In the structures of the title salts, two independent cations (Rb, Cs) are situated on a crystallographic twofold axis and on a center of inversion, respectively. Mutual intermolecular hydrogen bonding between the conjugate 3,5-dinitopyrazole NH-donor and 3,5-dinitropyrazolate N-acceptor sites of the anions governs the self-assembly of the translation-related anions in a predictable fashion. The anionic chains are further linked by multiple ion–dipole interactions involving the 12-coordinate cations bonded to two pyrazole N-atoms and all of the eight nitro O-atoms. The resulting ionic networks follow the CsCl topological archetype, with either metal or organic ions residing in an environment of eight counter-ions. Weak lone pair–π-hole interactions are also relevant to the packing.
Keywords: crystal structure, rubidium, caesium, nitropyrazoles, energetic materials, hydrogen bonding
Abstract
In the structures of the title salts, poly[[μ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium], [Rb(C6HN8O8)] n , (1), and its isostructural caesium analogue [Cs(C6HN8O8) n , (2), two independent cations M1 and M2 (M = Rb, Cs) are situated on a crystallographic twofold axis and on a center of inversion, respectively. Mutual intermolecular hydrogen bonding between the conjugate 3,5-dinitopyrazole NH-donor and 3,5-dinitropyrazole N-acceptor sites of the anions [N⋯N = 2.785 (2) Å for (1) and 2.832 (3) Å for (2)] governs the self-assembly of the translation-related anions in a predictable fashion. Such one-component modular construction of the organic subtopology supports the utility of the crystal-engineering approach towards designing the structures of polynitro energetic materials. The anionic chains are further linked by multiple ion–dipole interactions involving the 12-coordinate cations bonded to two pyrazole N-atoms [Rb—N = 3.1285 (16), 3.2261 (16) Å; Cs—N = 3.369 (2), 3.401 (2) Å] and all of the eight nitro O-atoms [Rb—O = 2.8543 (15)–3.6985 (16) Å; Cs—O = 3.071 (2)–3.811 (2) Å]. The resulting ionic networks follow the CsCl topological archetype, with either metal or organic ions residing in an environment of eight counter-ions. Weak lone pair–π-hole interactions [pyrazole-N atoms to NO2 groups; N⋯N = 2.990 (3)–3.198 (3) Å] are also relevant to the packing. The Hirshfeld surfaces and percentage two-dimensional fingerprint plots for (1) and (2) are described.
Chemical context
Many issues of crystal engineering, in regard to control over bonding patterns, supramolecular topologies, molecular packing, and crystal morphologies are highly relevant to the area of energetic materials. In particular, non-covalent contacts involving common explosophore nitro groups (Bauzá et al., 2017 ▸) establish pathways to transmit intermolecular interactions and they are often responsible for higher densities of the solids (Zhang et al., 2000 ▸). The layered architectures of the energetic solids provide better buffering against external mechanical stimuli, which is essential for developing insensitive materials (Zhang et al., 2008 ▸). At the same time, incorporation of specific coordination geometries for the assembly of metal–organic solids offers potential for the synthesis of new perchlorate-free flame colorants and pyrotechnics (Glück et al., 2017 ▸). However, successful applications of the crystal-engineering methodology toward designing the structures of polynitro compounds are relatively rare, so far (Domasevitch et al., 2020 ▸). This is predetermined by a lack of reliable supramolecular synthons comprising the nitro groups, which are only weak acceptors of conventional hydrogen bonds (Robinson et al., 2000 ▸) and are only weak donors with respect to the metal ions. A more severe limitation is associated with the need for direct bonding between the nitro-rich functionalities only, since the incorporation of any low-energetic component or solvent molecules is an inevitable penalty to the performance. Such dilution of the energetic moieties in the crystals is relevant, for example, to a series of hydrogen-bonded solids prepared by Aakeröy et al. (2015 ▸) with acidic ethylenedinitramine and common bitopic pyridine-N bases.
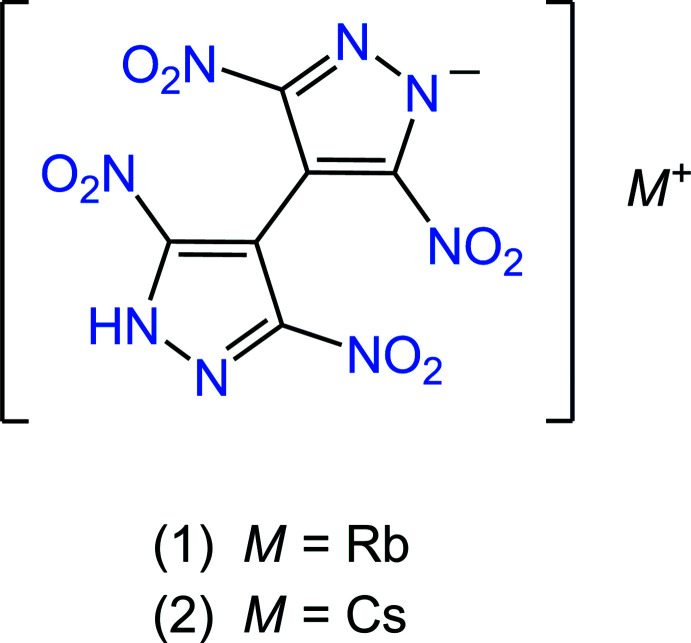
Recently, we have reported a new strategy for the construction of energetic salts, which offers higher degree of control over the structure. Double functionality of the well-performing material 3,3′,5,5′-tetranitro-4,4′-bipyrazole [H2(TNBP)] (Domasevitch et al., 2019 ▸) grants synthetic access either to singly or doubly anionic species [{H(TNBP)}− and {TNBP}2−, respectively]. The former combine conjugate dinitropyrazole donor and dinitropyrazolate acceptor sites for sustaining particularly strong N—H⋯N bonding. In fact, such bonding of two explosophores dominated the self-assembly in a very predictable fashion and it was traced in all of the previously examined salts with a range of nitrogen-rich cations (Gospodinov et al., 2020 ▸). That the resulting networks are ionic may find further applications to the synthesis of inorganic nitro-rich salts, based upon Li+, Rb+, Cs+, Sr2+, Ba2+ and other s- and p-block cations, which are a new generation of ‘green’ pyrotechnic formulations (Steinhauser & Klapötke, 2008 ▸).
Following the above findings, we now describe the synthesis and structure of rubidium and caesium 4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazolates M{H(TNBP)} [M = Rb (1) and Cs (2)], incorporating the peculiar half-deprotonated bipyrazole tectons. These materials may give an insight into the development of flame colorants in pyrotechnics: rubidium and caesium compounds exhibit, respectively, purple and orange colors when burned.
Structural commentary
The title compounds are isostructural, crystallizing in space group C2/c. The molecular structure of the rubidium salt (1) is shown in Fig. 1 ▸, with the unique part comprising one organic anion {H(TNBP)}− (or C6HN8O8 −) and two cations situated on a crystallographic twofold axis [Rb1] or on a center of inversion [Rb2]. The easy formation of such salts is conditioned by the appreciable acidity of polynitropyrazoles, c.f. pK a = 3.14 for 3,5-dinitropyrazole versus 14.63 for the parent pyrazole (Janssen et al., 1973 ▸), while for the crystallization of singly charged hydrogen bipyrazolate derivatives, the weakly polarizing, large Rb+ and Cs+ cations are important.
Figure 1.
The molecular structure and the atom-labeling scheme for (1) [the atom labeling for (2) is identical, with Cs1 and Cs2 instead of Rb1 and Rb2], with displacement ellipsoids drawn at the 50% probability level and the N—H⋯N hydrogen bond shown as a dashed line. [Symmetry code: (i) x, y + 1, z.]
Both unique metal ions exhibit exceptionally high coordination numbers of twelve, which are completed with ten O atoms [Rb—O = 2.8543 (15)–3.6985 (16) Å; Cs—O 3.071 (2)–3.811 (2) Å] and two N atoms of the pyrazole rings [Rb—N = 3.1285 (16) and 3.2261 (16) Å; Cs—N = 3.369 (2) and 3.401 (2) Å] (Tables 1 ▸ and 2 ▸). Most of these separations slightly exceed the sum of the corresponding ionic radii [which are M—O = 3.13 and 3.28 Å; M—N = 3.18 and 3.34 Å for 12-coordinate Rb and Cs ions, respectively (Shannon, 1976 ▸)], indicating the weakness of these relatively distal ion–dipole interactions. This may be best related to the bonding in the ionic salts with polynitro anions lacking conventional donor sites. For example, in caesium picrate, the cations display a comparable 12-fold coordination and a wide spread of Cs—O separations of 3.028 (3)–3.847 (2) Å (Schouten et al., 1990 ▸). The coordination polyhedra of the two unique cations are very similar and represent essentially distorted icosahedra (Fig. 2 ▸). These are completed with a twofold axis [for M1] or inversion [for M2] related pairs of chelating nitropyrazole-N,O groups, pseudo-chelating NO2 groups and two singly coordinated NO2 groups. Both kinds of cations reside in a closest environment of eight {H(TNBP)}− anions, which maintain supramolecular boxes with a small internal cavity for the cation (Fig. 3 ▸). It is notable that all of the eight O atoms present and the two pyrazole N atoms coordinate to the metal ions.
Table 1. Selected bond lengths (Å) for (1) .
| Rb1—O1i | 2.8543 (15) | Rb2—O5iii | 2.9616 (17) |
| Rb1—O5 | 2.9673 (16) | Rb2—O7 | 2.9690 (15) |
| Rb1—N3 | 3.1285 (16) | Rb2—O3i | 3.0743 (17) |
| Rb1—O8ii | 3.3074 (16) | Rb2—N2i | 3.2261 (16) |
| Rb1—O3iii | 3.424 (2) | Rb2—O6iii | 3.2275 (16) |
| Rb1—O4iii | 3.4942 (16) | Rb2—O2ii | 3.6985 (16) |
Symmetry codes: (i) x, y+1, z; (ii) -x+{\script{1\over 2}}, -y+{\script{1\over 2}}, -z; (iii) -x+{\script{1\over 2}}, y+{\script{1\over 2}}, -z+{\script{1\over 2}}.
Table 2. Selected bond lengths (Å) for (2) .
| Cs1—O1i | 3.071 (2) | Cs2—O7 | 3.109 (2) |
| Cs1—O5 | 3.177 (2) | Cs2—O5ii | 3.159 (2) |
| Cs1—O3ii | 3.351 (3) | Cs2—O3i | 3.297 (2) |
| Cs1—N3 | 3.369 (2) | Cs2—O6ii | 3.396 (2) |
| Cs1—O4ii | 3.464 (2) | Cs2—N2i | 3.401 (2) |
| Cs1—O8iii | 3.514 (2) | Cs2—O2iv | 3.811 (2) |
Symmetry codes: (i) x, y+1, z; (ii) -x+{\script{1\over 2}}, y+{\script{1\over 2}}, -z+{\script{1\over 2}}; (iii) -x+{\script{1\over 2}}, -y+{\script{1\over 2}}, -z; (iv) x-{\script{1\over 2}}, y+{\script{1\over 2}}, z.
Figure 2.
