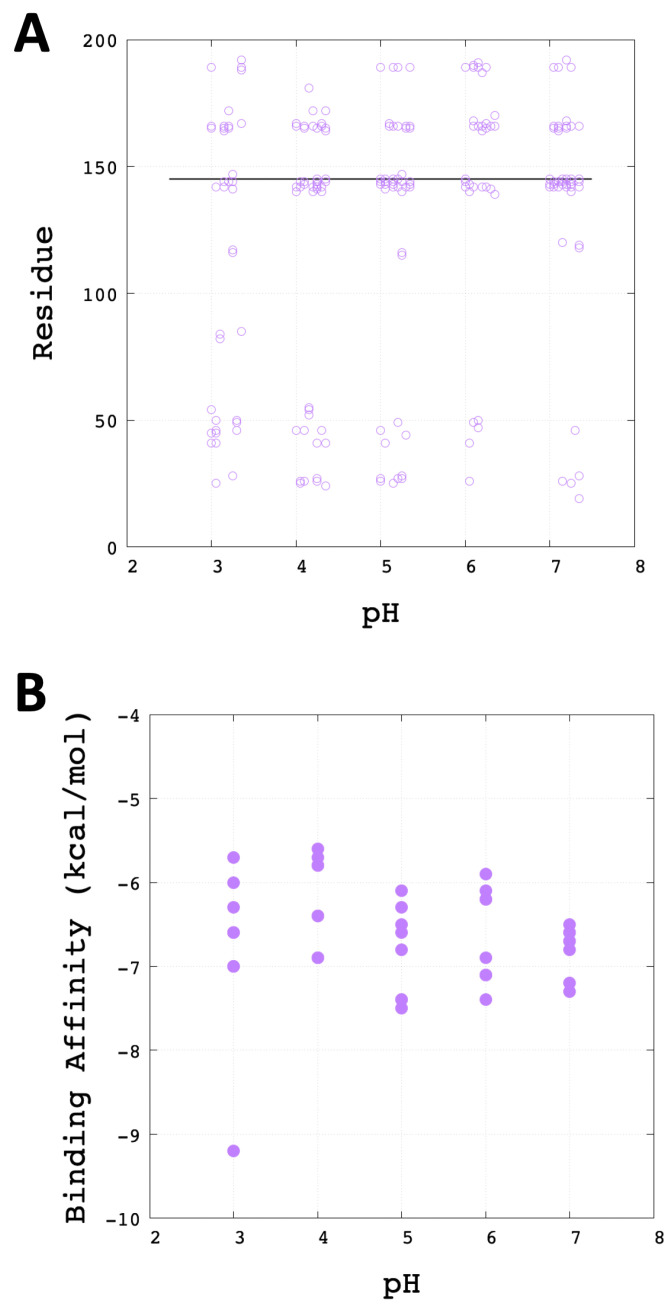
In AutoDock Vina the electrostatic interactions were mainly determined through hydrogen bonding terms. Figure 10A shows the residues of Mpro that establish hydrogen bonds with the PF-00835231 drug at different pHs (Figure S5 shows the 2D representation of the best coupling of each pH). Furthermore, at pH3, an additional pattern of interaction could be seen that shows the interaction of the drug with residues Met82, Asn84 and Cys85. On the other hand, Figure 10B shows the binding affinity of the best docking poses for each microstate. The binding mode of the drug close to the catalytic Cys145 shows a similar range of affinity (−7 to −6 kcal/mol) among all studied pHs. However, the more acidic pH shows an additional mode of interaction, being more separated from the catalytic Cys145 with the highest affinity.
Figure 10.
Binding analysis. (A) Mpro residues that interact with the drug PF-00835231 through hydrogen bonds at different pH (3, 4, 5, 6 and 7). For each pH, the best docking of each of the 8 microstates is analysed. To distinguish between the different docking poses, each is shifted slightly on the pH axis. A horizontal line is also depicted at 145 to help visualise the interaction with the catalytic Cys145. (B) The predicted binding affinity of drug PF-00835231 with Mpro at different pH. For each pH, the best binding affinity of the 8 different microstates is shown.
