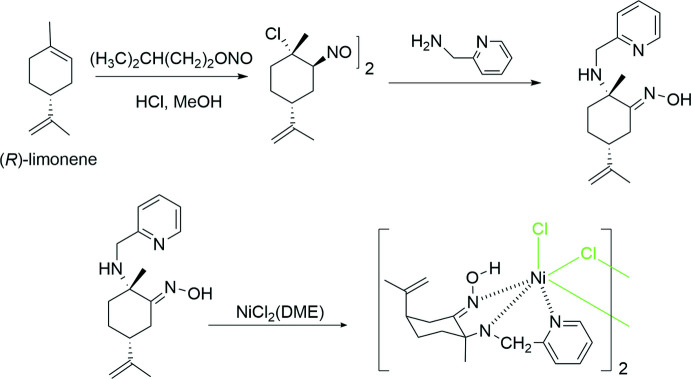
The reaction of a nickel precursor with an enantiomerically pure amino-oxime issued from (R)-limonene led to the formation of bis[κ3 N,N,N-(aminooxime)-μ-chlorido]dichlorodinickel as a new dinuclear nickel complex.
Keywords: Nickel, α-aminooxime, (R)-limonene, crystal structure
Abstract
A dinuclear nickel complex with (S)-limonene based aminooxime ligand has been isolated and its crystal structure determined. The resolved structure of dichloridobis{(2S,5R)-2-methyl-5-(prop-1-en-2-yl)-2-[(pyridin-2-yl)methylamino]cyclohexan-1-one oxime}dinickel(II), [Ni2Cl2(C16H23ClN3O)2], at 100 K has monoclinic (P21) symmetry. The two NiII ions in the dinuclear complex are each coordinated in a distorted octahedral environment by three nitrogen atoms, a terminal chloride and two μ bridging chlorides. Each oxime ligand is coordinated to nickel(II) by the three nitrogen atoms, leading to two five-membered chelate rings, each displaying an envelope conformation. In the crystal, numerous intermolecular and intramolecular hydrogen bonds lead to the formation of a three-dimensional network structure.
Chemical context
Asymmetric synthesis allows the preparation of enantiomerically enriched compounds either by using a chiral auxiliary, which will be temporarily introduced, or by using catalytic procedures (Gawley & Aubé, 2012 ▸). This latter method is particularly attractive as it contributes to the development of green chemistry, which maximizes efficiency and minimizes hazardous effects on human health and the environment (Anastas & Zimmerman, 2013 ▸). Thus, asymmetric catalysis avoids synthetic steps and only catalytic amounts of the optically pure auxiliary are needed (Ojima, 2010 ▸). As part of the development of this chemistry, the synthesis of new chiral organometallic complexes is always challenging. The pivotal point is then the synthesis of optically pure ligands, which will be coordinated to the metal center. In terms of sustainable chemistry, using the chiral pool to develop new ligands is most interesting (Elalami et al., 2015 ▸). Coordination metal complexes containing terpenoid fragments are widely used in the pharmaceutical field and in catalysis. We have therefore developed ligands based on terpenes such as pinene and limonene (El Alami et al., 2009 ▸, 2015 ▸; Chahboun et al., 2012 ▸). In particular, the synthesis of optically pure amino-oxime ligands has been performed successfully from (R)-limonene (El Alami et al., 2012 ▸). These compounds possess structures with two or three nitrogen atoms as donor heteroatoms that could coordinate to the metal center. They have advantageously replaced phosphine ligands, which are generally unstable under air. Ruthenium (Benabdelouahab et al., 2015 ▸) and palladium (de la Cueva-Alique et al., 2019 ▸) complexes have already been synthezised with these ligands. Here we report the first synthesis of a limonene-based α-aminooxime nickel complex and its crystal structure. In the dinuclear title complex, each nickel ion is coordinated by (1S,4R)-1-picolylamino-p-menth-8-en-2-one oxime. The ligand was first synthesized from (R)-limonene through the addition of nitrosyl chloride, NOCl, to a picolylamine moiety, allowing the formation of the oxime moiety.
Structural commentary
The title compound (Fig. 1 ▸) crystallizes in the monoclinic space group P21 with two chiral molecules per unit cell. The two NiII ions in the dinuclear complex are each coordinated by three nitrogen atoms, a terminal chloride and two μ bridging chlorides. The environment around each metal center can then be described as a distorted octahedron with N1—Ni1—N2 and Cl1—Ni1—Cl3 angles of 79.91 (13) and 91.99 (4)°, respectively, together with Cl1—Ni1—N2 and Cl2—Ni1—N1 angles of 165.04 (11) and 88.69 (10)°, respectively. A similar arrangement can be found around the Ni2 atom [N4—Ni2—N5, Cl2—Ni2—Cl4, Cl4—Ni2—N5 and Cl4—Ni2—N4 = 79.7 (2), 99.38 (4), 166.04 (12) and 93.24 (16)°, respectively].
Figure 1.
Displacement ellipsoid plot at the 50% probability level for Ni2(amino-oxime)2Cl4. H atoms are omitted for clarity.
Each aminooxime ligand is coordinated to nickel(II) by the three nitrogen atoms, leading to two five-membered chelate rings, each displaying an envelope conformation (with N2 as the flap for Ni1/N1/C5/C6/N2 and N5 for Ni2/N4/C21/C22/N5). The six-membered carbocycles of the limonene units adopt a chair conformation. The lengths of the Ni1—N1, Ni1—N2 and Ni1—N3 bonds are 2.077 (3), 2.126 (4) and 2.041 (3) Å, respectively, while Ni2—N4, Ni2—N5 and Ni2—N6 are 2.095 (4), 2.103 (4) and 2.027 (3) Å. Atoms Cl1 and Cl4 are in a trans-position at distances of 2.4408 (12) and 2.4077 (14) Å from the metal centers Ni1 and Ni2, respectively. The two metal centers are linked by two bridging Cl atoms with an average Ni—Cl distance of 2.42 Å, which is normal for these bond lengths. All these values compare well with literature values. The two nickel ions are separated by a distance of 3.5198 (7) Å, which is similar to average values (Zheng et al., 2010 ▸; Cheng et al., 2012 ▸).