Twelvefold coordination environments adopted by the Rb1 and Rb2 ions in (1), in the form of distorted icosahedra. The coordination of the two Cs ions in (2) is almost identical. [Symmetry codes: (i) x, y + 1, z; (ii) −x +
 , y +
, y +
 , −z +
, −z +
 ; (iii) −x +
; (iii) −x +
 , −y +
, −y +
 , −z; (iv) x −
, −z; (iv) x −
 , y +
, y +
 , z; (v) −x + 1, y + 1, −z +
, z; (v) −x + 1, y + 1, −z +
 ; (vi) −x + 1, y, −z +
; (vi) −x + 1, y, −z +
 ; (vii) x +
; (vii) x +
 , y +
, y +
 , z; (viii) x +
, z; (viii) x +
 , −y +
, −y +
 , z +
, z +
 ; (ix) −x, −y + 1, −z; (x) x −
; (ix) −x, −y + 1, −z; (x) x −
 , −y +
, −y +
 , z −
, z −
 ; (xi) −x, −y, −z.]
; (xi) −x, −y, −z.]
Figure 3.
The Rb2 ion resides inside a supramolecular prism (represented here as a gray box) adopted by eight anions, which complete the coordination environment. The vertices of the prism are built through the mid-points of the central C—C bonds of the molecules. The environments of Rb1 and the respective Cs ions in the structure of (2) are similar. [Symmetry codes: (i) x, y + 1, z; (ii) −x +
 , y +
, y +
 , −z +
, −z +
 ; (iv) x −
; (iv) x −
 , y +
, y +
 , z; (ix) −x, −y + 1, −z.]
, z; (ix) −x, −y + 1, −z.]
The main geometrical parameters of the organic anions are very similar to those of the parent [H2(TNBP)] (Domasevitch et al., 2019 ▸). In the case of (1), the protolytic inequivalency of two pyrazole halves is reflected by the ring C—N distances, which are almost the same for anionic ring A (atoms C4/C5/C6/N3/N4) [N3—C4 = 1.343 (2) and N4—C6 1.348 (2) Å] and are slightly differentiated for the neutral ring B (C1/C2/C3/N1/N2) [N1—C1 1.348 (2) and N2—C3 1.331 (2) Å] (Fig. 1 ▸). In addition, the deprotonation causes slight elongation of the N—N bond, which is 1.336 (2) Å for ring B and 1.347 (2) Å for ring A. Even more sensitive parameters are the bond angles at the N atoms, which are perceptibly different for the former fragment [N2—N1—C1 = 110.67 (15); C3—N2—N1 = 104.29 (15)°], being much closer for the latter [106.38 (15) and 107.59 (15)°]. In the case of (2), the corresponding geometries are nearly identical for rings A and B [C—N = 1.340 (3)–1.346 (3) Å; N2—N1—C1 = 109.8 (2); N3—N4—C6 = 109.6 (2)° and N1—N2—C3 = 104.9 (2); N4—N3—C4 = 105.1 (2)°]. This situation agrees with the disorder of the H atoms between two positions [at the N1 or N4 carrier atoms] within the N—H⋯N hydrogen bond in (2) as discussed below.
In both structures, the {H(TNBP)}− anions display twisted conformations, with the dihedral angles between the rings being 42.99 (8) and 44.86 (10)° for (1) and (2), respectively. These angles, however, are unusually small. For example, typical parameters for the structurally similar 3,3′,5,5′-tetramethyl-4,4′-bipyrazole unit are 65–90° (Ponomarova et al., 2013 ▸). The flattening of the {H(TNBP)}− anion suggests certain attractivity in steric interactions of the NO2 groups, which generates a set of short intramolecular O⋯N contacts, the shortest being O2⋯N7 at 2.786 (3) Å observed for (2). Indeed, the nitro/nitro stackings are energetically favorable, as a special kind of lone pair–π-hole bond (Bauzá et al., 2017 ▸).
Supramolecular features
The ionic structures of the title compounds may be regarded as three-dimensional networks, which are related to the structure of CsCl. The metal ions themselves constitute a distorted primitive cubic framework with the cells representing elongated prisms [the M⋯M edges are 5.2560 (3), 6.5962 (3), 8.8395 (8) and 5.4775 (4), 6.3932 (5), 9.1482 (12) Å for (1) and (2), respectively]. Every such cell is populated with the organic anion and, conversely, every cation resides inside the distorted prismatic box of eight anions (Figs. 3 ▸ and 4 ▸).
Figure 4.
Fragment of the crystal structure of (1), showing the polar hydrogen-bonded anionic chains propagating along the b-axis direction, in the environment of the Rb cations. Blue lines indicate a pseudo-primitive cubic net arrangement of the cations, with every cell populated by a single anion (c.f. the structure of CsCl). [Symmetry codes: (i) x, y + 1, z; (vi) −x + 1, y, −z +
 ; (xiv) x, y − 1, z.]
; (xiv) x, y − 1, z.]
Beyond Coulombic attraction, the principal supramolecular interaction is strong and directional N—H⋯N hydrogen bonding between the pyrazole and pyrazolate halves of translation-related anions [N1⋯N4xiv = 2.785 (2) and 2.832 (3) Å; H⋯Nxiv = 1.93 and 1.99 Å; N1H⋯N4xiv = 166 and 163° for (1) and (2), respectively; symmetry code: (xiv) x, y − 1, z], arranging the latter into linear polar chains propagating along the b-axis direction (Fig. 4 ▸). Such bonding involving the conjugate acid (pyrazole-NH) and base (pyrazolate-N) sites is a very rare, if not the only, example of a highly reliable supramolecular synthon for crystal engineering with energetic polynitro derivatives. In fact, the conjugate interactions are relevant for many organic species, e.g. carboxylates (Speakman, 1972 ▸) and oximes (Domasevitch et al., 1998 ▸), being often the most crucial bonding for the crystal patterns. With the aid of such a synthon, the assembly of the organic subtopology of lower dimensionality is possible in a very rational and predictable fashion and the title structures exactly follow the motifs of previously examined NH3OH+ and 3,3′,5,5′-tetramethyl-4,4′-bipyrazolium {H(TNBP)}− salts (Gospodinov et al., 2020 ▸).
The above hydrogen-bonded chains associate to yield layers lying parallel to the ac plane and the latter are separated by the layers of metal cations (Fig. 5 ▸). There are two kinds of weaker interactions, which facilitate close packing of the chains. The first of these is identified by close N3⋯N6ii and N2⋯N7xii contacts [the shortest of 2.990 (3) Å] originating in situation of the pyrazole N atoms almost exactly above the NO2 N atoms (Table 3 ▸). This peculiar lone pair–π-hole interaction occurs instead of the more common NO2/NO2 bonding (Bauzá et al., 2017 ▸), which is also relevant for the structure of [H2(TNBP)] itself (Domasevitch et al., 2019 ▸). One can note that extensive ion-dipole interactions M⋯O2N in (1) and (2) mitigate against mutual interactions of nitro groups, which are totally eliminated from the suite of supramolecular bonds. The second type of interchain interaction is stacking between pairs of inversion-related pyrazole and pyrazolate rings (Fig. 6 ▸), with the O7 and N5 atoms situated nearly above the centroids of the rings A
iii and B
xiii, respectively [symmetry codes: (iii) −x +
 , −y +
, −y +
 , −z; (xiii) −x +
, −z; (xiii) −x +
 , −y −
, −y −
 , −z.] (Table 4 ▸). As a result of the inversion symmetry of the stacks, the alignment of two polar hydrogen-bonded chains in (1) is antiparallel, while the above lone pair–π-hole interactions support coherent alignment of the contributing chains (Fig. 6 ▸). This results in pairing of the chains possessing identical polarities (Fig. 5 ▸). In the structure of (2), the polarity of the chains is eliminated because of the disorder of the H atoms in the N—H⋯N/N⋯H—N bonds.
, −z.] (Table 4 ▸). As a result of the inversion symmetry of the stacks, the alignment of two polar hydrogen-bonded chains in (1) is antiparallel, while the above lone pair–π-hole interactions support coherent alignment of the contributing chains (Fig. 6 ▸). This results in pairing of the chains possessing identical polarities (Fig. 5 ▸). In the structure of (2), the polarity of the chains is eliminated because of the disorder of the H atoms in the N—H⋯N/N⋯H—N bonds.
Figure 5.
Structure of (1) viewed in projection on the ac plane (down the direction of the anionic chains) showing the organic layers, which are separated by layers of the cations. The chains of opposite polarity are identified by blue and red colors. [Symmetry codes: (iv) x −
 , y +
, y +
 , z; (v) −x + 1, y + 1, −z +
, z; (v) −x + 1, y + 1, −z +
 .]
.]
Table 3. Geometry of lone pair–π-hole interactions (Å, °) in (1) and (2).
N⋯plane is a distance of an N-donor to the mean plane of a nitro group and φ is an angle of the N⋯N axis to the plane of the nitro group.
| Compound | N-Donor | Group | N⋯N | N⋯plane | φ |
|---|---|---|---|---|---|
| (1) | N2 | (C4N7O5O6)xii | 2.997 (3) | 2.980 (2) | 83.9 (2) |
| N3 | (C3N6O3O4)ii | 3.198 (3) | 3.093 (2) | 75.3 (2) | |
| (2) | N2 | (C4N7O5O6)xii | 2.990 (3) | 2.976 (3) | 84.5 (2) |
| N3 | (C3N6O3O4)ii | 3.186 (3) | 3.083 (3) | 75.4 (2) |
Symmetry codes: (ii) −x + {1\over 2}, y + {1\over 2}, −z + {1\over 2}; (xii) −x + {1\over 2}, y − {1\over 2}, −z + {1\over 2}.
Figure 6.
A suite of non-covalent interactions of the {H(TNBP)}− anions, with two kinds of lone pair–π-hole bonds (marked in red) and two kinds of nitro/pyrazole stacks (marked in blue) complementing the conventional hydrogen bonding. [Symmetry codes: (ii) −x +
 , y +
, y +
 , −z +
, −z +
 ; (v) −x + 1, y + 1, −z +
; (v) −x + 1, y + 1, −z +
 ; (vi) −x + 1, y, −z +
; (vi) −x + 1, y, −z +
 ; (xiii) −x +
; (xiii) −x +
 , −y −
, −y −
 , −z; (xiv) x, y − 1, z.]
, −z; (xiv) x, y − 1, z.]
Table 4. Geometry of stacking interactions involving nitro and pyrazole groups (Å, °) in (1) and (2).
Atom⋯Cg is the shortest distance from the nitro group atom to the centroid of the ring; Atom⋯plane is the deviation of the given atom from the mean plane of the ring and φ is the angle of the atom⋯Cg axis to the plane of the ring.
| Compound | Atom | Ring | Atom⋯Cg | Atom⋯plane | φ |
|---|---|---|---|---|---|
| (1) | O7 | (C4C5C6N3N4)iii | 3.265 (3) | 3.262 (2) | 87.5 (2) |
| N5 | (C1C2C3N1N2)xiii | 3.541 (3) | 3.526 (3) | 84.7 (2) | |
| (2) | O7 | (C4C5C6N3N4)iii | 3.240 (3) | 3.232 (3) | 86.0 (3) |
| N5 | (C1C2C3N1N2)xiii | 3.448 (3) | 3.389 (3) | 79.4 (3) |
Symmetry codes: (iii) −x + {1\over 2}, −y + {1\over 2}, −z; (xiii) −x + {1\over 2}, −y − {1\over 2}, −z.
