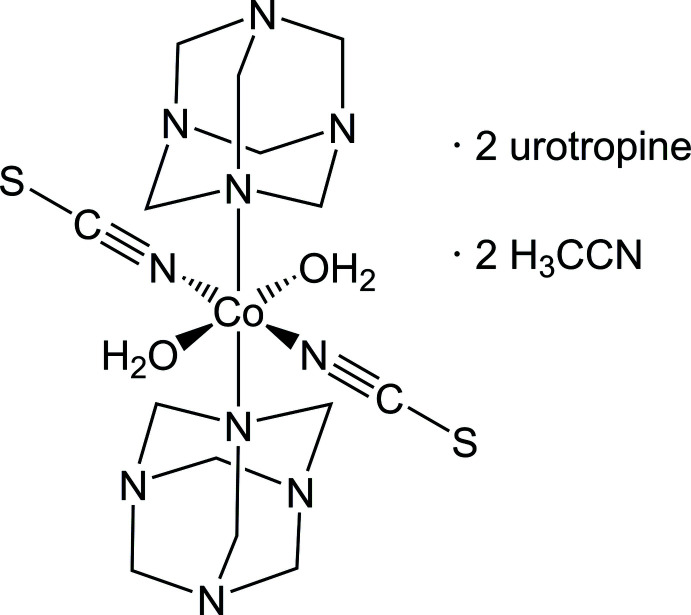
The crystal structure of the title compound consists of discrete neutral complexes in which the cobalt cations are octahedrally coordinated by two N-bonded thiocyanate anions, two hexamethylenetetraamine ligands and two water molecules with additional acetonitrile and hexamethylenetetramine solvate molecules, which are hydrogen bonded to the complexes.
Keywords: crystal structure, cobalt thiocyanate, hexamethylenetetramine, discrete complex, hydrogen bonding
Abstract
The crystal structure of the title solvated coordination compound, [Co(NCS)2(C6H12N4)2(H2O)2]·2C6H12N4·2C2H3N, consists of discrete complexes in which the Co2+ cations (site symmetry
 ) are sixfold coordinated by two N-bonded thiocyanate anions, two water molecules and two hexamethylenetetramine (HMT) molecules to generate distorted trans-CoN4O2 octahedra. The discrete complexes are each connected by two HMT solvate molecules into chains via strong O—H⋯N hydrogen bonds. These chains are further linked by additional O—H⋯N and C—H⋯N and C—H⋯S hydrogen bonds into a three-dimensional network. Within this network, channels are formed that propagate along the c-axis direction and in which additional acetonitrile solvent molecules are embedded, which are hydrogen bonded to the network. The CN stretching vibration of the thiocyanate ion occurs at 2062 cm−1, which is in agreement with the presence of N-bonded anionic ligands. XRPD investigations prove the formation of the title compound as the major phase accompanied by a small amount of a second unknown phase.
) are sixfold coordinated by two N-bonded thiocyanate anions, two water molecules and two hexamethylenetetramine (HMT) molecules to generate distorted trans-CoN4O2 octahedra. The discrete complexes are each connected by two HMT solvate molecules into chains via strong O—H⋯N hydrogen bonds. These chains are further linked by additional O—H⋯N and C—H⋯N and C—H⋯S hydrogen bonds into a three-dimensional network. Within this network, channels are formed that propagate along the c-axis direction and in which additional acetonitrile solvent molecules are embedded, which are hydrogen bonded to the network. The CN stretching vibration of the thiocyanate ion occurs at 2062 cm−1, which is in agreement with the presence of N-bonded anionic ligands. XRPD investigations prove the formation of the title compound as the major phase accompanied by a small amount of a second unknown phase.
Chemical context
For several years, we have been interested in the synthesis of coordination compounds based on cobalt thiocyanate and additional co-ligands that in most cases consist of N-donor ligands. As is the case for, e.g. cyanides and azides, even this anionic ligand is able to mediate reasonable magnetic exchange (Mekuimemba et al., 2018 ▸; Mousavi et al., 2020 ▸; Palion-Gazda et al., 2015 ▸). Therefore, we have focused especially on compounds in which the metal cations are linked by anionic ligands into coordination polymers. Most of the compounds with monocoordinating co-ligands consist of linear chains and show antiferromagnetic or ferromagnetic ordering or are single-chain magnets (Shi et al., 2006 ▸; Jin et al., 2007 ▸; Prananto et al., 2017 ▸; Mautner et al., 2018 ▸; Rams et al., 2020 ▸; Ceglarska et al., 2021 ▸; Werner et al., 2014 ▸, 2015 ▸), whereas in compounds with non-linear chains the magnetic exchange is completely suppressed (Böhme et al., 2020 ▸). In some cases, layered compounds are obtained, that are exclusively ferromagnets (Suckert et al., 2016 ▸; Wellm et al., 2020 ▸). All these compounds have in common that only monocoordinating co-ligands are used, which means that the thiocyanate substructures are not additionally connected into structures of higher dimensionality. We have therefore tried to link the Co(NCS)2 chains or layers by bridging co-ligands.
In this context, we became interested in urotropine, C6H12N4 (also called hexamethylenetetramine or 1,3,5,7-tetraazaadamantane), as a co-ligand. On one hand, this ligand is magnetically silent and on the other hand it is able to form tetrahedral networks and some examples have been reported in the literature (Czubacka et al., 2012 ▸; Li et al., 2012 ▸). It is noted that some compounds with this ligand and Co(NCS)2 have already been reported in the literature. In all cases, discrete complexes are formed in which the cobalt cations are octahedrally coordinated by two thiocyanate anions and some water, methanol or urotropine ligands (see Database survey). Compounds with urotropine in which the cobalt cations are linked by bridging thiocyanate anions have not been reported.
