The title coordination polymer consists of parallel linear chains built up of macrocyclic cations possessing a slightly tetragonally distorted NiN6 octahedral coordination geometry formed by four N atoms of the azamacrocyclic ligand in the equatorial plane and two trans N atoms of the cyanide groups of the bridging tetracyanonickelate anion in the axial positions. In the crystal, two independent [1
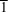 0] polymeric chains are cross-linked by N—H⋯Ow (w = water) and Ow—H⋯Nc (c = cyanide) hydrogen bonds into a three-dimensional network.
0] polymeric chains are cross-linked by N—H⋯Ow (w = water) and Ow—H⋯Nc (c = cyanide) hydrogen bonds into a three-dimensional network.
Keywords: crystal structure, coordination polymer, cyclam, nickel, hydrogen bonds
Abstract
The asymmetric unit of the title compound, catena-poly[[[(1,4,8,11-tetraazacyclotetradecane-κ4
N
1,N
4,N
8,N
11)nickel(II)]-μ-cyanido-κ2
N:C-[bis(cyanido-κC)nickel(II)]-μ-cyanido-κ2
C:N] dihydrate], {[Ni2(CN)4(C10H24N4)]·2H2O]
n
or [{[Ni(C10H24N4)][Ni(CN)4]}·2H2O]
n
, consists of a pair of crystallographically non-equivalent macrocyclic cations and anions. The nickel(II) ions (all with site symmetry
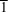 ) are coordinated by the four secondary N atoms of the macrocyclic ligands, which adopt the most energetically stable trans-III conformation, and the mutually trans N atoms of the tetracyanonickelate anion in a slightly tetragonally distorted NiN6 octahedral coordination geometry. The [Ni(CN)4)]2– anion exhibits a bridging function, resulting in the formation of parallel polymeric chains running along the [1
) are coordinated by the four secondary N atoms of the macrocyclic ligands, which adopt the most energetically stable trans-III conformation, and the mutually trans N atoms of the tetracyanonickelate anion in a slightly tetragonally distorted NiN6 octahedral coordination geometry. The [Ni(CN)4)]2– anion exhibits a bridging function, resulting in the formation of parallel polymeric chains running along the [1
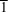 0] direction. The water molecules of crystallization play a pivotal role in the three-dimensional supramolecular organization of the crystal. Acting as acceptors, they form N—H⋯Ow (w = water) hydrogen bonds with the secondary amino groups of the macrocycles, forming layers oriented parallel to the (001) plane. At the same time, as donors, they interact with the non-coordinated cyano groups of the anion via Ow—H⋯Nc (c = cyanide) hydrogen bonds, giving two-dimensional layers oriented parallel to the (100) plane and thus generating a three-dimensional network.
0] direction. The water molecules of crystallization play a pivotal role in the three-dimensional supramolecular organization of the crystal. Acting as acceptors, they form N—H⋯Ow (w = water) hydrogen bonds with the secondary amino groups of the macrocycles, forming layers oriented parallel to the (001) plane. At the same time, as donors, they interact with the non-coordinated cyano groups of the anion via Ow—H⋯Nc (c = cyanide) hydrogen bonds, giving two-dimensional layers oriented parallel to the (100) plane and thus generating a three-dimensional network.
Chemical context
Transition-metal complexes of tetraazamacrocyclic ligands, in particular of 1,4,8,11-tetraazacyclotetradecane (cyclam, L), have been intensively studied for decades. This is explained by their unique properties, in particular, exceptionally high thermodynamic stability, kinetic inertness and the ability to stabilize uncommon oxidation states of coordinated metals (Melson, 1979 ▸; Yatsimirskii & Lampeka, 1985 ▸). Because of their conformational rigidity during chemical transformation (preservation of two vacant or labile trans axial positions in the coordination sphere of the metal ion), these complexes are also promising secondary building units for the construction of metal–organic frameworks (MOFs) (Lampeka & Tsymbal, 2004 ▸; Suh & Moon, 2007 ▸; Suh et al., 2012 ▸; Stackhouse & Ma, 2018 ▸), which possess great potential for applications in different areas including gas storage, separation, catalysis, sensing, etc (MacGillivray & Lukehart, 2014 ▸; Kaskel, 2016 ▸).
Cyanometallate anions refer to a type of bridging ligands for the creation of MOFs of different topologies possessing promising magnetic and electronic properties (Ohkoshi et al., 2019 ▸). Among such linkers, the tetracyanonickelate(II) dianion has attracted less attention compared to hexa- and octacyanometallates and only one work describing the structure of the coordination polymer formed by the metal(cyclam) complex and this anion, i.e., {Cu(L)[Ni(CN)4]}
n
, has been published to date (Černák et al., 2010 ▸). Interestingly, despite the diamagnetic nature of the bridging fragment, this complex displays a weak antiferromagnetic exchange coupling between the paramagnetic copper(II) centres.

We report herein the synthesis and crystal structure of the coordination polymer built up of the nickel(II) complex of L and the tetracyanonickelate(II) dianion, namely, catena-[bis(μ2-cyano-κ2C,N)-dicyano-(1,4,8,11-tetraazacyclotetradecane-κ4N1,N4,N8,N11)-dinickel(II) dihydrate], [{[Ni(L)][Ni(CN)4]}·2H2O] n , (I).