Hirshfeld analysis
The supramolecular interactions in the title structures were also assessed by Hirshfeld surface analysis (Spackman & Byrom, 1997 ▸; McKinnon et al., 2004 ▸; Hirshfeld, 1977 ▸; Spackman & McKinnon, 2002 ▸) performed with CrystalExplorer17 (Turner et al., 2017 ▸). The contributions of different kinds of interatomic contacts to the Hirshfeld surfaces of the individual anions are listed in Table 5 ▸ and the fingerprint plots for (1) are shown in Fig. 7 ▸. The most significant contributors are O⋯O contacts (37.4%), while the fraction of O,N⋯Rb (15.4%) is relatively modest due to the larger lengths of the ion–dipole interactions. The shortest O⋯O separation on the plot of ∼2.8 Å corresponds to the contact O1⋯O8xiii = 2.741 (2) Å [2.732 (3) Å in (2); symmetry code (xiii) −x +
 , −y −
, −y −
 , −z]. We note that slight contraction of the O⋯M fraction in the case of M = Rb [13.6% for (1) and 13.0% for (2)] coincides with a larger contribution of less favorable O⋯O contacts [37.4% for (1) and 35.5% for (2)]. This may be an additional factor destabilizing the structure: the crystals of (1) eventually decompose under the mother solution, unlike the stable Cs analogue. The lone pair–π-hole pyrazole-NO2 interactions generate 5.3% (1) and 6.3% (2) of the contacts of the Hirshfeld surfaces, with the shortest N⋯N = 2.9 Å. The nature of the O⋯N/N⋯O and N⋯C/C⋯N contacts [in total 23.3% (1) and 22.8% (2)] is similar, since they correspond to the stacking of pyrazole and NO2 groups with shortest O⋯N and N⋯C distances of 3.2 and 3.3 Å, respectively. However, there are no pairs of the features that are characteristic for the mutual O⋯N/N⋯O interactions of NO2 groups themselves (Domasevitch et al., 2020 ▸). The contributions of the O⋯H/H⋯O and N⋯H/H⋯N contacts are comparable and perceptible [5.4 and 6.9% for (1) and 5.2 and 6.6% for (2)], but only the latter correspond to hydrogen bonding, as is reflected in the plots. These bonds are responsible for a pair of very sharp features pointing to the lower left, with a shortest contact of 1.9 Å, whereas O⋯H/H⋯O contacts are identified only with a diffuse collection of points between the above features and with a shortest contact of 2.8 Å.
, −z]. We note that slight contraction of the O⋯M fraction in the case of M = Rb [13.6% for (1) and 13.0% for (2)] coincides with a larger contribution of less favorable O⋯O contacts [37.4% for (1) and 35.5% for (2)]. This may be an additional factor destabilizing the structure: the crystals of (1) eventually decompose under the mother solution, unlike the stable Cs analogue. The lone pair–π-hole pyrazole-NO2 interactions generate 5.3% (1) and 6.3% (2) of the contacts of the Hirshfeld surfaces, with the shortest N⋯N = 2.9 Å. The nature of the O⋯N/N⋯O and N⋯C/C⋯N contacts [in total 23.3% (1) and 22.8% (2)] is similar, since they correspond to the stacking of pyrazole and NO2 groups with shortest O⋯N and N⋯C distances of 3.2 and 3.3 Å, respectively. However, there are no pairs of the features that are characteristic for the mutual O⋯N/N⋯O interactions of NO2 groups themselves (Domasevitch et al., 2020 ▸). The contributions of the O⋯H/H⋯O and N⋯H/H⋯N contacts are comparable and perceptible [5.4 and 6.9% for (1) and 5.2 and 6.6% for (2)], but only the latter correspond to hydrogen bonding, as is reflected in the plots. These bonds are responsible for a pair of very sharp features pointing to the lower left, with a shortest contact of 1.9 Å, whereas O⋯H/H⋯O contacts are identified only with a diffuse collection of points between the above features and with a shortest contact of 2.8 Å.
Table 5. Contributions of the different kinds of the contacts (%) to the Hirshfeld surfaces of individual anions in (1) a and (2).
M = Rb (1) and Cs (2)
| Contact | (1) | (2) |
|---|---|---|
| O⋯M | 13.0 | 13.6 |
| N⋯M | 2.4 | 2.2 |
| O⋯O | 37.4 | 35.5 |
| N⋯N | 5.3 | 6.3 |
| C⋯C | 1.0 | 0.7 |
| O⋯N/N⋯O | 15.8 | 16.2 |
| O⋯C/C⋯O | 3.8 | 5.4 |
| N⋯C/C⋯N | 7.5 | 6.6 |
| N⋯H/H⋯N | 6.9 | 6.6 |
| O⋯H/H⋯O | 5.4 | 5.2 |
| C⋯H/H⋯C | 1.5 | 1.7 |
Note: (a) For the two-dimensional plots for (1), see Fig. 7 ▸.
Figure 7.
Two-dimensional fingerprint plots for the individual anions in (1), and delineated into the principal contributions of O,N⋯Rb, O⋯O, N⋯N, O⋯N/N⋯O, O⋯C/C⋯O, N⋯C/C⋯C, N⋯H/H⋯N and O⋯H/H⋯O contacts. Other contacts are C⋯H/H⋯C (1.5%) and C⋯C (1.0%).
Synthesis and crystallization
3,3′,5,5′-Tetranitro-4,4′-bipyrazole [H2(TNBP)] was synthesized in 92% yield by nitration of 4,4′-bipyrazole in mixed acids and then crystallized from water as a monohydrate (Domasevitch et al., 2019 ▸).
To prepare the Rb salt (1), 0.332 g (1.0 mmol) of H2(TNBP)·H2O was added to a solution of 0.116 g (0.5 mmol) of Rb2CO3 in 8 ml of water and the mixture was heated at 353–363 K until total dissolution was observed. The solution was cooled to room temperature and left for a few hours for crystallization. Pale-yellow crystals of Rb{H(TNBP)} were isolated in a yield of 0.325 g (82%) and dried in air. The compound is unstable when stored under the reaction solution as the initially formed crystals dissolve in a period of 10–15 d and colorless H2(TNBP)·H2O deposits. In a similar way, the reaction of 0.332 g (1.0 mmol) of H2(TNBP)·H2O and 0.163 g (0.5 mmol) of Cs2CO3 in 8 ml of water gives 0.415 g (93%) of pale-yellow Cs{H(TNBP)} (2). Unlike (1), this material is stable under the mother solution. Similar reactions with Na2CO3 and K2CO3 did not afford any hydrogen bipyrazolates and led to soluble M 2{TNBP} (M = Na, K) and precipitation of the excess amount of H2(TNBP)·H2O.
Analysis (%) calculated for (1), C6HN8O8Rb: C 18.08, H 0.25, N 28.12; found: C 17.93, H 0.44, N 28.49. IR (KBr, cm−1): 590 w, 708 w, 838 m, 854 s, 996 m, 1024 m, 1308 s, 1352 vs, 1398 vs, 1432 m, 1490 vs, 1500 m, 1556 vs, 1636 w, 3448 br.
Analysis (%) calculated for (2), C6HCsN8O8: C 16.15, H 0.23, N 25.13; found: C 16.01, H 0.38, N 28.11. IR (KBr, cm−1): 516 w, 586 m, 708 m, 838 s, 852 s, 994 s, 1022 m, 1170 w, 1306 s, 1324 s, 1350 vs, 1396 vs, 1432 s, 1488 vs, 1512 vs, 1544 vs, 1634 m, 3024 br, 3442 br.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 6 ▸. The hydrogen atoms were located and then refined as riding with N—H = 0.87 Å and U iso(H) = 1.5U eq(N). For (2), the H atom is equally disordered over two positions corresponding to the N1 and N4 carrier atoms.
Table 6. Experimental details.
