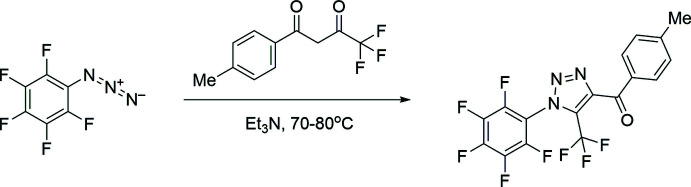
The title compound was obtained via the reaction of 1-azido-2,3,4,5,6-pentafluorobenzene with 4,4,4-trifluoro-1-(p-tolyl)butane-1,3-dione using triethylamine as a base catalyst and solvent. In the crystal, the molecules are linked by C—H⋯F and C—H⋯O hydrogen bonds as well as by aromatic π–π stacking interactions into a three-dimensional network.
Keywords: crystal structure; 1,2,3-triazole; Hirshfeld surface analysis
Abstract
The title compound, C17H7F8N3O, was obtained via the reaction of 1-azido-2,3,4,5,6-pentafluorobenzene with 4,4,4-trifluoro-1-(p-tolyl)butane-1,3-dione using triethylamine as a base catalyst and solvent. The dihedral angles between the pentafluorophenyl (A), triazole (B) and p-tolyl (C) rings are A/B = 62.3 (2), B/C = 43.9 (3) and A/C = 19.1 (3)°. In the crystal, the molecules are linked by C—H⋯F and C—H⋯O hydrogen bonds as well as by aromatic π–π stacking interactions into a three-dimensional network. To further analyse the intermolecular interactions, a Hirshfeld surface analysis was performed.
Chemical context
Compounds with perfluoroaromatic motifs are of interest for the design of fluorescence materials, including their application in optoelectronic devices (Funabiki et al., 2021 ▸; Feng et al., 2021 ▸; Moseev et al., 2019 ▸; Kandhadi et al., 2018 ▸; Lukeš et al., 2016 ▸; Wang et al., 2013 ▸; Matsui et al., 2008 ▸). For instance, the perfluorobiphenyl moiety was used as an electron acceptor for new donor–acceptor compounds with thermally activated delayed fluorescence (TADF) applied for the fabrication of TADF-based OLEDs (Danyliv et al., 2021 ▸; Hladka et al., 2018 ▸). On the other hand, 1,2,3-triazoles, as a result of their electron-accepting properties, are widely used in the design of organic phosphors (Gavlik et al., 2017 ▸; Fernández-Hernández et al., 2013 ▸; Tomkute-Luksiene et al., 2013 ▸; Ichikawa et al., 2011 ▸). Recently, a series of 3,6-bis(4-triazolyl)pyridazines equipped with terminal phenyl substituents with varying degrees of fluorination were synthesized and proposed to be used as electron-transporting/hole-blocking materials in organic electronics (Birkenfelder et al., 2017 ▸). In view of this, we decided to combine these two fragments in order to construct new molecular scaffolds of compounds that have the potential for use in optoelectronic devices.
Despite the prospects of using 1-(perfluorophenyl)-1H-1,2,3-triazole in the creation of phosphor materials, the paths of their synthesis are poorly studied. It is known that azides are convenient precursors of 1,2,3-triazoles. A literature survey showed limited data on the reaction of perfluorophenylazide in the synthesis of 1,2,3-triazoles. The reactions of such azides with acetylenes, which occur as a 1,3-dipolar cycloaddition, are primarily studied. For example, perfluorophenylazide was studied in the copper-catalysed azide–alkyne cycloaddition (CuAAC) click reaction with propargyl alcohol (Lavoie et al., 2017 ▸), 5-chloropent-1-yne and 6-chlorohex-1-yne (Berry et al., 2014 ▸), trimethyl[(perfluorophenyl)ethynyl]silane (Lu et al., 2012 ▸), iodoethynylarenes (Maugeri et al., 2016 ▸), 4-ethynylphospholo[3,2-b:4,5-b′]dithiophene 4-oxide (He, Zhang et al., 2013 ▸), [4-(iodoethynyl)phenyl]diphenylphosphine oxide (Maugeri et al., 2017 ▸), 2-ethynylpyridine (Liu et al., 2011 ▸), bis-alkynes (Milo et al., 2015 ▸) and 2,8-diethynyl-5-phenyl-4H-phosphepino[4,3-b:5,6-b′]dithiophene-4,6(5H)-dione (He, Borau-Garcia et al., 2013 ▸). The CuAAC reaction of perfluorophenylazide was used for the synthesis and bioactivity of phthalimide analogues as potential drugs to treat schistosomiasis (Singh et al., 2020 ▸) and for identification of sialoside analogues for siglec-based cell targeting (Rillahan et al., 2012 ▸). Moreover, the 1,3-dipolar cycloaddition of perfluorinated aryl azides with enamines and strained dipolarophiles has been studied (Xie et al., 2015 ▸). Additionally, non-catalytic Huisgen (3 + 2) cycloaddition of perfluorophenylazide with ethyl propiolate and a one-pot tandem Sonogashira cross-coupling/CuAAC reaction were studied (Kloss et al.. 2011 ▸). Conversely, for the synthesis of fully substituted 1,2,3-triazoles, Dimroth-type reactions are the most convenient. However, there is only one example of base-promoted cyclization of perfluorophenylazide with methylene active ketones (Dimroth-type reaction) in the triazole synthesis. Thus, by the reaction of perfluorophenylazide with acetylacetone in CHCl3 under Et3N and DBU catalysis, 1,2,3-triazoles were formed in 57% yield (Shafran et al., 2019 ▸). It should be noted that the classical conditions of the Dimroth reaction are MeONa/MeOH (Krivopalov et al., 2005 ▸). Such conditions are suitable for the rapid formation of polyheterocyclic 1,2,3-triazole derivatives via a domino reaction (Pokhodylo & Shyyka, 2017 ▸p; Pokhodylo et al., 2014 ▸), but for reagents with labile functional groups (Pokhodylo et al., 2018 ▸, 2020 ▸) or to avoid concurrent Regitz diazotransfer reaction (Pokhodylo & Obushak, 2019 ▸), mild bases such as K2CO3 (Pokhodylo et al., 2017 ▸) or organic bases (Et3N, DBU, pyrrolidine) are more suitable (Blastik et al., 2018 ▸; Ramachary et al., 2008 ▸; Danence et al., 2011 ▸). Furthermore, it has been shown that mild bases Et3N could be used for regioselective introduction of strongly electron-withdrawing groups such as trifluoroiodomethyl (CF3) in the 1,2,3-triazole ring in the reaction with asymmetric 1,3-diketones (Rozin et al., 2012 ▸).