In the course of this project, we reacted Co(NCS)2 with urotropine in acetonitrile, resulting in the formation of a light-yellow-colored crystalline phase, for which IR spectroscopic investigations revealed the CN stretching vibration to be 2062 cm−1. This indicates the presence of only N-bonded thiocyanate anions (see Fig. S1 in the supporting information). To identify this compound, a single-crystal structure analysis was performed, which proves that a discrete complex has formed. Comparison of the X-ray powder pattern of this crystalline phase with that calculated from single-crystal data reveals that the title compound has formed as the major phase, but that there are still some reflections indicating the formation of an additional and unknown crystalline phase (Fig. S2).
Structural commentary
In the crystal structure of the title compound, [Co(NCS)2(H2O)2(C6H12N4)2]·(C6H12N4)2(C2H3N)2, the cobalt cations are each octahedrally coordinated by two N-bonded thiocyanate anions, two urotropine molecules and two water molecules to form discrete complexes that are located on centers of inversion (Fig. 1 ▸). The Co1—O1 and the thiocyanate Co1—N1 bond lengths are similar, whereas the Co1—N11 bond length to the neutral co-ligand is significantly longer (Table 1 ▸). The cis-angles around the Co centers deviate from ideal values, showing that the octahedra are slightly distorted [range of cis bond angles = 87.51 (4)–92.49 (4)°]. This is also apparent from the value of the octahedral angle variance of 2.540°2 and the mean octahedral quadratic elongation of 1.006 calculated by the method of Robinson et al. (1971 ▸).
Figure 1.
Crystal structure of the title compound with atom labeling and displacement ellipsoids drawn at the 50% probability level. Symmetry operation for the generation of equivalent atoms: (A) −x + 1, −y + 1, −z + 1.
Table 1. Selected geometric parameters (Å, °).
| Co1—N1 | 2.0744 (10) | Co1—N11 | 2.3112 (9) |
| Co1—O1 | 2.0661 (8) | ||
| C1—N1—Co1 | 161.53 (9) |
Supramolecular features
The crystal structure of the title compound is dominated by a variety of intermolecular O—H⋯N, C—H⋯N and C—H⋯S hydrogen bonds (Table 2 ▸). Each complex molecule is connected to two adjacent non-coordinating urotropine solvate molecules via O—H⋯N hydrogen bonds from one of the water H atoms. The O—H⋯N angle is close to linear and the N⋯H distance amounts to 1.85 (2) Å, which indicates a very strong interaction (Fig. 2 ▸). The complex molecules are linked by the urotropine solvate molecules into chains (Fig. 3 ▸). The chains are further connected by an O—H⋯N hydrogen bond arising from the second water hydrogen atom into layers, and these layers are further linked into a three-dimensional network by a number of weak C—H⋯N and C—H⋯S hydrogen bonds. In this way, channels are formed along the crystallographic c-axis direction in which additional acetonitrile molecules are located (Fig. 4 ▸). These molecules are linked to the main network via C—H⋯N interactions (Table 2 ▸).
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1A⋯N21 | 0.88 (2) | 1.85 (2) | 2.7298 (13) | 171.8 (18) |
| O1—H1B⋯N22i | 0.88 (2) | 2.01 (2) | 2.8759 (13) | 167.7 (19) |
| C12—H12B⋯O1 | 0.97 | 2.60 | 3.0752 (14) | 111 |
| C13—H13A⋯S1i | 0.97 | 2.95 | 3.8089 (12) | 148 |
| C13—H13B⋯N24ii | 0.97 | 2.66 | 3.5045 (16) | 146 |
| C16—H16A⋯O1iii | 0.97 | 2.52 | 3.0571 (14) | 115 |
| C16—H16B⋯N1 | 0.97 | 2.70 | 3.2713 (15) | 118 |
| C21—H21A⋯S1 | 0.97 | 3.00 | 3.9471 (12) | 165 |
| C23—H23B⋯S1iv | 0.97 | 2.89 | 3.6683 (12) | 138 |
| C26—H26A⋯N31iii | 0.97 | 2.56 | 3.4794 (17) | 158 |
| C32—H32A⋯S1v | 0.96 | 3.02 | 3.9560 (15) | 166 |
| C32—H32B⋯N23vi | 0.96 | 2.58 | 3.4685 (16) | 154 |
| C32—H32C⋯N14v | 0.96 | 2.61 | 3.4750 (17) | 149 |
Symmetry codes: (i) x, -y+{\script{3\over 2}}, z-{\script{1\over 2}}; (ii) -x+1, y+{\script{1\over 2}}, -z+{\script{3\over 2}}; (iii) -x+1, -y+1, -z+1; (iv) -x+1, -y+1, -z+2; (v) -x, -y+1, -z+1; (vi) x-1, -y+{\script{3\over 2}}, z-{\script{1\over 2}}.
Figure 2.
View of a discrete complex that is connected to two hexamethylenetramine solvent molecules via O—H⋯N hydrogen bonds (dashed lines).
Figure 3.
Part of the crystal structure of the title compound showing the connection of discrete complexes by hexamethylenetramine solvate molecules via O—H⋯N hydrogen bonds (dashed lines).
Figure 4.
Crystal structure of the title compound viewed along the c-axis.
