Abstract
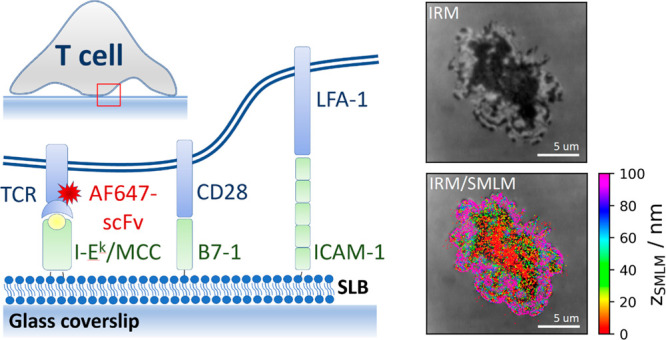
T-cells engage with antigen-presenting cells in search for antigenic peptides and form transient interfaces termed immunological synapses. Synapse topography affects receptor binding rates and the mutual segregation of proteins due to size exclusion effects. It is hence important to determine the 3D topography of the immunological synapse at high precision. Current methods provide only rather coarse images of the protein distribution within the synapse. Here, we applied supercritical angle fluorescence microscopy combined with defocused imaging, which allows three-dimensional single molecule localization microscopy (3D-SMLM) at an isotropic localization precision below 15 nm. Experiments were performed on hybrid synapses between primary T-cells and functionalized glass-supported lipid bilayers. We used 3D-SMLM to quantify the cleft size within the synapse by mapping the position of the T-cell receptor (TCR) with respect to the supported lipid bilayer, yielding average distances of 18 nm up to 31 nm for activating and nonactivating bilayers, respectively.
Keywords: T-cells, Immunological synapse, Interference Reflection Microscopy, T-cell receptor, 3D Single Molecule Localization Microscopy
Introduction
Understanding the topography of cellular interfaces is central for addressing many cell biological questions. The distance between the two juxtaposing cell surfaces not only regulates the affinity of protein–protein transinteractions,1,2 but the extension of the intercellular cleft also affects the spatial distribution of membrane proteins with differently sized ectodomains.3 A prominent example is the size-exclusion of the large phosphatase CD45 upon contact formation between a T-cell and an antigen-presenting cell (APC), which is suspected to represent an important regulatory mechanism for the phosphorylation of the T-cell receptor (TCR);4 according to this model, shifting the balance between Lck-mediated phosphorylation and CD45-mediated dephosphorylation induces downstream signaling.
A common way to study antigen-specific T-cell activation involves the use of functionalized glass-supported lipid bilayers (SLBs) as surrogates of APCs.5−7 This experimental design has several advantages when it comes to the application of high-resolution microscopy techniques while still preserving the essential hallmarks of T-cell signaling including the formation of an immunological synapse, the recruitment of the kinase ZAP-70 and other downstream signaling effectors, the increase in intracellular calcium, and the release of cytokines.
First, interfacing cells with a glass coverslip further allows for exploiting the interference of light reflected from the glass–water interface and light reflected from the cell membrane for imaging purposes.8,9 This technique, termed interference reflection microscopy (IRM), yields high precision information on the separation of the cell surface from the surface of the glass coverslip, eventually limited only by the signal-to-noise ratio of the data and by the knowledge of the interference model.9 IRM has been frequently applied to qualitatively assess the homogeneity of T-cell adhesion to activating or inert surfaces,5,10−12 often yielding patches of close contact next to areas of substantial elevation of the T-cell surface. Quantitative interpretation of the data, however, is often hampered by unknowns of the interference model.9 For example, tilted membranes or the presence of second or third order interferences are difficult to account for. In addition, multiple reflections from different layers of varying refractive index induce phase shifts in the IRM intensity profiles, thereby impeding absolute distance measurements. In fact, protein ectodomains and the glycocalyx contribute to the change in refractive index between the aqueous environment and the cell, rendering the plane of reflection rather poorly defined. Finally, IRM images are of diffraction-limited spatial resolution and thereby yield averages of the interference contrast over a few 100 nm. Fluctuations at smaller scales would hence be averaged out.
As a second advantage of hybrid synapses, they facilitate the application of total internal reflection (TIR) excitation to accentuate the signal of dyes proximal to the glass surface over intracellular background.13 Using TIR excitation, researchers discovered, for example, the formation of TCR microclusters upon T-cell activation.14−16
A third advantage relates to the use of single molecule localization microscopy (SMLM) for studying the organization of signaling molecules in the course of T-cell activation.17−22 Briefly, SMLM achieves superior spatial resolution by precisely localizing well-separated single molecule signals that can be obtained from blinking chromophores.23 Although a two-dimensional localization precision below 20 nm was frequently reported, it is difficult to achieve similar precision along the optical axis.24 In this context, the presence of the glass coverslip in the vicinity of the fluorophores of interest allows for using the supercritical angle fluorescence as a parameter for determining the distance between the dye molecule and the glass surface.25,26 Of note, the dye’s distance from the glass surface affects the shape of the recorded point spread functions due to different supercritical angle contributions. We have recently demonstrated that when combined with defocused imaging supercritical angle three-dimensional SMLM (3D-SMLM) achieves isotropic localization precision down to ∼10 nm in all three dimensions.27
Here, we used 3D-SMLM based on supercritical angle microscopy to study the topography of the immunological synapse formed between primary murine CD4+ T-cells and functionalized SLBs at isotropic localization precision below 15 nm. The obtained TCR localization maps were correlated with IRM images of the same synapse, thereby allowing not only cross-validation of the two approaches but also the identification of artifacts inherent to IRM images. From the TCR z-coordinates, we quantified the roughness of the T-cell surface within the synapse, as well as its separation from the SLB: Both for activating and nonactivating conditions we observed multiple TCR-proximal spots of close contact, which would qualify for CD45 exclusion. We finally quantitatively compared 3D-SMLM images with diffraction-limited TIR fluorescence microscopy of T-cell synapses to disentangle different contributions to the appearance of TCR microclusters.