Taking into account the above facts, in this work, the title compound, (I), was obtained and its crystal structure determined.

Structural commentary
The title compound crystallizes in the non-centrosymmetric space group P212121, with one molecule in the asymmetric unit. As shown in Fig. 1 ▸, it is constructed from three aromatic rings (C10–C15 4-methylphenyl, C1–C6 pentafluorophenyl and C7/C8/N1/N2/N3 triazole rings). The pentafluorophenyl ring and the heterocyclic ring are twisted relative to each other by 62.3 (2)° because of the significant steric hindrance of the trifluoromethyl group attached to C7. This dihedral angle is significantly smaller than the angle of 87.1° between the 4-nitrophenyl and triazole rings in the structure of 1-[5-methyl-1-(4-nitrophenyl)-1H-1,2,3-triazol-4-yl]ethanone (VI) (Vinutha et al., 2013 ▸) but considerably larger than the analogous angle between aromatic rings in the structures of 3-(4-fluorophenyl)-1-[1-(4-fluorophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]prop-2-en-1-one (39.6°; El-Hiti et al., 2018 ▸), (4-methylphenyl)(5-methyl-1-phenyl-1H-1,2,3-triazol-4-yl)methanone (44.5°; Li et al., 2014 ▸), 1-[1-(4-chlorophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]ethanone (45.6°) and 1-[1-(4-bromophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]ethanone (47.1°) (Zeghada et al., 2011 ▸). The carbonyl group of the title compound is not in the plane of the adjacent aromatic rings: the C7—C8—C9—O1 and C15—C10—C9—O1 torsion angles are −25.4 (9) and −16.8 (9)°, respectively]. The 4-methylphenyl and triazole rings are twisted relative to each other by 43.9 (2)° and the 4-methylphenyl and pentafluorophenyl rings by 19.1 (3)°.
Figure 1.
The molecular structure of (I) with displacement ellipsoids drawn at the 50% probability level.
Supramolecular features
As shown in Fig. 2 ▸ and listed in Table 1 ▸, the crystal structure of (I) features several weak intermolecular interactions. The hydrogen atoms of the 4-methylphenyl ring are involved in C—H⋯F hydrogen bonding with the trifluoromethyl substituents of adjacent molecules, while a hydrogen atom of the methyl group forms a C—H⋯O hydrogen bond with the carbonyl O atom of another adjacent molecule. The 4-methylphenyl and pentafluorophenyl rings of adjacent molecules are also involved into face-to-face π–π stacking interaction with a centroid–centroid separation of 3.783 (6) Å, while at the same time the triazole rings are involved into edge-to-face aromatic interactions at 3.218 (6) Å. The molecules are linked by the above-mentioned intermolecular interactions into a three-dimensional network (Fig. 3 ▸).
Figure 2.
The hydrogen bonding of molecules in (I). Hydrogen bonds are shown as dashed lines. The symmetry codes are as in Table 1 ▸.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C11—H11⋯F3i | 0.95 | 2.49 | 3.155 (5) | 127 |
| C15—H15⋯F3ii | 0.95 | 2.61 | 3.463 (6) | 149 |
| C16—H16A⋯F2iii | 0.98 | 2.57 | 3.054 (6) | 111 |
| C16—H16A⋯O1iii | 0.98 | 2.54 | 3.505 (6) | 167 |
Symmetry codes: (i) x+{\script{1\over 2}}, -y+{\script{1\over 2}}, -z+1; (ii) -x+{\script{1\over 2}}, -y+1, z+{\script{1\over 2}}; (iii) -x+1, y-{\script{1\over 2}}, -z+{\script{3\over 2}}.
Figure 3.
A view along the b-axis direction of the crystal packing of (I).
Hirshfeld surface analysis
Hirshfeld surface analysis was used to analyse the various intermolecular interactions in (I), through mapping the normalized contact distance (d norm) using CrystalExplorer (Turner et al., 2017 ▸; Spackman & Jayatilaka, 2009 ▸). The most prominent interactions (bifurcated interactions of atom H16A of the methyl group with the carbonyl group O atom and the fluorine atom of the trifluoromethyl substituent of neighbouring molecules, as well as the F⋯F interaction between neighbouring pentafluorophenyl rings) can be seen in the Hirshfeld surface plot as red areas (Fig. 4 ▸). Fingerprint plots were produced to show the intermolecular surface bond distances with the regions highlighted for F⋯H/H⋯F and F⋯F contacts interactions (Fig. 4 ▸). The contribution to the surface area for such contacts are 36.6% and 13.6%, respectively. The contribution to the surface area for O⋯H/H⋯O and H⋯H contacts are 4.6% and 5.7%, respectively.
Figure 4.
(a) Hirshfeld surface for (I) mapped with d norm over the range −0.12 to 1.53 a.u. showing C—H⋯O and C—H⋯F hydrogen-bonded contacts as well as F⋯F contacts. Fingerprint plots resolved into (b) F⋯H/H⋯F and (c) F⋯F contacts. Neighbouring molecules associated with close contacts are also shown.