Database survey
Some crystal structures have already been deposited in the Cambridge Structure Database (CSD version 5.42, last update November 2020; Groom et al., 2016 ▸) that contain cobalt cations, thiocyanate anions and urotropine molecules. These include [Co(NCS)2(C6H12N4)(CH3OH)2(H2O)] (refcode: POFGAT; Shang et al., 2008 ▸), which consists of neutral complexes in which the cobalt cations are octahedrally coordinated by the N atoms of two thiocyanate anions, two methanol, one water and one urotropine ligand to generate a mer-CoN3O3 coordination polyhedron. [Co(NCS)2(H2O)4]·2C6H12N4 (XILXOG; Li et al., 2007 ▸) is a discrete complex with a cobalt cation coordinated octahedrally by two thiocyanate anions and four water ligands (as a trans-CoN2O4 octahedron) with two additional urotropine solvent molecules. The structure of [Co(NCS)2(C6H12N4)2(H2O)2][Co(NCS)2(H2O)4]·2H2O has been determined several times (MOTNIS; Liu et al., 2002 ▸; MOTNIS01; Zhang et al., 1999 ▸; MOTNIS02; Chakraborty et al., 2006 ▸; MOTNIS03; Lu et al., 2010 ▸) and contains two discrete octahedral cobalt complexes: one metal ion is coordinated by two thiocyanate anions, two water molecules and two urotropine molecules (trans-CoN4O2 octahedron) and the other by two thiocyanate anions and four water molecules (trans-CoO4N2 octahedron); two water molecules of crystallization complete the structure.
Synthesis and crystallization
Synthesis
Co(NCS)2 and urotropine were purchased from Merck. All chemicals were used without further purification.
Light-yellow-colored single crystals suitable for single crystal X-ray analysis were obtained after heating 0.15 mmol Co(NCS)2 (26.3 mg) and 0.30 mmol urotropine (42.1 mg) in 0.5 ml MeCN up to 353 K and then storing the mixture at 333 K overnight.
Since it was not possible to obtain a crystalline powder of the title component from solution, a sample was taken from the single crystal batch, crushed and measured.
IR: ν = 2967 (w), 2958 (sh), 2930 (sh), 2920 (w), 2881 (w), 2309 (vw), 2281 (w), 2252 (vw), 2234 (vw), 2185 (vw), 2168 (vw), 2062 (s), 1952 (vw), 1684–1560 (vw), 1461 (m), 1417 (sh), 1378 (w), 1372 (w), 1363 (w), 1325 (vw), 1231 (s), 1049 (w), 994 (vs), 935 (w), 917 (m), 825 (m), 800 (m), 782 (m), 770 (m), 731 (sh), 690 (s), 662 (s), 506 (m) cm−1.
Experimental details
The data collection for the single-crystal structure analysis was performed using an XtaLAB Synergy, Dualflex, HyPix diffractometer from Rigaku with Cu Kα radiation.
The PXRD measurement was performed with Cu Kα1 radiation (λ = 1.540598 Å) using a Stoe transmission powder diffraction system (STADI P) equipped with a MYTHEN 1K detector and a Johansson-type Ge(111) monochromator.
The IR spectrum was measured using an ATI Mattson Genesis Series FTIR spectrometer, control software: WINFIRST, from ATI Mattson.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All non-hydrogen atoms were refined anisotropically. Water O atoms were freely refined. The C-bound H atoms were located in a difference map but positioned with idealized geometry (C—H = 0.96–0.97 Å, methyl H atoms allowed to rotate but not to tip) and were refined isotropically with U iso(H) = 1.2U eq(C) (1.5 for methyl H atoms) using a riding model.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [Co(NCS)2(C6H12N4)2(H2O)2]·2C6H12N4·2C2H3N |
| M r | 854.01 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 13.0008 (2), 12.5903 (2), 12.9988 (2) |
| β (°) | 114.899 (2) |
| V (Å3) | 1929.93 (6) |
| Z | 2 |
| Radiation type | Cu Kα |
| μ (mm−1) | 4.99 |
| Crystal size (mm) | 0.20 × 0.04 × 0.03 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Dualflex, HyPix |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2021 ▸) |
| T min, T max | 0.779, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 25388, 4088, 3972 |
| R int | 0.021 |