| (1) | (2) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | [Rb(C6HN8O8)] | [Cs(C6HN8O8)] |
| M r | 398.62 | 446.06 |
| Crystal system, space group | Monoclinic, C2/c | Monoclinic, C2/c |
| Temperature (K) | 213 | 213 |
| a, b, c (Å) | 19.4400 (15), 8.6070 (4), 16.0977 (10) | 19.944 (2), 8.6307 (7), 16.2083 (17) |
| β (°) | 115.264 (7) | 113.766 (8) |
| V (Å3) | 2435.8 (3) | 2553.4 (5) |
| Z | 8 | 8 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 4.13 | 2.97 |
| Crystal size (mm) | 0.20 × 0.16 × 0.14 | 0.20 × 0.16 × 0.14 |
| Data collection | ||
| Diffractometer | Stoe IPDS | Stoe IPDS |
| Absorption correction | Numerical [X-RED (Stoe & Cie, 2001 ▸) and X-SHAPE (Stoe & Cie, 1999 ▸)] | Numerical [X-RED (Stoe & Cie, 2001 ▸) and X-SHAPE (Stoe & Cie, 1999 ▸)] |
| T min, T max | 0.672, 0.789 | 0.677, 0.772 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 9925, 2890, 2186 | 9014, 2990, 2686 |
| R int | 0.033 | 0.042 |
| (sin θ/λ)max (Å−1) | 0.658 | 0.656 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.026, 0.055, 0.89 | 0.027, 0.060, 1.21 |
| No. of reflections | 2890 | 2990 |
| No. of parameters | 210 | 211 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.37, −0.39 | 0.67, −0.77 |
Supplementary Material
Crystal structure: contains datablock(s) global, 1, 2. DOI: 10.1107/S2056989021010227/hb7988sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989021010227/hb79881sup2.hkl
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2056989021010227/hb79882sup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Crystal data
| [Rb(C6HN8O8)] | F(000) = 1552 |
| Mr = 398.62 | Dx = 2.174 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 19.4400 (15) Å | Cell parameters from 8000 reflections |
| b = 8.6070 (4) Å | θ = 2.3–27.9° |
| c = 16.0977 (10) Å | µ = 4.13 mm−1 |
| β = 115.264 (7)° | T = 213 K |
| V = 2435.8 (3) Å3 | Prism, yellow |
| Z = 8 | 0.20 × 0.16 × 0.14 mm |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Data collection
| Stoe IPDS diffractometer | 2186 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.033 |
| φ oscillation scans | θmax = 27.9°, θmin = 2.3° |
| Absorption correction: numerical [X-RED (Stoe & Cie, 2001) and X-SHAPE (Stoe & Cie, 1999)] | h = −25→25 |
| Tmin = 0.672, Tmax = 0.789 | k = −10→11 |
| 9925 measured reflections | l = −20→20 |
| 2890 independent reflections |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.026 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.055 | H-atom parameters constrained |
| S = 0.89 | w = 1/[σ2(Fo2) + (0.0326P)2] where P = (Fo2 + 2Fc2)/3 |
| 2890 reflections | (Δ/σ)max < 0.001 |
| 210 parameters | Δρmax = 0.37 e Å−3 |
| 0 restraints | Δρmin = −0.39 e Å−3 |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Rb1 | 0.500000 | 0.39279 (3) | 0.250000 | 0.02840 (9) | |
| Rb2 | 0.000000 | 0.500000 | 0.000000 | 0.03230 (9) | |
| O1 | 0.38330 (9) | −0.38304 (18) | 0.16106 (14) | 0.0422 (5) | |
| O2 | 0.38489 (9) | −0.14611 (18) | 0.11639 (13) | 0.0344 (4) | |
| O3 | 0.03697 (9) | −0.16032 (19) | 0.06215 (16) | 0.0490 (5) | |
| O4 | 0.10681 (9) | 0.03244 (17) | 0.13792 (13) | 0.0347 (4) | |
| O5 | 0.45319 (8) | 0.09854 (18) | 0.30718 (12) | 0.0329 (4) | |
| O6 | 0.37797 (9) | −0.09294 (16) | 0.29580 (11) | 0.0301 (4) | |
| O7 | 0.13123 (9) | 0.34370 (17) | −0.01293 (13) | 0.0344 (4) | |
| O8 | 0.12218 (8) | 0.09417 (17) | −0.03095 (11) | 0.0280 (3) | |
| N1 | 0.23222 (9) | −0.36612 (17) | 0.10737 (13) | 0.0199 (4) | |
| H1 | 0.247444 | −0.461154 | 0.107308 | 0.030* | |
| N2 | 0.16218 (9) | −0.32601 (19) | 0.09532 (13) | 0.0210 (4) | |
| N3 | 0.33470 (9) | 0.27100 (18) | 0.19547 (13) | 0.0207 (4) | |
| N4 | 0.26946 (9) | 0.31935 (17) | 0.12643 (13) | 0.0200 (4) | |
| N5 | 0.35387 (9) | −0.25662 (18) | 0.13349 (12) | 0.0212 (4) | |
| N6 | 0.09790 (10) | −0.0935 (2) | 0.10011 (14) | 0.0266 (4) | |
| N7 | 0.39083 (9) | 0.03467 (19) | 0.27152 (13) | 0.0218 (4) | |
| N8 | 0.15509 (9) | 0.21089 (19) | 0.01062 (13) | 0.0206 (4) | |
| C1 | 0.27606 (10) | −0.2391 (2) | 0.11965 (14) | 0.0175 (4) | |
| C2 | 0.23545 (10) | −0.1061 (2) | 0.11814 (14) | 0.0161 (4) | |
| C3 | 0.16475 (10) | −0.1719 (2) | 0.10251 (15) | 0.0181 (4) | |
| C4 | 0.32927 (10) | 0.1159 (2) | 0.19966 (14) | 0.0177 (4) | |
| C5 | 0.26131 (10) | 0.0553 (2) | 0.13416 (14) | 0.0158 (4) | |
| C6 | 0.22651 (10) | 0.1926 (2) | 0.08941 (14) | 0.0167 (4) |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Rb1 | 0.01707 (13) | 0.01962 (14) | 0.0442 (2) | 0.000 | 0.00897 (13) | 0.000 |
| Rb2 | 0.02226 (15) | 0.03355 (17) | 0.0307 (2) | 0.00090 (12) | 0.00132 (12) | −0.01165 (14) |
| O1 | 0.0311 (8) | 0.0188 (8) | 0.0702 (14) | 0.0128 (6) | 0.0153 (9) | 0.0038 (8) |
| O2 | 0.0284 (8) | 0.0279 (8) | 0.0533 (12) | −0.0005 (6) | 0.0234 (8) | 0.0034 (8) |
| O3 | 0.0212 (8) | 0.0283 (9) | 0.0941 (17) | −0.0011 (7) | 0.0214 (9) | 0.0036 (9) |
| O4 | 0.0411 (9) | 0.0214 (8) | 0.0519 (12) | 0.0071 (7) | 0.0299 (9) | −0.0003 (7) |
| O5 | 0.0206 (7) | 0.0336 (8) | 0.0327 (10) | −0.0044 (6) | 0.0001 (7) | 0.0048 (7) |
| O6 | 0.0375 (8) | 0.0155 (7) | 0.0272 (10) | −0.0001 (6) | 0.0040 (7) | 0.0046 (6) |
| O7 | 0.0313 (8) | 0.0190 (7) | 0.0470 (12) | 0.0121 (6) | 0.0111 (8) | 0.0127 (7) |
| O8 | 0.0225 (7) | 0.0231 (8) | 0.0305 (10) | −0.0046 (6) | 0.0037 (6) | 0.0008 (7) |
| N1 | 0.0231 (8) | 0.0089 (7) | 0.0267 (11) | 0.0004 (6) | 0.0098 (7) | −0.0010 (7) |
| N2 | 0.0222 (8) | 0.0127 (8) | 0.0283 (11) | −0.0004 (6) | 0.0109 (7) | 0.0020 (7) |
| N3 | 0.0220 (8) | 0.0141 (8) | 0.0230 (10) | −0.0016 (6) | 0.0067 (7) | −0.0010 (7) |
| N4 | 0.0213 (8) | 0.0103 (7) | 0.0277 (11) | −0.0001 (6) | 0.0098 (7) | 0.0009 (7) |
| N5 | 0.0211 (8) | 0.0164 (8) | 0.0230 (10) | 0.0031 (6) | 0.0064 (7) | −0.0051 (7) |
| N6 | 0.0256 (9) | 0.0174 (9) | 0.0423 (13) | 0.0030 (7) | 0.0198 (8) | 0.0077 (8) |
| N7 | 0.0241 (9) | 0.0176 (9) | 0.0196 (10) | 0.0010 (6) | 0.0053 (7) | −0.0014 (7) |
| N8 | 0.0187 (8) | 0.0180 (8) | 0.0263 (11) | 0.0031 (6) | 0.0108 (7) | 0.0057 (7) |
| C1 | 0.0203 (9) | 0.0115 (9) | 0.0193 (12) | 0.0001 (7) | 0.0070 (8) | 0.0000 (7) |
| C2 | 0.0187 (9) | 0.0104 (8) | 0.0188 (11) | 0.0010 (7) | 0.0075 (8) | 0.0008 (8) |
| C3 | 0.0203 (9) | 0.0122 (9) | 0.0223 (12) | 0.0008 (7) | 0.0095 (8) | 0.0019 (8) |
| C4 | 0.0206 (9) | 0.0119 (8) | 0.0192 (11) | 0.0007 (7) | 0.0070 (8) | −0.0003 (7) |
| C5 | 0.0177 (9) | 0.0109 (8) | 0.0202 (12) | 0.0014 (7) | 0.0094 (8) | 0.0003 (7) |
| C6 | 0.0185 (9) | 0.0126 (8) | 0.0198 (11) | 0.0016 (7) | 0.0089 (8) | 0.0011 (7) |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Geometric parameters (Å, º)
| Rb1—O1i | 2.8543 (15) | O1—N5 | 1.221 (2) |
| Rb1—O1ii | 2.8543 (15) | O2—N5 | 1.219 (2) |
| Rb1—O5 | 2.9673 (16) | O3—N6 | 1.220 (2) |
| Rb1—O5iii | 2.9673 (16) | O4—N6 | 1.219 (2) |
| Rb1—N3 | 3.1285 (16) | O5—N7 | 1.228 (2) |
| Rb1—N3iii | 3.1285 (16) | O6—N7 | 1.227 (2) |
| Rb1—O8iv | 3.3074 (16) | O7—N8 | 1.231 (2) |
| Rb1—O8v | 3.3074 (16) | O8—N8 | 1.225 (2) |
| Rb1—O3vi | 3.424 (2) | N1—N2 | 1.336 (2) |
| Rb1—O3vii | 3.424 (2) | N1—C1 | 1.348 (2) |
| Rb1—O4vi | 3.4942 (16) | N1—H1 | 0.8700 |
| Rb1—O4vii | 3.4942 (16) | N2—C3 | 1.331 (2) |
| Rb2—O5viii | 2.9616 (17) | N3—C4 | 1.343 (2) |
| Rb2—O5vii | 2.9616 (17) | N3—N4 | 1.347 (2) |
| Rb2—O7ix | 2.9690 (15) | N4—C6 | 1.348 (2) |
| Rb2—O7 | 2.9690 (15) | N5—C1 | 1.440 (2) |
| Rb2—O3x | 3.0743 (17) | N6—C3 | 1.450 (2) |
| Rb2—O3ii | 3.0743 (17) | N7—C4 | 1.441 (2) |
| Rb2—N2x | 3.2261 (16) | N8—C6 | 1.435 (3) |
| Rb2—N2ii | 3.2261 (16) | C1—C2 | 1.384 (3) |
| Rb2—O6viii | 3.2275 (16) | C2—C3 | 1.406 (3) |
| Rb2—O6vii | 3.2275 (16) | C2—C5 | 1.462 (2) |
| Rb2—O2v | 3.6985 (16) | C4—C5 | 1.393 (3) |
| Rb2—O2xi | 3.6985 (16) | C5—C6 | 1.399 (3) |
| O1i—Rb1—O1ii | 94.94 (7) | O5vii—Rb2—O6viii | 139.46 (4) |
| O1i—Rb1—O5 | 134.80 (5) | O7ix—Rb2—O6viii | 71.28 (5) |
