Abstract
The routine diagnosis of Neospora caninum abortion is based upon histopathologic changes in fetal tissues and identification of tissue parasites by immunohistochemistry. Confirmation of N. caninum infection by immunohistochemistry has low sensitivity. In the present study, we examined the utility of PCR in detecting N. caninum infection in fetal tissues from spontaneous bovine abortion. DNA was obtained from fresh and formalin-fixed tissues from 61 bovine fetuses submitted for abortion diagnosis. Histopathology and immunohistochemistry determined the true status of N. caninum infection in each fetus. In formalin-fixed paraffin-embedded tissues, PCR detected N. caninum DNA in 13 of 13 true-positive fetuses (100%) and in 1 of 16 true-negative fetuses (6%). In fresh or frozen tissues, PCR detected N. caninum DNA in 10 of 13 true-positive fetuses (77%) and 0 of 11 true-negative fetuses (0%). PCR also detected N. caninum DNA in 6 of 8 fetuses that had typical lesions of N. caninum but were immunohistochemistry negative, indicating a higher sensitivity of PCR in comparison to that of immunohistochemistry. N. caninum DNA was amplified most consistently from brain tissue. PCR detection of N. caninum DNA in formalin-fixed, paraffin-embedded tissues was superior to that in fresh tissues, presumably because of the increased accuracy of sample selection inherent in histologic specimens.
Bovine neosporosis, caused by the apicomplexan protozoan parasite Neospora caninum, was initially recognized in 1989 (35) and is now reported as a leading infectious cause of reproductive failure in dairy cattle in countries worldwide (2, 3, 14, 29, 36, 40). The principle method of diagnosing Neospora caninum infection in aborted fetuses is by histopathology (HP) of fetal tissues, followed by specific identification of parasites within tissue lesions by immunohistochemistry (IHC) (5, 13, 24). Typical fetal lesions, which are not pathognomonic, include multifocal nonsuppurative necrotizing encephalitis and nonsuppurative myocarditis with or without focal necrosis in the liver (8, 39). IHC is relatively insensitive as a confirmatory test for neosporosis because parasite numbers in infected tissue can be very low, possibly leading to false negatives (12, 14, 18). Fetal serology has been used to confirm N. caninum abortion in individual fetuses, but the assay is not highly sensitive, as demonstrated by two separate studies where N. caninum-specific antibodies were present in only 50 to 65% of confirmed N. caninum-infected fetuses (6, 38). Maternal serology also is not consistently useful to confirm N. caninum abortion in individual cows (28, 31, 33). Thus, there is a practical need for a fast and reliable method to confirm N. caninum infection in tissues from aborted fetuses.
A sensitive and specific PCR detection assay for N. caninum DNA would be useful to augment the diagnosis of N. caninum abortion where pathologic changes in fetal tissues are consistent with neosporosis but cannot be consistently confirmed by IHC or serology. Although multiple PCR methods have been described for detection of N. caninum DNA in bovine tissues (16, 18, 19, 21, 23, 32, 41), N. caninum PCR has been tested infrequently for the routine diagnosis of naturally occurring N. caninum abortion. Gottstein et al. (18) used pNC-5 Neospora PCR to define the N. caninum infection status of 83 aborted bovine fetuses and identified a poor correlation between N. caninum PCR-positive status and the presence of N. caninum-compatible histologic lesions (nonsuppurative encephalitis and myocarditis) or N. caninum-positive serology (18), suggesting that the true status of the examined fetuses was not clearly identified. Ellis et al. (15) reported only 16 of 40 positive cases identified by ITS1 Neospora PCR in fetuses with brain and heart lesions compatible with N. caninum abortion (15), indicating a poor sensitivity of ITS1 PCR for clinical material. Thus, there is a need for further studies using defined N. caninum-positive and N. caninum-negative fetal populations to investigate the utility of PCR for the routine diagnosis of naturally occurring N. caninum abortion in cattle.