Results
Correlative 3D-Single Molecule Localization Microscopy and Interference Reflection Microscopy
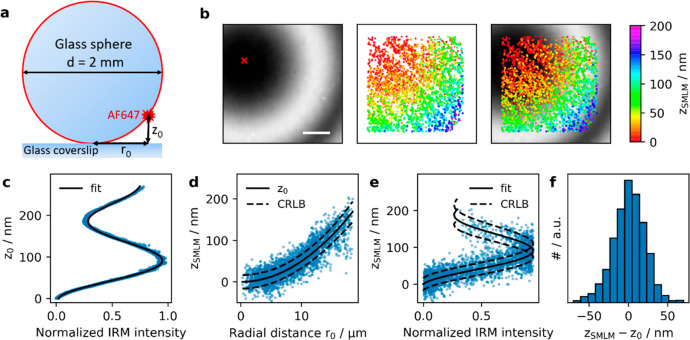
In order to evaluate the correlation between 3D-SMLM and IRM data, we sought a system with known separation of the detected single dye molecules from the glass surface. We opted for an AlexaF647-coated glass sphere of 1 mm radius adhered to a glass coverslip, which yielded z-distances of up to 150 nm within the field of view of 22 × 22 μm2 (Figure 1a). Figure 1b shows an IRM image recorded next to the contact point between the sphere and the glass surface. Concentric interference fringes are clearly visible. When plotting the recorded IRM intensity values versus the distance of the according pixels from the glass surface, z0, we observed the characteristic cosine-dependence (Figure 1c). In this case, three branches of the IRM intensity can be distinguished, corresponding to different orders of the interference pattern. The first branch covers zSMLM values up to 90 nm, the second values of 90–180 nm, and the third values above 180 nm. A slight decrease in amplitude and wavelength of the recorded IRM curve is noted for increasing IRM interference orders, which is a consequence of reflections on the curved surface.9 In Figure 1b, we also included the recorded 3D-SMLM data, with the color-code indicating the calculated displacement from the glass surface, zSMLM. When plotted against the radial distance from the sphere’s contact point, r0, the measured single molecule displacements zSMLM follow closely the surface of the sphere. (Figure 1d). We further correlated zSMLM with the IRM intensity values recorded on the corresponding pixels (Figure 1e), yielding very good agreement of the two data sets. The solid black line shows the calibration curve obtained in Figure 1c, and the dashed lines indicate the expected Cramér-Rao lower bound (CRLB).27,28 To obtain a quantitative measure of the method’s z-precision, we calculated for each localization the difference between zSMLM and the theoretical z0 (Figure 1f). The determined standard deviation of 21 nm agrees well with the Cramér-Rao lower bound of 17 nm.
Figure 1.
Control experiment on a fluorescently labeled glass sphere. A glass sphere of 2 mm diameter was labeled with BSA, Biotin–Streptavidin-AF647. On the basis of the known size of the sphere, the distance of the AF647 to the coverslip z0 was calculated from the measured radial distance r0 from the contact point of the sphere with the coverslip (a) (see SI Materials and Methods eq 1). (b) IRM and SMLM images were recorded next to the contact point of the sphere with the coverslip (red cross). (c) We plotted the z0 corresponding to the center of the respective pixel against the normalized IRM intensity and fitted with SI Materials and Methods eq 2. (d) The measured distances zSMLM of all localizations were plotted against their radial distance r0. The solid line indicates the expected behavior for a spherical surface (SI Materials and Methods eq 1), dashed lines indicate the square root of the Cramér-Rao lower bound. (e) Dependence of zSMLM on the normalized IRM intensity. Solid line indicates fit results from panel (c), dashed lines indicate the square root of the Cramér-Rao lower bound. (f) The difference between the measured values of zSMLM and the calculated values of z0 showed a standard deviation of 21 nm. Scale bar 5 μm.
Correlative IRM, TIR, and 3D-SMLM of the Immunological Synapse
Next, we applied the method to image the three-dimensional topography of the immunological synapse formed between CD4+ murine 5c.c7 TCR-transgenic T-cells and stimulatory or inert surfaces. To visualize the position of the TCR, T-cells were labeled with an AlexaF647-conjugated single chain antibody fragment6 against the TCR β subunit. Before analysis, all 3D-SMLM images were corrected for overcounts, which eventually allowed us to obtain a valid estimation of the surface roughness (see Supporting Information (SI) Materials and Methods). An isotropic mean precision of 12.7, 12.0, and 14.6 nm was determined for single molecule localization along the x-, y-, and z-axis, respectively (SI Figure 1).