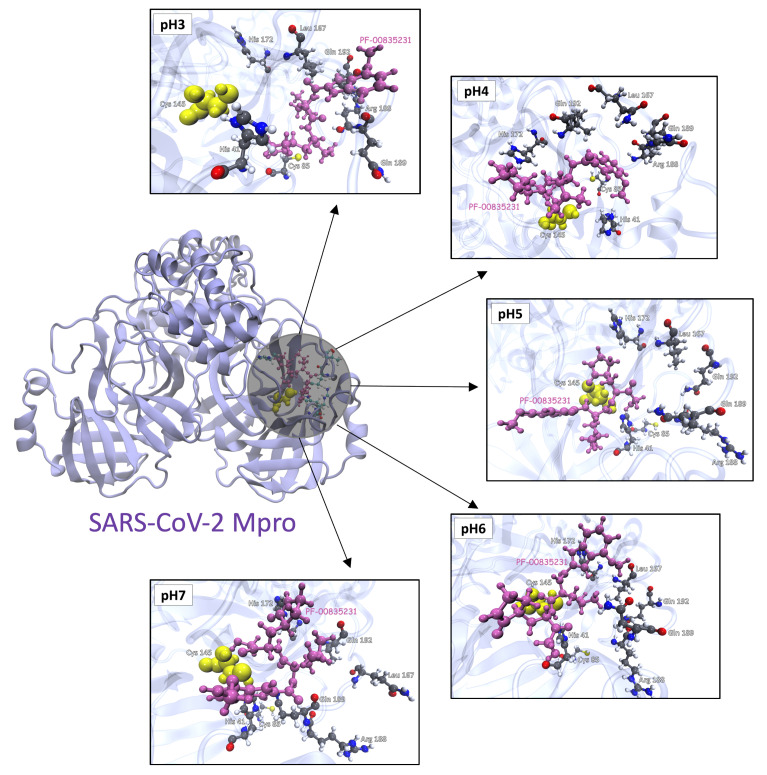
According to the docking results, the Figure 11 reports the best coupling energies for each system studied. Where the His41 and Cys145 residues remain very close to PF-00835231 at different pH conditions.
Figure 11.
Docking snapshot of the best binding affinity at different pHs between PF-0085231 and SARS-CoV-2 Mpro. The Cys145 is highlighted in yellow and the PF-00835231 drug is highlighted in pink. The ligand is located close to the SARS-CoV-2 Mpro active site at different conditions of pHs.
4. Conclusions
The SARS-CoV-2 Mpro is an exciting target gaining the attention of many research groups globally. Its primary importance lies in the replication of the virus, and its inhibition could be the key to stopping the virus from spreading. However, one of the parameters in which is compromised the activity of this enzyme is the pH-dependence.
Although from the point of view of experimental analysis, the study of proteins under low pH conditions is a great challenge. The computational chemistry approach looks the way to study the effect of pH on many important mechanisms such as explored herein.
The program was developed to define a total charge distribution for a specific pH, developed under the SGCMC method. This work showed the distribution of protonation/deprotonation states of the titratable residues with a total net positive charge in pH 3 until pH 5 and a total net negative charge at pH 6 and 7. Moreover, this analysis was able to estimate the isoelectric point (pI = 5.5) computationally. The conventional all-atom MD showed us that Mpro has a stable at low pH structure.
Indeed, Mpro at pH 4 to 7 showed high structural stability of the whole protein, active site, permitting the promoters’ dimerisation. For the case of microstates at pH 3, slight conformational changes were observed. This pH-dependent computational approach revealed that Met49 showed the most significant fluctuations at pH 3 and increased the Mpro pocket volume. Identifying these mechanisms could be used for target selectivity validation.
The last part of the paper elucidates the druggability interface for protonation-dependent Mpro binding sides for a drug candidate in the clinical trial phase. The protein surface was explored using a molecular docking-based screen for a new drug candidate (PF-00835231). The predicted binding modes and affinity for the drug candidate were found in the range from pH 4 to pH 7. While at pH 3, an additional mode of binding was observed; however, more studies could be necessary to elucidate its relevance in Mpro inhibition at very acidic pH.