In the present study, our laboratory extended the PCR methodology for detection of the pNC-5 gene of N. caninum (41). PCR and seminested-PCR procedures were optimized by using primers pairs Np4-Np7 and Np6-Np7, and testing was done on groups of fetuses whose N. caninum infection status was defined by HP and IHC. PCR assays were developed as a multiplex procedure and included the use of primer pairs to the bovine prolactin (PRL) gene (27, 34) to exclude false-negative results due to poor-quality DNA or unknown PCR inhibitors in the clinical samples. The purposes of the study were (i) to determine the utility of PCR for the identification of N. caninum infection in defined N. caninum-aborted bovine fetuses; (ii) to determine the optimal fetal tissue to be analyzed by PCR; (iii) to determine whether detection of DNA by N. caninum PCR was more sensitive than detection of tachyzoites by IHC; (iv) to compare the utility of PCR assay of formalin-fixed, paraffin-embedded tissues with that of PCR assay of fresh tissues; and (v) to determine whether detection of N. caninum infection in clinical fetal tissues requires nested PCR procedures.
MATERIALS AND METHODS
Clinical samples.
Whole cadavers or tissues from 61 naturally aborted bovine fetuses were submitted to the Washington Animal Disease Diagnostic Laboratory for routine abortion diagnosis. The fetuses originated from commercial dairy and beef herds in the Pacific Northwest region of the United States (Washington, Idaho, and Oregon). Abortion diagnosis examinations were part of an abortion diagnostic kit that included examination of fetal tissues by histopathology, bacterial culture, virus isolation, and examination of maternal serum for antibodies to abortofacient pathogens (N. caninum, infectious bovine rhinotracheitis herpesvirus [IBR], bovine virus diarrhea virus [BVDV], leptospires [Leptospira icterohemorrhagica, L. hardjo, L. canicola, L. pamona, L. bratislava, and L. grippotyphosa], and Brucella abortus). N. caninum antibodies were detected in the dams by competitive enzyme-linked immunosorbent assay (ELISA; VMRD Inc., Pullman, Wash.) modified from a previously published procedure (10). A total of 162 fetal tissue samples were examined by N. caninum PCR; these included samples from brain, heart, kidney, liver, lung, spleen, and placenta. Not all tissues were available from all fetuses.
Experimental design.
Fetuses were grouped as outlined in Table 1 by tissue treatment (formalin-fixed, paraffin-embedded versus fresh or frozen) and N. caninum infection status (true status) as determined by HP and IHC. Fetuses classified as N. caninum true positive (group 1) had histopathologic changes consistent with N. caninum infection and tachyzoites within affected tissues detectable by IHC (HP+ IHC+). For the purposes of the present study, histopathologic changes compatible with N. caninum abortion had to be present, at a minimum, in the brain and heart (4, 39). The targeted lesions consisted of moderate or severe multifocal necrosis and gliosis in the brain associated with nonsuppurative encephalitis and moderate or severe nonsuppurative myocarditis. Fetuses classified as N. caninum true negative (group 2) had no lesions compatible with N. caninum infection and no detectable N. caninum tachyzoites by IHC (HP− IHC−) and served as the uninfected negative control group. The N. caninum-negative group contained fetuses diagnosed as resulting from idiopathic abortion, sporadic bacterial abortion, BVDV abortion, and IBR virus abortion. A third group of fetuses had histopathologic changes of N. caninum abortion but no tachyzoites were found by IHC (HP+ IHC−); these fetuses were used to compare the abilities of PCR and IHC to detect N. caninum infection.
TABLE 1.
Fetuses and fetal tissues examined by N. caninum PCR
| Group | N. caninum statusa | Tissue type | No. of fetuses | No. of tissue samples |
|---|---|---|---|---|
| 1 | HP+ IHC+ | Formalin fixed | 13 | 80 |
| 2 | HP− IHC− | Formalin fixed | 16 | 32 |
| 3 | HP+ IHC− | Formalin fixed | 8 | 8 |
| Total fixed | 37 | 120 | ||
| 4 | HP+ IHC+ | Fresh or frozen | 13 | 31 |
| 5 | HP− IHC− | Fresh or frozen | 11 | 11 |
| Total fresh | 24 | 42 |
HP, histopathologic changes consistent with N. caninum infection; IHC, N. caninum specific immunohistochemistry.