Supramolecular features
The crystal structure is stabilized by numerous intermolecular and intramolecular hydrogen bonds (Table 1 ▸), which link the component into a three-dimensional network (Figs. 2 ▸ and 3 ▸). In particular, the two {Ni(aminoxime)μ-Cl}Cl units are slightly asymmetrical with the existence of a hydrogen-bonding interaction between the amine N2—H2 linked to Ni1 and the chlorine atom Cl4 linked to Ni2. In addition, the two oxygen atoms O1 and O2 of the oxime groups are involved in intramolecular O1—H1⋯Cl1 and O2—H2A⋯Cl4 hydrogen bonds and in intermolecular C3—H3⋯O1 and C26—H26⋯O2 interactions.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯Cl1 | 0.85 (7) | 2.32 (6) | 3.009 (4) | 139 (6) |
| N2—H2⋯Cl4 | 0.77 (5) | 2.46 (5) | 3.209 (4) | 166 (5) |
| O2—H2A⋯Cl4 | 0.76 (8) | 2.31 (7) | 2.978 (4) | 147 (7) |
| C3—H3⋯O1i | 0.95 | 2.58 | 3.432 (5) | 149 |
| C1—H1A⋯Cl1 | 0.95 | 2.75 | 3.369 (5) | 124 |
| C6—H6A⋯Cl2 | 0.99 | 2.76 | 3.309 (5) | 115 |
| C11—H11B⋯Cl3ii | 0.99 | 2.64 | 3.573 (5) | 156 |
| C17—H17⋯Cl4 | 0.95 | 2.69 | 3.327 (6) | 125 |
| C26—H26⋯O2iii | 1.00 | 2.56 | 3.489 (6) | 154 |
| C22—H22B⋯Cl2 | 0.99 | 2.81 | 3.352 (6) | 115 |
| C19—H19⋯Cl1iv | 0.95 | 2.64 | 3.570 (7) | 167 |
Symmetry codes: (i) -x+2, y-{\script{1\over 2}}, -z+1; (ii) -x+1, y-{\script{1\over 2}}, -z+1; (iii) -x, y+{\script{1\over 2}}, -z; (iv) -x+1, y-{\script{1\over 2}}, -z.
Figure 2.
Intermolecular and intramolecular hydrogen bonds in the structure, shown as dashed lines.
Figure 3.
Packing diagram.
Database survey
The aminooxime ligand used in this study was previously reacted with palladium and platinum precursors, generating three N-coordinated cationic complexes as enantiopure compounds (de la Cueva-Alique et al., 2019 ▸). A heteronuclear TiIV/PdII complex has also been described. The compounds were studied to assess their potential biological activity, a high anticancer activity (de la Cueva-Alique et al., 2019 ▸).
Synthesis and crystallization
To a solution of NiII chloride ethylene glycol dimethyl ether (0.15 g, 1.48 mmol) in MeOH (5 mL) was added (1S,4R)-1-picolylamino-p-menth-8-en-2-one-oxime (0.101 g, 0.36 mmol) dissolved in MeOH (3 mL). The solution turned green. The mixture was stirred overnight at room temperature during which time the mixture changed color to blue–green. The solvent was then evaporated to produce a crude solid that was washed with diethyl ether before crystallization. Single crystals were grown by slow diffusion at room temperature of diethyl ether into a dichloromethane solution. Elemental analysis calculated for C32H46Cl4N6Ni2O2: C, 46.33; H, 5.54; N, 9.65. Found: C, 46.35; H, 5.672; N, 9.77.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. N- and O-bound atoms were refined with the restraint U iso(H) = 1.2U eq(N) or 1.5U eq(O). H atoms were positioned geometrically(C—H = 0.95–1.00 Å) and refined as riding with U iso(H) = 1.2U eq(C) or 1.5U eq(C-methyl)
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Ni2Cl2(C16H23ClN3O)2] |
| M r | 805.97 |
| Crystal system, space group | Monoclinic, P21 |
| Temperature (K) | 100 |
| a, b, c (Å) | 13.3729 (9), 8.9363 (7), 16.4248 (16) |
| β (°) | 114.014 (2) |
| V (Å3) | 1792.9 (3) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.39 |
| Crystal size (mm) | 0.21 × 0.17 × 0.12 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.669, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 42747, 10769, 9436 |
| R int | 0.037 |
| (sin θ/λ)max (Å−1) | 0.714 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.043, 0.109, 1.05 |
| No. of reflections | 10769 |
| No. of parameters | 431 |
| No. of restraints | 13 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 1.50, −1.19 |
| Absolute structure | Flack x determined using 3850 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | −0.009 (4) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021010537/ex2048sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010537/ex2048Isup2.hkl