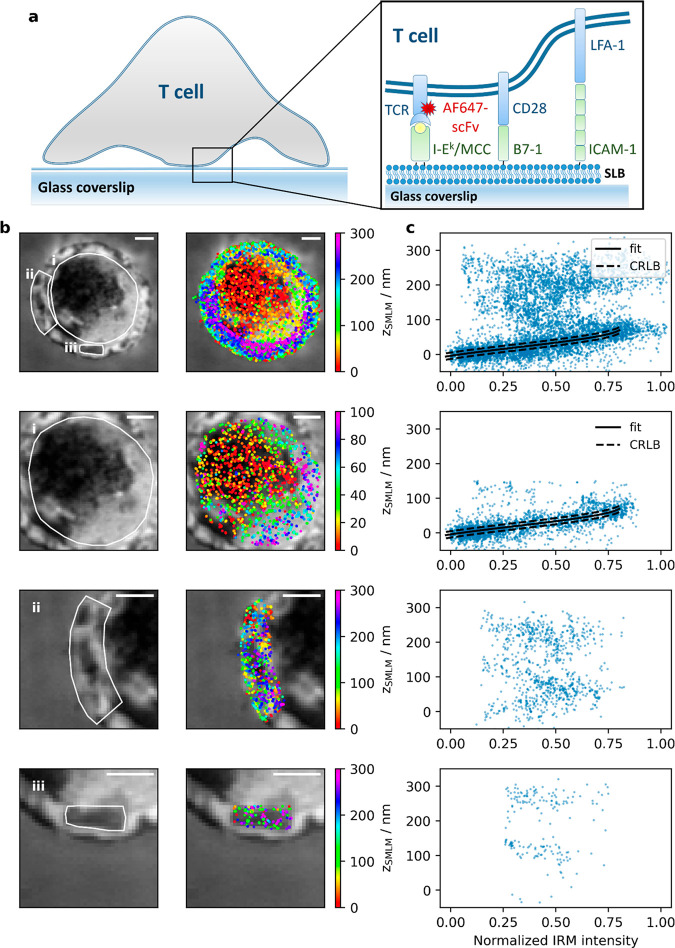
We first addressed the three-dimensional organization of the TCR in activated T-cells. To this end, T-cells were seeded onto fluid SLBs functionalized with MCC-loaded I-Ek at a surface density of 100 ± 30 molecules/μm2, which is known to stimulate intracellular calcium release;29 in addition, bilayers contained the adhesion molecule ICAM-1 and the costimulatory molecule B7-1 (Figure 2a). For all applied conditions, T-cell activation was controlled via ratiometric calcium imaging (SI Figure 3). Prior to conducting imaging experiments, T-cells were fixed at specific time points (here 5 min) after their seeding onto the SLBs and imaged by both via IRM and 3D-SMLM. The IRM images show substantial contrast fluctuations (Figure 2b), indicating corresponding fluctuations in the distance of the T-cell membrane from the SLB. This is supported by the 3D-SMLM data, where the determined TCR z-positions spread between 0 and 300 nm. Plotting zSMLM versus the IRM intensity revealed a good correlation between the two data sets for the first branch of the IRM signal (Figure 2c, solid black line). This correlation vanished, however, for higher order IRM branches. We attribute this lack of correlation to additional parameters affecting the measured IRM contrast such as unknown angles of the reflecting surfaces or the occurrence of multiple interferences, which particularly disturbs IRM signals originating from reflections at larger distances from the glass surface. We therefore did not fit those regions of the IRM curves. In addition, different resolutions of the two methods impede direct comparison of the two data sets: while 3D-SMLM data report on z-distances specific for 2D coordinates that can be determined with a precision below the diffraction-limit, IRM images are limited by diffraction and hence provide average values over areas given by the size of the 2D point spread function.
Figure 2.
Correlative 3D-SMLM and IRM within the immunological synapse. (a) Experiments were performed on a T-cell adhering to an SLB functionalized with I-Ek/MCC, B7-1 and ICAM-1; the TCR was labeled via AF647-conjugated H57-scFv. (b) IRM (left) and SMLM/IRM overlay images (right) of the immunological synapse. The areas i, ii, and iii are shown in magnification below. (c) Correlation plots between zSMLM and the normalized IRM intensity for the areas indicated in panel b. Data points with zSMLM < 100 nm were fitted with SI Materials and Methods eq 3 (solid line). The dashed lines indicate the square root of the Cramér-Rao lower bound. Scale bars 2 μm.
For detailed assessment, we selected in region (i) of Figure 2b the cell center in which the left half of the region was well adhered to the SLB surface while the right half featured a separation of ∼60 nm from the SLB. Here, results obtained with the two imaging modalities are in very good agreement. In region (ii) showing the cell edge, where the T-cell formed a narrow lamellipodium, we observed two distinct clusters in the zSMLM data, one reflecting the bottom, the other reflecting the top membrane of the lamellipodium. The two clusters were separated by ∼160 nm (SI Figure 2), which corresponded to the thickness of the lamellipodium.30 Interestingly, the z-positions of this particular region hardly correlated with the IRM patterns, likely due to high inclinations of the reflecting membrane at the lamellipodium edge possibly causing additional interferences in the IRM image. In region (iii), another feature of membrane topography became apparent: the central dark IRM area did not indicate an area of close contact but instead showed rather distal lamellipodium regions reflected by zSMLM coordinates more than 100 nm away from the glass surface. Also in this region, the separation of the two lamellipodia membranes of ∼160 nm became apparent from two well-separated localization clusters in the zSMLM data.