| (sin θ/λ)max (Å−1) | 0.635 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.025, 0.071, 1.09 |
| No. of reflections | 4088 |
| No. of parameters | 259 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.39 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021010033/hb7982sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010033/hb7982Isup2.hkl
Fig. S1. Experimental (top) and calculated X-ray powder pattern (bottom) of the title compound. The title compound is contaminated with at least one additional unknown phase. For the calculation of the powder pattern the data obtained from a a single crystal measured at 24 C was used. DOI: 10.1107/S2056989021010033/hb7982sup3.png
Fig. S2. IR spectra of the title compound. The value of the CN stretching vibration of the thiocyanat anions is given. DOI: 10.1107/S2056989021010033/hb7982sup4.png
CCDC reference: 2112185
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Co(NCS)2(C6H12N4)2(H2O)2]·2C6H12N4·2C2H3N | F(000) = 906 |
| Mr = 854.01 | Dx = 1.470 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54178 Å |
| a = 13.0008 (2) Å | Cell parameters from 20611 reflections |
| b = 12.5903 (2) Å | θ = 3.7–77.9° |
| c = 12.9988 (2) Å | µ = 4.99 mm−1 |
| β = 114.899 (2)° | T = 100 K |
| V = 1929.93 (6) Å3 | Needle, light yellow |
| Z = 2 | 0.20 × 0.04 × 0.03 mm |
Data collection
| XtaLAB Synergy, Dualflex, HyPix diffractometer | 4088 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, PhotonJet (Cu) X-ray Source | 3972 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.021 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 78.0°, θmin = 3.8° |
| ω scans | h = −12→16 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2021) | k = −15→15 |
| Tmin = 0.779, Tmax = 1.000 | l = −16→16 |
| 25388 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.025 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.071 | w = 1/[σ2(Fo2) + (0.0427P)2 + 0.5759P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max = 0.001 |
| 4088 reflections | Δρmax = 0.20 e Å−3 |
| 259 parameters | Δρmin = −0.39 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Co1 | 0.500000 | 0.500000 | 0.500000 | 0.01162 (8) | |
| N1 | 0.49434 (8) | 0.46285 (8) | 0.65301 (8) | 0.01532 (19) | |
| C1 | 0.46823 (9) | 0.46250 (9) | 0.72848 (10) | 0.0135 (2) | |
| S1 | 0.43052 (3) | 0.46382 (2) | 0.83365 (3) | 0.02096 (8) | |
| O1 | 0.62345 (7) | 0.61232 (7) | 0.58179 (7) | 0.01480 (16) | |
| H1A | 0.6441 (16) | 0.6265 (15) | 0.6543 (17) | 0.034 (5)* | |
| H1B | 0.6416 (17) | 0.6677 (18) | 0.5509 (18) | 0.046 (6)* | |
| N11 | 0.36295 (8) | 0.62692 (8) | 0.47457 (8) | 0.01335 (19) | |
| N12 | 0.31137 (9) | 0.78613 (8) | 0.55340 (9) | 0.0178 (2) | |
| N13 | 0.23595 (9) | 0.76282 (8) | 0.34775 (9) | 0.0188 (2) | |
| C11 | 0.32652 (10) | 0.68571 (9) | 0.36510 (10) | 0.0175 (2) | |
| H11A | 0.391355 | 0.722732 | 0.363717 | 0.021* | |
| H11B | 0.300127 | 0.635132 | 0.303039 | 0.021* | |
| C12 | 0.40035 (10) | 0.70880 (9) | 0.56706 (10) | 0.0163 (2) | |
| H12A | 0.423172 | 0.673366 | 0.639612 | 0.020* | |
| H12B | 0.465940 | 0.746048 | 0.567719 | 0.020* | |
| C13 | 0.27803 (11) | 0.83883 (9) | 0.44318 (11) | 0.0194 (2) | |
| H13A | 0.342809 | 0.876660 | 0.442661 | 0.023* | |
| H13B | 0.219187 | 0.890515 | 0.432994 | 0.023* | |
| C14 | 0.21257 (10) | 0.72912 (10) | 0.55236 (11) | 0.0194 (2) | |
| H14A | 0.153212 | 0.779834 | 0.543355 | 0.023* | |
| H14B | 0.233623 | 0.693517 | 0.624588 | 0.023* | |
| C15 | 0.13874 (10) | 0.70598 (10) | 0.35186 (10) | 0.0196 (2) | |
| H15A | 0.110306 | 0.654944 | 0.290289 | 0.023* | |
| H15B | 0.078532 | 0.756406 | 0.340611 | 0.023* | |
| C16 | 0.25985 (9) | 0.57494 (9) | 0.47493 (10) | 0.0156 (2) | |
| H16A | 0.231942 | 0.522809 | 0.414419 | 0.019* | |
| H16B | 0.280830 | 0.537541 | 0.546167 | 0.019* | |
| N14 | 0.16833 (8) | 0.64994 (8) | 0.46020 (9) | 0.0173 (2) | |