CCDC reference: 2115017
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We would like to thank Céline Delabre for the elemental analysis.
supplementary crystallographic information
Crystal data
| [Ni2Cl2(C16H23ClN3O)2] | F(000) = 840 |
| Mr = 805.97 | Dx = 1.493 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 13.3729 (9) Å | Cell parameters from 9996 reflections |
| b = 8.9363 (7) Å | θ = 2.7–30.0° |
| c = 16.4248 (16) Å | µ = 1.39 mm−1 |
| β = 114.014 (2)° | T = 100 K |
| V = 1792.9 (3) Å3 | Block, green |
| Z = 2 | 0.21 × 0.17 × 0.12 mm |
Data collection
| Bruker APEXII CCD diffractometer | 9436 reflections with I > 2σ(I) |
| Radiation source: microfocus sealed X-ray tube | Rint = 0.037 |
| φ and ω scans | θmax = 30.5°, θmin = 1.4° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −17→19 |
| Tmin = 0.669, Tmax = 0.746 | k = −12→12 |
| 42747 measured reflections | l = −23→21 |
| 10769 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.109 | w = 1/[σ2(Fo2) + (0.0581P)2 + 0.9636P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 10769 reflections | Δρmax = 1.50 e Å−3 |
| 431 parameters | Δρmin = −1.18 e Å−3 |
| 13 restraints | Absolute structure: Flack x determined using 3850 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| Primary atom site location: dual | Absolute structure parameter: −0.009 (4) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni1 | 0.66455 (4) | 0.50226 (6) | 0.35327 (3) | 0.01526 (11) | |
| Ni2 | 0.41917 (4) | 0.48960 (6) | 0.15985 (3) | 0.01813 (12) | |
| Cl2 | 0.61155 (8) | 0.45928 (14) | 0.19208 (7) | 0.0259 (2) | |
| Cl3 | 0.48342 (7) | 0.60270 (12) | 0.30567 (7) | 0.0212 (2) | |
| Cl1 | 0.74487 (8) | 0.75170 (12) | 0.37010 (7) | 0.0249 (2) | |
| Cl4 | 0.38501 (9) | 0.25191 (13) | 0.21270 (9) | 0.0336 (3) | |
| O1 | 0.7388 (3) | 0.6301 (4) | 0.5389 (2) | 0.0244 (7) | |
| H1 | 0.740 (5) | 0.705 (7) | 0.507 (4) | 0.037* | |
| N1 | 0.8111 (3) | 0.3926 (4) | 0.3786 (2) | 0.0182 (7) | |
| N3 | 0.6939 (2) | 0.5043 (5) | 0.4853 (2) | 0.0183 (6) | |
| N2 | 0.6216 (3) | 0.2811 (4) | 0.3745 (2) | 0.0183 (7) | |
| H2 | 0.563 (4) | 0.289 (6) | 0.339 (3) | 0.022* | |
| O2 | 0.1908 (3) | 0.4533 (5) | 0.1497 (2) | 0.0339 (9) | |
| H2A | 0.226 (6) | 0.389 (9) | 0.176 (5) | 0.051* | |
| C8 | 0.6846 (3) | 0.3877 (5) | 0.5258 (3) | 0.0203 (8) | |
| C5 | 0.8008 (3) | 0.2431 (5) | 0.3699 (3) | 0.0202 (8) | |
| N6 | 0.2619 (3) | 0.5471 (4) | 0.1318 (2) | 0.0212 (8) | |
| N5 | 0.4081 (3) | 0.6943 (5) | 0.0936 (3) | 0.0394 (12) | |
| H5 | 0.468 (5) | 0.736 (8) | 0.140 (4) | 0.047* | |
| C12 | 0.7001 (3) | 0.1123 (6) | 0.5107 (3) | 0.0250 (9) | |
| H12A | 0.660298 | 0.022653 | 0.478112 | 0.030* | |
| H12B | 0.770464 | 0.117488 | 0.504159 | 0.030* | |
| C14 | 0.9033 (3) | 0.2356 (6) | 0.6758 (3) | 0.0257 (9) | |
| C3 | 0.9925 (4) | 0.2136 (6) | 0.4131 (3) | 0.0301 (11) | |
| H3 | 1.054899 | 0.151943 | 0.425362 | 0.036* | |
| C9 | 0.7192 (4) | 0.3765 (6) | 0.6253 (3) | 0.0279 (10) | |
| H9A | 0.653362 | 0.378843 | 0.638523 | 0.034* | |
| H9B | 0.764996 | 0.464264 | 0.654444 | 0.034* | |
| C1 | 0.9109 (3) | 0.4531 (5) | 0.4044 (3) | 0.0220 (9) | |
| H1A | 0.918464 | 0.558677 | 0.410777 | 0.026* | |
| C30 | 0.0369 (4) | 0.6416 (7) | −0.1114 (4) | 0.0357 (12) | |
| N4 | 0.3669 (3) | 0.4143 (6) | 0.0282 (3) | 0.0361 (11) | |
| C2 | 1.0041 (3) | 0.3660 (6) | 0.4222 (3) | 0.0277 (10) | |
| H2B | 1.074096 | 0.411370 | 0.440161 | 0.033* | |
| C25 | 0.1029 (4) | 0.7072 (7) | 0.0546 (4) | 0.0384 (13) | |
| H25A | 0.095090 | 0.786671 | 0.093629 | 0.046* | |
| H25B | 0.059972 | 0.619493 | 0.058673 | 0.046* | |
| C7 | 0.6314 (3) | 0.2540 (5) | 0.4678 (3) | 0.0206 (8) | |
| C4 | 0.8897 (4) | 0.1499 (6) | 0.3860 (3) | 0.0278 (10) | |
| H4 | 0.880183 | 0.044699 | 0.378533 | 0.033* | |
| C24 | 0.2208 (4) | 0.6641 (5) | 0.0869 (3) | 0.0258 (10) | |