Abbreviations
The following abbreviations are used in this manuscript:
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| Mpro | Main Protease |
| SGCMC | Semi-Grand Canonical Monte Carlo |
| MD | Molecular Dynamics |
| RMSD | Root-Mean Squared Deviation |
| RMSF | Root-Mean Squared Fluctuation |
| RG | Radius of Gyration |
| HB | Number of Hydrogen Bonds |
| SASA | Solvent Accessible Surface Area |
| RDF | Radial Distribution Function |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/polym13213823/s1, Figure S1: Plot of hydrogen bonds and SASA during the time of MD simulations at different pHs. (a) Solvent Accessible Surface Area. (b) Intramolecular Hydrogen bonds between protein-protein. (c) Intermolecular Hydrogen bonds between protein-waters, Figure S2: Distances between C-atoms from residues of the active site (Cys145 and His41) and S1 pocket in Mpro. (a) Average distance Cys145-His41. (b) Average distance of Met49-Gln189. (c) Average distance of His172-Phe140, Figure S3: Radial Distribution of active site residues against Mpro. (a) Sγ–ions of Cys145. (b) Nδ1–ions of His41. (c) Sγ–ions of Met49, Figure S4: 2D representation of PF-00835231, Figure S5: 2D representation of the best binding affinity Mpro–PF-00835231. (a) pH3. (b) pH4. (c) pH5. (d) pH6. (e) pH7, Table S1: Total charge of each titratable amino acids, Table S2: Protonate/deprotonate microstate of His41 and His172. The symbol P and Ddefine the protonated and deprotonated states, respectively, Table S3: Values of the MD was fitted against the average structure of the last 100 ns of the dynamics was used, Table S4: Distances between amino acids from the catalytic dy-ad for each microstate, Table S5: Numerical values of Ramachandran plot of the last frame of each microstate determined by Molprobity server, Table S6: Docking results of each microstate at different pHs.
Author Contributions
Conceptualisation, H.L.B.-C., S.M., F.M. and M.N.; methodology, H.L.B.-C., S.M., F.M. and M.N.; software, H.L.B.-C. and S.M.; formal analysis, H.L.B.-C., S.M. and M.N.; investigation, H.L.B.-C., S.M., F.M. and M.N.; resources, S.M. and F.M.; data curation, H.L.B.-C.; writing—original draft preparation, H.L.B.-C.; writing—review and editing, S.M., F.M. and M.N.; supervision, S.M. and F.M.; funding acquisition, S.M. and F.M. All authors have read and agreed to the published version of the manuscript.
Funding
S.M. and F.M. acknowledge the financial support from Generalitat de Catalunya (Grant 2017SGR1033). H.L.B.-C., S.M. and F.M. acknowledge the Spanish Structures of Excellence María de Maeztu program through (Grant MDM-2017(0767). M.N. gratefully acknowledged for the funding of the EOSCsecretariat.eu—funding from the European Union’s Horizon Programme call H2020-INFRAEOSC-05-2018-2019, grant agreement number 831644. The authors thankfully acknowledge the computer resources at Pirineus (CSUC from Generalitat de Catalunya) and the technical support provided by Marenostrum at Barcelona Supercomputing centre (RES-QSB-2020-2-0013). The author M.N. is gratefully acknowledged for the computing time granted through Nestum Cluster funded by the ERDF Project BG161PO003-1.2.05-0001-C0001 at Sofia Tech Park, Sofia, Bulgaria.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The access code of the Mpro SARS-CoV-2 crystal structure of this study is described in the Computational Methods Section. The homemade developed code is freely available at GitHub https://github.com/smadurga/Protein-Protonation.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawase M., Shirato K., Matsuyama S., Taguchi F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 2009;83:712–721. doi: 10.1128/JVI.01933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stauffer S., Feng Y., Nebioglu F., Heilig R., Picotti P., Helenius A. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J. Virol. 2014;88:13029–13046. doi: 10.1128/JVI.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S., Sieben C., Ludwig K., Höfer C.T., Chiantia S., Herrmann A., Eghiaian F., Schaap I.A. pH-Controlled two-step uncoating of influenza virus. Biophys. J. 2014;106:1447–1456. doi: 10.1016/j.bpj.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/S0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 9.Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto H.H., Schirmeister T. Cysteine proteases and their inhibitors. Chem. Rev. 1997;97:133–172. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- 11.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 12.Tang J., Wong R.N. Evolution in the structure and function of aspartic proteases. J. Cell. Biochem. 1987;33:53–63. doi: 10.1002/jcb.240330106. [DOI] [PubMed] [Google Scholar]