| N21 | 0.70755 (8) | 0.64919 (8) | 0.80957 (8) | 0.01507 (19) | |
| N22 | 0.71117 (9) | 0.72608 (8) | 0.98413 (9) | 0.0167 (2) | |
| N23 | 0.87597 (9) | 0.74634 (8) | 0.93941 (9) | 0.0184 (2) | |
| N24 | 0.83066 (8) | 0.57193 (8) | 0.99309 (8) | 0.0165 (2) | |
| C21 | 0.64056 (10) | 0.70037 (9) | 0.86374 (10) | 0.0166 (2) | |
| H21A | 0.579657 | 0.653203 | 0.858804 | 0.020* | |
| H21B | 0.606714 | 0.765088 | 0.823090 | 0.020* | |
| C22 | 0.75820 (10) | 0.55052 (9) | 0.87319 (10) | 0.0159 (2) | |
| H22A | 0.697996 | 0.502245 | 0.867830 | 0.019* | |
| H22B | 0.802836 | 0.516073 | 0.838924 | 0.019* | |
| C23 | 0.76143 (11) | 0.62520 (10) | 1.04253 (10) | 0.0184 (2) | |
| H23A | 0.808248 | 0.639798 | 1.121947 | 0.022* | |
| H23B | 0.700979 | 0.577825 | 1.038421 | 0.022* | |
| C24 | 0.80542 (11) | 0.79507 (10) | 0.98977 (11) | 0.0195 (2) | |
| H24A | 0.774373 | 0.861136 | 0.950611 | 0.023* | |
| H24B | 0.852716 | 0.811921 | 1.068513 | 0.023* | |
| C25 | 0.92101 (10) | 0.64522 (10) | 0.99882 (10) | 0.0197 (2) | |
| H25A | 0.966605 | 0.611322 | 0.965300 | 0.024* | |
| H25B | 0.969896 | 0.659937 | 1.077698 | 0.024* | |
| C26 | 0.80131 (10) | 0.72159 (10) | 0.82069 (10) | 0.0180 (2) | |
| H26A | 0.845870 | 0.689137 | 0.785169 | 0.022* | |
| H26B | 0.769710 | 0.787150 | 0.780642 | 0.022* | |
| N31 | 0.02532 (10) | 0.44658 (10) | 0.24906 (11) | 0.0297 (3) | |
| C31 | −0.03061 (10) | 0.46842 (10) | 0.29362 (11) | 0.0198 (2) | |
| C32 | −0.10137 (13) | 0.49834 (10) | 0.35086 (13) | 0.0242 (3) | |
| H32A | −0.179624 | 0.497188 | 0.297589 | 0.036* | |
| H32B | −0.081381 | 0.568548 | 0.381669 | 0.036* | |
| H32C | −0.089767 | 0.449026 | 0.411080 | 0.036* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Co1 | 0.01283 (13) | 0.01259 (14) | 0.01083 (13) | −0.00162 (9) | 0.00635 (10) | −0.00092 (9) |
| N1 | 0.0171 (5) | 0.0161 (5) | 0.0139 (5) | −0.0003 (4) | 0.0077 (4) | 0.0002 (4) |
| C1 | 0.0131 (5) | 0.0120 (5) | 0.0148 (5) | −0.0004 (4) | 0.0052 (4) | −0.0002 (4) |
| S1 | 0.02595 (16) | 0.02425 (17) | 0.01980 (15) | 0.00071 (11) | 0.01660 (12) | −0.00011 (11) |
| O1 | 0.0176 (4) | 0.0156 (4) | 0.0124 (4) | −0.0034 (3) | 0.0074 (3) | −0.0018 (3) |
| N11 | 0.0151 (4) | 0.0126 (4) | 0.0136 (4) | −0.0011 (3) | 0.0072 (4) | −0.0004 (4) |
| N12 | 0.0189 (5) | 0.0153 (5) | 0.0207 (5) | 0.0013 (4) | 0.0099 (4) | −0.0024 (4) |
| N13 | 0.0229 (5) | 0.0147 (5) | 0.0199 (5) | 0.0035 (4) | 0.0099 (4) | 0.0022 (4) |
| C11 | 0.0226 (6) | 0.0159 (5) | 0.0164 (5) | 0.0025 (4) | 0.0104 (5) | 0.0022 (4) |
| C12 | 0.0164 (5) | 0.0145 (5) | 0.0178 (5) | 0.0002 (4) | 0.0071 (4) | −0.0040 (4) |
| C13 | 0.0232 (6) | 0.0125 (5) | 0.0245 (6) | 0.0008 (4) | 0.0120 (5) | 0.0004 (4) |
| C14 | 0.0208 (6) | 0.0197 (6) | 0.0220 (6) | 0.0026 (5) | 0.0133 (5) | −0.0008 (5) |
| C15 | 0.0186 (5) | 0.0175 (6) | 0.0197 (6) | 0.0023 (4) | 0.0051 (5) | 0.0012 (4) |
| C16 | 0.0140 (5) | 0.0133 (5) | 0.0201 (5) | 0.0002 (4) | 0.0078 (4) | 0.0008 (4) |
| N14 | 0.0157 (5) | 0.0156 (5) | 0.0216 (5) | 0.0024 (4) | 0.0088 (4) | 0.0013 (4) |
| N21 | 0.0186 (5) | 0.0135 (4) | 0.0142 (4) | 0.0001 (4) | 0.0080 (4) | −0.0003 (4) |
| N22 | 0.0219 (5) | 0.0151 (5) | 0.0170 (5) | −0.0011 (4) | 0.0119 (4) | −0.0011 (4) |
| N23 | 0.0202 (5) | 0.0178 (5) | 0.0195 (5) | −0.0041 (4) | 0.0107 (4) | −0.0024 (4) |
| N24 | 0.0174 (5) | 0.0173 (5) | 0.0153 (5) | −0.0001 (4) | 0.0074 (4) | 0.0009 (4) |
| C21 | 0.0172 (5) | 0.0160 (5) | 0.0178 (6) | 0.0015 (4) | 0.0084 (4) | 0.0002 (4) |
| C22 | 0.0194 (5) | 0.0129 (5) | 0.0159 (5) | −0.0001 (4) | 0.0079 (4) | −0.0011 (4) |
| C23 | 0.0240 (6) | 0.0190 (6) | 0.0159 (5) | 0.0006 (5) | 0.0120 (5) | 0.0017 (4) |
| C24 | 0.0252 (6) | 0.0159 (5) | 0.0203 (6) | −0.0051 (5) | 0.0124 (5) | −0.0059 (4) |
| C25 | 0.0162 (5) | 0.0235 (6) | 0.0189 (6) | −0.0017 (5) | 0.0067 (4) | −0.0007 (5) |