| C6 | 0.6855 (3) | 0.1848 (5) | 0.3402 (3) | 0.0218 (8) | |
| H6A | 0.649955 | 0.182503 | 0.274260 | 0.026* | |
| H6B | 0.687313 | 0.081400 | 0.362311 | 0.026* | |
| C10 | 0.7844 (4) | 0.2319 (6) | 0.6644 (3) | 0.0281 (10) | |
| H10 | 0.785669 | 0.219265 | 0.725250 | 0.034* | |
| C23 | 0.2999 (4) | 0.7724 (5) | 0.0732 (4) | 0.0365 (12) | |
| C15 | 0.9419 (4) | 0.3220 (6) | 0.6296 (3) | 0.0301 (10) | |
| H15A | 1.016603 | 0.314817 | 0.638711 | 0.036* | |
| H15B | 0.894916 | 0.391186 | 0.587469 | 0.036* | |
| C31 | 0.0663 (3) | 0.5006 (7) | −0.0931 (3) | 0.0322 (10) | |
| H31A | 0.053020 | 0.430999 | −0.140077 | 0.039* | |
| H31B | 0.100801 | 0.468485 | −0.032826 | 0.039* | |
| C13 | 0.5139 (3) | 0.2394 (7) | 0.4628 (3) | 0.0314 (11) | |
| H13A | 0.471161 | 0.327732 | 0.433110 | 0.047* | |
| H13B | 0.516636 | 0.231714 | 0.523147 | 0.047* | |
| H13C | 0.479378 | 0.149469 | 0.428806 | 0.047* | |
| C11 | 0.7236 (4) | 0.0951 (6) | 0.6094 (3) | 0.0306 (11) | |
| H11A | 0.768517 | 0.004325 | 0.633040 | 0.037* | |
| H11B | 0.653697 | 0.081821 | 0.615749 | 0.037* | |
| C16 | 0.9776 (4) | 0.1259 (6) | 0.7427 (3) | 0.0313 (11) | |
| H16A | 0.980940 | 0.149758 | 0.802043 | 0.047* | |
| H16B | 1.051172 | 0.132186 | 0.743621 | 0.047* | |
| H16C | 0.948965 | 0.024270 | 0.726130 | 0.047* | |
| C18 | 0.2741 (6) | 0.2441 (10) | −0.0937 (4) | 0.0582 (18) | |
| H18 | 0.234602 | 0.153747 | −0.115022 | 0.070* | |
| C17 | 0.3151 (5) | 0.2832 (8) | −0.0055 (4) | 0.0493 (16) | |
| H17 | 0.306660 | 0.213712 | 0.035033 | 0.059* | |
| C26 | 0.0565 (4) | 0.7638 (7) | −0.0421 (4) | 0.0423 (14) | |
| H26 | −0.016160 | 0.810447 | −0.054167 | 0.051* | |
| C21 | 0.3804 (4) | 0.5139 (10) | −0.0254 (4) | 0.0504 (17) | |
| C28 | 0.2479 (5) | 0.8323 (7) | −0.0237 (4) | 0.0486 (16) | |
| H28A | 0.293285 | 0.915184 | −0.030027 | 0.058* | |
| H28B | 0.247256 | 0.751483 | −0.065083 | 0.058* | |
| C32 | −0.0176 (7) | 0.6926 (9) | −0.2067 (4) | 0.068 (2) | |
| H32A | 0.032890 | 0.756035 | −0.220891 | 0.101* | |
| H32B | −0.083660 | 0.749858 | −0.215141 | 0.101* | |
| H32C | −0.037751 | 0.605256 | −0.246168 | 0.101* | |
| C22 | 0.4373 (5) | 0.6511 (10) | 0.0162 (4) | 0.064 (2) | |
| H22A | 0.416191 | 0.732765 | −0.028357 | 0.077* | |
| H22B | 0.517365 | 0.635850 | 0.038173 | 0.077* | |
| C27 | 0.1312 (6) | 0.8883 (7) | −0.0493 (4) | 0.0552 (18) | |
| H27A | 0.101814 | 0.926492 | −0.111198 | 0.066* | |
| H27B | 0.132010 | 0.972060 | −0.009604 | 0.066* | |
| C29 | 0.3272 (7) | 0.8968 (8) | 0.1403 (5) | 0.068 (2) | |
| H29A | 0.368891 | 0.856492 | 0.200143 | 0.102* | |
| H29B | 0.259425 | 0.942465 | 0.137844 | 0.102* | |
| H29C | 0.371164 | 0.972534 | 0.126705 | 0.102* | |
| C19 | 0.2927 (7) | 0.3391 (10) | −0.1474 (5) | 0.069 (2) | |
| H19 | 0.270015 | 0.312455 | −0.208304 | 0.082* | |
| C20 | 0.3427 (6) | 0.4734 (12) | −0.1196 (4) | 0.070 (2) | |
| H20 | 0.353114 | 0.539853 | −0.160608 | 0.084* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.01393 (19) | 0.0171 (2) | 0.0136 (2) | 0.0000 (2) | 0.00431 (17) | 0.0017 (2) |
| Ni2 | 0.0158 (2) | 0.0210 (3) | 0.0153 (2) | −0.0019 (2) | 0.00406 (17) | −0.0011 (2) |
| Cl2 | 0.0180 (4) | 0.0431 (7) | 0.0155 (4) | 0.0016 (4) | 0.0057 (3) | 0.0037 (4) |
| Cl3 | 0.0178 (4) | 0.0225 (5) | 0.0194 (5) | 0.0017 (4) | 0.0034 (4) | −0.0031 (4) |
| Cl1 | 0.0240 (5) | 0.0198 (5) | 0.0244 (5) | −0.0048 (4) | 0.0031 (4) | 0.0052 (4) |
| Cl4 | 0.0301 (5) | 0.0180 (5) | 0.0367 (7) | −0.0042 (4) | −0.0027 (5) | 0.0021 (5) |
| O1 | 0.0275 (15) | 0.0233 (17) | 0.0205 (16) | 0.0014 (13) | 0.0077 (13) | −0.0051 (13) |
| N1 | 0.0159 (14) | 0.0249 (19) | 0.0133 (16) | 0.0017 (13) | 0.0054 (13) | 0.0017 (14) |
| N3 | 0.0163 (13) | 0.0206 (17) | 0.0170 (15) | 0.0001 (15) | 0.0056 (12) | 0.0000 (16) |
| N2 | 0.0122 (13) | 0.0204 (19) | 0.0200 (18) | 0.0019 (13) | 0.0044 (13) | 0.0019 (14) |
| O2 | 0.0186 (14) | 0.052 (3) | 0.0312 (19) | −0.0003 (14) | 0.0099 (13) | 0.0128 (17) |