Database survey
The most closely related compounds, containing a similar 1-aryl-1H-1,2,3-triazole-4-carbonyl skeleton to the title compound but with different substituents on the carbonyl group are: 2,2′-(quinoxaline-2,3-diyldisulfanediyl)bis{1-[5-methyl-1-(4-methylphenyl)-1H-1,2,3-triazol-4-yl]ethan-1-one} (II) [Cambridge Structural Database (Version 2021.1; Groom et al., 2016 ▸) refcode ETUVEX; Mohamed et al., 2021 ▸], 4-(4-acetyl-5-methyl-1H-1,2,3-triazol-1-yl)benzonitrile (III) (SILBOH; Zukerman-Schpector et al., 2018 ▸), 1-[5-methyl-1-(4-methylphenyl)-1H-1,2,3-triazol-4-yl]ethan-1-one (IV) (LEMSUU; El-Hiti et al., 2017 ▸), 3-(4-fluorophenyl)-1-[1-(4-fluorophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]prop-2-en-1-one (V) (MESTAI; El-Hiti et al., 2018 ▸), 1-[5-methyl-1-(4-nitrophenyl)-1H-1,2,3-triazol-4-yl]ethanone (VI) (QIRQOZ; Vinutha et al., 2013 ▸), 2-bromo-1-[1-(4-bromophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]ethanone (VII) (XODSAM; Bunev et al., 2014 ▸), (4-methylphenyl)(5-methyl-1-phenyl-1H-1,2,3-triazol-4-yl)methanone (VIII) (COCYAW; Li et al., 2014 ▸), (2E)-3-(4-fluorophenyl)-1-[5-methyl-1-(4-methylphenyl)-1H-1,2,3-triazol-4-yl]prop-2-en-1-one (IX) (IDITUM; Abdel-Wahab et al., 2013 ▸), 1-[1-(4-chlorophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]ethanone (X) (ISOBUO; Zeghada et al., 2011 ▸) and 1-[1-(4-bromophenyl)-5-methyl-1H-1,2,3-triazol-4-yl]ethanone (XI) (ISOCAV; Zeghada et al., 2011 ▸).
Compounds (V), (IX), (X) and (XI) crystallize in the triclinic crystal system in space group P
 . Compounds (II), (III) and (IV), (VIII) crystallize in the monoclinic crystal system with space groups P21/n and P21/c, respectively, while compound (VII) is found in the monoclinic crystal system, space group Pn. Compound (VI) crystallizes in the orthorhombic crystal system in non-centrosymmetric space group Pca21. Structures (V), (VI) and (VII) contain two crystallographically independent molecules. The aryl and triazole rings in (VI) are twisted relative to each other by 87.1 and 38.2° in the two crystallographically independent molecules. In compounds (III), (IV) and (IX), the analogous angles between the aromatic rings are 54.7, 50.1 and 51.8°, respectively.
. Compounds (II), (III) and (IV), (VIII) crystallize in the monoclinic crystal system with space groups P21/n and P21/c, respectively, while compound (VII) is found in the monoclinic crystal system, space group Pn. Compound (VI) crystallizes in the orthorhombic crystal system in non-centrosymmetric space group Pca21. Structures (V), (VI) and (VII) contain two crystallographically independent molecules. The aryl and triazole rings in (VI) are twisted relative to each other by 87.1 and 38.2° in the two crystallographically independent molecules. In compounds (III), (IV) and (IX), the analogous angles between the aromatic rings are 54.7, 50.1 and 51.8°, respectively.
Synthesis and crystallization
A number of experimental conditions described previously were investigated for the synthesis of the title compound (Shafran et al., 2019 ▸; Pokhodylo et al., 2017 ▸; Blastik et al., 2018 ▸; Rozin et al., 2012 ▸). However, it was possible to obtain the target product only in the case of the protocol proposed by Rozin et al. (2012 ▸). The synthesis scheme is shown in Fig. 5 ▸.
Figure 5.
Synthesis scheme for (I)
(4-Methylphenyl)[1-(pentafluorophenyl)-5-(trifluoromethyl)-1 H -1,2,3-triazol-4-yl]methanone: A mixture of the corresponding 4,4,4-trifluoro-1-(p-tolyl)butane-1,3-dione 230 mg (1.00 mmol), 1-azido-2,3,4,5,6-pentafluorobenzene 209 mg (1.00 mmol), and triethylamine (0.43 ml, 3.00 mmol) was heated at 343–353 K for 3 h. Volatiles were evaporated in vacuo and the residue was purified by column chromatography on silica gel using dichloromethane as an eluent. Colourless crystals were grown by slow evaporation of a dichloromethane solution, yield 21%; m.p. 391–394 K; 1H NMR (500 MHz, DMSO-d 6) δ 8.02 (d, J = 7.9 Hz, 2H, HAr-2,6), 7.44 (d, J = 7.8 Hz, 2H, HAr-3,5), 2.43 (s, 3H); 13C NMR (126 MHz, DMSO-d 6) δ 183.74 (CO), 145.73 (CTol-4), 145.57 (CTriazole-4), 143.61 (m), 141.68 (m), 138.84 (m), 136.79 (m), 132.61 (CTol-1), 130.67 (q, J = 41.7 Hz, CTriazole-5), 130.63 (2 × CTol-2,6), 129.49 (2 × CTol-3,5), 118.36 (q, 1 J C–F = 270.9 Hz, CF3), 109.59 (m), 21.37 (CH3); 19F NMR (376 MHz, DMSO-d 6) δ −58.46 (CF3), −146.39 (d, J = 21.5 Hz, 2 × F-2,6), −146.53 (t, J = 23.4 Hz, F-4), −159.61 (t, J = 23.3 Hz, 2 × F-3,5); MS, m/z = 422 (M + + 1). Calculated for C17H7F8N3O, (%): C, 48.47; H, 1.68; N, 9.98. Found (%): C, 48.55; H, 1.67; N, 9.91.