| O1ii—Rb1—O5 | 116.69 (4) | O7—Rb2—O6viii | 108.72 (5) |
| O1i—Rb1—O5iii | 116.69 (4) | O3x—Rb2—O6viii | 86.34 (5) |
| O1ii—Rb1—O5iii | 134.80 (5) | O3ii—Rb2—O6viii | 93.66 (5) |
| O5—Rb1—O5iii | 62.80 (7) | N2x—Rb2—O6viii | 59.09 (4) |
| O1i—Rb1—N3 | 150.29 (5) | N2ii—Rb2—O6viii | 120.91 (4) |
| O1ii—Rb1—N3 | 65.44 (5) | O5viii—Rb2—O6vii | 139.46 (4) |
| O5—Rb1—N3 | 52.18 (4) | O5vii—Rb2—O6vii | 40.54 (4) |
| O5iii—Rb1—N3 | 92.36 (4) | O7ix—Rb2—O6vii | 108.72 (5) |
| O1i—Rb1—N3iii | 65.44 (5) | O7—Rb2—O6vii | 71.28 (5) |
| O1ii—Rb1—N3iii | 150.29 (5) | O3x—Rb2—O6vii | 93.66 (5) |
| O5—Rb1—N3iii | 92.36 (4) | O3ii—Rb2—O6vii | 86.34 (5) |
| O5iii—Rb1—N3iii | 52.18 (4) | N2x—Rb2—O6vii | 120.91 (4) |
| N3—Rb1—N3iii | 140.85 (6) | N2ii—Rb2—O6vii | 59.09 (4) |
| O1i—Rb1—O8iv | 52.19 (5) | O6viii—Rb2—O6vii | 180.00 (7) |
| O1ii—Rb1—O8iv | 124.56 (5) | O5viii—Rb2—O2v | 63.14 (4) |
| O5—Rb1—O8iv | 82.76 (4) | O5vii—Rb2—O2v | 116.86 (4) |
| O5iii—Rb1—O8iv | 100.60 (4) | O7ix—Rb2—O2v | 127.24 (4) |
| N3—Rb1—O8iv | 119.65 (4) | O7—Rb2—O2v | 52.75 (4) |
| N3iii—Rb1—O8iv | 61.84 (4) | O3x—Rb2—O2v | 105.45 (5) |
| O1i—Rb1—O8v | 124.56 (5) | O3ii—Rb2—O2v | 74.55 (5) |
| O1ii—Rb1—O8v | 52.19 (5) | N2x—Rb2—O2v | 126.63 (4) |
| O5—Rb1—O8v | 100.60 (4) | N2ii—Rb2—O2v | 53.37 (4) |
| O5iii—Rb1—O8v | 82.76 (4) | O6viii—Rb2—O2v | 74.96 (4) |
| N3—Rb1—O8v | 61.84 (4) | O6vii—Rb2—O2v | 105.04 (4) |
| N3iii—Rb1—O8v | 119.65 (4) | O5viii—Rb2—O2xi | 116.86 (4) |
| O8iv—Rb1—O8v | 176.11 (5) | O5vii—Rb2—O2xi | 63.14 (4) |
| O1i—Rb1—O3vi | 96.42 (5) | O7ix—Rb2—O2xi | 52.76 (4) |
| O1ii—Rb1—O3vi | 93.94 (5) | O7—Rb2—O2xi | 127.25 (4) |
| O5—Rb1—O3vi | 111.58 (4) | O3x—Rb2—O2xi | 74.55 (5) |
| O5iii—Rb1—O3vi | 53.44 (4) | O3ii—Rb2—O2xi | 105.45 (5) |
| N3—Rb1—O3vi | 106.56 (4) | N2x—Rb2—O2xi | 53.37 (4) |
| N3iii—Rb1—O3vi | 68.00 (4) | N2ii—Rb2—O2xi | 126.63 (4) |
| O8iv—Rb1—O3vi | 128.33 (4) | O6viii—Rb2—O2xi | 105.04 (4) |
| O8v—Rb1—O3vi | 52.33 (4) | O6vii—Rb2—O2xi | 74.96 (4) |
| O1i—Rb1—O3vii | 93.94 (5) | O2v—Rb2—O2xi | 180.00 (3) |
| O1ii—Rb1—O3vii | 96.42 (5) | N5—O1—Rb1xii | 158.96 (13) |
| O5—Rb1—O3vii | 53.44 (4) | N6—O3—Rb2xii | 130.77 (13) |
| O5iii—Rb1—O3vii | 111.58 (4) | N6—O3—Rb1xiii | 91.12 (15) |
| N3—Rb1—O3vii | 68.00 (4) | Rb2xii—O3—Rb1xiii | 107.84 (5) |
| N3iii—Rb1—O3vii | 106.56 (4) | N6—O4—Rb1xiii | 87.86 (12) |
| O8iv—Rb1—O3vii | 52.33 (4) | N7—O5—Rb2xiv | 99.33 (12) |
| O8v—Rb1—O3vii | 128.33 (4) | N7—O5—Rb1 | 127.55 (12) |
| O3vi—Rb1—O3vii | 164.66 (6) | Rb2xiv—O5—Rb1 | 124.88 (5) |
| O1i—Rb1—O4vi | 60.41 (5) | N7—O6—Rb2xiv | 86.64 (11) |
| O1ii—Rb1—O4vi | 91.65 (5) | N8—O7—Rb2 | 128.40 (13) |
| O5—Rb1—O4vi | 141.47 (4) | N8—O8—Rb1v | 121.64 (12) |
| O5iii—Rb1—O4vi | 78.75 (4) | N2—N1—C1 | 110.67 (15) |
| N3—Rb1—O4vi | 137.39 (5) | N2—N1—H1 | 124.7 |
| N3iii—Rb1—O4vi | 59.64 (4) | C1—N1—H1 | 124.7 |
| O8iv—Rb1—O4vi | 102.96 (4) | C3—N2—N1 | 104.29 (15) |
| O8v—Rb1—O4vi | 75.66 (4) | C3—N2—Rb2xii | 119.80 (12) |
| O3vi—Rb1—O4vi | 36.38 (4) | N1—N2—Rb2xii | 132.66 (12) |
| O3vii—Rb1—O4vi | 153.75 (4) | C4—N3—N4 | 106.38 (15) |
| O1i—Rb1—O4vii | 91.65 (5) | C4—N3—Rb1 | 114.35 (12) |
| O1ii—Rb1—O4vii | 60.41 (5) | N4—N3—Rb1 | 128.37 (12) |
| O5—Rb1—O4vii | 78.75 (4) | N3—N4—C6 | 107.59 (15) |
| O5iii—Rb1—O4vii | 141.47 (4) | O2—N5—O1 | 125.25 (17) |
| N3—Rb1—O4vii | 59.64 (4) | O2—N5—C1 | 118.13 (15) |
| N3iii—Rb1—O4vii | 137.39 (5) | O1—N5—C1 | 116.61 (16) |
| O8iv—Rb1—O4vii | 75.66 (4) | O4—N6—O3 | 124.73 (18) |
| O8v—Rb1—O4vii | 102.96 (4) | O4—N6—C3 | 117.74 (17) |
| O3vi—Rb1—O4vii | 153.75 (4) | O3—N6—C3 | 117.50 (18) |
| O3vii—Rb1—O4vii | 36.38 (4) | O4—N6—Rb1xiii | 72.68 (11) |
| O4vi—Rb1—O4vii | 139.76 (5) | O3—N6—Rb1xiii | 69.40 (14) |
| O5viii—Rb2—O5vii | 180.00 (8) | C3—N6—Rb1xiii | 132.87 (13) |
| O5viii—Rb2—O7ix | 108.32 (4) | O6—N7—O5 | 123.15 (18) |
| O5vii—Rb2—O7ix | 71.68 (4) | O6—N7—C4 | 118.54 (16) |
| O5viii—Rb2—O7 | 71.68 (4) | O5—N7—C4 | 118.29 (16) |
| O5vii—Rb2—O7 | 108.32 (4) | O6—N7—Rb2xiv | 72.15 (11) |
| O7ix—Rb2—O7 | 180.0 | O5—N7—Rb2xiv | 59.69 (11) |
| O5viii—Rb2—O3x | 57.45 (5) | C4—N7—Rb2xiv | 146.67 (12) |
| O5vii—Rb2—O3x | 122.55 (5) | O8—N8—O7 | 123.57 (18) |
| O7ix—Rb2—O3x | 111.42 (4) | O8—N8—C6 | 118.40 (15) |
| O7—Rb2—O3x | 68.58 (4) | O7—N8—C6 | 118.00 (17) |
| O5viii—Rb2—O3ii | 122.55 (5) | N1—C1—C2 | 110.36 (16) |
| O5vii—Rb2—O3ii | 57.45 (5) | N1—C1—N5 | 119.62 (15) |
| O7ix—Rb2—O3ii | 68.58 (4) | C2—C1—N5 | 130.01 (16) |
| O7—Rb2—O3ii | 111.42 (4) | C1—C2—C3 | 100.20 (16) |
| O3x—Rb2—O3ii | 180.00 (10) | C1—C2—C5 | 129.17 (17) |
| O5viii—Rb2—N2x | 64.39 (4) | C3—C2—C5 | 130.53 (17) |
| O5vii—Rb2—N2x | 115.61 (4) | N2—C3—C2 | 114.47 (16) |
| O7ix—Rb2—N2x | 63.20 (4) | N2—C3—N6 | 117.47 (16) |
| O7—Rb2—N2x | 116.80 (4) | C2—C3—N6 | 127.90 (17) |
| O3x—Rb2—N2x | 50.09 (4) | N3—C4—C5 | 113.82 (17) |
| O3ii—Rb2—N2x | 129.91 (4) | N3—C4—N7 | 117.80 (17) |
| O5viii—Rb2—N2ii | 115.61 (4) | C5—C4—N7 | 128.31 (17) |
| O5vii—Rb2—N2ii | 64.39 (4) | C4—C5—C6 | 99.63 (15) |
| O7ix—Rb2—N2ii | 116.80 (4) | C4—C5—C2 | 129.27 (17) |
| O7—Rb2—N2ii | 63.20 (4) | C6—C5—C2 | 131.10 (17) |
| O3x—Rb2—N2ii | 129.91 (4) | N4—C6—C5 | 112.57 (17) |
| O3ii—Rb2—N2ii | 50.09 (4) | N4—C6—N8 | 119.01 (16) |
| N2x—Rb2—N2ii | 180.0 | C5—C6—N8 | 128.38 (16) |
| O5viii—Rb2—O6viii | 40.54 (4) | ||
| C1—N1—N2—C3 | −1.3 (2) | C1—C2—C3—N2 | 0.0 (2) |
| C1—N1—N2—Rb2xii | 157.43 (14) | C5—C2—C3—N2 | −176.6 (2) |
| C4—N3—N4—C6 | −1.0 (2) | C1—C2—C3—N6 | 175.2 (2) |
| Rb1—N3—N4—C6 | 140.39 (14) | C5—C2—C3—N6 | −1.4 (4) |
| Rb1xii—O1—N5—O2 | 26.7 (6) | O4—N6—C3—N2 | 154.3 (2) |
| Rb1xii—O1—N5—C1 | −153.8 (4) | O3—N6—C3—N2 | −24.1 (3) |
| Rb1xiii—O4—N6—O3 | 48.3 (2) | Rb1xiii—N6—C3—N2 | 62.6 (3) |
| Rb1xiii—O4—N6—C3 | −129.88 (17) | O4—N6—C3—C2 | −20.8 (3) |
| Rb2xii—O3—N6—O4 | −164.56 (17) | O3—N6—C3—C2 | 160.8 (2) |
| Rb1xiii—O3—N6—O4 | −49.6 (2) | Rb1xiii—N6—C3—C2 | −112.5 (2) |
| Rb2xii—O3—N6—C3 | 13.7 (3) | N4—N3—C4—C5 | 0.3 (2) |
| Rb1xiii—O3—N6—C3 | 128.59 (17) | Rb1—N3—C4—C5 | −147.15 (14) |
| Rb2xii—O3—N6—Rb1xiii | −114.92 (19) | N4—N3—C4—N7 | −177.06 (17) |
| Rb2xiv—O6—N7—O5 | 32.5 (2) | Rb1—N3—C4—N7 | 35.5 (2) |
| Rb2xiv—O6—N7—C4 | −145.49 (16) | O6—N7—C4—N3 | 156.77 (19) |
| Rb2xiv—O5—N7—O6 | −36.3 (2) | O5—N7—C4—N3 | −21.3 (3) |
| Rb1—O5—N7—O6 | 174.67 (14) | Rb2xiv—N7—C4—N3 | 55.7 (3) |
| Rb2xiv—O5—N7—C4 | 141.67 (15) | O6—N7—C4—C5 | −20.2 (3) |
| Rb1—O5—N7—C4 | −7.3 (3) | O5—N7—C4—C5 | 161.7 (2) |
| Rb1—O5—N7—Rb2xiv | −149.01 (17) | Rb2xiv—N7—C4—C5 | −121.3 (2) |
| Rb1v—O8—N8—O7 | 22.7 (3) | N3—C4—C5—C6 | 0.4 (2) |
| Rb1v—O8—N8—C6 | −155.32 (13) | N7—C4—C5—C6 | 177.5 (2) |
| Rb2—O7—N8—O8 | 72.4 (3) | N3—C4—C5—C2 | −179.85 (19) |
| Rb2—O7—N8—C6 | −109.63 (17) | N7—C4—C5—C2 | −2.8 (4) |
| N2—N1—C1—C2 | 1.4 (2) | C1—C2—C5—C4 | −41.0 (4) |
| N2—N1—C1—N5 | −179.72 (17) | C3—C2—C5—C4 | 134.7 (2) |
| O2—N5—C1—N1 | 157.8 (2) | C1—C2—C5—C6 | 138.7 (2) |
| O1—N5—C1—N1 | −21.7 (3) | C3—C2—C5—C6 | −45.6 (4) |
| O2—N5—C1—C2 | −23.6 (3) | N3—N4—C6—C5 | 1.3 (2) |
| O1—N5—C1—C2 | 156.9 (2) | N3—N4—C6—N8 | −176.64 (16) |
| N1—C1—C2—C3 | −0.8 (2) | C4—C5—C6—N4 | −1.0 (2) |
| N5—C1—C2—C3 | −179.5 (2) | C2—C5—C6—N4 | 179.2 (2) |
| N1—C1—C2—C5 | 175.8 (2) | C4—C5—C6—N8 | 176.7 (2) |
| N5—C1—C2—C5 | −2.9 (4) | C2—C5—C6—N8 | −3.1 (4) |
| N1—N2—C3—C2 | 0.8 (2) | O8—N8—C6—N4 | 168.87 (18) |
| Rb2xii—N2—C3—C2 | −161.31 (14) | O7—N8—C6—N4 | −9.2 (3) |
| N1—N2—C3—N6 | −174.94 (18) | O8—N8—C6—C5 | −8.7 (3) |
| Rb2xii—N2—C3—N6 | 23.0 (2) | O7—N8—C6—C5 | 173.2 (2) |
Symmetry codes: (i) −x+1, y+1, −z+1/2; (ii) x, y+1, z; (iii) −x+1, y, −z+1/2; (iv) x+1/2, −y+1/2, z+1/2; (v) −x+1/2, −y+1/2, −z; (vi) x+1/2, y+1/2, z; (vii) −x+1/2, y+1/2, −z+1/2; (viii) x−1/2, −y+1/2, z−1/2; (ix) −x, −y+1, −z; (x) −x, −y, −z; (xi) x−1/2, y+1/2, z; (xii) x, y−1, z; (xiii) x−1/2, y−1/2, z; (xiv) −x+1/2, y−1/2, −z+1/2.