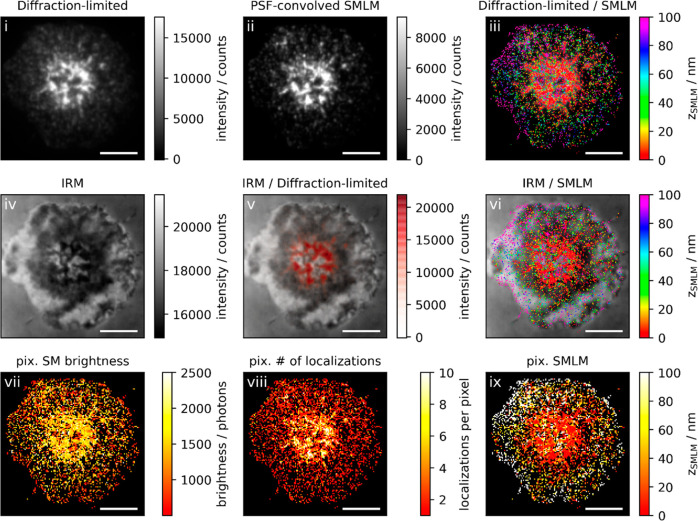
It is also instructive to compare the obtained 3D SMLM and IRM images with conventional diffraction-limited TIR fluorescence microscopy images (Figure 3; see also SI supporting gallery Figures 1 and 2 for additional examples). In this particular example, the cell had been fixed just before the formation of the central supramolecular activation cluster (cSMAC), when TCR microclusters were observed in a ringlike structure around the cell center (Figure 3i). This image hence presumably reflects a snapshot of the directional microcluster transport toward the cSMAC.31 While TCRs in the ring itself showed tight contact with the glass surface, likely due to binding to the cognate pMHC, 3D-SMLM revealed that the engulfed circular membrane patch contained TCRs substantially elevated by ∼100 nm (Figure 3iii and Figure 3ix), in agreement with the observation that the cSMAC is a site of TCR endocytosis.32 In addition, some of the SLB engaged TCR in the cSMAC reside in ∼100 nm extracellular microvesicles that elevate the nonengaged TCR bearing plasma membrane by ∼100 nm above the SLB,33 potentially contributing to the two layers of TCR in the cSMAC.
Figure 3.
Correlative 3D-SMLM, IRM, and diffraction-limited TIR microscopy of the immunological synapse. T-cells were activated on a SLB functionalized with I-Ek/MCC, B7-1, and high densities of ICAM-1 and fixed 10 min post seeding. The T-cell was imaged with IRM and fluorescence microscopy. (i) Diffraction-limited TIR image of the T-cell. (ii) Reconstruction of the diffraction-limited image by convolving the 3D-SMLM image with the corresponding psf. (iii) Overlay of the diffraction-limited TIR image with the 3D-SMLM image. The color-code indicates distances to the coverslip zSMLM. (iv) IRM image. (v) Overlay of the IRM image with the diffraction-limited image. (vi) Overlay of the IRM and the 3D-SMLM image. Bottom row images were generated by calculating the pixel-wise average of the 3D-SMLM images (pixel size of 146 nm is consistent with diffraction-limited image) according to pixelated mean single molecule (SM) brightness (vii), pixelated number of localizations (viii), and pixelated mean zSMLM (ix). Scale bars: 5 μm.
We next analyzed TCR microclusters in more detail. In principle, TCR microcluster contrast is not only determined by protein enrichment but also by the closer proximity of the fluorophores to the glass surface, yielding both increased excitation intensity in the evanescent field as well as increased detection efficiency due to the collection of supercritical angle fluorescence. Because our method allows for disentangling single molecule fluorescence brightness, z-position, and local clustering, we addressed the different contributions to the diffraction-limited TIR images. For this, we compared the diffraction-limited TIR images (Figure 3i) with data obtained from 3D-SMLM. The single molecule brightness indeed showed some correlation with the positions of microclusters in the diffraction-limited image (Figure 3vii), which was also reflected in a map of the single molecule z-positions (Figure 3ix). Furthermore, single molecule localizations were strongly clustered at the positions of the microclusters (Figure 3viii). We sought to reconstruct the diffraction-limited image by convolving the single molecule localization map with the brightness-weighted point spread function (psf) (Figure 3ii). The reconstructed image agreed well with the original diffraction-limited image, down to the level of the individual TCR microclusters.
To quantitatively disentangle the different contributions, we identified TCR microclusters by intensity-thresholding the diffraction-limited images (SI Figure 6) and analyzed the single molecule properties separately for localizations coinciding with the TCR microcluster regions versus localizations outside of TCR microclusters. Figure 4 shows the ratios of single molecule brightness and the number of single molecule localizations per pixel, which were obtained from multiple cells. While the brightness levels in microclusters increased only by 20%, we observed about 3-fold enrichment of localizations. The product of the two ratios quantitatively matched the average brightness ratios of pixels corresponding to microclusters versus pixels of outside regions obtained from the diffraction-limited images. Importantly, as this analysis was based on pixel-wise discrimination of microclusters in the diffraction-limited images, it was not affected by residual overcounts in the SMLM images. Taken together, we conclude that the increased brightness of TCR microclusters in diffraction-limited images is mainly explained by enrichment of TCR molecules. Of note, we observed the expected increased brightness of microclusters fixed at late time-points (red data points) compared to those fixed at early time-points (orange data points);16 also, this effect is explained by an increase in the number of single molecule localizations.
Figure 4.
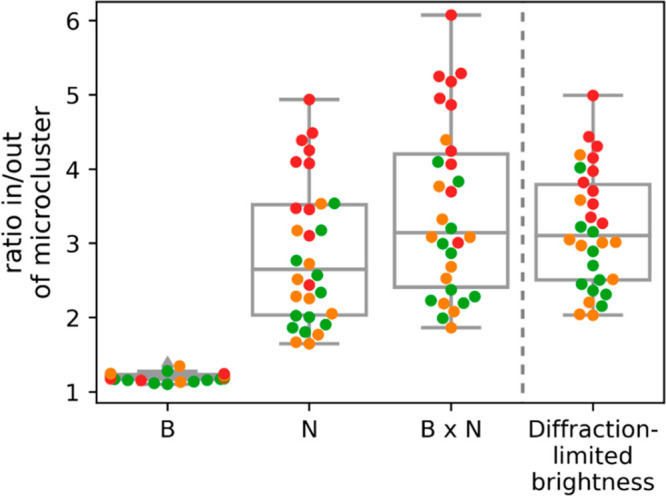
Disentangling single molecule brightness and molecular enrichment in TCR microclusters. We quantified the average single molecule brightness, B, and the number of localizations, N, in pixels corresponding to TCR microclusters (“in”) and the complementary regions of the synapse (“out”). Plotted are the ratios in/out for B, N, the product B × N, and the average diffraction-limited brightness per pixel. Each data point corresponds to the average ratio per cell. Colors indicate the time point of fixation post seeding (orange, 5–10 min; green, 10 min; red, 10–15 min). (n = 30 cells).