- 13.Szecsi P.B. The aspartic proteases. Scand. J. Clin. Lab. Investig. 1992;52:5–22. doi: 10.1080/00365519209104650. [DOI] [PubMed] [Google Scholar]
- 14.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2021;39:3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanhed A.M., Patel D.V., Teli D.M., Patel N.R., Chhabria M.T., Yadav M.R. Identification of potential Mpro inhibitors for the treatment of COVID-19 by using systematic virtual screening approach. Mol. Divers. 2021;25:383–401. doi: 10.1007/s11030-020-10130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhary J., Dheeman S., Sharma V., Katiyar P., Karn S.K., Sarangi M.K., Chauhan A.K., Verma G., Baliyan N. Insights of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) pandemic: A current review. Biol. Proced. Online. 2021;23:1–22. doi: 10.1186/s12575-020-00141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL M pro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorganic Med. Chem. Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Römer R.A., Römer N.S., Wallis A.K. Flexibility and mobility of SARS-CoV-2-related protein structures. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-82849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 24.Sharma A., Gupta S.P. Viral Proteases and Their Inhibitors. Elsevier; Amsterdam, The Netherlands: 2017. Fundamentals of viruses and their proteases; pp. 1–24. [Google Scholar]
- 25.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 26.Tan J., Verschueren K.H., Anand K., Shen J., Yang M., Xu Y., Rao Z., Bigalke J., Heisen B., Mesters J.R., et al. pH-dependent conformational flexibility of the SARS-CoV main proteinase (Mpro) dimer: Molecular dynamics simulations and multiple X-ray structure analyses. J. Mol. Biol. 2005;354:25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., et al. Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: A potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 29.Talley K., Alexov E. On the pH-optimum of activity and stability of proteins. Proteins Struct. Funct. Bioinform. 2010;78:2699–2706. doi: 10.1002/prot.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 32.Chang H.P., Chou C.Y., Chang G.G. Reversible unfolding of the severe acute respiratory syndrome coronavirus main protease in guanidinium chloride. Biophys. J. 2007;92:1374–1383. doi: 10.1529/biophysj.106.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilgenfeld R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan P., Yang Z., Song H., Wang K., Yang Y., Xie L., Huang S., Liu J., Ran L., Song Z. Three Main Inducers of Alphacoronavirus Infection of Enterocytes: Sialic Acid, Proteases, and Low pH. Intervirology. 2018;61:53–63. doi: 10.1159/000492424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suárez D., Díaz N. SARS-CoV-2 Main Protease: A Molecular Dynamic Study. J. Chem. Inf. Model. 2020;60:5815–5831. doi: 10.1021/acs.jcim.0c00575. [DOI] [PubMed] [Google Scholar]
- 36.Díaz N., Suárez D. Influence of charge configuration on substrate binding to SARS-CoV-2 main protease. Chem. Commun. 2021;57:5314–5317. doi: 10.1039/D1CC01449H. [DOI] [PubMed] [Google Scholar]
- 37.Sharma S., Deep S. pH Effect on the Dynamics of SARS-CoV-2 Main Protease (Mpro) bioRxiv. 2020 doi: 10.1101/2020.11.30.404384. [DOI] [Google Scholar]