Structural commentary
The molecular structure of I is shown in Fig. 1 ▸. It represents a one-dimensional coordination polymer built up from two crystallographically independent centrosymmetric tetragonal macrocyclic [Ni(L)]2+ cations and tetracyanonickelate anions [Ni(CN)4]2–. The coordination of the trans cyanide groups of the anions in the axial positions of the coordination sphere of the metal ions in cations results in the formation of two structurally non-equivalent parallel polymeric chains (Ni1/Ni3 and Ni2/Ni4) running along the [1
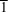 0] direction.
0] direction.
Figure 1.
The extended asymmetric unit in I showing the coordination environment of the Ni atoms and the atom-labelling scheme (displacement ellipsoids are drawn at the 40% probability level). C-bound H atoms are omitted for clarity. Dotted lines represent hydrogen-bonding interactions. Symmetry codes: (i) −x + 1, −y + 1, −z; (ii) −x, −y + 2, −z; (iii) x − 1, y + 1, z; (iv) −x + 1, −y, −z + 1; (v) −x, −y + 1, −z + 1.
The location of the metal ions on inversion centres enforces strict planarity of the Ni(N4) and Ni(C4) coordination moieties. The macrocyclic ligand in the complex cations adopts the most common and energetically favorable trans-III (R,R,S,S) conformation (Bosnich et al., 1965 ▸) with almost equal Ni—N bond lengths (Table 1 ▸). The five-membered chelate rings are present in gauche (bite angles ca 85.5°) and the six-membered in chair (bite angles ca 94.5°) conformations (Table 1 ▸). The geometric parameters observed are characteristic of high-spin d 8 nickel(II) complexes with macrocyclic 14-membered tetraamine ligands (Lampeka & Tsymbal, 2004 ▸; Tsymbal et al., 2021 ▸). The axial Ni—N(CN) bond lengths are somewhat longer than the Ni—N(amine) ones, resulting in a slight tetragonal distortion of the trans-NiN4N2 coordination polyhedron.
Table 1. Selected bond lengths (Å).
| Ni1—N5 | 2.100 (4) | Ni2—N4 | 2.079 (4) |
| Ni1—N2 | 2.070 (4) | Ni3—C11 | 1.874 (6) |
| Ni1—N1 | 2.082 (4) | Ni3—C12 | 1.857 (6) |
| Ni2—N3 | 2.069 (4) | Ni4—C13 | 1.866 (6) |
| Ni2—N7 | 2.095 (4) | Ni4—C14 | 1.863 (6) |
The Ni—C—N angles in the anion deviate only slightly (less than 4°) from linearity. In I, each tetracyanonickelate unit uses two trans cyanide groups for coordination to two macrocyclic moieties in a bent fashion [Ni—N—C = 166.1 (4)°], giving rise to a linear polymeric chain, whereas the two remaining trans CN− groups are monodentate. The adjacent Ni⋯Ni distance in the chain is 5.0558 (5) Å, and the shortest interchain Ni⋯Ni distance is 6.6159 (5) Å.
Supramolecular features
The crystals of I are composed of linear polymeric chains of [Ni(L)]2+ cations bridged by the [Ni(CN)4]2− anions, which propagate along the [1
 0] direction. There are no direct contacts between the chains and the water molecules of crystallization play a key role in assembling them into a three-dimensional supramolecular network. In particular, serving as the acceptor for N—H⋯O hydrogen bonds arising from the secondary amino groups of different macrocyclic ligands in the crystallographically equivalent chains (O1W for Ni1/Ni3, O2W for Ni2/Ni4), the water molecules link them in two-dimensional layers oriented parallel to the (001) plane (Table 2 ▸, Fig. 2 ▸
a). At the same time, acting as the donors in O—H⋯N hydrogen-bonding interactions with the nitrogen atoms of the non-coordinating cyanide groups of the anions belonging to crystallographically non-equivalent polymeric chains, they form two-dimensional layers oriented parallel to the (100) plane (Table 2 ▸, Fig. 2 ▸
b) thus realizing a three-dimensional system of hydrogen bonds in the crystal.