The following comparisons were analyzed by chi-square analysis using 2 × 2 contingency tables: (i) PCR detection of N. caninum infection in formalin-fixed tissues versus fresh tissues (Table 1, group 1 versus group 4), and (ii) PCR detection of N. caninum infection versus IHC detection of N. caninum infection (Table 1, group 1 versus group 3). The specificity and sensitivity of detection of N. caninum infection by PCR were determined for formalin-fixed and fresh tissues by using the HP+ IHC+ groups as true positives and the HP− IHC− groups as true negatives. Sensitivity was defined in the HP+ IHC+ groups (Table 1, groups 1 and 4) by the following formula: sensitivity = (PCR-positive fetuses/true-positive fetuses) × 100. Specificity was defined in the HP− IHC− groups (Table 1, groups 2 and 5) by the following formula: specificity = PCR-negative fetuses/true-negative fetuses) × 100. Agreement (concordance) between PCR detection of N. caninum DNA and true status (as determined by HP and IHC) was defined by the following formula: agreement = [(PCR-positive fetuses + PCR-negative fetuses)/(true-positive fetuses + true-negative fetuses)] × 100.
Histopathology and immunohistochemistry.
Fetal tissues were fixed in 10% neutral buffered formalin, paraffin embedded, and stained with hematoxylin and eosin for routine histologic examination. A second set of paraffin sections were mounted on positive-charged glass slides (Probe-On Plus; Fisher Scientific) and processed for IHC as previously described (24, 26) with avidin-biotin-complex (ABC) immunoperoxidase methods (Vector Elite ABC-peroxidase) with an automated capillary action immunostainer (Vantana Inc.). Sections were dehydrated, enzymatically treated with 0.1% protease XIV (Sigma Chemical Co., St. Louis, Mo.) for antigen retrieval, and incubated with 5% normal horse serum (Vector Laboratories, Burlingame, Calif.) to block nonspecific immunoglobulin binding. The primary antibody was anti-N. caninum hyperimmune goat serum (VMRD Inc.) diluted 1:2,000. Immunostaining was visualized with amino-ethyl-carbazol substrate (Dako Inc.), and sections were counterstained with Mayer's hematoxylin (Sigma Diagnostics, St. Louis, Mo.) and examined microscopically. Positive control tissue consisted of formalin-fixed brain tissue from BALB/c mice experimentally inoculated with the NC-1 strain of N. caninum (25). Negative controls consisted of (one) replacement of the primary antibody with a similar dilution of normal goat serum on all examined tissues. The specificity of the anti-N. caninum goat serum was confirmed in-house by the absence of immunoreactivity on archived tissue sections containing previously confirmed Toxoplasma gondii tachyzoites (cat with systemic toxoplasmosis) or Sarcocystis cruzi tachyzoites (bovine sarcocystosis abortion).
PCR.
PCR analysis was done by using primer pairs Np4-Np7 and Np6-Np7 of the pNC-5 gene of N. caninum (41) in standard or seminested PCR procedures. The Np4-Np7 primer pair is one of several pairs previously shown to be specific for N. caninum when tested against Toxoplasma, Sarcocystis, and Hammondia spp. (41). Preliminary studies in our laboratory using spiked bovine blood or spiked bovine brain showed that primer pair Np4-Np7, when used in a standard procedure or when used before primer pairs Np6-Np7 in a seminested procedure, provided optimal results in bovine tissues (data not shown). The Np4-Np7 primer pair amplifies a DNA fragment of 275 bp, and the Np6-Np7 primer pair amplifies a DNA fragment of 227 bp. To exclude the possibility of false-negative PCR results due to poor-quality DNA or the presence of nonspecific PCR inhibitors in the clinical tissue samples, all samples negative when N. caninum primer pairs alone were used were retested with a multiplex PCR, using N. caninum primers and PCR primers for the PRL gene, a constitutive gene expressed in bovine cells (27, 34). The PRL HL033-HL035 primer pair amplifies a 156-bp DNA fragment. Any fetal tissue samples that were PCR negative for both PRL and N. caninum were excluded from the study because of the poor quality of DNA in the clinical sample.