| C8 | 0.0184 (18) | 0.026 (2) | 0.020 (2) | 0.0079 (16) | 0.0113 (16) | 0.0058 (17) |
| C5 | 0.0221 (18) | 0.025 (2) | 0.0146 (19) | 0.0034 (17) | 0.0086 (15) | 0.0006 (17) |
| N6 | 0.0182 (15) | 0.028 (2) | 0.0163 (17) | 0.0011 (13) | 0.0055 (14) | −0.0007 (14) |
| N5 | 0.029 (2) | 0.040 (3) | 0.033 (2) | −0.0178 (19) | −0.0040 (18) | 0.020 (2) |
| C12 | 0.0200 (18) | 0.025 (2) | 0.027 (2) | −0.0009 (17) | 0.0064 (17) | 0.0088 (19) |
| C14 | 0.0249 (19) | 0.033 (3) | 0.016 (2) | 0.0063 (18) | 0.0045 (16) | 0.0044 (19) |
| C3 | 0.025 (2) | 0.036 (3) | 0.029 (3) | 0.0125 (19) | 0.0111 (19) | 0.004 (2) |
| C9 | 0.032 (2) | 0.037 (3) | 0.018 (2) | 0.011 (2) | 0.0145 (18) | 0.006 (2) |
| C1 | 0.0194 (17) | 0.028 (2) | 0.019 (2) | 0.0006 (16) | 0.0077 (15) | 0.0024 (17) |
| C30 | 0.035 (2) | 0.041 (3) | 0.033 (3) | 0.006 (2) | 0.015 (2) | 0.000 (2) |
| N4 | 0.0223 (18) | 0.061 (3) | 0.021 (2) | 0.0157 (19) | 0.0044 (16) | −0.005 (2) |
| C2 | 0.0149 (17) | 0.041 (3) | 0.028 (2) | 0.0019 (18) | 0.0097 (17) | 0.007 (2) |
| C25 | 0.033 (2) | 0.047 (3) | 0.031 (3) | 0.022 (2) | 0.009 (2) | −0.001 (2) |
| C7 | 0.0174 (16) | 0.025 (2) | 0.022 (2) | 0.0014 (16) | 0.0105 (15) | 0.0077 (18) |
| C4 | 0.025 (2) | 0.032 (3) | 0.027 (2) | 0.0106 (18) | 0.0114 (18) | 0.002 (2) |
| C24 | 0.029 (2) | 0.025 (2) | 0.017 (2) | 0.0080 (18) | 0.0041 (18) | −0.0052 (18) |
| C6 | 0.0243 (19) | 0.017 (2) | 0.024 (2) | 0.0046 (16) | 0.0094 (17) | −0.0026 (17) |
| C10 | 0.030 (2) | 0.034 (3) | 0.024 (2) | 0.007 (2) | 0.0142 (18) | 0.012 (2) |
| C23 | 0.048 (3) | 0.015 (2) | 0.032 (3) | −0.003 (2) | 0.001 (2) | 0.006 (2) |
| C15 | 0.0204 (19) | 0.036 (3) | 0.028 (2) | 0.0014 (18) | 0.0032 (18) | 0.008 (2) |
| C31 | 0.0267 (19) | 0.033 (2) | 0.032 (2) | −0.005 (2) | 0.0058 (18) | 0.001 (3) |
| C13 | 0.0198 (19) | 0.040 (3) | 0.034 (3) | −0.001 (2) | 0.0108 (18) | 0.010 (2) |
| C11 | 0.026 (2) | 0.033 (3) | 0.035 (3) | 0.0008 (19) | 0.015 (2) | 0.018 (2) |
| C16 | 0.033 (2) | 0.037 (3) | 0.024 (2) | 0.009 (2) | 0.0120 (19) | 0.011 (2) |
| C18 | 0.059 (4) | 0.067 (4) | 0.032 (3) | 0.026 (4) | 0.003 (3) | −0.011 (3) |
| C17 | 0.040 (3) | 0.057 (4) | 0.032 (3) | 0.025 (3) | −0.004 (2) | −0.020 (3) |
| C26 | 0.043 (3) | 0.047 (4) | 0.028 (3) | 0.027 (3) | 0.005 (2) | 0.002 (2) |
| C21 | 0.030 (2) | 0.091 (5) | 0.031 (3) | 0.024 (3) | 0.013 (2) | 0.016 (3) |
| C28 | 0.053 (3) | 0.029 (3) | 0.043 (3) | −0.005 (3) | −0.002 (3) | 0.018 (3) |
| C32 | 0.115 (7) | 0.050 (4) | 0.026 (3) | 0.028 (4) | 0.016 (4) | 0.002 (3) |
| C22 | 0.033 (3) | 0.113 (7) | 0.049 (4) | 0.010 (3) | 0.020 (3) | 0.057 (4) |
| C27 | 0.069 (4) | 0.029 (3) | 0.041 (3) | 0.017 (3) | −0.004 (3) | 0.007 (3) |
| C29 | 0.091 (5) | 0.027 (3) | 0.051 (4) | 0.000 (3) | −0.007 (4) | −0.001 (3) |
| C19 | 0.093 (6) | 0.071 (5) | 0.055 (4) | 0.014 (4) | 0.044 (4) | −0.020 (4) |
| C20 | 0.068 (4) | 0.104 (6) | 0.040 (3) | 0.030 (4) | 0.025 (3) | 0.035 (4) |
Geometric parameters (Å, º)
| Ni1—Cl2 | 2.4762 (11) | C2—H2B | 0.9500 |
| Ni1—Cl3 | 2.3964 (10) | C25—H25A | 0.9900 |
| Ni1—Cl1 | 2.4408 (12) | C25—H25B | 0.9900 |
| Ni1—N1 | 2.077 (3) | C25—C24 | 1.495 (6) |
| Ni1—N3 | 2.041 (3) | C25—C26 | 1.536 (8) |
| Ni1—N2 | 2.126 (4) | C7—C13 | 1.545 (5) |
| Ni2—Cl2 | 2.4216 (10) | C4—H4 | 0.9500 |
| Ni2—Cl3 | 2.4128 (12) | C24—C23 | 1.516 (7) |
| Ni2—Cl4 | 2.4077 (14) | C6—H6A | 0.9900 |
| Ni2—N6 | 2.027 (3) | C6—H6B | 0.9900 |
| Ni2—N5 | 2.103 (4) | C10—H10 | 1.0000 |
| Ni2—N4 | 2.095 (4) | C10—C11 | 1.540 (8) |
| O1—H1 | 0.85 (7) | C23—C28 | 1.550 (8) |
| O1—N3 | 1.403 (5) | C23—C29 | 1.502 (9) |
| N1—C5 | 1.345 (6) | C15—H15A | 0.9500 |
| N1—C1 | 1.338 (5) | C15—H15B | 0.9500 |
| N3—C8 | 1.269 (6) | C31—H31A | 0.9500 |
| N2—H2 | 0.77 (5) | C31—H31B | 0.9500 |
| N2—C7 | 1.503 (5) | C13—H13A | 0.9800 |
| N2—C6 | 1.477 (5) | C13—H13B | 0.9800 |
| O2—H2A | 0.76 (8) | C13—H13C | 0.9800 |
| O2—N6 | 1.385 (5) | C11—H11A | 0.9900 |