0] direction. There are no direct contacts between the chains and the water molecules of crystallization play a key role in assembling them into a three-dimensional supramolecular network. In particular, serving as the acceptor for N—H⋯O hydrogen bonds arising from the secondary amino groups of different macrocyclic ligands in the crystallographically equivalent chains (O1W for Ni1/Ni3, O2W for Ni2/Ni4), the water molecules link them in two-dimensional layers oriented parallel to the (001) plane (Table 2 ▸, Fig. 2 ▸
a). At the same time, acting as the donors in O—H⋯N hydrogen-bonding interactions with the nitrogen atoms of the non-coordinating cyanide groups of the anions belonging to crystallographically non-equivalent polymeric chains, they form two-dimensional layers oriented parallel to the (100) plane (Table 2 ▸, Fig. 2 ▸
b) thus realizing a three-dimensional system of hydrogen bonds in the crystal.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1W i | 0.98 | 2.28 | 3.115 (6) | 143 |
| N2—H2⋯O1W ii | 0.98 | 2.10 | 3.020 (6) | 156 |
| N3—H3⋯O2W i | 0.98 | 2.15 | 3.083 (7) | 159 |
| N4—H4⋯O2W iii | 0.98 | 2.26 | 3.080 (6) | 140 |
| O1W—H1WA⋯N8 | 0.85 | 2.03 | 2.872 (7) | 173 |
| O1W—H1WB⋯N6iv | 0.85 | 2.27 | 3.112 (7) | 171 |
| O2W—H2WA⋯N6 | 0.85 | 2.03 | 2.853 (6) | 164 |
| O2W—H2WB⋯N8i | 0.85 | 2.30 | 3.149 (7) | 175 |
Symmetry codes: (i) -x, -y+1, -z+1; (ii) x, y, z-1; (iii) x, y-1, z; (iv) -x, -y+2, -z+1.
Figure 2.
View of the sheets of polymeric chains formed due to the hydrogen-bond acceptor (a) and donor (b) properties of the water molecules of crystallization. The macrocyclic ligands in the crystallographically non-equivalent nickel ions are shown in violet (Ni1/Ni3) and green (Ni2/Ni4) colors.
Database survey
A search of the Cambridge Structural Database (CSD, version 5.42, last update February 2021; Groom et al., 2016 ▸) indicated that several one-dimensional coordination polymers formed by diazacyclam nickel(II) cations (diazacyclam = 1,3,5,8,10,12-hexaazacyclotetradecane) and the tetracyanonickelate anion have been characterized structurally. They include compounds with monomacrocylic (refcode MIMJIB; Kou et al., 2002 ▸) and macrotricyclic [NADVOE (Zhou et al., 2004 ▸), YUBHEK, YUBHIO and YUBHOU (Jiang et al., 2015 ▸)] tetradentate ligands. The structures of the polymeric chains in these compounds are very similar. In particular, because of comparable Ni—N(CN) bond lengths and Ni—N—C bond angles, the interchain Ni⋯Ni distances fall in the range 5.07–5.15 Å and are slightly longer than that observed in I. Surprisingly, a similar value for this parameter (5.056 Å) is also observed in the complex of the [Cu(L)]2+ cation with [Ni(CN)4]2− (XABGEO; Černák et al., 2010 ▸), despite the substantially longer Cu—N(CN) distance (2.532 Å). This feature is explained by the considerable bending of the Cu—N—C (133.0°) angle as compared the nickel analogues.
Synthesis and crystallization
All reagents and solvents used in this work were analytical grade and were used without further purification. The macrocyclic nickel(II) complex Ni(L)(ClO4)2 was prepared according to procedures described previously (Barefield et al., 1976 ▸).