PCR sensitivity.
The methodological sensitivity of the standard PCR procedure was determined from fresh bovine brain spiked with purified cell culture-derived N. caninum tachyzoites (NC-1 isolate). Parasites were grown in Vero cells and purified by centrifugation in Percoll as described previously (10) and then counted with a hemocytometer. Zero, 10, 20, 30, 40, 50, 500, and 5,000 purified tachyzoites were diluted in culture medium, mixed with 20 mg of homogenized brain tissue, and DNA extracted for PCR. Sensitivity was expressed as organism equivalents as determined by microgram of DNA in the PCR by the following formula: tachyzoite equivalents = [(150 ng/total nanograms of DNA extracted in sample) × 100] × number of tachyzoites spiked in sample.
DNA extraction from fresh and formalin-fixed, paraffin-embedded tissue.
DNA was extracted from fresh or frozen and from formalin-fixed, paraffin-embedded tissues by using proteinase K digestion followed by ethanol precipitation without phenol-chloroform extraction (11, 22). Twenty milligrams of tissue was used for DNA extraction from fresh tissues. For formalin-fixed tissues, four 10-μm paraffin sections were cut with a standard microtome, placed in a 1.5-ml microcentrifuge tube with a sterile forceps, and dewaxed with xylene and ethanol washes. The microtome blade was cleaned between blocks with a xylene substitute (Histoclear) and 70% ethanol to prevent carryover. An empty microcentrifuge tube (lacking paraffin sections similarly processed) was included every 10 tubes as a paraffin negative control for contamination during sectioning. DNA was precipitated from digested tissue by using an equal volume of 4 M ammonium acetate followed by 2 volumes of isopropanol. The concentration of DNA was determined by spectrophotometric analysis at A260/280. Only DNA with A260/280 ratios of >1.0 were kept for PCR analysis. Tissue DNA was stored at −80°C.
The PCR mixture of 50 μl contained 150 ng of target DNA, 2 mM MgCl2, 10× reaction buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.1% Triton X-100), 1 mg of gelatin per ml, 10 pmol of each PCR primer, 200 μM each dNTP, and 2 U of Taq DNA polymerase. PCRs using Np4 (5′CCTCCCAATGCGAACGAAA3′) and Np7 (5′GGGTGAACCGAGGGAGTTG3′) were performed in a thermocycler (GeneAmp 2400; Perkin-Elmer) for 35 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 60 s. For multiplex PCR, similar concentrations of both Np4-Np7 and PRL primers HL033 (5′CGAGTCCTTATGAGCTTGATTCTT3′) and HL035 (5′GCCTTCCAGAAGTCGTTTGTTTTC3′) were simultaneously added to the PCR mixture described above. For seminested PCR, second-round primers Np6 (5′CAGTCAACCTACGTCTTCT3′) and Np7 (5′GGGTGAACCGAGGGAGTTG3′) used 2 μl of amplicon solution from first-round Np4-Np7 PCR amplification as target DNA with the same PCR mixture with 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 60 s. Amplicons were resolved on a 1.8% agarose gel stained with ethidium bromide and photographed under UV light. Positive controls (purified N. caninum tachyzoite DNA) and negative controls (DNA from normal bovine brain, paraffin control, and no DNA (double-distilled water) were included in each PCR run. PCR products were sequenced and compared to known published sequences to confirm that the correct DNA targets were being amplified.
RESULTS
Methodological sensitivity of Neospora PCR in spiked bovine brain.