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]rubidium] (1) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N4xii | 0.87 | 1.93 | 2.785 (2) | 166 |
Symmetry code: (xii) x, y−1, z.
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Crystal data
| [Cs(C6HN8O8)] | F(000) = 1696 |
| Mr = 446.06 | Dx = 2.321 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 19.944 (2) Å | Cell parameters from 8000 reflections |
| b = 8.6307 (7) Å | θ = 2.2–27.8° |
| c = 16.2083 (17) Å | µ = 2.97 mm−1 |
| β = 113.766 (8)° | T = 213 K |
| V = 2553.4 (5) Å3 | Prism, yellow |
| Z = 8 | 0.20 × 0.16 × 0.14 mm |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Data collection
| Stoe IPDS diffractometer | 2686 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.042 |
| φ oscillation scans | θmax = 27.8°, θmin = 2.2° |
| Absorption correction: numerical [X-RED (Stoe & Cie, 2001) and X-SHAPE (Stoe & Cie, 1999)] | h = −20→26 |
| Tmin = 0.677, Tmax = 0.772 | k = −11→11 |
| 9014 measured reflections | l = −21→21 |
| 2990 independent reflections |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.027 | H-atom parameters constrained |
| wR(F2) = 0.060 | w = 1/[σ2(Fo2) + (0.0214P)2 + 3.932P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.21 | (Δ/σ)max < 0.001 |
| 2990 reflections | Δρmax = 0.67 e Å−3 |
| 211 parameters | Δρmin = −0.77 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2018/1 (Sheldrick 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00151 (14) |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cs1 | 0.500000 | 0.42703 (3) | 0.250000 | 0.03031 (9) | |
| Cs2 | 0.000000 | 0.500000 | 0.000000 | 0.04080 (10) | |
| O1 | 0.36906 (14) | −0.3754 (3) | 0.1294 (2) | 0.0536 (7) | |
| O2 | 0.38087 (12) | −0.1274 (2) | 0.12176 (15) | 0.0369 (5) | |
| O3 | 0.04269 (12) | −0.1420 (3) | 0.07349 (18) | 0.0460 (6) | |
| O4 | 0.11168 (14) | 0.0488 (2) | 0.14521 (17) | 0.0391 (5) | |
| O5 | 0.44367 (12) | 0.1085 (2) | 0.29706 (15) | 0.0379 (5) | |
| O6 | 0.36857 (13) | −0.0765 (2) | 0.29226 (14) | 0.0351 (5) | |
| O7 | 0.13940 (12) | 0.3610 (2) | −0.01257 (15) | 0.0391 (5) | |
| O8 | 0.12142 (13) | 0.1123 (2) | −0.02282 (14) | 0.0370 (5) | |
| N1 | 0.22944 (13) | −0.3511 (2) | 0.10259 (15) | 0.0260 (5) | |
| H1A | 0.243307 | −0.445633 | 0.099377 | 0.039* | 0.5 |
| N2 | 0.16260 (13) | −0.3095 (2) | 0.09520 (15) | 0.0267 (5) | |
| N3 | 0.32972 (13) | 0.2858 (2) | 0.19322 (15) | 0.0266 (5) | |
| N4 | 0.26741 (13) | 0.3309 (2) | 0.12597 (15) | 0.0252 (5) | |
| H1B | 0.256042 | 0.426103 | 0.108253 | 0.038* | 0.5 |
| N5 | 0.34581 (13) | −0.2427 (2) | 0.12310 (15) | 0.0260 (5) | |
| N6 | 0.10201 (14) | −0.0772 (3) | 0.10736 (17) | 0.0304 (5) | |
| N7 | 0.38266 (14) | 0.0482 (3) | 0.26693 (15) | 0.0268 (5) | |
| N8 | 0.15723 (13) | 0.2273 (2) | 0.01333 (15) | 0.0253 (5) | |
| C1 | 0.27188 (15) | −0.2249 (3) | 0.11572 (17) | 0.0228 (5) | |
| C2 | 0.23398 (15) | −0.0903 (3) | 0.11868 (16) | 0.0216 (5) | |
| C3 | 0.16581 (15) | −0.1553 (3) | 0.10518 (17) | 0.0236 (5) | |
| C4 | 0.32386 (15) | 0.1316 (3) | 0.19768 (17) | 0.0230 (5) | |
| C5 | 0.25873 (14) | 0.0703 (3) | 0.13417 (16) | 0.0208 (5) | |
| C6 | 0.22504 (15) | 0.2064 (3) | 0.08996 (17) | 0.0230 (5) |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cs1 | 0.02186 (13) | 0.02958 (13) | 0.03945 (15) | 0.000 | 0.01232 (10) | 0.000 |
| Cs2 | 0.02484 (14) | 0.04935 (19) | 0.03776 (16) | −0.00107 (11) | 0.00175 (11) | −0.01535 (12) |
| O1 | 0.0407 (13) | 0.0213 (10) | 0.095 (2) | 0.0120 (10) | 0.0230 (14) | −0.0019 (12) |
| O2 | 0.0366 (12) | 0.0257 (10) | 0.0524 (13) | 0.0006 (9) | 0.0220 (10) | 0.0023 (9) |
| O3 | 0.0277 (11) | 0.0353 (11) | 0.0727 (16) | −0.0002 (10) | 0.0177 (11) | 0.0033 (11) |
| O4 | 0.0456 (13) | 0.0234 (10) | 0.0604 (14) | 0.0040 (9) | 0.0339 (12) | −0.0020 (9) |
| O5 | 0.0276 (11) | 0.0354 (11) | 0.0418 (12) | −0.0014 (8) | 0.0047 (9) | 0.0046 (9) |
| O6 | 0.0440 (13) | 0.0198 (9) | 0.0305 (10) | −0.0020 (8) | 0.0035 (9) | 0.0055 (8) |
| O7 | 0.0395 (12) | 0.0224 (9) | 0.0488 (12) | 0.0106 (9) | 0.0107 (10) | 0.0125 (9) |
| O8 | 0.0369 (12) | 0.0276 (10) | 0.0369 (11) | −0.0045 (9) | 0.0048 (9) | 0.0035 (8) |
| N1 | 0.0305 (11) | 0.0154 (9) | 0.0310 (11) | 0.0029 (9) | 0.0113 (9) | 0.0008 (9) |
| N2 | 0.0312 (12) | 0.0172 (10) | 0.0313 (11) | −0.0004 (9) | 0.0121 (10) | 0.0011 (9) |
| N3 | 0.0305 (12) | 0.0187 (10) | 0.0282 (11) | −0.0016 (9) | 0.0095 (9) | −0.0018 (9) |
| N4 | 0.0311 (12) | 0.0127 (9) | 0.0306 (11) | 0.0031 (8) | 0.0111 (9) | 0.0014 (8) |
| N5 | 0.0272 (12) | 0.0207 (10) | 0.0283 (11) | 0.0053 (9) | 0.0094 (9) | −0.0017 (8) |
| N6 | 0.0314 (13) | 0.0229 (11) | 0.0427 (14) | 0.0025 (10) | 0.0208 (11) | 0.0070 (10) |
| N7 | 0.0299 (12) | 0.0211 (10) | 0.0254 (11) | 0.0028 (9) | 0.0069 (9) | −0.0004 (8) |
| N8 | 0.0268 (12) | 0.0206 (10) | 0.0296 (11) | 0.0045 (9) | 0.0123 (9) | 0.0048 (9) |
| C1 | 0.0282 (13) | 0.0143 (10) | 0.0232 (12) | 0.0035 (9) | 0.0076 (10) | 0.0018 (9) |
| C2 | 0.0288 (13) | 0.0131 (10) | 0.0214 (11) | 0.0013 (10) | 0.0085 (10) | 0.0011 (8) |
| C3 | 0.0284 (13) | 0.0163 (11) | 0.0279 (12) | 0.0015 (10) | 0.0134 (11) | 0.0023 (9) |
| C4 | 0.0274 (13) | 0.0171 (11) | 0.0227 (11) | 0.0022 (10) | 0.0082 (10) | 0.0010 (9) |
| C5 | 0.0267 (13) | 0.0130 (10) | 0.0232 (11) | 0.0020 (9) | 0.0105 (10) | 0.0021 (9) |
| C6 | 0.0265 (13) | 0.0162 (11) | 0.0260 (12) | 0.0052 (9) | 0.0101 (10) | 0.0026 (9) |
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Geometric parameters (Å, º)
| Cs1—O1i | 3.071 (2) | O2—N5 | 1.222 (3) |
| Cs1—O1ii | 3.071 (2) | O3—N6 | 1.221 (3) |
| Cs1—O5iii | 3.177 (2) | O4—N6 | 1.225 (3) |
| Cs1—O5 | 3.177 (2) | O5—N7 | 1.229 (3) |
| Cs1—O3iv | 3.351 (3) | O6—N7 | 1.224 (3) |
| Cs1—O3v | 3.351 (3) | O7—N8 | 1.230 (3) |
| Cs1—N3 | 3.369 (2) | O8—N8 | 1.225 (3) |
| Cs1—N3iii | 3.369 (2) | N1—N2 | 1.338 (3) |