[{[Ni( L )][Ni(CN)4]}·2H2O] n , (I): A solution of 121 mg (0.50 mmol) of K2[Ni(CN)4] in 15 ml of water was added under stirring to a solution of 290 mg (0.50 mmol) Ni(L)(ClO4)2 in 10 ml of dimethylformamide. Filtration and slow evaporation of the resulting solution gave after several days a light-yellow crystalline precipitate, which was washed with DMF, methanol and dried in air. Yield 160 mg (35%). Analysis calculated for C14H28N8Ni2O2: C, 36.72; H, 6.16; N, 24.47%. Found: C, 36.62; H, 6.26; N, 24.19%. Single crystals suitable for X-ray diffraction analysis were selected from the sample resulting from the synthesis. Safety note: perchlorate salts of metal complexes are potentially explosive and should be handled with care.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All H atoms in I were placed in geometrically idealized positions and constrained to ride on their parent atoms, with C—H = 0.97 Å, N—H = 0.98 Å and water O—H = 0.85 Å, with U iso(H) values of 1.2 or 1.5Ueq of the parent atoms.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [Ni2(CN)4(C10H24N4)]·2H2O |
| M r | 457.86 |
| Crystal system, space group | Triclinic, P\overline{1} |
| Temperature (K) | 180 |
| a, b, c (Å) | 7.7325 (6), 8.8809 (7), 15.7780 (12) |
| α, β, γ (°) | 88.673 (6), 85.682 (7), 74.623 (7) |
| V (Å3) | 1041.74 (15) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.83 |
| Crystal size (mm) | 0.30 × 0.20 × 0.06 |
| Data collection | |
| Diffractometer | Rigaku Xcalibur, Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2018 ▸) |
| T min, T max | 0.852, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7144, 3673, 2417 |
| R int | 0.036 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.059, 0.159, 1.07 |
| No. of reflections | 3673 |
| No. of parameters | 247 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 1.31, −0.42 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021010902/hb7992sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010902/hb7992Isup2.hkl
CCDC reference: 2114932
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Ni2(CN)4(C10H24N4)]·2H2O | Z = 2 |
| Mr = 457.86 | F(000) = 480 |
| Triclinic, P1 | Dx = 1.460 Mg m−3 |
| a = 7.7325 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.8809 (7) Å | Cell parameters from 2303 reflections |
| c = 15.7780 (12) Å | θ = 2.7–28.8° |
| α = 88.673 (6)° | µ = 1.83 mm−1 |
| β = 85.682 (7)° | T = 180 K |
| γ = 74.623 (7)° | Block, colourless |
| V = 1041.74 (15) Å3 | 0.30 × 0.20 × 0.06 mm |
Data collection
| Rigaku Xcalibur, Eos diffractometer | 3673 independent reflections |
| Radiation source: fine-focus sealed X-ray tube, Enhance (Mo) X-ray Source | 2417 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.036 |
| Detector resolution: 8.0797 pixels mm-1 | θmax = 25.0°, θmin = 2.4° |
| ω scans | h = −9→9 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2018) | k = −9→10 |
| Tmin = 0.852, Tmax = 1.000 | l = −17→18 |
| 7144 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.059 | H-atom parameters constrained |
| wR(F2) = 0.159 | w = 1/[σ2(Fo2) + (0.064P)2 + 1.1599P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 3673 reflections | Δρmax = 1.31 e Å−3 |
| 247 parameters | Δρmin = −0.42 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni4 | 0.000000 | 0.500000 | 0.500000 | 0.0201 (3) | |
| Ni3 | 0.000000 | 1.000000 | 0.000000 | 0.0195 (3) | |
| Ni1 | 0.500000 | 0.500000 | 0.000000 | 0.0172 (3) | |
| Ni2 | 0.500000 | 0.000000 | 0.500000 | 0.0177 (3) | |
| C13 | 0.1682 (7) | 0.3061 (6) | 0.4882 (3) | 0.0190 (11) | |