| C26 | 0.0247 (6) | 0.0171 (5) | 0.0167 (6) | −0.0022 (4) | 0.0130 (5) | 0.0000 (4) |
| N31 | 0.0242 (6) | 0.0341 (6) | 0.0311 (6) | −0.0005 (5) | 0.0119 (5) | −0.0095 (5) |
| C31 | 0.0180 (6) | 0.0164 (6) | 0.0216 (6) | 0.0005 (5) | 0.0051 (5) | −0.0009 (5) |
| C32 | 0.0226 (7) | 0.0262 (7) | 0.0267 (7) | 0.0014 (5) | 0.0131 (6) | 0.0011 (5) |
Geometric parameters (Å, º)
| Co1—N1i | 2.0744 (10) | C16—H16B | 0.9700 |
| Co1—N1 | 2.0744 (10) | C16—N14 | 1.4670 (14) |
| Co1—O1i | 2.0661 (8) | N21—C21 | 1.4786 (14) |
| Co1—O1 | 2.0661 (8) | N21—C22 | 1.4840 (14) |
| Co1—N11 | 2.3112 (9) | N21—C26 | 1.4797 (15) |
| Co1—N11i | 2.3112 (9) | N22—C21 | 1.4788 (15) |
| N1—C1 | 1.1654 (16) | N22—C23 | 1.4827 (15) |
| C1—S1 | 1.6352 (12) | N22—C24 | 1.4784 (15) |
| O1—H1A | 0.88 (2) | N23—C24 | 1.4669 (15) |
| O1—H1B | 0.88 (2) | N23—C25 | 1.4763 (16) |
| N11—C11 | 1.4931 (14) | N23—C26 | 1.4695 (15) |
| N11—C12 | 1.5008 (14) | N24—C22 | 1.4673 (15) |
| N11—C16 | 1.4934 (14) | N24—C23 | 1.4697 (15) |
| N12—C12 | 1.4645 (14) | N24—C25 | 1.4707 (15) |
| N12—C13 | 1.4690 (16) | C21—H21A | 0.9700 |
| N12—C14 | 1.4665 (15) | C21—H21B | 0.9700 |
| N13—C11 | 1.4685 (15) | C22—H22A | 0.9700 |
| N13—C13 | 1.4773 (16) | C22—H22B | 0.9700 |
| N13—C15 | 1.4727 (16) | C23—H23A | 0.9700 |
| C11—H11A | 0.9700 | C23—H23B | 0.9700 |
| C11—H11B | 0.9700 | C24—H24A | 0.9700 |
| C12—H12A | 0.9700 | C24—H24B | 0.9700 |
| C12—H12B | 0.9700 | C25—H25A | 0.9700 |
| C13—H13A | 0.9700 | C25—H25B | 0.9700 |
| C13—H13B | 0.9700 | C26—H26A | 0.9700 |
| C14—H14A | 0.9700 | C26—H26B | 0.9700 |
| C14—H14B | 0.9700 | N31—C31 | 1.1377 (18) |
| C14—N14 | 1.4764 (16) | C31—C32 | 1.4554 (18) |
| C15—H15A | 0.9700 | C32—H32A | 0.9600 |
| C15—H15B | 0.9700 | C32—H32B | 0.9600 |
| C15—N14 | 1.4740 (15) | C32—H32C | 0.9600 |
| C16—H16A | 0.9700 | ||
| N1i—Co1—N1 | 180.0 | N11—C16—H16B | 108.9 |
| N1—Co1—N11i | 92.49 (4) | H16A—C16—H16B | 107.7 |
| N1i—Co1—N11i | 87.51 (4) | N14—C16—N11 | 113.40 (9) |
| N1—Co1—N11 | 87.51 (4) | N14—C16—H16A | 108.9 |
| N1i—Co1—N11 | 92.49 (4) | N14—C16—H16B | 108.9 |
| O1—Co1—N1i | 90.32 (4) | C15—N14—C14 | 107.93 (9) |
| O1i—Co1—N1i | 89.68 (4) | C16—N14—C14 | 108.13 (9) |
| O1—Co1—N1 | 89.68 (4) | C16—N14—C15 | 107.69 (9) |
| O1i—Co1—N1 | 90.32 (4) | C21—N21—C22 | 108.15 (9) |
| O1i—Co1—O1 | 180.0 | C21—N21—C26 | 108.13 (9) |
| O1—Co1—N11 | 89.16 (3) | C26—N21—C22 | 107.90 (9) |
| O1i—Co1—N11 | 90.84 (3) | C21—N22—C23 | 107.32 (9) |
| O1—Co1—N11i | 90.84 (3) | C24—N22—C21 | 108.26 (9) |
| O1i—Co1—N11i | 89.16 (3) | C24—N22—C23 | 107.49 (9) |
| N11i—Co1—N11 | 180.0 | C24—N23—C25 | 108.18 (9) |
| C1—N1—Co1 | 161.53 (9) | C24—N23—C26 | 107.26 (9) |
| N1—C1—S1 | 179.08 (11) | C26—N23—C25 | 107.92 (9) |
| Co1—O1—H1A | 120.6 (12) | C22—N24—C23 | 108.11 (9) |
| Co1—O1—H1B | 127.1 (14) | C22—N24—C25 | 108.12 (9) |
| H1A—O1—H1B | 107.8 (18) | C23—N24—C25 | 108.33 (9) |
| C11—N11—Co1 | 113.28 (7) | N21—C21—N22 | 111.82 (9) |
| C11—N11—C12 | 106.78 (9) | N21—C21—H21A | 109.3 |
| C11—N11—C16 | 107.17 (9) | N21—C21—H21B | 109.3 |
| C12—N11—Co1 | 112.96 (7) | N22—C21—H21A | 109.3 |
| C16—N11—Co1 | 109.60 (7) | N22—C21—H21B | 109.3 |
| C16—N11—C12 | 106.68 (8) | H21A—C21—H21B | 107.9 |
| C12—N12—C13 | 108.19 (9) | N21—C22—H22A | 109.2 |
| C12—N12—C14 | 108.65 (9) | N21—C22—H22B | 109.2 |
| C14—N12—C13 | 108.33 (10) | N24—C22—N21 | 111.96 (9) |
| C11—N13—C13 | 108.05 (9) | N24—C22—H22A | 109.2 |
| C11—N13—C15 | 108.54 (9) | N24—C22—H22B | 109.2 |
| C15—N13—C13 | 107.73 (9) | H22A—C22—H22B | 107.9 |
| N11—C11—H11A | 109.0 | N22—C23—H23A | 109.1 |
| N11—C11—H11B | 109.0 | N22—C23—H23B | 109.1 |
| N13—C11—N11 | 112.72 (9) | N24—C23—N22 | 112.71 (9) |
| N13—C11—H11A | 109.0 | N24—C23—H23A | 109.1 |
| N13—C11—H11B | 109.0 | N24—C23—H23B | 109.1 |
| H11A—C11—H11B | 107.8 | H23A—C23—H23B | 107.8 |