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were positioned geometrically with C—H = 0.95–0.98 Å and refined as riding atoms. The constraint U iso(H) = 1.2U eq(carrier) or 1.5U eq(C-methyl carrier) was applied in all cases.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C17H7F8N3O |
| M r | 421.26 |
| Crystal system, space group | Orthorhombic, P212121 |
| Temperature (K) | 150 |
| a, b, c (Å) | 6.7605 (6), 15.065 (1), 16.0849 (9) |
| V (Å3) | 1638.2 (2) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.55 |
| Crystal size (mm) | 0.43 × 0.12 × 0.08 |
| Data collection | |
| Diffractometer | New Gemini, Dual, Cu at home/near, Atlas |
| Absorption correction | Analytical (CrysAlis PRO; Rigaku OD, 2021 ▸) |
| T min, T max | 0.320, 0.678 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 15249, 3187, 2311 |
| R int | 0.088 |
| (sin θ/λ)max (Å−1) | 0.618 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.047, 0.109, 1.03 |
| No. of reflections | 3187 |
| No. of parameters | 264 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.18, −0.18 |
| Absolute structure | Refined as an inversion twin. |
| Absolute structure parameter | 0.2 (3) |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989021010070/hb7985sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010070/hb7985Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021010070/hb7985Isup4.cml
NMR spectra. DOI: 10.1107/S2056989021010070/hb7985sup3.pdf
CCDC reference: 2112438
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C17H7F8N3O | Dx = 1.708 Mg m−3 |
| Mr = 421.26 | Cu Kα radiation, λ = 1.54184 Å |
| Orthorhombic, P212121 | Cell parameters from 2646 reflections |
| a = 6.7605 (6) Å | θ = 4.0–72.2° |
| b = 15.065 (1) Å | µ = 1.55 mm−1 |
| c = 16.0849 (9) Å | T = 150 K |
| V = 1638.2 (2) Å3 | Irregular, colourless |
| Z = 4 | 0.43 × 0.12 × 0.08 mm |
| F(000) = 840 |
Data collection
| New Gemini, Dual, Cu at home/near, Atlas diffractometer | 2311 reflections with I > 2σ(I) |
| Detector resolution: 10.6426 pixels mm-1 | Rint = 0.088 |
| ω scans | θmax = 72.3°, θmin = 4.0° |
| Absorption correction: analytical (CrysalisPro; Rigaku OD, 2021) | h = −7→8 |
| Tmin = 0.320, Tmax = 0.678 | k = −18→18 |
| 15249 measured reflections | l = −19→19 |
| 3187 independent reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.047 | w = 1/[σ2(Fo2) + (0.031P)2 + 0.1487P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.109 | (Δ/σ)max < 0.001 |
| S = 1.03 | Δρmax = 0.18 e Å−3 |
| 3187 reflections | Δρmin = −0.18 e Å−3 |
| 264 parameters | Absolute structure: Refined as an inversion twin. |
| 0 restraints | Absolute structure parameter: 0.2 (3) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component inversion twin. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.0932 (5) | 0.4231 (2) | 0.51007 (19) | 0.0563 (9) | |
| F2 | 0.2755 (6) | 0.52826 (18) | 0.4634 (2) | 0.0575 (10) | |
| F3 | 0.1757 (5) | 0.42585 (19) | 0.38129 (17) | 0.0456 (8) | |
| F4 | 0.5987 (6) | 0.50612 (18) | 0.33789 (17) | 0.0565 (9) | |
| F5 | 0.6160 (6) | 0.5157 (2) | 0.16979 (18) | 0.0609 (10) | |
| F6 | 0.5614 (6) | 0.3674 (3) | 0.07762 (16) | 0.0612 (10) | |
| F7 | 0.4985 (6) | 0.2089 (2) | 0.15264 (18) | 0.0566 (9) | |
| F8 | 0.4847 (5) | 0.19857 (17) | 0.32018 (16) | 0.0456 (8) | |
| O1 | 0.3633 (7) | 0.4664 (2) | 0.6435 (2) | 0.0496 (10) | |
| N1 | 0.5368 (7) | 0.3474 (2) | 0.4207 (2) | 0.0351 (10) | |
| N2 | 0.6809 (7) | 0.2985 (3) | 0.4582 (2) | 0.0418 (11) | |
| N3 | 0.6565 (8) | 0.3072 (3) | 0.5387 (2) | 0.0410 (10) | |
| C1 | 0.5367 (8) | 0.3525 (3) | 0.3316 (3) | 0.0339 (11) | |
| C2 | 0.5726 (8) | 0.4330 (3) | 0.2930 (3) | 0.0396 (12) | |