| C8—C9 | 1.509 (6) | C11—H11B | 0.9900 |
| C8—C7 | 1.513 (7) | C16—H16A | 0.9800 |
| C5—C4 | 1.385 (6) | C16—H16B | 0.9800 |
| C5—C6 | 1.508 (6) | C16—H16C | 0.9800 |
| N6—C24 | 1.269 (6) | C18—H18 | 0.9500 |
| N5—H5 | 0.93 (7) | C18—C17 | 1.370 (8) |
| N5—C23 | 1.517 (7) | C18—C19 | 1.318 (12) |
| N5—C22 | 1.524 (9) | C17—H17 | 0.9500 |
| C12—H12A | 0.9900 | C26—H26 | 1.0000 |
| C12—H12B | 0.9900 | C26—C27 | 1.532 (10) |
| C12—C7 | 1.555 (6) | C21—C22 | 1.457 (12) |
| C12—C11 | 1.528 (7) | C21—C20 | 1.465 (10) |
| C14—C10 | 1.523 (6) | C28—H28A | 0.9900 |
| C14—C15 | 1.326 (7) | C28—H28B | 0.9900 |
| C14—C16 | 1.506 (7) | C28—C27 | 1.525 (9) |
| C3—H3 | 0.9500 | C32—H32A | 0.9800 |
| C3—C2 | 1.373 (8) | C32—H32B | 0.9800 |
| C3—C4 | 1.384 (7) | C32—H32C | 0.9800 |
| C9—H9A | 0.9900 | C22—H22A | 0.9900 |
| C9—H9B | 0.9900 | C22—H22B | 0.9900 |
| C9—C10 | 1.545 (7) | C27—H27A | 0.9900 |
| C1—H1A | 0.9500 | C27—H27B | 0.9900 |
| C1—C2 | 1.396 (6) | C29—H29A | 0.9800 |
| C30—C31 | 1.318 (8) | C29—H29B | 0.9800 |
| C30—C26 | 1.521 (8) | C29—H29C | 0.9800 |
| C30—C32 | 1.503 (8) | C19—H19 | 0.9500 |
| N4—C17 | 1.359 (8) | C19—C20 | 1.360 (13) |
| N4—C21 | 1.314 (8) | C20—H20 | 0.9500 |
| Cl3—Ni1—Cl2 | 84.13 (4) | C8—C7—C13 | 108.0 (4) |
| Cl3—Ni1—Cl1 | 91.99 (4) | C13—C7—C12 | 110.9 (4) |
| Cl1—Ni1—Cl2 | 100.61 (4) | C5—C4—H4 | 120.7 |
| N1—Ni1—Cl2 | 88.69 (10) | C3—C4—C5 | 118.5 (5) |
| N1—Ni1—Cl3 | 171.31 (10) | C3—C4—H4 | 120.7 |
| N1—Ni1—Cl1 | 94.14 (11) | N6—C24—C25 | 124.3 (5) |
| N1—Ni1—N2 | 79.91 (13) | N6—C24—C23 | 116.7 (4) |
| N3—Ni1—Cl2 | 170.10 (12) | C25—C24—C23 | 118.8 (4) |
| N3—Ni1—Cl3 | 94.30 (9) | N2—C6—C5 | 110.5 (4) |
| N3—Ni1—Cl1 | 89.21 (12) | N2—C6—H6A | 109.6 |
| N3—Ni1—N1 | 91.94 (13) | N2—C6—H6B | 109.6 |
| N3—Ni1—N2 | 77.38 (15) | C5—C6—H6A | 109.6 |
| N2—Ni1—Cl2 | 93.02 (10) | C5—C6—H6B | 109.6 |
| N2—Ni1—Cl3 | 95.56 (9) | H6A—C6—H6B | 108.1 |
| N2—Ni1—Cl1 | 165.04 (11) | C14—C10—C9 | 114.7 (4) |
| Cl3—Ni2—Cl2 | 84.97 (4) | C14—C10—H10 | 106.7 |
| Cl4—Ni2—Cl2 | 99.38 (4) | C14—C10—C11 | 111.4 (4) |
| Cl4—Ni2—Cl3 | 93.14 (5) | C9—C10—H10 | 106.7 |
| N6—Ni2—Cl2 | 171.72 (12) | C11—C10—C9 | 110.3 (4) |
| N6—Ni2—Cl3 | 92.13 (11) | C11—C10—H10 | 106.7 |
| N6—Ni2—Cl4 | 88.51 (11) | N5—C23—C28 | 112.0 (5) |
| N6—Ni2—N5 | 79.29 (16) | C24—C23—N5 | 109.5 (4) |
| N6—Ni2—N4 | 88.11 (15) | C24—C23—C28 | 108.9 (4) |
| N5—Ni2—Cl2 | 93.15 (13) | C29—C23—N5 | 104.7 (5) |
| N5—Ni2—Cl3 | 94.06 (15) | C29—C23—C24 | 109.9 (5) |
| N5—Ni2—Cl4 | 166.04 (12) | C29—C23—C28 | 111.8 (5) |
| N4—Ni2—Cl2 | 93.92 (11) | C14—C15—H15A | 120.0 |
| N4—Ni2—Cl3 | 173.62 (15) | C14—C15—H15B | 120.0 |
| N4—Ni2—Cl4 | 93.24 (16) | H15A—C15—H15B | 120.0 |
| N4—Ni2—N5 | 79.7 (2) | C30—C31—H31A | 120.0 |
| Ni2—Cl2—Ni1 | 91.88 (4) | C30—C31—H31B | 120.0 |
| Ni1—Cl3—Ni2 | 94.09 (4) | H31A—C31—H31B | 120.0 |
| N3—O1—H1 | 111 (4) | C7—C13—H13A | 109.5 |
| C5—N1—Ni1 | 113.6 (3) | C7—C13—H13B | 109.5 |
| C1—N1—Ni1 | 127.6 (3) | C7—C13—H13C | 109.5 |
| C1—N1—C5 | 118.8 (4) | H13A—C13—H13B | 109.5 |
| O1—N3—Ni1 | 121.4 (3) | H13A—C13—H13C | 109.5 |
| C8—N3—Ni1 | 122.2 (3) | H13B—C13—H13C | 109.5 |
| C8—N3—O1 | 115.9 (3) | C12—C11—C10 | 112.0 (4) |
| Ni1—N2—H2 | 93 (4) | C12—C11—H11A | 109.2 |
| C7—N2—Ni1 | 113.6 (3) | C12—C11—H11B | 109.2 |
| C7—N2—H2 | 116 (4) | C10—C11—H11A | 109.2 |
| C6—N2—Ni1 | 104.0 (2) | C10—C11—H11B | 109.2 |
| C6—N2—H2 | 109 (4) | H11A—C11—H11B | 107.9 |
| C6—N2—C7 | 118.1 (3) | C14—C16—H16A | 109.5 |
| N6—O2—H2A | 105 (5) | C14—C16—H16B | 109.5 |
| N3—C8—C9 | 124.7 (4) | C14—C16—H16C | 109.5 |
| N3—C8—C7 | 116.1 (4) | H16A—C16—H16B | 109.5 |
| C9—C8—C7 | 119.2 (4) | H16A—C16—H16C | 109.5 |
| N1—C5—C4 | 122.3 (4) | H16B—C16—H16C | 109.5 |
| N1—C5—C6 | 115.1 (4) | C17—C18—H18 | 121.9 |
| C4—C5—C6 | 122.6 (4) | C19—C18—H18 | 121.9 |
| O2—N6—Ni2 | 122.4 (3) | C19—C18—C17 | 116.1 (8) |
| C24—N6—Ni2 | 120.3 (3) | N4—C17—C18 | 124.7 (7) |