| N5 | 0.2721 (5) | 0.6907 (5) | 0.0165 (3) | 0.0232 (10) | |
| C8 | 0.3761 (7) | −0.2867 (6) | 0.4659 (4) | 0.0312 (14) | |
| H8A | 0.496131 | −0.353457 | 0.472014 | 0.037* | |
| H8B | 0.316556 | −0.338924 | 0.428528 | 0.037* | |
| N8 | −0.0359 (7) | 0.4416 (6) | 0.6889 (3) | 0.0424 (14) | |
| N3 | 0.3772 (6) | −0.0524 (5) | 0.6138 (3) | 0.0259 (11) | |
| H3 | 0.254063 | 0.015213 | 0.617490 | 0.031* | |
| N6 | −0.0089 (7) | 1.0667 (7) | 0.1866 (3) | 0.0459 (14) | |
| N2 | 0.4008 (6) | 0.4404 (5) | −0.1091 (3) | 0.0247 (11) | |
| H2 | 0.278118 | 0.507374 | −0.111458 | 0.030* | |
| C11 | 0.1664 (7) | 0.8059 (7) | 0.0125 (3) | 0.0205 (12) | |
| O2W | 0.0288 (6) | 0.9077 (5) | 0.3461 (2) | 0.0374 (10) | |
| H2WA | 0.036923 | 0.959680 | 0.300850 | 0.056* | |
| H2WB | 0.032480 | 0.814393 | 0.333553 | 0.056* | |
| C14 | −0.0241 (7) | 0.4667 (6) | 0.6168 (4) | 0.0273 (13) | |
| C10 | 0.5288 (8) | 0.0021 (7) | 0.3194 (4) | 0.0322 (14) | |
| H10A | 0.601399 | −0.007329 | 0.265908 | 0.039* | |
| H10B | 0.415596 | 0.079153 | 0.312246 | 0.039* | |
| N1 | 0.3727 (5) | 0.3666 (5) | 0.0802 (3) | 0.0224 (10) | |
| H1 | 0.247298 | 0.427127 | 0.090795 | 0.027* | |
| C3 | 0.3903 (7) | 0.2784 (7) | −0.1143 (4) | 0.0336 (14) | |
| H3A | 0.510949 | 0.209177 | −0.117835 | 0.040* | |
| H3B | 0.334252 | 0.265566 | −0.165612 | 0.040* | |
| N7 | 0.2769 (5) | 0.1898 (5) | 0.4838 (3) | 0.0231 (10) | |
| C4 | 0.5107 (7) | 0.4876 (7) | −0.1807 (3) | 0.0289 (14) | |
| H4A | 0.448916 | 0.495059 | −0.232483 | 0.035* | |
| H4B | 0.625088 | 0.409578 | −0.189027 | 0.035* | |
| C12 | −0.0032 (7) | 1.0368 (6) | 0.1155 (4) | 0.0273 (13) | |
| O1W | 0.0212 (6) | 0.5830 (5) | 0.8426 (3) | 0.0383 (11) | |
| H1WA | −0.004667 | 0.541681 | 0.798936 | 0.057* | |
| H1WB | 0.027685 | 0.675211 | 0.830710 | 0.057* | |
| C1 | 0.3698 (7) | 0.2153 (6) | 0.0457 (4) | 0.0300 (14) | |
| H1A | 0.303586 | 0.163745 | 0.086184 | 0.036* | |
| H1B | 0.491913 | 0.149679 | 0.037738 | 0.036* | |
| N4 | 0.3896 (5) | −0.1378 (5) | 0.4269 (3) | 0.0225 (10) | |
| H4 | 0.267023 | −0.077143 | 0.417091 | 0.027* | |
| C2 | 0.2837 (7) | 0.2331 (7) | −0.0382 (4) | 0.0373 (15) | |
| H2A | 0.169263 | 0.311348 | −0.031187 | 0.045* | |
| H2B | 0.258498 | 0.135002 | −0.050764 | 0.045* | |
| C6 | 0.3638 (7) | −0.2135 (7) | 0.6248 (4) | 0.0336 (15) | |
| H6A | 0.295970 | −0.222107 | 0.678108 | 0.040* | |
| H6B | 0.483387 | −0.283061 | 0.627483 | 0.040* | |
| C5 | 0.5424 (7) | 0.6439 (7) | −0.1614 (3) | 0.0295 (14) | |
| H5A | 0.619798 | 0.672475 | −0.206694 | 0.035* | |
| H5B | 0.428831 | 0.723764 | −0.157782 | 0.035* | |
| C9 | 0.4941 (7) | −0.1553 (7) | 0.3439 (3) | 0.0323 (14) | |
| H9A | 0.427445 | −0.187587 | 0.301267 | 0.039* | |
| H9B | 0.607290 | −0.234112 | 0.347689 | 0.039* | |
| C7 | 0.2724 (7) | −0.2638 (7) | 0.5524 (4) | 0.0364 (16) | |
| H7A | 0.244558 | −0.361196 | 0.568747 | 0.044* | |
| H7B | 0.159067 | −0.186226 | 0.546429 | 0.044* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni4 | 0.0180 (5) | 0.0115 (5) | 0.0275 (6) | 0.0020 (4) | −0.0016 (4) | −0.0006 (4) |
| Ni3 | 0.0170 (5) | 0.0117 (5) | 0.0265 (6) | 0.0015 (4) | −0.0005 (4) | −0.0005 (4) |
| Ni1 | 0.0152 (5) | 0.0111 (5) | 0.0234 (6) | −0.0001 (4) | −0.0012 (4) | −0.0006 (4) |
| Ni2 | 0.0177 (5) | 0.0099 (5) | 0.0228 (6) | 0.0010 (4) | −0.0005 (4) | −0.0019 (4) |