| C3 | 0.5793 (9) | 0.4382 (4) | 0.2066 (3) | 0.0451 (13) | |
| C4 | 0.5531 (9) | 0.3623 (4) | 0.1611 (3) | 0.0430 (13) | |
| C5 | 0.5215 (9) | 0.2822 (3) | 0.1983 (3) | 0.0405 (12) | |
| C6 | 0.5130 (8) | 0.2771 (3) | 0.2847 (3) | 0.0357 (11) | |
| C7 | 0.4210 (8) | 0.3878 (3) | 0.4781 (3) | 0.0319 (11) | |
| C8 | 0.4981 (8) | 0.3615 (3) | 0.5531 (3) | 0.0337 (11) | |
| C9 | 0.4375 (9) | 0.3934 (3) | 0.6379 (3) | 0.0385 (13) | |
| C10 | 0.4732 (8) | 0.3363 (3) | 0.7117 (3) | 0.0345 (11) | |
| C11 | 0.5049 (8) | 0.2451 (3) | 0.7063 (3) | 0.0340 (10) | |
| H11 | 0.509418 | 0.217046 | 0.653465 | 0.041* | |
| C12 | 0.5297 (8) | 0.1955 (3) | 0.7776 (3) | 0.0360 (11) | |
| H12 | 0.549256 | 0.133197 | 0.773331 | 0.043* | |
| C13 | 0.5267 (8) | 0.2352 (3) | 0.8560 (3) | 0.0374 (12) | |
| C14 | 0.4964 (10) | 0.3264 (3) | 0.8609 (3) | 0.0454 (14) | |
| H14 | 0.494442 | 0.354584 | 0.913774 | 0.054* | |
| C15 | 0.4693 (9) | 0.3765 (3) | 0.7897 (3) | 0.0438 (14) | |
| H15 | 0.447771 | 0.438645 | 0.794080 | 0.053* | |
| C16 | 0.5547 (10) | 0.1808 (3) | 0.9339 (3) | 0.0493 (16) | |
| H16A | 0.562191 | 0.117776 | 0.919256 | 0.074* | |
| H16B | 0.442677 | 0.190607 | 0.971434 | 0.074* | |
| H16C | 0.677506 | 0.198827 | 0.961531 | 0.074* | |
| C17 | 0.2417 (9) | 0.4415 (3) | 0.4588 (3) | 0.0405 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.045 (2) | 0.0728 (19) | 0.0511 (16) | 0.0166 (18) | 0.0065 (16) | 0.0039 (15) |
| F2 | 0.077 (3) | 0.0330 (13) | 0.0620 (18) | 0.0102 (16) | −0.0228 (18) | −0.0058 (14) |
| F3 | 0.049 (2) | 0.0459 (15) | 0.0416 (14) | 0.0086 (15) | −0.0134 (14) | −0.0077 (12) |
| F4 | 0.084 (3) | 0.0395 (15) | 0.0464 (15) | −0.0048 (17) | 0.0019 (16) | 0.0027 (13) |
| F5 | 0.073 (3) | 0.0606 (18) | 0.0492 (16) | −0.001 (2) | 0.0050 (17) | 0.0228 (15) |
| F6 | 0.061 (3) | 0.094 (2) | 0.0285 (13) | 0.005 (2) | −0.0016 (15) | 0.0040 (14) |
| F7 | 0.058 (2) | 0.0650 (18) | 0.0470 (15) | −0.001 (2) | −0.0009 (17) | −0.0199 (14) |
| F8 | 0.050 (2) | 0.0372 (13) | 0.0497 (15) | −0.0019 (16) | 0.0064 (15) | −0.0027 (11) |
| O1 | 0.072 (3) | 0.0354 (17) | 0.0412 (18) | 0.0107 (19) | 0.0010 (19) | 0.0006 (14) |
| N1 | 0.040 (3) | 0.0344 (18) | 0.0312 (17) | 0.005 (2) | −0.0025 (19) | 0.0008 (15) |
| N2 | 0.042 (3) | 0.049 (2) | 0.0345 (19) | 0.008 (2) | 0.0005 (19) | 0.0094 (19) |
| N3 | 0.042 (3) | 0.050 (2) | 0.0316 (19) | 0.004 (2) | 0.0010 (19) | 0.0040 (17) |
| C1 | 0.031 (3) | 0.041 (2) | 0.030 (2) | 0.004 (2) | −0.002 (2) | 0.0003 (18) |
| C2 | 0.042 (3) | 0.038 (2) | 0.039 (2) | 0.004 (3) | −0.001 (2) | 0.000 (2) |
| C3 | 0.046 (3) | 0.053 (3) | 0.037 (2) | 0.001 (3) | 0.003 (2) | 0.016 (2) |
| C4 | 0.037 (4) | 0.067 (3) | 0.026 (2) | 0.008 (3) | 0.002 (2) | 0.002 (2) |
| C5 | 0.030 (3) | 0.053 (3) | 0.038 (2) | 0.002 (3) | −0.003 (2) | −0.008 (2) |
| C6 | 0.026 (3) | 0.039 (2) | 0.041 (2) | 0.002 (2) | 0.002 (2) | 0.0021 (19) |
| C7 | 0.037 (3) | 0.0276 (19) | 0.031 (2) | −0.002 (2) | 0.000 (2) | 0.0012 (17) |
| C8 | 0.032 (3) | 0.034 (2) | 0.035 (2) | −0.002 (2) | 0.000 (2) | 0.0018 (17) |
| C9 | 0.046 (4) | 0.037 (2) | 0.032 (2) | 0.002 (2) | −0.003 (2) | 0.0021 (19) |
| C10 | 0.036 (3) | 0.039 (2) | 0.0285 (19) | 0.001 (2) | 0.000 (2) | 0.0009 (17) |
| C11 | 0.030 (3) | 0.039 (2) | 0.033 (2) | 0.002 (2) | 0.000 (2) | −0.0025 (17) |
| C12 | 0.034 (3) | 0.038 (2) | 0.036 (2) | 0.003 (2) | 0.001 (2) | 0.0006 (18) |
| C13 | 0.035 (3) | 0.045 (2) | 0.032 (2) | 0.009 (2) | −0.004 (2) | 0.0030 (18) |
| C14 | 0.058 (4) | 0.047 (3) | 0.031 (2) | 0.011 (3) | −0.001 (3) | −0.0058 (19) |
| C15 | 0.049 (4) | 0.045 (3) | 0.037 (2) | 0.012 (3) | −0.007 (3) | −0.005 (2) |
| C16 | 0.061 (5) | 0.052 (3) | 0.036 (2) | 0.014 (3) | −0.001 (3) | 0.005 (2) |
| C17 | 0.050 (4) | 0.038 (2) | 0.034 (2) | 0.007 (3) | −0.006 (2) | −0.004 (2) |
Geometric parameters (Å, º)
| F1—C17 | 1.328 (7) | C5—C6 | 1.393 (6) |
| F2—C17 | 1.329 (6) | C7—C8 | 1.373 (6) |
| F3—C17 | 1.345 (6) | C7—C17 | 1.490 (8) |
| F4—C2 | 1.328 (6) | C8—C9 | 1.502 (6) |
| F5—C3 | 1.332 (6) | C9—C10 | 1.485 (6) |
| F6—C4 | 1.346 (5) | C10—C11 | 1.393 (6) |
| F7—C5 | 1.336 (6) | C10—C15 | 1.394 (6) |
| F8—C6 | 1.328 (5) | C11—H11 | 0.9500 |