| C24—N6—O2 | 116.7 (4) | N4—C17—H17 | 117.7 |
| Ni2—N5—H5 | 94 (4) | C18—C17—H17 | 117.7 |
| C23—N5—Ni2 | 112.0 (3) | C30—C26—C25 | 114.3 (5) |
| C23—N5—H5 | 115 (4) | C30—C26—H26 | 107.1 |
| C23—N5—C22 | 118.6 (4) | C30—C26—C27 | 112.5 (5) |
| C22—N5—Ni2 | 102.9 (4) | C25—C26—H26 | 107.1 |
| C22—N5—H5 | 110 (4) | C27—C26—C25 | 108.5 (5) |
| H12A—C12—H12B | 107.8 | C27—C26—H26 | 107.1 |
| C7—C12—H12A | 109.0 | N4—C21—C22 | 116.4 (5) |
| C7—C12—H12B | 109.0 | N4—C21—C20 | 117.2 (8) |
| C11—C12—H12A | 109.0 | C22—C21—C20 | 126.3 (7) |
| C11—C12—H12B | 109.0 | C23—C28—H28A | 109.2 |
| C11—C12—C7 | 113.0 (4) | C23—C28—H28B | 109.2 |
| C15—C14—C10 | 124.9 (4) | H28A—C28—H28B | 107.9 |
| C15—C14—C16 | 120.2 (4) | C27—C28—C23 | 112.2 (6) |
| C16—C14—C10 | 114.9 (4) | C27—C28—H28A | 109.2 |
| C2—C3—H3 | 120.1 | C27—C28—H28B | 109.2 |
| C2—C3—C4 | 119.7 (4) | C30—C32—H32A | 109.5 |
| C4—C3—H3 | 120.1 | C30—C32—H32B | 109.5 |
| C8—C9—H9A | 109.2 | C30—C32—H32C | 109.5 |
| C8—C9—H9B | 109.2 | H32A—C32—H32B | 109.5 |
| C8—C9—C10 | 112.2 (4) | H32A—C32—H32C | 109.5 |
| H9A—C9—H9B | 107.9 | H32B—C32—H32C | 109.5 |
| C10—C9—H9A | 109.2 | N5—C22—H22A | 109.7 |
| C10—C9—H9B | 109.2 | N5—C22—H22B | 109.7 |
| N1—C1—H1A | 119.0 | C21—C22—N5 | 110.0 (4) |
| N1—C1—C2 | 122.1 (4) | C21—C22—H22A | 109.7 |
| C2—C1—H1A | 119.0 | C21—C22—H22B | 109.7 |
| C31—C30—C26 | 124.8 (5) | H22A—C22—H22B | 108.2 |
| C31—C30—C32 | 120.1 (5) | C26—C27—H27A | 109.3 |
| C32—C30—C26 | 115.1 (5) | C26—C27—H27B | 109.3 |
| C17—N4—Ni2 | 126.7 (4) | C28—C27—C26 | 111.5 (5) |
| C21—N4—Ni2 | 113.2 (4) | C28—C27—H27A | 109.3 |
| C21—N4—C17 | 119.9 (5) | C28—C27—H27B | 109.3 |
| C3—C2—C1 | 118.6 (4) | H27A—C27—H27B | 108.0 |
| C3—C2—H2B | 120.7 | C23—C29—H29A | 109.5 |
| C1—C2—H2B | 120.7 | C23—C29—H29B | 109.5 |
| H25A—C25—H25B | 107.9 | C23—C29—H29C | 109.5 |
| C24—C25—H25A | 109.2 | H29A—C29—H29B | 109.5 |
| C24—C25—H25B | 109.2 | H29A—C29—H29C | 109.5 |
| C24—C25—C26 | 112.1 (4) | H29B—C29—H29C | 109.5 |
| C26—C25—H25A | 109.2 | C18—C19—H19 | 118.5 |
| C26—C25—H25B | 109.2 | C18—C19—C20 | 123.0 (7) |
| N2—C7—C8 | 109.8 (3) | C20—C19—H19 | 118.5 |
| N2—C7—C12 | 112.5 (3) | C21—C20—H20 | 120.5 |
| N2—C7—C13 | 107.1 (3) | C19—C20—C21 | 119.0 (7) |
| C8—C7—C12 | 108.5 (3) | C19—C20—H20 | 120.5 |
| Ni1—N1—C5—C4 | −178.5 (3) | N4—C21—C20—C19 | 2.4 (9) |
| Ni1—N1—C5—C6 | 2.4 (4) | C2—C3—C4—C5 | −0.9 (7) |
| Ni1—N1—C1—C2 | 177.8 (3) | C25—C24—C23—N5 | −168.7 (4) |
| Ni1—N3—C8—C9 | 171.5 (3) | C25—C24—C23—C28 | −45.9 (6) |
| Ni1—N3—C8—C7 | −10.6 (5) | C25—C24—C23—C29 | 76.9 (6) |
| Ni1—N2—C7—C8 | −7.7 (4) | C25—C26—C27—C28 | 59.6 (6) |
| Ni1—N2—C7—C12 | −128.6 (3) | C7—N2—C6—C5 | −83.5 (4) |
| Ni1—N2—C7—C13 | 109.3 (4) | C7—C8—C9—C10 | 47.9 (5) |
| Ni1—N2—C6—C5 | 43.4 (4) | C7—C12—C11—C10 | −57.9 (5) |
| Ni2—N6—C24—C25 | 173.2 (4) | C4—C5—C6—N2 | 148.6 (4) |
| Ni2—N6—C24—C23 | −12.5 (6) | C4—C3—C2—C1 | 0.6 (7) |
| Ni2—N5—C23—C24 | −13.2 (5) | C24—C25—C26—C30 | 73.6 (6) |
| Ni2—N5—C23—C28 | −134.2 (4) | C24—C25—C26—C27 | −52.8 (6) |
| Ni2—N5—C23—C29 | 104.5 (5) | C24—C23—C28—C27 | 49.5 (6) |
| Ni2—N5—C22—C21 | 44.6 (5) | C6—N2—C7—C8 | 114.5 (4) |
| Ni2—N4—C17—C18 | 174.2 (4) | C6—N2—C7—C12 | −6.5 (5) |
| Ni2—N4—C21—C22 | 5.4 (6) | C6—N2—C7—C13 | −128.6 (4) |
| Ni2—N4—C21—C20 | −178.0 (4) | C6—C5—C4—C3 | 180.0 (4) |
| O1—N3—C8—C9 | −0.7 (6) | C23—N5—C22—C21 | −79.6 (6) |
| O1—N3—C8—C7 | 177.3 (3) | C23—C28—C27—C26 | −59.7 (7) |
| N1—C5—C4—C3 | 1.0 (7) | C15—C14—C10—C9 | −24.5 (7) |
| N1—C5—C6—N2 | −32.3 (5) | C15—C14—C10—C11 | 101.6 (6) |
| N1—C1—C2—C3 | −0.2 (7) | C31—C30—C26—C25 | −6.7 (8) |
| N3—C8—C9—C10 | −134.2 (4) | C31—C30—C26—C27 | 117.5 (6) |
| N3—C8—C7—N2 | 11.5 (5) | C11—C12—C7—N2 | 172.3 (3) |
| N3—C8—C7—C12 | 134.8 (4) | C11—C12—C7—C8 | 50.6 (4) |
| N3—C8—C7—C13 | −104.9 (4) | C11—C12—C7—C13 | −67.8 (5) |
| O2—N6—C24—C25 | 1.3 (6) | C16—C14—C10—C9 | 158.2 (4) |