| C13 | 0.017 (3) | 0.020 (3) | 0.020 (3) | −0.006 (2) | 0.002 (2) | −0.001 (2) |
| N5 | 0.015 (2) | 0.012 (3) | 0.037 (3) | 0.005 (2) | −0.0038 (19) | −0.002 (2) |
| C8 | 0.024 (3) | 0.020 (3) | 0.051 (4) | −0.007 (3) | −0.007 (3) | −0.009 (3) |
| N8 | 0.058 (4) | 0.029 (3) | 0.037 (3) | −0.005 (3) | −0.004 (3) | −0.001 (3) |
| N3 | 0.022 (2) | 0.025 (3) | 0.029 (3) | −0.002 (2) | −0.0016 (19) | 0.002 (2) |
| N6 | 0.055 (4) | 0.045 (4) | 0.037 (4) | −0.012 (3) | 0.000 (3) | −0.002 (3) |
| N2 | 0.020 (2) | 0.023 (3) | 0.028 (3) | −0.001 (2) | −0.0050 (18) | −0.003 (2) |
| C11 | 0.024 (3) | 0.019 (3) | 0.020 (3) | −0.007 (2) | −0.006 (2) | −0.001 (2) |
| O2W | 0.041 (2) | 0.030 (3) | 0.039 (3) | −0.004 (2) | −0.0061 (19) | 0.005 (2) |
| C14 | 0.020 (3) | 0.014 (3) | 0.044 (4) | 0.004 (2) | −0.005 (2) | −0.002 (3) |
| C10 | 0.035 (3) | 0.029 (4) | 0.029 (3) | −0.001 (3) | −0.002 (2) | −0.005 (3) |
| N1 | 0.022 (2) | 0.018 (3) | 0.026 (3) | −0.0045 (19) | 0.0040 (18) | 0.000 (2) |
| C3 | 0.032 (3) | 0.025 (4) | 0.046 (4) | −0.008 (3) | −0.008 (3) | −0.011 (3) |
| N7 | 0.017 (2) | 0.014 (3) | 0.034 (3) | 0.002 (2) | −0.0014 (18) | 0.003 (2) |
| C4 | 0.028 (3) | 0.034 (4) | 0.023 (3) | −0.006 (3) | −0.001 (2) | −0.003 (3) |
| C12 | 0.027 (3) | 0.015 (3) | 0.039 (4) | −0.005 (2) | 0.003 (2) | 0.005 (3) |
| O1W | 0.038 (2) | 0.033 (3) | 0.042 (3) | −0.003 (2) | −0.0109 (19) | −0.006 (2) |
| C1 | 0.019 (3) | 0.012 (3) | 0.057 (4) | −0.004 (2) | 0.008 (2) | 0.007 (3) |
| N4 | 0.021 (2) | 0.013 (2) | 0.034 (3) | −0.0037 (19) | −0.0065 (18) | −0.006 (2) |
| C2 | 0.025 (3) | 0.026 (4) | 0.064 (4) | −0.011 (3) | −0.003 (3) | −0.010 (3) |
| C6 | 0.027 (3) | 0.027 (4) | 0.045 (4) | −0.006 (3) | 0.003 (3) | 0.010 (3) |
| C5 | 0.024 (3) | 0.030 (4) | 0.028 (3) | 0.002 (3) | 0.003 (2) | 0.009 (3) |
| C9 | 0.038 (3) | 0.030 (4) | 0.026 (3) | −0.002 (3) | −0.004 (2) | −0.012 (3) |
| C7 | 0.018 (3) | 0.027 (4) | 0.064 (5) | −0.008 (3) | −0.002 (3) | 0.010 (3) |
Geometric parameters (Å, º)
| Ni1—N5 | 2.100 (4) | N2—C4 | 1.482 (7) |
| Ni1—N5i | 2.100 (4) | O2W—H2WA | 0.8491 |
| Ni1—N2i | 2.070 (4) | O2W—H2WB | 0.8493 |
| Ni1—N2 | 2.070 (4) | C10—H10A | 0.9700 |
| Ni1—N1i | 2.082 (4) | C10—H10B | 0.9700 |
| Ni1—N1 | 2.082 (4) | C10—C9 | 1.528 (8) |
| Ni2—N3ii | 2.069 (4) | N1—H1 | 0.9800 |
| Ni2—N3 | 2.069 (4) | N1—C1 | 1.468 (7) |
| Ni2—N7 | 2.095 (4) | N1—C5i | 1.474 (6) |
| Ni2—N7ii | 2.095 (4) | C3—H3A | 0.9700 |
| Ni2—N4 | 2.079 (4) | C3—H3B | 0.9700 |
| Ni2—N4ii | 2.079 (4) | C3—C2 | 1.512 (8) |
| Ni3—C11 | 1.874 (6) | C4—H4A | 0.9700 |
| Ni3—C11iii | 1.874 (6) | C4—H4B | 0.9700 |
| Ni3—C12iii | 1.857 (6) | C4—C5 | 1.514 (8) |
| Ni3—C12 | 1.857 (6) | O1W—H1WA | 0.8499 |
| Ni4—C13iv | 1.866 (6) | O1W—H1WB | 0.8492 |
| Ni4—C13 | 1.866 (6) | C1—H1A | 0.9700 |
| Ni4—C14iv | 1.863 (6) | C1—H1B | 0.9700 |
| Ni4—C14 | 1.863 (6) | C1—C2 | 1.511 (8) |
| C13—N7 | 1.145 (7) | N4—H4 | 0.9800 |
| N5—C11 | 1.132 (7) | N4—C9 | 1.475 (7) |
| C8—H8A | 0.9700 | C2—H2A | 0.9700 |
| C8—H8B | 0.9700 | C2—H2B | 0.9700 |
| C8—N4 | 1.470 (6) | C6—H6A | 0.9700 |
| C8—C7 | 1.521 (8) | C6—H6B | 0.9700 |
| N8—C14 | 1.156 (7) | C6—C7 | 1.521 (8) |
| N3—H3 | 0.9800 | C5—H5A | 0.9700 |
| N3—C10ii | 1.466 (7) | C5—H5B | 0.9700 |
| N3—C6 | 1.468 (7) | C9—H9A | 0.9700 |
| N6—C12 | 1.154 (7) | C9—H9B | 0.9700 |
| N2—H2 | 0.9800 | C7—H7A | 0.9700 |