- 38.White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan N., Chen X., Geiger J.D. Role of endolysosomes in severe acute respiratory syndrome coronavirus-2 infection and coronavirus disease 2019 pathogenesis: Implications for potential treatments. Front. Pharmacol. 2020;11:1739. doi: 10.3389/fphar.2020.595888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jimenez L., Campos Codo A., Sampaio V.d.S., Oliveira A.E., Ferreira L.K.K., Davanzo G.G., Brito Monteiro L.d., Victor Virgilio-da Silva J., Borba M.G.S., Fabiano de Souza G., et al. Acid pH Increases SARS-CoV-2 Infection and the Risk of Death by COVID-19. Front. Med. 2021:1358. doi: 10.3389/fmed.2021.637885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nechipurenko Y.D., Semyonov D.A., Lavrinenko I.A., Lagutkin D.A., Generalov E.A., Zaitceva A.Y., Matveeva O.V., Yegorov Y.E. The Role of Acidosis in the Pathogenesis of Severe Forms of COVID-19. Biology. 2021;10:852. doi: 10.3390/biology10090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henry B.M., Aggarwal G., Wong J., Benoit S., Vikse J., Plebani M., Lippi G. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: A pooled analysis. Am. J. Emerg. Med. 2020;38:1722–1726. doi: 10.1016/j.ajem.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira J.C., Rabeh W.M. Biochemical and biophysical characterization of the main protease, 3-chymotrypsin-like protease (3CLpro) from the novel coronavirus SARS-CoV 2. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-020-79357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamdullah K.S., Tanzila A., ZainaSher M., Iqra A., Mohtasheemul H. pH Dependent Differential Binding Behavior of Prtotease Inhibitor Molecular Drugs for SARS-CoV-2. 2020. [(accessed on 10 January 2021)]. Available online: https://pesquisa.bvsalud.org/global-literature-on-novel-coronavirus-2019-ncov/resource/pt/ppcovidwho-148.
- 47.Vatansever E.C., Yang K.S., Drelich A.K., Kratch K.C., Cho C.C., Kempaiah K.R., Hsu J.C., Mellott D.M., Xu S., Tseng C.T.K., et al. Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. USA. 2021;118:e2012201118. doi: 10.1073/pnas.2012201118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen T.T.H., Jung J.H., Kim M.K., Lim S., Choi J.M., Chung B., Kim D.W., Kim D. The Inhibitory Effects of Plant Derivate Polyphenols on the Main Protease of SARS Coronavirus 2 and Their Structure–Activity Relationship. Molecules. 2021;26:1924. doi: 10.3390/molecules26071924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamdi M., Abdel-Bar H.M., Elmowafy E., El-Khouly A., Mansour M., Awad G.A. Investigating the Internalization and COVID-19 Antiviral Computational Analysis of Optimized Nanoscale Zinc Oxide. ACS Omega. 2021;6:6848–6860. doi: 10.1021/acsomega.0c06046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dražić T., Kühl N., Leuthold M.M., Behnam M.A., Klein C.D. Efficiency Improvements and Discovery of New Substrates for a SARS-CoV-2 Main Protease FRET Assay. Slas Discov. Adv. Sci. Drug Discov. 2021:24725552211020681. doi: 10.1177/24725552211020681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., et al. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graziano V., McGrath W.J., DeGruccio A.M., Dunn J.J., Mangel W.F. Enzymatic activity of the SARS coronavirus main proteinase dimer. FEBS Lett. 2006;580:2577–2583. doi: 10.1016/j.febslet.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S., Chen L.L., Luo H.B., Sun T., Chen J., Ye F., Cai J.H., Shen J.K., Shen X., Jiang H.L. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique 1. Acta Pharmacol. Sin. 2005;26:99–106. doi: 10.1111/j.1745-7254.2005.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verma N., Henderson J.A., Shen J. Proton-Coupled Conformational Activation of SARS Coronavirus Main Proteases and Opportunity for Designing Small-Molecule Broad-Spectrum Targeted Covalent Inhibitors. J. Am. Chem. Soc. 2020;142:21883–21890. doi: 10.1021/jacs.0c10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavlova A., Lynch D.L., Smith M.D., Smith J.D., Gumbart J.C. Inhibitor Binding Influences the Protonation State of Histidines in SARS-CoV-2 Main Protease. Biophys. J. 2021;120:204a–205a. doi: 10.1016/j.bpj.2020.11.