| O2—N6—C24—C23 | 175.6 (4) | C16—C14—C10—C11 | −75.7 (5) |
| C8—C9—C10—C14 | 77.6 (5) | C18—C19—C20—C21 | 1.5 (12) |
| C8—C9—C10—C11 | −49.1 (5) | C17—N4—C21—C22 | −179.7 (5) |
| C5—N1—C1—C2 | 0.3 (6) | C17—N4—C21—C20 | −3.0 (7) |
| N6—C24—C23—N5 | 16.7 (6) | C17—C18—C19—C20 | −4.4 (11) |
| N6—C24—C23—C28 | 139.5 (5) | C26—C25—C24—N6 | −136.7 (5) |
| N6—C24—C23—C29 | −97.7 (5) | C26—C25—C24—C23 | 49.2 (7) |
| N5—C23—C28—C27 | 170.8 (5) | C21—N4—C17—C18 | 0.0 (8) |
| C14—C10—C11—C12 | −72.9 (5) | C32—C30—C26—C25 | 174.8 (6) |
| C9—C8—C7—N2 | −170.4 (3) | C32—C30—C26—C27 | −60.9 (7) |
| C9—C8—C7—C12 | −47.1 (5) | C22—N5—C23—C24 | 106.3 (6) |
| C9—C8—C7—C13 | 73.2 (5) | C22—N5—C23—C28 | −14.6 (6) |
| C9—C10—C11—C12 | 55.6 (5) | C22—N5—C23—C29 | −135.9 (6) |
| C1—N1—C5—C4 | −0.6 (6) | C22—C21—C20—C19 | 178.6 (6) |
| C1—N1—C5—C6 | −179.7 (4) | C29—C23—C28—C27 | −72.1 (7) |
| C30—C26—C27—C28 | −67.9 (6) | C19—C18—C17—N4 | 3.8 (9) |
| N4—C21—C22—N5 | −35.0 (7) | C20—C21—C22—N5 | 148.7 (6) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···Cl1 | 0.85 (7) | 2.32 (6) | 3.009 (4) | 139 (6) |
| N2—H2···Cl4 | 0.77 (5) | 2.46 (5) | 3.209 (4) | 166 (5) |
| O2—H2A···Cl4 | 0.76 (8) | 2.31 (7) | 2.978 (4) | 147 (7) |
| C3—H3···O1i | 0.95 | 2.58 | 3.432 (5) | 149 |
| C1—H1A···Cl1 | 0.95 | 2.75 | 3.369 (5) | 124 |
| C6—H6A···Cl2 | 0.99 | 2.76 | 3.309 (5) | 115 |
| C11—H11B···Cl3ii | 0.99 | 2.64 | 3.573 (5) | 156 |
| C17—H17···Cl4 | 0.95 | 2.69 | 3.327 (6) | 125 |
| C26—H26···O2iii | 1.00 | 2.56 | 3.489 (6) | 154 |
| C22—H22B···Cl2 | 0.99 | 2.81 | 3.352 (6) | 115 |
| C19—H19···Cl1iv | 0.95 | 2.64 | 3.570 (7) | 167 |
Symmetry codes: (i) −x+2, y−1/2, −z+1; (ii) −x+1, y−1/2, −z+1; (iii) −x, y+1/2, −z; (iv) −x+1, y−1/2, −z.
Funding Statement
This work was funded by Ministère de l’Enseignement Supérieur de la Recherche et de l’Innovation (France); Ministère de la Recherche (Maroc); Institut Chevreul; Région Hauts-de-France; FEDER.
References
- Anastas, P. T. & Zimmerman, J. B. (2013). Environ. Sci. Technol. 37, 95A–101A. [DOI] [PubMed]
- Benabdelouahab, Y., Muñoz-Moreno, L., Frik, M., de la Cueva-Alique, I., El Amrani, M. A., Contel, M., Bajo, A. M., Cuenca, T. & Royo, E. (2015). Eur. J. Inorg. Chem. pp. 2295–2307. [DOI] [PMC free article] [PubMed]
- Bruker (2019). APEX2 and SAINT. Bruker AXS Inc., Madison Wisconsin, USA.
- Chahboun, G., Brito, J. A., Royo, B., El Amrani, M. A., Gómez-Bengoa, E., Mosquera, M. E. G., Cuenca, T. & Royo, E. (2012). Eur. J. Inorg. Chem. pp. 2940–2949.
- Cheng, T.-P., Liao, B.-S., Liu, Y.-H., Peng, S.-M. & Liu, S.-T. (2012). Dalton Trans. 41, 3468–3473. [DOI] [PubMed]
- Cueva-Alique, I. de la, Muñoz-Moreno, L., de la Torre-Rubio, E., Bajo, A. M., Gude, L., Cuenca, T. & Royo, E. (2019). Dalton Trans. 48, 14279–14293. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Elalami, M. S., Dahdouh, A. A., Mansour, A. I., ElAmrani, M. A., Suisse, I., Mortreux, A. & Agbossou-Niedercorn, F. (2009). C. R. Chim. 12, 1253–1258.
- El Alami, M. S. I., El Amrani, M. A., Agbossou-Niedercorn, F., Suisse, I. & Mortreux, A. (2015). Chem. Eur. J. 21, 1398–1413. [DOI] [PubMed]
- Gawley, R. E. & Aubé, J. (2012). Principles and applications of asymmetric synthesis, 2nd ed. Amsterdam: Elsevier Science
- El Alami, M. S. I., El Amrani, M. A., Dahdouh, A., Roussel, P., Suisse, I. & Mortreux, A. (2012). Chirality, 24, 675–682. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Ojima, I. (2010). Catalytic asymmetric synthesis, 3rd ed. Hoboken: Wiley
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Zheng, L., Zhang, S., Li, K., Chen, W., Chen, Y., Xu, B., Hu, B., Li, Y. & Li, W. (2010). J. Mol. Struct. 984, 153–156.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021010537/ex2048sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021010537/ex2048Isup2.hkl
CCDC reference: 2115017
Additional supporting information: crystallographic information; 3D view; checkCIF report