| N2—C3 | 1.467 (7) | C7—H7B | 0.9700 |
| C13iv—Ni4—C13 | 180.0 | N3ii—C10—C9 | 109.3 (4) |
| C14iv—Ni4—C13iv | 89.7 (2) | H10A—C10—H10B | 108.3 |
| C14—Ni4—C13iv | 90.3 (2) | C9—C10—H10A | 109.8 |
| C14iv—Ni4—C13 | 90.3 (2) | C9—C10—H10B | 109.8 |
| C14—Ni4—C13 | 89.7 (2) | Ni1—N1—H1 | 107.1 |
| C14iv—Ni4—C14 | 180.0 | C1—N1—Ni1 | 115.1 (3) |
| C11—Ni3—C11iii | 180.0 | C1—N1—H1 | 107.1 |
| C12—Ni3—C11 | 90.1 (2) | C1—N1—C5i | 114.3 (4) |
| C12iii—Ni3—C11iii | 90.1 (2) | C5i—N1—Ni1 | 105.7 (3) |
| C12—Ni3—C11iii | 89.9 (2) | C5i—N1—H1 | 107.1 |
| C12iii—Ni3—C11 | 89.9 (2) | N2—C3—H3A | 109.2 |
| C12iii—Ni3—C12 | 180.0 | N2—C3—H3B | 109.2 |
| N5i—Ni1—N5 | 180.0 | N2—C3—C2 | 111.9 (5) |
| N2—Ni1—N5 | 89.31 (17) | H3A—C3—H3B | 107.9 |
| N2—Ni1—N5i | 90.69 (17) | C2—C3—H3A | 109.2 |
| N2i—Ni1—N5i | 89.31 (17) | C2—C3—H3B | 109.2 |
| N2i—Ni1—N5 | 90.69 (17) | C13—N7—Ni2 | 166.1 (4) |
| N2i—Ni1—N2 | 180.00 (11) | N2—C4—H4A | 109.8 |
| N2—Ni1—N1i | 85.61 (17) | N2—C4—H4B | 109.8 |
| N2i—Ni1—N1 | 85.61 (17) | N2—C4—C5 | 109.6 (5) |
| N2i—Ni1—N1i | 94.39 (17) | H4A—C4—H4B | 108.2 |
| N2—Ni1—N1 | 94.39 (17) | C5—C4—H4A | 109.8 |
| N1i—Ni1—N5 | 90.34 (17) | C5—C4—H4B | 109.8 |
| N1—Ni1—N5 | 89.66 (17) | N6—C12—Ni3 | 176.9 (5) |
| N1i—Ni1—N5i | 89.66 (17) | H1WA—O1W—H1WB | 109.4 |
| N1—Ni1—N5i | 90.34 (17) | N1—C1—H1A | 109.2 |
| N1i—Ni1—N1 | 180.0 | N1—C1—H1B | 109.2 |
| N3ii—Ni2—N3 | 180.0 | N1—C1—C2 | 111.9 (4) |
| N3ii—Ni2—N7ii | 89.44 (17) | H1A—C1—H1B | 107.9 |
| N3—Ni2—N7ii | 90.56 (17) | C2—C1—H1A | 109.2 |
| N3ii—Ni2—N7 | 90.56 (17) | C2—C1—H1B | 109.2 |
| N3—Ni2—N7 | 89.44 (17) | Ni2—N4—H4 | 107.0 |
| N3ii—Ni2—N4 | 85.36 (17) | C8—N4—Ni2 | 115.6 (3) |
| N3—Ni2—N4ii | 85.36 (17) | C8—N4—H4 | 107.0 |
| N3—Ni2—N4 | 94.64 (17) | C8—N4—C9 | 113.8 (4) |
| N3ii—Ni2—N4ii | 94.64 (17) | C9—N4—Ni2 | 105.9 (3) |
| N7ii—Ni2—N7 | 180.0 | C9—N4—H4 | 107.0 |
| N4ii—Ni2—N7ii | 89.87 (17) | C3—C2—H2A | 108.1 |
| N4ii—Ni2—N7 | 90.13 (17) | C3—C2—H2B | 108.1 |
| N4—Ni2—N7ii | 90.13 (17) | C1—C2—C3 | 116.7 (4) |
| N4—Ni2—N7 | 89.87 (17) | C1—C2—H2A | 108.1 |
| N4ii—Ni2—N4 | 180.0 | C1—C2—H2B | 108.1 |
| N7—C13—Ni4 | 176.4 (5) | H2A—C2—H2B | 107.3 |
| C11—N5—Ni1 | 166.1 (4) | N3—C6—H6A | 109.2 |
| H8A—C8—H8B | 107.9 | N3—C6—H6B | 109.2 |
| N4—C8—H8A | 109.2 | N3—C6—C7 | 111.9 (4) |
| N4—C8—H8B | 109.2 | H6A—C6—H6B | 107.9 |
| N4—C8—C7 | 112.1 (5) | C7—C6—H6A | 109.2 |
| C7—C8—H8A | 109.2 | C7—C6—H6B | 109.2 |
| C7—C8—H8B | 109.2 | N1i—C5—C4 | 109.3 (4) |
| Ni2—N3—H3 | 106.9 | N1i—C5—H5A | 109.8 |
| C10ii—N3—Ni2 | 105.8 (3) | N1i—C5—H5B | 109.8 |
| C10ii—N3—H3 | 106.9 | C4—C5—H5A | 109.8 |
| C10ii—N3—C6 | 113.3 (4) | C4—C5—H5B | 109.8 |
| C6—N3—Ni2 | 116.6 (4) | H5A—C5—H5B | 108.3 |
| C6—N3—H3 | 106.9 | C10—C9—H9A | 110.1 |
| Ni1—N2—H2 | 107.0 | C10—C9—H9B | 110.1 |
| C3—N2—Ni1 | 116.2 (3) | N4—C9—C10 | 108.2 (5) |
| C3—N2—H2 | 107.0 | N4—C9—H9A | 110.1 |
| C3—N2—C4 | 113.7 (4) | N4—C9—H9B | 110.1 |
| C4—N2—Ni1 | 105.5 (3) | H9A—C9—H9B | 108.4 |
| C4—N2—H2 | 107.0 | C8—C7—C6 | 117.0 (4) |
| N5—C11—Ni3 | 176.5 (5) | C8—C7—H7A | 108.1 |
| H2WA—O2W—H2WB | 109.4 | C8—C7—H7B | 108.1 |
| N8—C14—Ni4 | 178.0 (5) | C6—C7—H7A | 108.1 |
| N3ii—C10—H10A | 109.8 | C6—C7—H7B | 108.1 |
| N3ii—C10—H10B | 109.8 | H7A—C7—H7B | 107.3 |
| Ni1—N2—C3—C2 | 54.8 (5) | N2—C4—C5—N1i | 56.3 (6) |
| Ni1—N2—C4—C5 | −40.9 (5) | C10ii—N3—C6—C7 | −177.7 (5) |