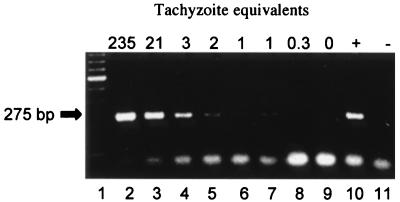
To determine the detection limit of the PCR assay in bovine tissues, PCR was performed on normal fresh bovine brain tissue spiked with N. caninum tachyzoites. Tachyzoite equivalents were calculated by the amount of target DNA in the PCR; generally, 150 ng of target DNA represented 3 to 6% of total DNA in the sample. Figure 1 shows a weak 275-bp band at 1 to 2 tachyzoite equivalents and a strong 275-bp band at ≥3 tachyzoite equivalents. The results indicated an adequate detection limit, in the range of 20 to 40 tachyzoites in 20 mg of bovine brain tissue, to pursue PCR detection of N. caninum in clinical samples.
FIG. 1.
Detection limit of N. caninum PCR in bovine brain spiked with tachyzoites. Total DNA was isolated from spiked bovine brain, and parasite DNA was detected by using the Np4-Np7 primer pair of the pNC-5 gene of N. caninum. PCR products were separated on agarose gel and stained with ethidium bromide. Lane 1, molecular mass ladder (100 kb); lanes 2 to 9, 20 mg of uninfected bovine brain spiked with 5,000, 500, 50, 40, 30, 20, 10, and 0 N. caninum tachyzoites, respectively (numbers at the tops of the lanes indicate N. caninum tachyzoite equivalents based upon micrograms of DNA loaded in the PCR as described in Materials and Methods); lane 10, positive control (N. caninum DNA from Vero cell cultures); lane 11, negative control (double-distilled water). Left arrow, specific 275-kb Np4-Np7 PCR product detected in lanes 2 to 7 and 10. The bottom bands are nonspecific unused PCR reagents and primer dimers.
Sensitivity and specificity of Neospora PCR in clinical samples.
The sensitivity and specificity were determined for PCR of fresh and paraffin-embedded tissues by using defined Neospora-positive and Neospora-negative fetuses (Table 1, groups 1, 2, 4, and 5). All fetal samples were initially tested with the Np4-Np7 primer pair only (Fig. 2). All PCR-negative samples were subsequently tested by using seminested PCR with Np4-Np7 followed by Np6-Np7 to increase the methodological sensitivity of the PCR assay. Finally, all samples negative by seminested PCR were tested with multiplex PCR using Neospora primer pair Np4-Np7 and primer pair HL033-HL035 to bovine PRL to identify false negatives due to poor-quality DNA or the presence of PCR inhibitors in the fetal tissue samples (Fig. 3). An N. caninum-positive PCR amplicon from any fetal tissue sample classified that particular fetus as PCR positive. The PCR results from the clinical samples are summarized in Table 2. Sensitivities of PCR were 100% for formalin-fixed, paraffin-embedded brain tissue (13 of 13 true-positive fetuses were PCR positive) and 77% for fresh brain (10 of 13 true-positive fetuses were PCR positive). Specificities of PCR were 94% for formalin-fixed paraffin-embedded brain tissue (1 to 16 true-negative fetuses were PCR positive) and 100% for fresh brain (0 of 13 true-negative fetuses were PCR positive). Agreement (concordance) rates between N. caninum PCR and true status were 97% for formalin-fixed, paraffin-embedded tissue and 88% for fresh tissue. Chi-square analysis comparing detection of N. caninum DNA in formalin-fixed tissue with that in fresh tissue samples revealed no significant difference (P > 0.05).
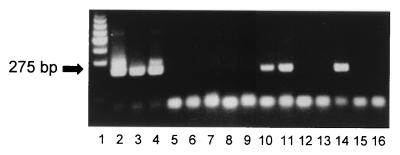
FIG. 2.