| N11—C12—H12A | 109.0 | N22—C24—H24A | 108.9 |
| N11—C12—H12B | 109.0 | N22—C24—H24B | 108.9 |
| N12—C12—N11 | 112.74 (9) | N23—C24—N22 | 113.15 (9) |
| N12—C12—H12A | 109.0 | N23—C24—H24A | 108.9 |
| N12—C12—H12B | 109.0 | N23—C24—H24B | 108.9 |
| H12A—C12—H12B | 107.8 | H24A—C24—H24B | 107.8 |
| N12—C13—N13 | 112.28 (10) | N23—C25—H25A | 109.1 |
| N12—C13—H13A | 109.1 | N23—C25—H25B | 109.1 |
| N12—C13—H13B | 109.1 | N24—C25—N23 | 112.47 (10) |
| N13—C13—H13A | 109.1 | N24—C25—H25A | 109.1 |
| N13—C13—H13B | 109.1 | N24—C25—H25B | 109.1 |
| H13A—C13—H13B | 107.9 | H25A—C25—H25B | 107.8 |
| N12—C14—H14A | 109.2 | N21—C26—H26A | 109.1 |
| N12—C14—H14B | 109.2 | N21—C26—H26B | 109.1 |
| N12—C14—N14 | 112.24 (9) | N23—C26—N21 | 112.70 (9) |
| H14A—C14—H14B | 107.9 | N23—C26—H26A | 109.1 |
| N14—C14—H14A | 109.2 | N23—C26—H26B | 109.1 |
| N14—C14—H14B | 109.2 | H26A—C26—H26B | 107.8 |
| N13—C15—H15A | 109.1 | N31—C31—C32 | 178.95 (14) |
| N13—C15—H15B | 109.1 | C31—C32—H32A | 109.5 |
| N13—C15—N14 | 112.61 (10) | C31—C32—H32B | 109.5 |
| H15A—C15—H15B | 107.8 | C31—C32—H32C | 109.5 |
| N14—C15—H15A | 109.1 | H32A—C32—H32B | 109.5 |
| N14—C15—H15B | 109.1 | H32A—C32—H32C | 109.5 |
| N11—C16—H16A | 108.9 | H32B—C32—H32C | 109.5 |
| Co1—N11—C11—N13 | 177.50 (7) | C16—N11—C12—N12 | −56.82 (12) |
| Co1—N11—C12—N12 | −177.30 (7) | C21—N21—C22—N24 | −58.62 (12) |
| Co1—N11—C16—N14 | 179.51 (7) | C21—N21—C26—N23 | 59.00 (12) |
| N11—C16—N14—C14 | −58.13 (12) | C21—N22—C23—N24 | 58.82 (12) |
| N11—C16—N14—C15 | 58.26 (12) | C21—N22—C24—N23 | −58.16 (13) |
| N12—C14—N14—C15 | −58.00 (12) | C22—N21—C21—N22 | 58.99 (12) |
| N12—C14—N14—C16 | 58.24 (12) | C22—N21—C26—N23 | −57.76 (12) |
| N13—C15—N14—C14 | 58.10 (12) | C22—N24—C23—N22 | −58.82 (12) |
| N13—C15—N14—C16 | −58.43 (12) | C22—N24—C25—N23 | 59.03 (12) |
| C11—N11—C12—N12 | 57.52 (12) | C23—N22—C21—N21 | −58.65 (12) |
| C11—N11—C16—N14 | −57.18 (12) | C23—N22—C24—N23 | 57.48 (12) |
| C11—N13—C13—N12 | −58.92 (12) | C23—N24—C22—N21 | 58.22 (12) |
| C11—N13—C15—N14 | 58.66 (12) | C23—N24—C25—N23 | −57.90 (12) |
| C12—N11—C11—N13 | −57.52 (12) | C24—N22—C21—N21 | 57.10 (12) |
| C12—N11—C16—N14 | 56.90 (12) | C24—N22—C23—N24 | −57.44 (12) |
| C12—N12—C13—N13 | 58.99 (12) | C24—N23—C25—N24 | 57.61 (12) |
| C12—N12—C14—N14 | −58.86 (12) | C24—N23—C26—N21 | −58.72 (12) |
| C13—N12—C12—N11 | −58.73 (12) | C25—N23—C24—N22 | −57.80 (13) |
| C13—N12—C14—N14 | 58.46 (12) | C25—N23—C26—N21 | 57.63 (12) |
| C13—N13—C11—N11 | 58.71 (12) | C25—N24—C22—N21 | −58.86 (12) |
| C13—N13—C15—N14 | −58.10 (12) | C25—N24—C23—N22 | 58.11 (12) |
| C14—N12—C12—N11 | 58.67 (12) | C26—N21—C21—N22 | −57.60 (12) |
| C14—N12—C13—N13 | −58.62 (12) | C26—N21—C22—N24 | 58.13 (11) |
| C15—N13—C11—N11 | −57.84 (12) | C26—N23—C24—N22 | 58.38 (13) |
| C15—N13—C13—N12 | 58.16 (12) | C26—N23—C25—N24 | −58.14 (12) |
| C16—N11—C11—N13 | 56.49 (12) |
Symmetry code: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···N21 | 0.88 (2) | 1.85 (2) | 2.7298 (13) | 171.8 (18) |
| O1—H1B···N22ii | 0.88 (2) | 2.01 (2) | 2.8759 (13) | 167.7 (19) |
| C12—H12B···O1 | 0.97 | 2.60 | 3.0752 (14) | 111 |
| C13—H13A···S1ii | 0.97 | 2.95 | 3.8089 (12) | 148 |
| C13—H13B···N24iii | 0.97 | 2.66 | 3.5045 (16) | 146 |
| C16—H16A···O1i | 0.97 | 2.52 | 3.0571 (14) | 115 |
| C16—H16B···N1 | 0.97 | 2.70 | 3.2713 (15) | 118 |
| C21—H21A···S1 | 0.97 | 3.00 | 3.9471 (12) | 165 |
| C23—H23B···S1iv | 0.97 | 2.89 | 3.6683 (12) | 138 |
| C26—H26A···N31i | 0.97 | 2.56 | 3.4794 (17) | 158 |
| C32—H32A···S1v | 0.96 | 3.02 | 3.9560 (15) | 166 |
| C32—H32B···N23vi | 0.96 | 2.58 | 3.4685 (16) | 154 |
| C32—H32C···N14v | 0.96 | 2.61 | 3.4750 (17) | 149 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, −y+3/2, z−1/2; (iii) −x+1, y+1/2, −z+3/2; (iv) −x+1, −y+1, −z+2; (v) −x, −y+1, −z+1; (vi) x−1, −y+3/2, z−1/2.