We further studied T-cells contacting SLBs that were functionalized with the adhesion molecule ICAM-1 only, so that no calcium signal was triggered (see SI Figure 3 for ratiometric calcium analysis and SI supporting gallery Figures 3 and 4 for exemplary images). As expected, we did not observe the formation of TCR microclusters. While in general the cells spread well on such substrates, areas of close contact appeared more fragmented than in the activated situation. This effect was more pronounced when we reduced the density of the adhesion molecule ICAM-1 in the SLB from 125 ± 22 to 4 ± 2 molecules/μm 2.
Quantitative Analysis of T-cell Surface Topography within the Immunological Synapse
For quantification, we determined the
roughness of the T-cell surface within the immunological synapse.
To prevent the inclusion of data originating from the top membrane
of lamellipodia we only considered localizations with zSMLM < 100 nm. At nonactivating conditions and low
ICAM-1 densities, we observed substantial fluctuations of the recorded z-positions (Figure 5a). For quantitative determination of the surface roughness,
we compared the obtained variances σz,TCR2 with values obtained
for fluorescently labeled SLB-anchored I-Ek/MCC, σz,MHC, according to 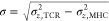 , yielding a standard deviation
of σ
= 37 nm. Fluctuations decreased to 29 nm when we increased the density
of the adhesion molecule ICAM-1 in the lipid bilayer. Upon activation
via higher densities of I-Ek/MCC, T-cells adhered more
smoothly to the surface, as indicated by reduced overall z-fluctuations of 19 nm. Also, in the case of activation we observed
the T-cell surface flattening out to a considerable extent with ICAM-1
present at higher densities.
, yielding a standard deviation
of σ
= 37 nm. Fluctuations decreased to 29 nm when we increased the density
of the adhesion molecule ICAM-1 in the lipid bilayer. Upon activation
via higher densities of I-Ek/MCC, T-cells adhered more
smoothly to the surface, as indicated by reduced overall z-fluctuations of 19 nm. Also, in the case of activation we observed
the T-cell surface flattening out to a considerable extent with ICAM-1
present at higher densities.
Figure 5.
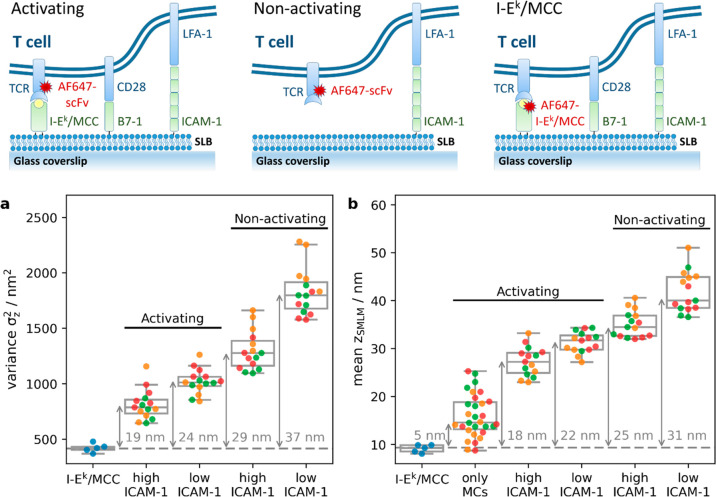
Contact analysis for T-cells recorded under activating and nonactivating conditions: T-cells were seeded on SLBs either functionalized with I-Ek/MCC, B7-1, and ICAM-1 (termed “activating”) or with ICAM-1 only (termed “non-activating”). To vary adhesive strength, we used 125 ICAM-1 molecules per μm2 (termed “high ICAM-1”) or 4 ICAM-1 molecules per μm2 (termed “low ICAM-1”). T-cells were allowed to spread on SLBs and were fixed after 5–10 (orange), 10 (green), or 10–15 (red) minutes post seeding. As a control, unlabeled T-cells were seeded on SLBs containing I-Ek/MCC labeled with AF647. The recorded localizations were filtered for zSMLM < 100 nm in order to exclude contributions from the upper surface of lamellipodia. (a) The variance of zSMLM per cell was plotted for different conditions. Arrows indicate the difference in variance to the control I-Ek/MCC data, numbers indicate the corresponding square root. (b) Mean zSMLM per cell was plotted for different conditions. Arrows indicate the difference with the control I-Ek/MCC data. To calculate the mean distance of TCR microclusters from the glass surface we considered only localizations within microclusters (termed “only MCs”) in experiments performed both at high and low densities of ICAM-1 (n = 15 cells per condition).