| Ni1—N1—C1—C2 | −56.5 (5) | N1—C1—C2—C3 | 71.8 (7) |
| Ni2—N3—C6—C7 | −54.5 (5) | C3—N2—C4—C5 | −169.4 (4) |
| Ni2—N4—C9—C10 | −40.9 (5) | C4—N2—C3—C2 | 177.5 (4) |
| C8—N4—C9—C10 | −169.0 (4) | N4—C8—C7—C6 | −70.7 (6) |
| N3ii—C10—C9—N4 | 57.3 (6) | C5i—N1—C1—C2 | −179.1 (4) |
| N3—C6—C7—C8 | 69.7 (6) | C7—C8—N4—Ni2 | 55.4 (5) |
| N2—C3—C2—C1 | −70.5 (7) | C7—C8—N4—C9 | 178.2 (4) |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) −x+1, −y, −z+1; (iii) −x, −y+2, −z; (iv) −x, −y+1, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1Wiv | 0.98 | 2.28 | 3.115 (6) | 143 |
| N2—H2···O1Wv | 0.98 | 2.10 | 3.020 (6) | 156 |
| N3—H3···O2Wiv | 0.98 | 2.15 | 3.083 (7) | 159 |
| N4—H4···O2Wvi | 0.98 | 2.26 | 3.080 (6) | 140 |
| O1W—H1WA···N8 | 0.85 | 2.03 | 2.872 (7) | 173 |
| O1W—H1WB···N6vii | 0.85 | 2.27 | 3.112 (7) | 171 |
| O2W—H2WA···N6 | 0.85 | 2.03 | 2.853 (6) | 164 |
| O2W—H2WB···N8iv | 0.85 | 2.30 | 3.149 (7) | 175 |
Symmetry codes: (iv) −x, −y+1, −z+1; (v) x, y, z−1; (vi) x, y−1, z; (vii) −x, −y+2, −z+1.
References
- Barefield, E. K., Wagner, F., Herlinger, A. W. & Dahl, A. R. (1976). Inorg. Synth. 16, 220–224.
- Bosnich, B., Poon, C. K. & Tobe, M. C. (1965). Inorg. Chem. 4, 1102–1108.
- Černák, J., Kuchár, J., Stolárová, M., Kajňaková, M., Vavra, M., Potočňák, I., Falvello, L. R. & Tomás, M. (2010). Transition Met. Chem. 35, 737–744.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jiang, X., Tao, B., Yu, X., Wang, Y. & Xia, H. (2015). RSC Adv. 5, 19034–19040.
- Kaskel, S. (2016). Editor. The Chemistry of Metal–Organic Frameworks: Synthesis, Characterization, and Applications, 2 volumes. Weinheim: Wiley-VCH.
- Kou, H.-Z., Si, S.-F., Gao, S., Liao, D.-Z., Jiang, Z.-H., Yan, S.-P., Fan, Y.-G. & Wang, G.-L. (2002). Eur. J. Inorg. Chem. pp. 699–702.
- Lampeka, Ya. D. & Tsymbal, L. V. (2004). Theor. Exp. Chem. 40, 345–371.
- MacGillivray, L. R. & Lukehart, C. M. (2014). Editors. Metal–Organic Framework Materials. Hoboken: John Wiley and Sons.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Melson, G. A. (1979). Editor. Coordination Chemistry of Macrocyclic Compounds. New York: Plenum Press.
- Ohkoshi, S., Namai, A. & Tokoro, H. (2019). Coord. Chem. Rev. 380, 572–583.
- Rigaku OD (2018). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Stackhouse, C. A. & Ma, S. (2018). Polyhedron, 145, 154–165.
- Suh, M. P. & Moon, H. R. (2007). Advances in Inorganic Chemistry, Vol. 59, edited by R. van Eldik & K. Bowman-James, pp. 39–79. San Diego: Academic Press.
- Suh, M. P., Park, H. J., Prasad, T. K. & Lim, D.-W. (2012). Chem. Rev. 112, 782–835. [DOI] [PubMed]
- Tsymbal, L. V., Andriichuk, I. L., Shova, S., Trzybiński, D., Woźniak, K., Arion, V. B. & Lampeka, Ya. D. (2021). Cryst. Growth Des. 21, 2355–2370.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yatsimirskii, K. B. & Lampeka, Ya. D. (1985). Physicochemistry of Metal Complexes with Macrocyclic Ligands. Kiev: Naukova Dumka. (In Russian)
- Zhou, H.-B., Dong, W., Zhu, L.-N., Yu, L.-H., Wang, Q.-L., Liao, D.-Z., Jiang, Z.-H., Yan, S.-P. & Cheng, P. (2004). J. Mol. Struct. 703, 103–106.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021010902/hb7992sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010902/hb7992Isup2.hkl
CCDC reference: 2114932
Additional supporting information: crystallographic information; 3D view; checkCIF report