| O1—C9 | 1.213 (6) | C11—C12 | 1.380 (6) |
| N1—N2 | 1.362 (6) | C12—H12 | 0.9500 |
| N1—C1 | 1.435 (5) | C12—C13 | 1.395 (6) |
| N1—C7 | 1.355 (6) | C13—C14 | 1.392 (7) |
| N2—N3 | 1.313 (6) | C13—C16 | 1.508 (6) |
| N3—C8 | 1.368 (7) | C14—H14 | 0.9500 |
| C1—C2 | 1.384 (7) | C14—C15 | 1.384 (7) |
| C1—C6 | 1.373 (7) | C15—H15 | 0.9500 |
| C2—C3 | 1.393 (7) | C16—H16A | 0.9800 |
| C3—C4 | 1.369 (8) | C16—H16B | 0.9800 |
| C4—C5 | 1.363 (8) | C16—H16C | 0.9800 |
| N2—N1—C1 | 118.1 (4) | C10—C9—C8 | 119.7 (4) |
| C7—N1—N2 | 110.8 (4) | C11—C10—C9 | 123.1 (4) |
| C7—N1—C1 | 131.0 (4) | C11—C10—C15 | 119.1 (4) |
| N3—N2—N1 | 107.1 (4) | C15—C10—C9 | 117.7 (4) |
| N2—N3—C8 | 109.0 (4) | C10—C11—H11 | 119.9 |
| C2—C1—N1 | 119.6 (4) | C12—C11—C10 | 120.1 (4) |
| C6—C1—N1 | 120.4 (4) | C12—C11—H11 | 119.9 |
| C6—C1—C2 | 119.9 (4) | C11—C12—H12 | 119.4 |
| F4—C2—C1 | 120.4 (4) | C11—C12—C13 | 121.1 (4) |
| F4—C2—C3 | 119.4 (5) | C13—C12—H12 | 119.4 |
| C1—C2—C3 | 120.1 (5) | C12—C13—C16 | 121.0 (4) |
| F5—C3—C2 | 119.9 (5) | C14—C13—C12 | 118.5 (4) |
| F5—C3—C4 | 121.2 (4) | C14—C13—C16 | 120.5 (4) |
| C4—C3—C2 | 118.9 (5) | C13—C14—H14 | 119.6 |
| F6—C4—C3 | 118.7 (5) | C15—C14—C13 | 120.7 (4) |
| F6—C4—C5 | 119.7 (5) | C15—C14—H14 | 119.6 |
| C5—C4—C3 | 121.6 (4) | C10—C15—H15 | 119.8 |
| F7—C5—C4 | 120.6 (4) | C14—C15—C10 | 120.4 (4) |
| F7—C5—C6 | 119.8 (5) | C14—C15—H15 | 119.8 |
| C4—C5—C6 | 119.6 (5) | C13—C16—H16A | 109.5 |
| F8—C6—C1 | 121.1 (4) | C13—C16—H16B | 109.5 |
| F8—C6—C5 | 119.0 (4) | C13—C16—H16C | 109.5 |
| C1—C6—C5 | 119.9 (4) | H16A—C16—H16B | 109.5 |
| N1—C7—C8 | 104.5 (4) | H16A—C16—H16C | 109.5 |
| N1—C7—C17 | 124.9 (4) | H16B—C16—H16C | 109.5 |
| C8—C7—C17 | 130.4 (4) | F1—C17—F2 | 107.5 (4) |
| N3—C8—C7 | 108.7 (4) | F1—C17—F3 | 106.8 (5) |
| N3—C8—C9 | 123.9 (4) | F1—C17—C7 | 111.9 (4) |
| C7—C8—C9 | 127.0 (5) | F2—C17—F3 | 106.3 (4) |
| O1—C9—C8 | 118.0 (4) | F2—C17—C7 | 112.5 (5) |
| O1—C9—C10 | 122.2 (4) | F3—C17—C7 | 111.6 (4) |
| F4—C2—C3—F5 | 1.5 (9) | C2—C1—C6—F8 | 177.8 (5) |
| F4—C2—C3—C4 | 179.7 (5) | C2—C1—C6—C5 | −1.2 (9) |
| F5—C3—C4—F6 | −1.2 (9) | C2—C3—C4—F6 | −179.4 (5) |
| F5—C3—C4—C5 | 178.0 (6) | C2—C3—C4—C5 | −0.3 (10) |
| F6—C4—C5—F7 | −0.1 (9) | C3—C4—C5—F7 | −179.3 (6) |
| F6—C4—C5—C6 | 180.0 (5) | C3—C4—C5—C6 | 0.9 (10) |
| F7—C5—C6—F8 | 0.9 (9) | C4—C5—C6—F8 | −179.2 (5) |
| F7—C5—C6—C1 | −180.0 (5) | C4—C5—C6—C1 | −0.1 (9) |
| O1—C9—C10—C11 | 161.5 (6) | C6—C1—C2—F4 | −179.0 (5) |
| O1—C9—C10—C15 | −16.8 (9) | C6—C1—C2—C3 | 1.8 (9) |
| N1—N2—N3—C8 | −0.2 (6) | C7—N1—N2—N3 | 0.4 (6) |
| N1—C1—C2—F4 | −2.9 (9) | C7—N1—C1—C2 | 61.7 (8) |
| N1—C1—C2—C3 | 177.9 (5) | C7—N1—C1—C6 | −122.2 (6) |
| N1—C1—C6—F8 | 1.8 (9) | C7—C8—C9—O1 | −25.4 (9) |
| N1—C1—C6—C5 | −177.3 (5) | C7—C8—C9—C10 | 155.8 (5) |
| N1—C7—C8—N3 | 0.3 (5) | C8—C7—C17—F1 | −37.5 (7) |
| N1—C7—C8—C9 | 174.0 (5) | C8—C7—C17—F2 | 83.6 (7) |
| N1—C7—C17—F1 | 136.6 (5) | C8—C7—C17—F3 | −157.0 (5) |
| N1—C7—C17—F2 | −102.3 (5) | C8—C9—C10—C11 | −19.9 (9) |
| N1—C7—C17—F3 | 17.1 (7) | C8—C9—C10—C15 | 161.9 (5) |
| N2—N1—C1—C2 | −114.2 (6) | C9—C10—C11—C12 | −177.5 (5) |
| N2—N1—C1—C6 | 61.9 (7) | C9—C10—C15—C14 | 178.3 (6) |
| N2—N1—C7—C8 | −0.4 (5) | C10—C11—C12—C13 | −0.9 (9) |
| N2—N1—C7—C17 | −175.8 (4) | C11—C10—C15—C14 | 0.0 (9) |
| N2—N3—C8—C7 | −0.1 (6) | C11—C12—C13—C14 | 0.5 (9) |
| N2—N3—C8—C9 | −174.1 (5) | C11—C12—C13—C16 | −179.7 (6) |
| N3—C8—C9—O1 | 147.4 (5) | C12—C13—C14—C15 | 0.2 (10) |
| N3—C8—C9—C10 | −31.3 (8) | C13—C14—C15—C10 | −0.5 (10) |
| C1—N1—N2—N3 | 177.1 (4) | C15—C10—C11—C12 | 0.7 (9) |
| C1—N1—C7—C8 | −176.5 (5) | C16—C13—C14—C15 | −179.5 (6) |
| C1—N1—C7—C17 | 8.1 (8) | C17—C7—C8—N3 | 175.3 (5) |
| C1—C2—C3—F5 | −179.3 (5) | C17—C7—C8—C9 | −11.0 (9) |
| C1—C2—C3—C4 | −1.1 (10) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C11—H11···F3i | 0.95 | 2.49 | 3.155 (5) | 127 |
| C15—H15···F3ii | 0.95 | 2.61 | 3.463 (6) | 149 |
| C16—H16A···F2iii | 0.98 | 2.57 | 3.054 (6) | 111 |
| C16—H16A···O1iii | 0.98 | 2.54 | 3.505 (6) | 167 |
Symmetry codes: (i) x+1/2, −y+1/2, −z+1; (ii) −x+1/2, −y+1, z+1/2; (iii) −x+1, y−1/2, −z+3/2.