PCR detection of N. caninum in formalin-fixed paraffin-embedded tissues from aborted bovine fetuses. DNA was isolated as described in Materials and Methods, and parasite DNA was detected by using the Np4-Np7 primer pair of the pNC-5 gene of N. caninum. A representative illustration of PCR products separated on agarose gels and stained with ethidium bromide is shown. Lane 1, molecular mass ladder (100 kb); lane 2, positive control (N. caninum DNA); lanes 3 to 5, kidney (positive for N. caninum detection), brain (positive), and liver (negative) tissue from N. caninum-positive fetus 97-8417; lane 6, paraffin negative control; lanes 7 to 11, kidney (negative), lung (negative), liver (negative), heart (positive), and brain (positive) tissues from N. caninum-positive fetus 97-4707; lane 12, paraffin negative control; lane 13, brain tissue (negative) from N. caninum-negative fetus (IBR abortion) 96-713; lanes 14 and 15, brain (positive) and heart (negative) tissue from N. caninum-positive fetus 92-9309; lane 16, PCR-negative control (double-distilled water). Left arrow, specific 275-kb Np4-Np7 PCR product. Bottom bands are nonspecific unused PCR reagents and primer dimers.
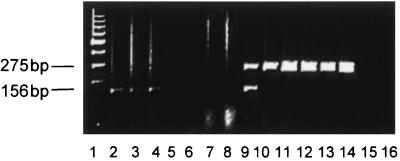
FIG. 3.
Multiplex PCR of N. caninum DNA and PRL DNA in formalin-fixed paraffin-embedded tissues from aborted bovine fetuses. Parasite DNA was detected by using the Np4-Np7 primer pair of the pNC-5 gene of N. caninum, and bovine PRL DNA was detected by using primer pair HL033-HL035. A representative illustration of PCR products separated on agarose gels and stained with ethidium bromide is shown. Lane 1, molecular mass ladder (100 kb); lanes 2 to 4, liver, heart, and brain tissue from N. caninum-negative fetus 96-10792 with detection of PRL DNA only; lanes 5 and 6, brain and kidney tissue from fetus 96-6445 with no detectable N. caninum or PRL DNA; lanes 7 and 8, brain and lung tissue from fetus 96-6445 with no detectable N. caninum or PRL DNA; lane 9, brain tissue from N. caninum-positive fetus 94-4569 with detectable N. caninum and PRL DNA; lanes 10 to 14, N. caninum-infected Vero cell cultures with detectable N. caninum DNA only (bovine PRL-negative control); lanes 15 and 16, PCR-negative control (double-distilled water). Left markers indicate a specific 275-kb Np4-Np7 PCR product detected in lanes 9 to 14 and a specific 156-kb HL033-HL035 PCR product detected in lanes 2 to 4 and 9.
TABLE 2.
Summary of N. caninum PCR results with Np4-Np7 on defined N. caninum-positive and -negative fetuses
| Type of tissue samplesa | Np4-Np7 PCR status | No. of samples
|
|
|---|---|---|---|
| True positive | True negative | ||
| Formalin fixed | Positive | 13 | 1 |
| Negative | 0 | 15 | |
| Fresh | Positive | 10 | 0 |
| Negative | 3 | 11 | |
Sensitivities, specificities, and agreements were 100, 94, and 97% for formalin-fixed tissue samples and 77, 100, and 88% for fresh tissues, respectively.
Tissue distribution of N. caninum DNA detected by PCR in N. caninum-infected fetuses.
The distribution and frequency of N. caninum infection in tissue were determined by Np4-Np7 PCR by using both formalin-fixed, paraffin-embedded tissues and fresh tissues (Table 3). With formalin-fixed, paraffin-embedded tissues, brain tissue was optimal for standard PCR detection of N. caninum, with 100% of defined positive cases being detected. The detection rate of standard PCR in fixed brain tissue was far superior to that in all other fixed tissues examined, with the detection rate for other tissues ranging from 17 to 50%. With fresh tissues, kidney and brain were optimal for standard PCR detection of N. caninum infection, with detection rates of 100 and 88%, respectively. Detection rates were much lower in other fresh tissues and ranged from 0 (spleen and placenta) to 60% in heart tissue.
TABLE 3.