According to the kinetic segregation model, the axial dimension of the intercellular cleft determines the accessibility of the large phosphatase CD45. We hence quantified the absolute distance of the TCR from the SLB, when compared to fluorescently labeled SLB-anchored I-Ek/MCC. Generally, we observed similar trends as for the standard deviations: with increasing densities of I-Ek/MCC and ICAM-1 the TCR was observed to be closer to the SLB surface (Figure 5b). The separation varied between 18 nm at high densities of I-Ek/MCC and ICAM-1 up to 31 nm for scanning T-cells recorded at low densities of ICAM-1. When selecting only signals corresponding to TCR microclusters for our distance analysis, we observed the expected close contact between TCR and I-Ek/MCC with a calculated separation of 5 nm. The residual separation reflected in all likelihood the distance separating the dye site-specifically conjugated to the single chain antibody fragment and the dye coupled to the MCC peptide’s C-terminus as presented by I-Ek.6
Discussion
We applied here a novel 3D-SMLM method to map and analyze the position of the TCR within the immunological synapse at isotropic localization precision below 15 nm and put it in the context of IRM imaging. IRM contrast arises from interferences due to optical path-length differences between the beam reflected at the glass–water interface and beams reflected from surfaces within the sample. Recording 3D-SMLM localization maps is fundamentally different from IRM. Using supercritical angle detection combined with defocused imaging, our method essentially determines the three-dimensional position of all visible dye molecules. We employed here an AlexaF647-conjugated single chain antibody fragment that specifically recognizes the TCR β subunit.6 In previous studies, we have shown that labeling neither activates T-cells nor impedes specific antigen recognition.6 Of note, we used TIR excitation in order to confine imaging to the synapse region. While the evanescent field was narrow enough to prevent contributions from TCRs at the top of the T-cell, we could observe both the bottom and the top membranes of lamellipodia.
Indeed, there was in general a good qualitative agreement between the two imaging modalities. Substantial differences, however, arose when distal membrane regions contributed to the IRM contrast values. Then, contrast values did not unequivocally correspond to the axial membrane separation. This was especially observed in lamellipodia, where both the bottom and the top membrane contributed to the signal.
Two aspects of our study contain important immunological implications for understanding T-cell antigen recognition
i. What Is the Physical Reason for the Observation of TCR Microclusters?
In TIR excitation, fluorescent molecules close to the glass coverslip naturally contribute with higher brightness than distal molecules. This effect is further amplified by the collection of supercritical angle fluorescence when using high NA objectives.25 Taken together, what appears as a bright spot in diffraction-limited fluorescence microscopy could be the consequence of increased density or of increased brightness of the fluorophores. Given the close contact between TCR microclusters and the surface,12 researchers became concerned whether microclusters actually reflect TCR enrichment.
Three-dimensional SMLM provides ground truth information on the origin of apparent TCR microclusters in diffraction-limited TIR microscopy, as it allows for disentangling molecular enrichment from brightness changes. We observed only marginal contributions from single molecule brightness increase, as brightness was largely attributable to molecular enrichment. This finding is in accord with our observation of a rather smooth interface between the SLB and the T-cell membrane under activating conditions, so that there are globally only minor variations in the single molecule’s z-distance in comparison to the TIR penetration depth.
ii. Does the Observed Cleft Size Support CD45 Segregation?
A prominent model for TCR triggering involves the balance in the activities of the kinase lck and the phosphatase CD45 for ITAM phosphorylation. In this kinetic segregation model, proteins with bulky extracellular domains, such as the large phosphatase CD45, are proposed to be segregated locally from the comparably short pMHC-TCR complexes.4 Although there are ample reports that would be consistent with this model,34−38 the community has not reached a consensus yet.39−41
One difficulty has been the precise measurement of the cleft size between the T-cell surface and the opposing membrane. Given the dimension of the TCR-pMHC-CD4 ternary complex of 10 nm42 and our apparent distance measurements between TCR and I-Ek/MCC within TCR microclusters of 5 nm, the obtained distance undervalues the cleft size by 5 nm. Upon correcting for this effect, an average cleft size within the whole synapse ranging between 23 and 36 nm for the conditions shown in Figure 5b can be estimated. In addition to the average cleft size, however, distance fluctuations within the synapse were of the same order (Figure 5a), indicating the presence of multiple contact sites between the two membranes. Assuming an axial length of CD45R0, the smallest CD45 isoform of approximately 22 nm,34 our measurements hence indicate the existence of numerous membrane contact sites that would be too narrow to host CD45, both for resting and activating conditions. In particular, an average cleft size of 30 nm together with an average surface roughness of 29 nm, as observed for scanning T-cells at high ICAM-1 densities, render the presence of multiple CD45 exclusion zones likely. Similar data were recently reported for the tips of microvilli which showed segregation of the TCR and CD45 prior to T-cell activation.22 Given that none of these scenarios promoted T-cell activation, it is possible that the ITAMs of these segregated TCRs are not accessible to kinases.43 In addition, TCR-CD45 segregation may be too transient to trigger stable phosphorylation of TCR ITAMs. Kinetic data about protein mobility in conjunction with the superresolution images would be needed in order to obtain a quantitative understanding of these key aspects of the TCR triggering process.
Acknowledgments
The study was supported by the Austrian Science Fund (FWF) (P 30214-N36, P 32105-N28), by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement (721358), and by the Wellcome Trust 100262Z/12/Z and the Kennedy Trust for Rheumatology Research.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c03160.