Funding Statement
This work was funded by Ministry of Education and Science of Ukraine.
References
- Abdel-Wahab, B. F., Mohamed, H. A., Ng, S. W. & Tiekink, E. R. T. (2013). Acta Cryst. E69, o638. [DOI] [PMC free article] [PubMed]
- Berry, M. T., Castrejon, D. & Hein, J. E. (2014). Org. Lett. 16, 3676–3679. [DOI] [PubMed]
- Birkenfelder, I., Gurke, J., Grubert, L., Hecht, S. & Schmidt, B. M. (2017). Chem. Asian J. 12, 3156–3161. [DOI] [PubMed]
- Blastik, Z. E., Klepetářová, B. & Beier, P. (2018). ChemistrySelect, 3, 7045–7048.
- Bunev, A. S., Troshina, M. A., Ostapenko, G. I., Pavlova, A. P. & Khrustalev, V. N. (2014). Acta Cryst. E70, o818. [DOI] [PMC free article] [PubMed]
- Danence, L. J. T., Gao, Y., Li, M., Huang, Y. & Wang, J. (2011). Chem. Eur. J. 17, 3584–3587. [DOI] [PubMed]
- Danyliv, I., Danyliv, Y., Lytvyn, R., Bezvikonnyi, O., Volyniuk, D., Simokaitiene, J., Ivaniuk, K., Tsiko, U., Tomkeviciene, A., Dabulienė, A., Skuodis, E., Stakhira, P. & Grazulevicius, J. V. (2021). Dyes Pigments, 193, 109493.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- El-Hiti, G. A., Abdel-Wahab, B. F., Alotaibi, M. H., Hegazy, A. S. & Kariuki, B. M. (2017). IUCrData, 2, x171782.
- El-Hiti, G. A., Abdel-Wahab, B. F., Alotaibi, M. H., Hegazy, A. S. & Kariuki, B. M. (2018). IUCrData, 3, x171841.
- Feng, Z., Chong, Y., Tang, S., Ruan, H., Fang, Y., Zhao, Y., Jiang, J. & Wang, X. (2021). Chin. J. Chem. 39, 1297–1302.
- Fernández-Hernández, J. M., Beltrán, J. I., Lemaur, V., Gálvez-López, M. D., Chien, C. H., Polo, F., Orselli, E., Fröhlich, R., Cornil, J. & De Cola, L. (2013). Inorg. Chem. 52, 1812–1824. [DOI] [PubMed]
- Funabiki, K., Yamada, K., Matsueda, H., Arisawa, Y., Agou, T., Kubota, Y., Inuzuka, T. & Wasada, H. (2021). Eur. J. Org. Chem. pp. 1344–1350.
- Gavlik, K. D., Sukhorukova, E. S., Shafran, Y. M., Slepukhin, P. A., Benassi, E. & Belskaya, N. P. (2017). Dyes Pigments, 136, 229–242.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- He, X., Borau-Garcia, J., Woo, A. Y., Trudel, S. & Baumgartner, T. (2013). J. Am. Chem. Soc. 135, 1137–1147. [DOI] [PubMed]
- He, X., Zhang, P., Lin, J. B., Huynh, H. V., Navarro Muñoz, S. E., Ling, C. C. & Baumgartner, T. (2013). Org. Lett. 15, 5322–5325. [DOI] [PubMed]
- Hladka, I., Volyniuk, D., Bezvikonnyi, O., Kinzhybalo, V., Bednarchuk, T. J., Danyliv, Y., Lytvyn, R., Lazauskas, A. & Grazulevicius, J. V. (2018). J. Mater. Chem. C. 6, 13179–13189.
- Ichikawa, M., Mochizuki, S., Jeon, H. G., Hayashi, S., Yokoyama, N. & Taniguchi, Y. (2011). J. Mater. Chem. 21, 11791–11799.
- Kandhadi, J., Yan, W. C., Cheng, F., Wang, H. & Liu, H. Y. (2018). New J. Chem. 42, 9987–9999.
- Kloss, F., Köhn, U., Jahn, B. O., Hager, M. D., Görls, H. & Schubert, U. S. (2011). Chem. Asian J. 6, 2816–2824. [DOI] [PubMed]
- Krivopalov, V. P. & Shkurko, O. P. (2005). Russ. Chem. Rev. 74, 339–379.
- Lavoie, K. D., Frauhiger, B. E., White, P. S. & Templeton, J. L. (2017). J. Mol. Catal. A Chem. 426, 474–489.