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan S., Xiao D., Wang Y., Liu L., Zhou X., Zhong W. Research progress on repositioning drugs and specific therapeutic drugs for SARS-CoV-2. Future Med. Chem. 2020;12:1565–1578. doi: 10.4155/fmc-2020-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 58.Coelho C., Gallo G., Campos C.B., Hardy L., Würtele M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS ONE. 2020;15:e0240079. doi: 10.1371/journal.pone.0240079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reiner Ž., Hatamipour M., Banach M., Pirro M., Al-Rasadi K., Jamialahmadi T., Radenkovic D., Montecucco F., Sahebkar A. Statins and the COVID-19 main protease: In silico evidence on direct interaction. Arch. Med. Sci. AMS. 2020;16:490. doi: 10.5114/aoms.2020.94655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gimeno A., Mestres-Truyol J., Ojeda-Montes M.J., Macip G., Saldivar-Espinoza B., Cereto-Massagué A., Pujadas G., Garcia-Vallvé S. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int. J. Mol. Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar N., Awasthi A., Kumari A., Sood D., Jain P., Singh T., Sharma N., Grover A., Chandra R. Antitussive noscapine and antiviral drug conjugates as arsenal against COVID-19: A comprehensive chemoinformatics analysis. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1808072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moneriz C., Castro-Salguedo C. Fármacos prometedores y potenciales para el tratamiento de COVID-19. Rev. Chil. Infectología. 2020;37:205–215. doi: 10.4067/s0716-10182020000300205. [DOI] [PubMed] [Google Scholar]
- 63.Yang H., Yang J. A review of latest research on Mpro targeted SARS-COV inhibitors. RSC Med. Chem. 2021;12:1026–1036. doi: 10.1039/D1MD00066G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao T., Cui M., Zheng C., Wang M., Sun R., Gao D., Bao J., Ren S., Yang B., Lin J., et al. Myricetin Inhibits SARS-CoV-2 Viral Replication by Targeting Mpro and Ameliorates Pulmonary Inflammation. Front. Pharmacol. 2021;12:1012. doi: 10.3389/fphar.2021.669642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mulu A., Gajaa M., Woldekidan H.B. The impact of curcumin derived polyphenols on the structure and flexibility COVID-19 main protease binding pocket: A molecular dynamics simulation study. PeerJ. 2021;9:e11590. doi: 10.7717/peerj.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silvestrini L., Belhaj N., Comez L., Gerelli Y., Lauria A., Libera V., Mariani P., Marzullo P., Ortore M.G., Piccionello A.P., et al. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci. Rep. 2021;11:1–16. doi: 10.1038/s41598-021-88630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9:622898. doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vicidomini C., Roviello V., Roviello G.N. In Silico Investigation on the Interaction of Chiral Phytochemicals from Opuntia ficus-indica with SARS-CoV-2 Mpro. Symmetry. 2021;13:1041. doi: 10.3390/sym13061041. [DOI] [Google Scholar]
- 69.Roviello V., Musumeci D., Mokhir A., Roviello G.N. Evidence of protein binding by a nucleopeptide based on a thymine-decorated L-diaminopropanoic acid through CD and in silico studies. Curr. Med. Chem. 2021;28:5004–5015. doi: 10.2174/0929867328666210201152326. [DOI] [PubMed] [Google Scholar]
- 70.Goyzueta-Mamani L.D., Barazorda-Ccahuana H.L., Mena-Ulecia K., Chávez-Fumagalli M.A. Antiviral Activity of Metabolites from Peruvian Plants against SARS-CoV-2: An In Silico Approach. Molecules. 2021;26:3882. doi: 10.3390/molecules26133882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rizzuti B., Grande F., Conforti F., Jimenez-Alesanco A., Ceballos-Laita L., Ortega-Alarcon D., Vega S., Reyburn H.T., Abian O., Velazquez-Campoy A. Rutin is a low micromolar inhibitor of SARS-CoV-2 main protease 3CLpro: Implications for drug design of quercetin analogs. Biomedicines. 2021;9:375. doi: 10.3390/biomedicines9040375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madurga S., Garcés J.L., Companys E., Rey-Castro C., Salvador J., Galceran J., Vilaseca E., Puy J., Mas F. Ion binding to polyelectrolytes: Monte Carlo simulations versus classical mean field theories. Theor. Chem. Accounts. 2009;123:127–135. doi: 10.1007/s00214-009-0550-z. [DOI] [Google Scholar]
- 73.Madurga S., Rey-Castro C., Pastor I., Vilaseca E., David C., Garcés J.L., Puy J., Mas F. A semi-grand canonical Monte Carlo simulation model for ion binding to ionizable surfaces: Proton binding of carboxylated latex particles as a case study. J. Chem. Phys. 2011;135:184103. doi: 10.1063/1.3658484. [DOI] [PubMed] [Google Scholar]