Additional supporting Figures 1–6; materials and methods including a description of the T-cell imaging experiment using supported lipid bilayers, an animal model and ethical compliance statement, as well as a description of the microscopy system, the performed data analysis, and the simulations; supporting gallery (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hu J.; Lipowsky R.; Weikl T. R. Binding constants of membrane-anchored receptors and ligands depend strongly on the nanoscale roughness of membranes. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (38), 15283–8. 10.1073/pnas.1305766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M. L.; Golan D. E.; Zhu D. M.; Miller J. M.; Meier W.; Davies E. A.; van der Merwe P. A. Low affinity interaction of human or rat T cell adhesion molecule CD2 with its ligand aligns adhering membranes to achieve high physiological affinity. J. Biol. Chem. 1997, 272 (49), 30889–98. 10.1074/jbc.272.49.30889. [DOI] [PubMed] [Google Scholar]
- Cartwright A. N. R.; Griggs J.; Davis D. M. The immune synapse clears and excludes molecules above a size threshold. Nat. Commun. 2014, 5 (1), 5479. 10.1038/ncomms6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. J.; van der Merwe P. A. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 2006, 7 (8), 803–9. 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- Grakoui A.; Bromley S. K.; Sumen C.; Davis M. M.; Shaw A. S.; Allen P. M.; Dustin M. L. The immunological synapse: a molecular machine controlling T cell activation. Science 1999, 285 (5425), 221–7. 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Huppa J. B.; Axmann M.; Mortelmaier M. A.; Lillemeier B. F.; Newell E. W.; Brameshuber M.; Klein L. O.; Schütz G. J.; Davis M. M. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 2010, 463 (7283), 963–967. 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves J. T.; Dustin M. L. Supported planar bilayers in studies on immune cell adhesion and communication. J. Immunol. Methods 2003, 278 (1–2), 19–32. 10.1016/S0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- Curtis A. S. G. The Mechanism of Adhesion of Cells to Glass: A Study by Interference Reflection Microscopy. J. Cell Biol. 1964, 20 (2), 199–215. 10.1083/jcb.20.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limozin L.; Sengupta K. Quantitative Reflection Interference Contrast Microscopy (RICM) in Soft Matter and Cell Adhesion. ChemPhysChem 2009, 10 (16), 2752–2768. 10.1002/cphc.200900601. [DOI] [PubMed] [Google Scholar]
- Smith A.-S.; Sengupta K.; Goennenwein S.; Seifert U.; Sackmann E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (19), 6906–6911. 10.1073/pnas.0801706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L.; Sherman E.; Barr V. A.; Samelson L. E. Imaging techniques for assaying lymphocyte activation in action. Nat. Rev. Immunol. 2011, 11 (1), 21–33. 10.1038/nri2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai E.; Marchuk K.; Beemiller P.; Beppler C.; Rubashkin M. G.; Weaver V. M.; Gérard A.; Liu T.-L.; Chen B.-C.; Betzig E.; Bartumeus F.; Krummel M. F. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science 2017, 356 (6338), eaal3118 10.1126/science.aal3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D.; Burghardt T. P.; Thompson N. L. Total internal reflection fluorescence. Annu. Rev. Biophys. Bioeng. 1984, 13, 247–68. 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Tane A.; Saito T. Dynamic Regulation of TCR-Microclusters and the Microsynapse for T Cell Activation. Front. Immunol. 2016, 7, 255. 10.3389/fimmu.2016.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T.; Sakata-Sogawa K.; Kobayashi W.; Hiroshima M.; Hashimoto-Tane A.; Tokunaga M.; Dustin M. L.; Saito T. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 2005, 6 (12), 1253–1262. 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- Varma R.; Campi G.; Yokosuka T.; Saito T.; Dustin M. L. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 2006, 25 (1), 117–27. 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossy J.; Owen D. M.; Williamson D. J.; Yang Z.; Gaus K. Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat. Immunol. 2013, 14, 82–89. 10.1038/ni.2488. [DOI] [PubMed] [Google Scholar]
- Pageon S. V.; Tabarin T.; Yamamoto Y.; Ma Y.; Nicovich P. R.; Bridgeman J. S.; Cohnen A.; Benzing C.; Gao Y.; Crowther M. D.; Tungatt K.; Dolton G.; Sewell A. K.; Price D. A.; Acuto O.; Parton R. G.; Gooding J. J.; Rossy J.; Rossjohn J.; Gaus K. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (37), E5454–63. 10.1073/pnas.1607436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossboth B.; Arnold A. M.; Ta H.; Platzer R.; Kellner F.; Huppa J. B.; Brameshuber M.; Baumgart F.; Schütz G. J. TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat. Immunol. 2018, 19 (8), 821–827. 10.1038/s41590-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncelli S.; Griffié J.; Williamson D. J.; Bibby J.; Bray C.; Zamoyska R.; Cope A. P.; Owen D. M. Multi-color Molecular Visualization of Signaling Proteins Reveals How C-Terminal Src Kinase Nanoclusters Regulate T Cell Receptor Activation. Cell Rep. 2020, 33 (12), 108523. 10.1016/j.celrep.2020.108523. [DOI] [PubMed] [Google Scholar]
- Jung Y.; Riven I.; Feigelson S. W.; Kartvelishvily E.; Tohya K.; Miyasaka M.; Alon R.; Haran G. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (40), E5916–E5924. 10.1073/pnas.1605399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.; Wen L.; Altman A.; Ley K. CD45 pre-exclusion from the tips of T cell microvilli prior to antigen recognition. Nat. Commun. 2021, 12 (1), 3872. 10.1038/s41467-021-23792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M.; Heilemann M. Single-Molecule Localization Microscopy in Eukaryotes. Chem. Rev. 2017, 117 (11), 7478–7509. 10.1021/acs.chemrev.6b00667. [DOI] [PubMed] [Google Scholar]