- Li, W., Du, Z., Huang, J., Jia, Q., Zhang, K. & Wang, J. (2014). Green Chem. 16, 3003–3006.
- Liu, S., Müller, P., Takase, M. K. & Swager, T. M. (2011). Inorg. Chem. 50, 7598–7609. [DOI] [PubMed]
- Lu, B. Y., Li, Z. M., Zhu, Y. Y., Zhao, X. & Li, Z. T. (2012). Tetrahedron, 68, 8857–8862.
- Lukeš, V., Michalík, M., Poliak, P., Cagardová, D., Végh, D., Bortňák, D., Fronc, M. & Kožíšek, J. (2016). Synth. Met. 219, 83–92.
- Matsui, M., Suzuki, M., Nunome, I., Kubota, Y., Funabiki, K., Shiro, M., Matsumoto, S. & Shiozaki, H. (2008). Tetrahedron, 64, 8830–8836.
- Maugeri, L., Asencio-Hernández, J., Lébl, T., Cordes, D. B., Slawin, A. M., Delsuc, M. A. & Philp, D. (2016). Chem. Sci. 7, 6422–6428. [DOI] [PMC free article] [PubMed]
- Maugeri, L., Lébl, T., Cordes, D. B., Slawin, A. M. & Philp, D. (2017). J. Org. Chem. 82, 1986–1995. [DOI] [PubMed]
- Milo, A., Neel, A. J., Toste, F. D. & Sigman, M. S. (2015). Science, 347, 737–743. [DOI] [PMC free article] [PubMed]
- Mohamed, H. A., Alotaibi, H. A., Kariuki, B. M. & El-Hiti, G. A. (2021). CSD Communication (CCDC 1861196). CCDC, Cambridge, England. https://doi.org/10.5517/ccdc.csd.cc20gqlt.
- Moseev, T. D., Varaksin, M. V., Gorlov, D. A., Nikiforov, E. A., Kopchuk, D. S., Starnovskaya, E. S., Khasanov, A. F., Zyryanov, G. V., Charushin, V. N. & Chupakhin, O. N. (2019). J. Fluor. Chem. 224, 89–99.
- Pokhodylo, N. T. & Obushak, M. D. (2019). Russ. J. Org. Chem. 55, 1241–1243.
- Pokhodylo, N. T. & Shyyka, O. Y. (2017). Synth. Commun. 47, 1096–1101.
- Pokhodylo, N. T., Shyyka, O. Y., Goreshnik, E. A. & Obushak, M. D. (2020). ChemistrySelect, 5, 260–264.
- Pokhodylo, N. T., Shyyka, O. Y., Matiychuk, V. S., Obushak, M. D. & Pavlyuk, V. V. (2017). ChemistrySelect, 2, 5871–5876.
- Pokhodylo, N. T., Shyyka, O. Y. & Obushak, M. D. (2014). Synth. Commun. 44, 1002–1006.
- Pokhodylo, N. T., Shyyka, O. Y. & Obushak, M. D. (2018). Chem. Heterocycl. Compd, 54, 773–779.
- Ramachary, D. B., Ramakumar, K. & Narayana, V. V. (2008). Chem. Eur. J. 14, 9143–9147. [DOI] [PubMed]
- Rigaku OD (2021). CrysAlis PRO. Rigaku Oxford Diffraction, Tokyo, Japan.
- Rillahan, C. D., Schwartz, E., McBride, R., Fokin, V. V. & Paulson, J. C. (2012). Angew. Chem. Int. Ed. 51, 11014–11018. [DOI] [PMC free article] [PubMed]
- Rozin, Y. A., Leban, J., Dehaen, W., Nenajdenko, V. G., Muzalevskiy, V. M., Eltsov, O. S. & Bakulev, V. A. (2012). Tetrahedron, 68, 614–618.
- Shafran, Y. M., Beryozkina, T. V., Efimov, I. V. & Bakulev, V. A. (2019). Chem. Heterocycl. Compd, 55, 704–715.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Singh, S., El-Sakkary, N., Skinner, D. E., Sharma, P. P., Ottilie, S., Antonova-Koch, Y., Kumar, P., Winzeler, E., Caffrey, C. R. & Rathi, B. (2020). Pharmaceuticals, 13, 25. https://doi.org/10.3390/ph13020025. [DOI] [PMC free article] [PubMed]
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Tomkute-Luksiene, D., Keruckas, J., Malinauskas, T., Simokaitiene, J., Getautis, V., Grazulevicius, J. V., Volyniuk, D., Cherpak, V., Stakhira, P., Yashchuk, V., Kosach, V., Luka, G. & Sidaravicius, J. (2013). Dyes Pigments, 96, 278–286.
- Turner, M. J., Mckinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. The University of Western Australia. http://hirshfeldsurface.net
- Vinutha, N., Madan Kumar, S., Nithinchandra, Balakrishna, K., Lokanath, N. K. & Revannasiddaiah, D. (2013). Acta Cryst. E69, o1724. [DOI] [PMC free article] [PubMed]
- Wang, C., Li, G. & Zhang, Q. (2013). Tetrahedron Lett. 54, 2633–2636.
- Xie, S., Lopez, S. A., Ramström, O., Yan, M. & Houk, K. N. (2015). J. Am. Chem. Soc. 137, 2958–2966. [DOI] [PMC free article] [PubMed]
- Zeghada, S., Bentabed-Ababsa, G., Derdour, A., Abdelmounim, S., Domingo, L. R., Sáez, J. A., Roisnel, T., Nassar, E. & Mongin, F. (2011). Org. Biomol. Chem. 9, 4295–4305. [DOI] [PubMed]
- Zukerman-Schpector, J., Dias, C. da S., Schwab, R. S., Jotani, M. M. & Tiekink, E. R. T. (2018). Acta Cryst. E74, 1195–1200. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989021010070/hb7985sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010070/hb7985Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021010070/hb7985Isup4.cml
NMR spectra. DOI: 10.1107/S2056989021010070/hb7985sup3.pdf
CCDC reference: 2112438
Additional supporting information: crystallographic information; 3D view; checkCIF report