| Cs1—O4iv | 3.464 (2) | N1—C1 | 1.342 (3) |
| Cs1—O4v | 3.464 (2) | N1—H1A | 0.8700 |
| Cs1—O8vi | 3.514 (2) | N2—C3 | 1.340 (3) |
| Cs1—O8vii | 3.514 (2) | N3—N4 | 1.339 (3) |
| Cs2—O7viii | 3.109 (2) | N3—C4 | 1.340 (3) |
| Cs2—O7 | 3.109 (2) | N4—C6 | 1.346 (3) |
| Cs2—O5ix | 3.159 (2) | N4—H1B | 0.8700 |
| Cs2—O5v | 3.159 (2) | N5—C1 | 1.439 (4) |
| Cs2—O3x | 3.297 (2) | N6—C3 | 1.453 (3) |
| Cs2—O3ii | 3.297 (2) | N7—C4 | 1.447 (3) |
| Cs2—O6ix | 3.396 (2) | N8—C6 | 1.432 (3) |
| Cs2—O6v | 3.396 (2) | C1—C2 | 1.398 (3) |
| Cs2—N2x | 3.401 (2) | C2—C3 | 1.404 (4) |
| Cs2—N2ii | 3.401 (2) | C2—C5 | 1.458 (3) |
| Cs2—O2xi | 3.811 (2) | C4—C5 | 1.396 (4) |
| Cs2—O2vi | 3.811 (2) | C5—C6 | 1.399 (3) |
| O1—N5 | 1.224 (3) | ||
| O1i—Cs1—O1ii | 112.57 (9) | O5ix—Cs2—N2x | 60.69 (6) |
| O1i—Cs1—O5iii | 109.94 (6) | O5v—Cs2—N2x | 119.31 (6) |
| O1ii—Cs1—O5iii | 128.30 (6) | O3x—Cs2—N2x | 47.39 (5) |
| O1i—Cs1—O5 | 128.30 (6) | O3ii—Cs2—N2x | 132.61 (5) |
| O1ii—Cs1—O5 | 109.94 (6) | O6ix—Cs2—N2x | 55.26 (5) |
| O5iii—Cs1—O5 | 60.14 (9) | O6v—Cs2—N2x | 124.74 (5) |
| O1i—Cs1—O3iv | 101.47 (7) | O7viii—Cs2—N2ii | 119.87 (6) |
| O1ii—Cs1—O3iv | 89.92 (7) | O7—Cs2—N2ii | 60.13 (6) |
| O5iii—Cs1—O3iv | 53.49 (6) | O5ix—Cs2—N2ii | 119.31 (6) |
| O5—Cs1—O3iv | 106.69 (6) | O5v—Cs2—N2ii | 60.69 (6) |
| O1i—Cs1—O3v | 89.92 (7) | O3x—Cs2—N2ii | 132.61 (5) |
| O1ii—Cs1—O3v | 101.47 (7) | O3ii—Cs2—N2ii | 47.39 (5) |
| O5iii—Cs1—O3v | 106.69 (6) | O6ix—Cs2—N2ii | 124.74 (5) |
| O5—Cs1—O3v | 53.49 (6) | O6v—Cs2—N2ii | 55.26 (5) |
| O3iv—Cs1—O3v | 159.51 (8) | N2x—Cs2—N2ii | 180.0 |
| O1i—Cs1—N3 | 151.61 (7) | O7viii—Cs2—O2xi | 46.92 (5) |
| O1ii—Cs1—N3 | 61.45 (6) | O7—Cs2—O2xi | 133.09 (5) |
| O5iii—Cs1—N3 | 92.08 (6) | O5ix—Cs2—O2xi | 114.79 (5) |
| O5—Cs1—N3 | 48.49 (5) | O5v—Cs2—O2xi | 65.21 (5) |
| O3iv—Cs1—N3 | 106.11 (6) | O3x—Cs2—O2xi | 78.04 (6) |
| O3v—Cs1—N3 | 66.04 (6) | O3ii—Cs2—O2xi | 101.96 (6) |
| O1i—Cs1—N3iii | 61.45 (6) | O6ix—Cs2—O2xi | 100.16 (5) |
| O1ii—Cs1—N3iii | 151.61 (7) | O6v—Cs2—O2xi | 79.84 (5) |
| O5iii—Cs1—N3iii | 48.49 (5) | N2x—Cs2—O2xi | 54.28 (5) |
| O5—Cs1—N3iii | 92.08 (6) | N2ii—Cs2—O2xi | 125.72 (5) |
| O3iv—Cs1—N3iii | 66.03 (6) | O7viii—Cs2—O2vi | 133.08 (5) |
| O3v—Cs1—N3iii | 106.11 (6) | O7—Cs2—O2vi | 46.91 (5) |
| N3—Cs1—N3iii | 137.57 (8) | O5ix—Cs2—O2vi | 65.21 (5) |
| O1i—Cs1—O4iv | 66.05 (7) | O5v—Cs2—O2vi | 114.79 (5) |
| O1ii—Cs1—O4iv | 93.97 (7) | O3x—Cs2—O2vi | 101.96 (6) |
| O5iii—Cs1—O4iv | 77.60 (6) | O3ii—Cs2—O2vi | 78.04 (6) |
| O5—Cs1—O4iv | 137.70 (5) | O6ix—Cs2—O2vi | 79.84 (5) |
| O3iv—Cs1—O4iv | 36.92 (6) | O6v—Cs2—O2vi | 100.16 (5) |
| O3v—Cs1—O4iv | 155.15 (6) | N2x—Cs2—O2vi | 125.72 (5) |
| N3—Cs1—O4iv | 138.81 (6) | N2ii—Cs2—O2vi | 54.28 (5) |
| N3iii—Cs1—O4iv | 57.79 (6) | O2xi—Cs2—O2vi | 180.00 (6) |
| O1i—Cs1—O4v | 93.97 (7) | N5—O1—Cs1xii | 140.2 (2) |
| O1ii—Cs1—O4v | 66.05 (7) | N6—O3—Cs2xii | 130.86 (18) |
| O5iii—Cs1—O4v | 137.70 (6) | N6—O3—Cs1xiii | 93.91 (18) |
| O5—Cs1—O4v | 77.60 (6) | Cs2xii—O3—Cs1xiii | 110.96 (7) |
| O3iv—Cs1—O4v | 155.15 (6) | N6—O4—Cs1xiii | 88.50 (16) |
| O3v—Cs1—O4v | 36.92 (6) | N7—O5—Cs2xiv | 99.67 (16) |
| N3—Cs1—O4v | 57.79 (6) | N7—O5—Cs1 | 131.31 (16) |
| N3iii—Cs1—O4v | 138.81 (6) | Cs2xiv—O5—Cs1 | 119.65 (7) |
| O4iv—Cs1—O4v | 144.69 (7) | N7—O6—Cs2xiv | 88.38 (14) |
| O1i—Cs1—O8vi | 140.41 (6) | N8—O7—Cs2 | 118.85 (17) |
| O1ii—Cs1—O8vi | 48.43 (6) | N8—O8—Cs1vi | 127.02 (16) |
| O5iii—Cs1—O8vi | 79.97 (5) | N2—N1—C1 | 109.8 (2) |
| O5—Cs1—O8vi | 90.40 (5) | N2—N1—H1A | 125.1 |
| O3iv—Cs1—O8vi | 52.64 (6) | C1—N1—H1A | 125.1 |
| O3v—Cs1—O8vi | 124.92 (6) | N1—N2—C3 | 104.9 (2) |
| N3—Cs1—O8vi | 59.05 (6) | N1—N2—Cs2xii | 130.12 (15) |
| N3iii—Cs1—O8vi | 116.39 (6) | C3—N2—Cs2xii | 121.71 (17) |
| O4iv—Cs1—O8vi | 79.82 (6) | N4—N3—C4 | 105.1 (2) |
| O4v—Cs1—O8vi | 103.61 (6) | N4—N3—Cs1 | 128.32 (16) |
| O1i—Cs1—O8vii | 48.43 (6) | C4—N3—Cs1 | 116.41 (17) |
| O1ii—Cs1—O8vii | 140.41 (6) | N3—N4—C6 | 109.6 (2) |
| O5iii—Cs1—O8vii | 90.40 (5) | N3—N4—H1B | 125.2 |
| O5—Cs1—O8vii | 79.97 (5) | C6—N4—H1B | 125.2 |
| O3iv—Cs1—O8vii | 124.92 (6) | O2—N5—O1 | 124.3 (3) |
| O3v—Cs1—O8vii | 52.64 (6) | O2—N5—C1 | 119.1 (2) |
| N3—Cs1—O8vii | 116.39 (6) | O1—N5—C1 | 116.6 (2) |
| N3iii—Cs1—O8vii | 59.05 (6) | O3—N6—O4 | 124.1 (3) |
| O4iv—Cs1—O8vii | 103.61 (6) | O3—N6—C3 | 118.4 (2) |
| O4v—Cs1—O8vii | 79.82 (6) | O4—N6—C3 | 117.5 (2) |
| O8vi—Cs1—O8vii | 168.92 (7) | O3—N6—Cs1xiii | 66.57 (17) |
| O7viii—Cs2—O7 | 180.0 | O4—N6—Cs1xiii | 71.87 (15) |
| O7viii—Cs2—O5ix | 103.32 (6) | C3—N6—Cs1xiii | 137.60 (16) |
| O7—Cs2—O5ix | 76.68 (6) | O6—N7—O5 | 124.3 (2) |
| O7viii—Cs2—O5v | 76.68 (6) | O6—N7—C4 | 118.3 (2) |
| O7—Cs2—O5v | 103.32 (6) | O5—N7—C4 | 117.4 (2) |
| O5ix—Cs2—O5v | 180.00 (11) | O6—N7—Cs2xiv | 71.61 (14) |
| O7viii—Cs2—O3x | 106.12 (6) | O5—N7—Cs2xiv | 60.53 (14) |
| O7—Cs2—O3x | 73.88 (6) | C4—N7—Cs2xiv | 149.11 (16) |
| O5ix—Cs2—O3x | 54.16 (6) | O8—N8—O7 | 124.3 (2) |
| O5v—Cs2—O3x | 125.84 (6) | O8—N8—C6 | 118.5 (2) |
| O7viii—Cs2—O3ii | 73.88 (6) | O7—N8—C6 | 117.1 (2) |
| O7—Cs2—O3ii | 106.12 (6) | N1—C1—C2 | 111.4 (2) |
| O5ix—Cs2—O3ii | 125.84 (6) | N1—C1—N5 | 119.1 (2) |
| O5v—Cs2—O3ii | 54.16 (6) | C2—C1—N5 | 129.5 (2) |
| O3x—Cs2—O3ii | 180.00 (10) | C1—C2—C3 | 99.5 (2) |
| O7viii—Cs2—O6ix | 68.69 (6) | C1—C2—C5 | 130.2 (3) |
| O7—Cs2—O6ix | 111.31 (6) | C3—C2—C5 | 130.2 (2) |
| O5ix—Cs2—O6ix | 38.43 (5) | N2—C3—C2 | 114.3 (2) |
| O5v—Cs2—O6ix | 141.57 (5) | N2—C3—N6 | 117.7 (2) |
| O3x—Cs2—O6ix | 80.81 (5) | C2—C3—N6 | 127.8 (2) |
| O3ii—Cs2—O6ix | 99.19 (5) | N3—C4—C5 | 114.3 (2) |
| O7viii—Cs2—O6v | 111.31 (6) | N3—C4—N7 | 118.3 (2) |
| O7—Cs2—O6v | 68.69 (6) | C5—C4—N7 | 127.3 (2) |
| O5ix—Cs2—O6v | 141.57 (5) | C4—C5—C6 | 99.9 (2) |
| O5v—Cs2—O6v | 38.43 (5) | C4—C5—C2 | 129.6 (2) |
| O3x—Cs2—O6v | 99.19 (5) | C6—C5—C2 | 130.6 (2) |