- Hajj B.; El Beheiry M.; Izeddin I.; Darzacq X.; Dahan M. Accessing the third dimension in localization-based super-resolution microscopy. Phys. Chem. Chem. Phys. 2014, 16 (31), 16340–16348. 10.1039/C4CP01380H. [DOI] [PubMed] [Google Scholar]
- Oheim M.; Salomon A.; Brunstein M. Supercritical Angle Fluorescence Microscopy and Spectroscopy. Biophys. J. 2020, 118 (10), 2339–2348. 10.1016/j.bpj.2020.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourg N.; Mayet C.; Dupuis G.; Barroca T.; Bon P.; Lécart S.; Fort E.; Lévêque-Fort S. Direct optical nanoscopy with axially localized detection. Nat. Photonics 2015, 9 (9), 587–593. 10.1038/nphoton.2015.132. [DOI] [Google Scholar]
- Zelger P.; Bodner L.; Velas L.; Schütz G. J.; Jesacher A. Defocused imaging exploits supercritical-angle fluorescence emission for precise axial single molecule localization microscopy. Biomed. Opt. Express 2020, 11 (2), 775–790. 10.1364/BOE.375678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober R. J.; Ram S.; Ward E. S. Localization accuracy in single-molecule microscopy. Biophys. J. 2004, 86 (2), 1185–200. 10.1016/S0006-3495(04)74193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmeier J.; Platzer R.; Eklund A. S.; Schlichthaerle T.; Karner A.; Motsch V.; Schneider M. C.; Kurz E.; Bamieh V.; Brameshuber M.; Preiner J.; Jungmann R.; Stockinger H.; Schütz G. J.; Huppa J. B.; Sevcsik E. DNA origami demonstrate the unique stimulatory power of single pMHCs as T cell antigens. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (4), e2016857118 10.1073/pnas.2016857118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham V. C.; Krishnamurthi V.; Taylor D. L.; Lanni F. The Actin-Based Nanomachine at the Leading Edge of Migrating Cells. Biophys. J. 1999, 77 (3), 1721–1732. 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Tane A.; Yokosuka T.; Sakata-Sogawa K.; Sakuma M.; Ishihara C.; Tokunaga M.; Saito T. Dynein-Driven Transport of T Cell Receptor Microclusters Regulates Immune Synapse Formation and T Cell Activation. Immunity 2011, 34 (6), 919–931. 10.1016/j.immuni.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Onnis A.; Baldari C. T. Orchestration of Immunological Synapse Assembly by Vesicular Trafficking. Front. Cell Dev. Biol. 2019, 7, 110. 10.3389/fcell.2019.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K.; Llodra J.; Roth E. W.; Tsai J.; Gordo S.; Wucherpfennig K. W.; Kam L. C.; Stokes D. L.; Dustin M. L. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature 2014, 507, 118–123. 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang V. T.; Fernandes R. A.; Ganzinger K. A.; Lee S. F.; Siebold C.; McColl J.; Jonsson P.; Palayret M.; Harlos K.; Coles C. H.; Jones E. Y.; Lui Y.; Huang E.; Gilbert R. J. C.; Klenerman D.; Aricescu A. R.; Davis S. J. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 2016, 17, 574–582. 10.1038/ni.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. M.; Ponjavic A.; Fritzsche M.; Fernandes R. A.; de la Serna J. B.; Wilcock M. J.; Schneider F.; Urbančič I.; McColl J.; Anzilotti C.; Ganzinger K. A.; Aßmann M.; Depoil D.; Cornall R. J.; Dustin M. L.; Klenerman D.; Davis S. J.; Eggeling C.; Lee S. F. Capturing resting T cells: the perils of PLL. Nat. Immunol. 2018, 19, 203–205. 10.1038/s41590-018-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K.; Wiseman D.; Brown M. H.; Gould K.; van der Merwe P. A. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 2005, 436 (7050), 578–82. 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- Taylor M. J.; Husain K.; Gartner Z. J.; Mayor S.; Vale R. D. A DNA-Based T Cell Receptor Reveals a Role for Receptor Clustering in Ligand Discrimination. Cell 2017, 169 (1), 108–119. 10.1016/j.cell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes R. A.; Ganzinger K. A.; Tzou J. C.; Jönsson P.; Lee S. F.; Palayret M.; Santos A. M.; Carr A. R.; Ponjavic A.; Chang V. T.; Macleod C.; Lagerholm B. C.; Lindsay A. E.; Dushek O.; Tilevik A.; Davis S. J.; Klenerman D. A cell topography-based mechanism for ligand discrimination by the T cell receptor. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (28), 14002–14010. 10.1073/pnas.1817255116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Aghbar M. A.; Chu Y.-S.; Chen B.-M.; Roffler S. R. High-Affinity Ligands Can Trigger T Cell Receptor Signaling Without CD45 Segregation. Front. Immunol. 2018, 9, 713. 10.3389/fimmu.2018.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney A. H.; Shvets A. A.; Lu W.; Griffante G.; Mollenauer M.; Horkova V.; Lo W.-L.; Yu S.; Stepanek O.; Chakraborty A. K.; Weiss A. CD45 functions as a signaling gatekeeper in T cells. Sci. Signaling 2019, 12 (604), eaaw8151 10.1126/scisignal.aaw8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H.; Muller J.; Depoil D.; Mayya V.; Sheetz M. P.; Dustin M. L.; Wind S. J. Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat. Nanotechnol. 2018, 13, 610–617. 10.1038/s41565-018-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y.; Wang X. X.; Mariuzza R. A. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (14), 5405–10. 10.1073/pnas.1118801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz A.-L.; Masi G.; Porciello N.; Cohnen A.; Cipria D.; Prakaash D.; Bálint Š.; Raggiaschi R.; Galgano D.; Cole D. K.; Lepore M.; Dushek O.; Dustin M. L.; Sansom M. S. P.; Kalli A. C.; Acuto O. Allosteric activation of T cell antigen receptor signaling by quaternary structure relaxation. Cell Rep. 2021, 36 (2), 109531. 10.1016/j.celrep.2021.109531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.