- 74.Blanco P.M., Madurga S., Mas F., Garcés J.L. Coupling of charge regulation and conformational equilibria in linear weak polyelectrolytes: Treatment of long-range interactions via effective short-ranged and pH-dependent interaction parameters. Polymers. 2018;10:811. doi: 10.3390/polym10080811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanco P.M., Madurga S., Narambuena C.F., Mas F., Garcés J.L. Role of charge regulation and fluctuations in the conformational and mechanical properties of weak flexible polyelectrolytes. Polymers. 2019;11:1962. doi: 10.3390/polym11121962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barazorda-Ccahuana H.L., Gómez B., Mas F., Madurga S. Effect of pH on the Supramolecular Structure of Helicobacter pylori Urease by Molecular Dynamics Simulations. Polymers. 2020;12:2713. doi: 10.3390/polym12112713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., et al. Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. BioRxiv. 2021:2020-09. doi: 10.1101/2020.09.12.293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Vries M., Mohamed A., Prescott R.A., Valero-Jimenez A., Desvignes L., O’Connor R., Steppan C., Devlin J.C., Ivanova E., Herrera A., et al. A comparative analysis of SARS-CoV-2 antivirals in human airway models characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. bioRxiv. 2021:2020-08. doi: 10.1101/2020.08.28.272880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Owen D.R., Allerton C.M., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., et al. An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19. medRxiv. 2021 doi: 10.1101/2021.07.28.21261232. [DOI] [PubMed] [Google Scholar]
- 80.Pavan M., Bolcato G., Bassani D., Sturlese M., Moro S. Supervised Molecular Dynamics (SuMD) Insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332. J. Enzym. Inhib. Med. Chem. 2021;36:1646–1650. doi: 10.1080/14756366.2021.1954919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anirudhan V., Lee H., Cheng H., Cooper L., Rong L. Targeting SARS-CoV-2 viral proteases as a therapeutic strategy to treat COVID-19. J. Med. Virol. 2021;93:2722–2734. doi: 10.1002/jmv.26814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baig M.H., Sharma T., Ahmad I., Abohashrh M., Alam M.M., Dong J.J. Is PF-00835231 a Pan-SARS-CoV-2 Mpro Inhibitor? A Comparative Study. Molecules. 2021;26:1678. doi: 10.3390/molecules26061678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olsson M.H., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical p K a predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 84.Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct. Funct. Bioinform. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 85.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 86.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Makov G., Payne M. Periodic boundary conditions in ab initio calculations. Phys. Rev. B. 1995;51:4014. doi: 10.1103/PhysRevB.51.4014. [DOI] [PubMed] [Google Scholar]
- 88.Mahoney M.W., Jorgensen W.L. Diffusion constant of the TIP5P model of liquid water. J. Chem. Phys. 2001;114:363–366. doi: 10.1063/1.1329346. [DOI] [Google Scholar]
- 89.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 92.Racine J. gnuplot 4.0: A portable interactive plotting utility. J. Appl. Econom. 2006;21:133–141. doi: 10.1002/jae.885. [DOI] [Google Scholar]
- 93.Scheller C., Krebs F., Minkner R., Astner I., Gil-Moles M., Wätzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41:1137–1151. doi: 10.1002/elps.202000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L., Buck M. Molecular simulations of a dynamic protein complex: Role of salt-bridges and polar interactions in configurational transitions. Biophys. J. 2013;105:2412–2417. doi: 10.1016/j.bpj.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Świderek K., Moliner V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 M pro by QM/MM computational methods. Chem. Sci. 2020;11:10626–10630. doi: 10.1039/D0SC02823A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The access code of the Mpro SARS-CoV-2 crystal structure of this study is described in the Computational Methods Section. The homemade developed code is freely available at GitHub https://github.com/smadurga/Protein-Protonation.