Funding Statement
This work was funded by Deutsche Forschungsgemeinschaft grant NA720/5-2; State of Schleswig-Holstein.
References
- Böhme, M., Jochim, A., Rams, M., Lohmiller, T., Suckert, S., Schnegg, A., Plass, W. & Näther, C. (2020). Inorg. Chem. 59, 5325–5338. [DOI] [PubMed]
- Brandenburg, K. & Putz, H. (1999). DIAMOND. Crystal Impact GbR, Bonn, Germany.
- Ceglarska, M., Böhme, M., Neumann, T., Plass, W., Näther, C. & Rams, M. (2021). Phys. Chem. Chem. Phys. 23, 10281–10289. [DOI] [PubMed]
- Chakraborty, J., Samanta, B., Rosair, G., Gramlich, V., Salah El Fallah, M., Ribas, J., Matsushita, T. & Mitra, S. (2006). Polyhedron, 25, 3006–3016.
- Czubacka, E., Kruszynski, R. & Sieranski, T. (2012). Struct. Chem. 23, 451–459.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jin, Y., Che, Y. X. & Zheng, J. M. (2007). J. Coord. Chem. 60, 2067–2074.
- Li, J., Meng, S., Zhang, J., Song, Y., Huang, Z., Zhao, H., Wei, H., Huang, W., Cifuentes, M. P., Humphrey, M. G. & Zhang, C. (2012). CrystEngComm, 14, 2787–2796.
- Li, X.-L., Niu, D.-Z. & Lu, Z.-S. (2007). Acta Cryst. E63, m2478.
- Liu, Q., Xi, H.-T., Sun, X.-Q., Zhu, J.-F. & Yu, K.-B. (2002). Chin. J. Struct. Chem. 21, 355–359.
- Lu, J., Liu, H.-T., Zhang, X.-X., Wang, D.-Q. & Niu, M.-J. (2010). Z. Anorg. Allg. Chem. 636, 641–647.
- Mautner, F. A., Traber, M., Fischer, R. C., Torvisco, A., Reichmann, K., Speed, S., Vicente, R. & Massoud, S. S. (2018). Polyhedron, 154, 436–442.
- Mekuimemba, C. D., Conan, F., Mota, A. J., Palacios, M. A., Colacio, E. & Triki, S. (2018). Inorg. Chem. 57, 2184–2192. [DOI] [PubMed]
- Mousavi, M., Duhayon, C., Bretosh, K., Béreau, V. & Sutter, J. P. (2020). Inorg. Chem. 59, 7603–7613. [DOI] [PubMed]
- Palion-Gazda, J., Machura, B., Lloret, F. & Julve, M. (2015). Cryst. Growth Des. 15, 2380–2388.
- Prananto, Y. P., Urbatsch, A., Moubaraki, B., Murray, K. S., Turner, D. R., Deacon, G. B. & Batten, S. R. (2017). Aust. J. Chem. 70, 516–528.
- Rams, M., Jochim, A., Böhme, M., Lohmiller, T., Ceglarska, M., Rams, M. M., Schnegg, A., Plass, W. & Näther, C. (2020). Chem. Eur. J. 26, 2837–2851. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2021). CrysAlis PRO. Rigaku Oxford Diffraction.
- Robinson, K., Gibbs, G. V. & Ribbe, P. H. (1971). Science, 172, 567–570. [DOI] [PubMed]
- Shang, W.-L., Bai, Y., Ma, C.-Z. & Li, Z.-M. (2008). Acta Cryst. E64, m1184–m1185. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shi, J.-M., Chen, J.-N. & Liu, L.-D. (2006). Pol. J. Chem. 80, 1909–1913.
- Suckert, S., Rams, M., Böhme, M., Germann, L. S., Dinnebier, R. E., Plass, W., Werner, J. & Näther, C. (2016). Dalton Trans. 45, 18190–18201. [DOI] [PubMed]
- Wellm, C., Majcher-Fitas, A., Rams, M. & Näther, C. (2020). Dalton Trans. 49, 16707–16714. [DOI] [PubMed]
- Werner, J., Rams, M., Tomkowicz, Z. & Näther, C. (2014). Dalton Trans. 43, 17333–17342. [DOI] [PubMed]
- Werner, J., Tomkowicz, Z., Rams, M., Ebbinghaus, S. G., Neumann, T. & Näther, C. (2015). Dalton Trans. 44, 14149–14158. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zhang, Y., Li, J., Xu, H., Hou, H., Nishiura, M. & Imamoto, T. (1999). J. Mol. Struct. 510, 191–196.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021010033/hb7982sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010033/hb7982Isup2.hkl
Fig. S1. Experimental (top) and calculated X-ray powder pattern (bottom) of the title compound. The title compound is contaminated with at least one additional unknown phase. For the calculation of the powder pattern the data obtained from a a single crystal measured at 24 C was used. DOI: 10.1107/S2056989021010033/hb7982sup3.png
Fig. S2. IR spectra of the title compound. The value of the CN stretching vibration of the thiocyanat anions is given. DOI: 10.1107/S2056989021010033/hb7982sup4.png
CCDC reference: 2112185
Additional supporting information: crystallographic information; 3D view; checkCIF report