| O3ii—Cs2—O6v | 80.81 (5) | N4—C6—C5 | 111.1 (2) |
| O6ix—Cs2—O6v | 180.00 (9) | N4—C6—N8 | 119.0 (2) |
| O7viii—Cs2—N2x | 60.13 (6) | C5—C6—N8 | 129.8 (2) |
| O7—Cs2—N2x | 119.87 (6) | ||
| C1—N1—N2—C3 | −0.7 (3) | C1—C2—C3—N2 | −0.1 (3) |
| C1—N1—N2—Cs2xii | 158.78 (16) | C5—C2—C3—N2 | −178.3 (2) |
| C4—N3—N4—C6 | −0.6 (3) | C1—C2—C3—N6 | 175.2 (2) |
| Cs1—N3—N4—C6 | 142.57 (18) | C5—C2—C3—N6 | −3.1 (4) |
| Cs1xii—O1—N5—O2 | 53.4 (5) | O3—N6—C3—N2 | −22.2 (4) |
| Cs1xii—O1—N5—C1 | −127.5 (3) | O4—N6—C3—N2 | 156.2 (2) |
| Cs2xii—O3—N6—O4 | −167.5 (2) | Cs1xiii—N6—C3—N2 | 63.3 (3) |
| Cs1xiii—O3—N6—O4 | −45.3 (3) | O3—N6—C3—C2 | 162.7 (3) |
| Cs2xii—O3—N6—C3 | 10.7 (4) | O4—N6—C3—C2 | −18.9 (4) |
| Cs1xiii—O3—N6—C3 | 132.9 (2) | Cs1xiii—N6—C3—C2 | −111.8 (3) |
| Cs2xii—O3—N6—Cs1xiii | −122.2 (2) | N4—N3—C4—C5 | 0.4 (3) |
| Cs1xiii—O4—N6—O3 | 43.4 (3) | Cs1—N3—C4—C5 | −147.90 (19) |
| Cs1xiii—O4—N6—C3 | −134.9 (2) | N4—N3—C4—N7 | −178.0 (2) |
| Cs2xiv—O6—N7—O5 | 31.4 (3) | Cs1—N3—C4—N7 | 33.7 (3) |
| Cs2xiv—O6—N7—C4 | −148.0 (2) | O6—N7—C4—N3 | 155.1 (3) |
| Cs2xiv—O5—N7—O6 | −34.6 (3) | O5—N7—C4—N3 | −24.4 (4) |
| Cs1—O5—N7—O6 | −179.42 (19) | Cs2xiv—N7—C4—N3 | 53.4 (4) |
| Cs2xiv—O5—N7—C4 | 144.81 (19) | O6—N7—C4—C5 | −23.1 (4) |
| Cs1—O5—N7—C4 | 0.0 (4) | O5—N7—C4—C5 | 157.4 (3) |
| Cs1—O5—N7—Cs2xiv | −144.8 (2) | Cs2xiv—N7—C4—C5 | −124.8 (3) |
| Cs1vi—O8—N8—O7 | 26.7 (4) | N3—C4—C5—C6 | −0.1 (3) |
| Cs1vi—O8—N8—C6 | −152.15 (18) | N7—C4—C5—C6 | 178.2 (3) |
| Cs2—O7—N8—O8 | 66.2 (3) | N3—C4—C5—C2 | 179.4 (3) |
| Cs2—O7—N8—C6 | −114.9 (2) | N7—C4—C5—C2 | −2.3 (5) |
| N2—N1—C1—C2 | 0.7 (3) | C1—C2—C5—C4 | −43.6 (4) |
| N2—N1—C1—N5 | −178.4 (2) | C3—C2—C5—C4 | 134.1 (3) |
| O2—N5—C1—N1 | 169.4 (2) | C1—C2—C5—C6 | 135.7 (3) |
| O1—N5—C1—N1 | −9.8 (4) | C3—C2—C5—C6 | −46.5 (4) |
| O2—N5—C1—C2 | −9.5 (4) | N3—N4—C6—C5 | 0.5 (3) |
| O1—N5—C1—C2 | 171.4 (3) | N3—N4—C6—N8 | −176.5 (2) |
| N1—C1—C2—C3 | −0.4 (3) | C4—C5—C6—N4 | −0.3 (3) |
| N5—C1—C2—C3 | 178.5 (2) | C2—C5—C6—N4 | −179.8 (3) |
| N1—C1—C2—C5 | 177.9 (2) | C4—C5—C6—N8 | 176.4 (3) |
| N5—C1—C2—C5 | −3.2 (5) | C2—C5—C6—N8 | −3.1 (5) |
| N1—N2—C3—C2 | 0.5 (3) | O8—N8—C6—N4 | 175.1 (2) |
| Cs2xii—N2—C3—C2 | −161.16 (16) | O7—N8—C6—N4 | −3.9 (4) |
| N1—N2—C3—N6 | −175.3 (2) | O8—N8—C6—C5 | −1.4 (4) |
| Cs2xii—N2—C3—N6 | 23.1 (3) | O7—N8—C6—C5 | 179.6 (3) |
Symmetry codes: (i) −x+1, y+1, −z+1/2; (ii) x, y+1, z; (iii) −x+1, y, −z+1/2; (iv) x+1/2, y+1/2, z; (v) −x+1/2, y+1/2, −z+1/2; (vi) −x+1/2, −y+1/2, −z; (vii) x+1/2, −y+1/2, z+1/2; (viii) −x, −y+1, −z; (ix) x−1/2, −y+1/2, z−1/2; (x) −x, −y, −z; (xi) x−1/2, y+1/2, z; (xii) x, y−1, z; (xiii) x−1/2, y−1/2, z; (xiv) −x+1/2, y−1/2, −z+1/2.
Poly[[µ4-4-(3,5-dinitropyrazol-4-yl)-3,5-dinitropyrazol-1-ido]caesium] (2) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···N4xii | 0.87 | 1.99 | 2.832 (3) | 162 |
| N4—H1B···N1ii | 0.87 | 1.99 | 2.832 (3) | 163 |
Symmetry codes: (ii) x, y+1, z; (xii) x, y−1, z.
Funding Statement
This work was funded by Ministry of Education and Science of Ukraine grant 19BF037-05.
References
- Aakeröy, C. B., Wijethunga, T. K. & Desper, J. (2015). Chem. Eur. J. 21, 11029–11037. [DOI] [PubMed]
- Bauzá, A., Sharko, A. V., Senchyk, G. A., Rusanov, E. B., Frontera, A. & Domasevitch, K. V. (2017). CrystEngComm, 19, 1933–1937.
- Brandenburg, K. (1999). DIAMOND. Crystal Impact GbR, Bonn, Germany.
- Domasevitch, K. V., Gospodinov, I., Krautscheid, H., Klapötke, T. M. & Stierstorfer, J. (2019). New J. Chem. 43, 1305–1312.
- Domasevitch, K. V., Ponomareva, V. V., Rusanov, E. B., Gelbrich, T., Sieler, J. & Skopenko, V. V. (1998). Inorg. Chim. Acta, 268, 93–101.
- Domasevitch, K. V., Senchyk, G. A. & Krautscheid, H. (2020). Acta Cryst. C76, 598–604. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Glück, J., Klapötke, T. M., Rusan, M., Sabatini, J. J. & Stierstorfer, J. (2017). Angew. Chem. Int. Ed. 56, 16507–16509. [DOI] [PubMed]
- Gospodinov, I., Domasevitch, K. V., Unger, C. C., Klapötke, T. M. & Stierstorfer, J. (2020). Cryst. Growth Des. 20, 755–764.
- Hirshfeld, F. L. (1977). Theor. Chim. Acta, 44, 129–138.
- Janssen, J. W. A. M., Kruse, C. C., Koeners, H. J. & Habraken, C. (1973). J. Heterocycl. Chem. 10, 1055–1058.
- McKinnon, J. J., Spackman, M. A. & Mitchell, A. S. (2004). Acta Cryst. B60, 627–668. [DOI] [PubMed]
- Ponomarova, V. V., Komarchuk, V. V., Boldog, I., Krautscheid, H. & Domasevitch, K. V. (2013). CrystEngComm, 15, 8280–8287.
- Robinson, J. M. A., Philp, D., Harris, K. D. M. & Kariuki, B. M. (2000). New J. Chem. 24, 799–806.
- Schouten, A., Kanters, J. A. & Poonia, N. S. (1990). Acta Cryst. C46, 61–64.
- Shannon, R. D. (1976). Acta Cryst. A32, 751–767.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Byrom, P. G. A. (1997). Chem. Phys. Lett. 267, 215–220.
- Spackman, M. A. & McKinnon, J. J. (2002). CrystEngComm, 4, 378–392.
- Speakman, J. C. (1972). Structure and Bonding. Vol. 12, pp. 141–199. Berlin, Heidelberg: Springer.
- Steinhauser, G. & Klapötke, T. M. (2008). Angew. Chem. Int. Ed. 47, 3330–3347. [DOI] [PubMed]
- Stoe & Cie (1999). X-SHAPE. Stoe & Cie GmbH, Darmstadt, Germany.
- Stoe & Cie (2000). IPDS Software. Stoe & Cie GmbH, Darmstadt, Germany.
- Stoe & Cie (2001). X-RED. Stoe & Cie GmbH, Darmstadt, Germany.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://crystalexplorer.scb.uwa.edu.au/
- Zhang, C., Wang, X. & Huang, H. (2008). J. Am. Chem. Soc. 130, 8359–8365. [DOI] [PubMed]
- Zhang, M.-X., Eaton, P. E. & Gilardi, R. (2000). Angew. Chem. Int. Ed. 39, 401–404. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, 1, 2. DOI: 10.1107/S2056989021010227/hb7988sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989021010227/hb79881sup2.hkl
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2056989021010227/hb79882sup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report