Tissue distribution of parasite DNA and comparison of nested and standard PCR methods for detection of N. caninum PCR
| Tissue type | No. of tissue samples tested | No. (%) of samples positive by:
|
|
|---|---|---|---|
| PCR | Nested PCR | ||
| Fixed | |||
| Brain | 13 | 13 (100) | 13 (100) |
| Lung | 12 | 6 (50) | 11 (92) |
| Placenta | 5 | 2 (40) | 5 (100) |
| Kidney | 13 | 5 (38) | 11 (85) |
| Liver | 12 | 3 (25) | 10 (83) |
| Heart | 13 | 3 (23) | 8 (62) |
| Spleen | 12 | 2 (17) | 7 (58) |
| Fresh | |||
| Kidney | 3 | 3 (100) | 3 (100) |
| Brain | 8 | 7 (88) | 8 (100) |
| Heart | 5 | 3 (60) | 4 (80) |
| Lung | 5 | 2 (40) | 5 (100) |
| Liver | 7 | 2 (29) | 3 (43) |
| Placenta | 1 | 0 (0) | 1 (100) |
| Spleen | 2 | 0 (0) | 1 (50) |
Comparison of nested and standard PCRs in clinical samples.
All tissues from defined true-positive fetuses (Table 1, groups 1 and 4) were also tested by seminested PCR to determine whether nested PCR would increase the sensitivity of detecting N. caninum in an individual tissue over that of standard PCR (Table 3). Seminested PCR was superior to standard PCR for all fetal tissues tested except for formalin-fixed brain. Of both fresh and formalin-fixed tissues, five had greater than 75% detection rates by seminested PCR. However, the ability to identify an N. caninum-infected fetus was not improved by the seminested PCR procedure, compared to standard PCR, because standard PCR detected N. caninum DNA in at least one tissue (formalin-fixed brain) in 100% of the true-positive cases. In fresh tissues, detection of all true-positive fetuses by standard PCR required examination of at least two fetal tissues, brain and kidney. Thus, seminested PCR did not increase the sensitivity of detecting N. caninum infection in aborted bovine fetuses.
Comparison of PCR and IHC to detect Neospora infection in clinical samples.
Aborted fetuses from group 3 (Table 1) were tested to determine whether PCR was more sensitive than IHC in detecting N. caninum-infected fetuses. Brain tissue was chosen for the comparative analysis because the most consistent and diagnostic lesions of N. caninum infection occur in brain (4), and tachyzoites are most commonly associated with brain lesions in infected fetuses (5, 39). All PCR analyses were done on formalin-fixed, paraffin-embedded tissues. Both Np4-Np7 PCR and seminested PCR using Np4-Np7 followed by Np6-Np7 detected N. caninum DNA in six of eight (75%) fetuses that were IHC negative. Chi-square analysis comparing detection rates of N. caninum in formalin-fixed tissue samples by PCR and IHC revealed a significant difference (P < 0.01).
DISCUSSION
The goal of the present study was to apply PCR technology to the routine diagnosis of N. caninum-induced abortion in cattle. The exquisite sensitivity of PCR to detect small numbers of parasites in tissue together with the ability of HP to select appropriate tissue sections for PCR analysis (based upon the presence of tissue lesions) provide an ideal combination to reliably identify parasites and link them directly to areas of tissue damage. We determined the sensitivity and specificity of pNC-5 PCR by using fetuses defined as true N. caninum positive or true N. caninum negative. The “gold standard” criteria used to determine true N. caninum infection status were HP and IHC, a rationale supported by abundant literature showing that N. caninum infection in aborted bovine fetuses most consistently causes encephalitis and myocarditis (9, 14, 39). Chi-square analysis comparing fixed- and fresh-tissue PCRs revealed no statistically significant difference in detecting infection in 26 N. caninum-positive fetuses. However, when the 26 N. caninum-positive fetuses and 27 N. caninum-negative fetuses were analyzed, the sensitivity of fixed-tissue PCR (100%) and agreement with true status (97%) were superior to the sensitivity of the fresh-tissue PCR (77%) and agreement with true status (88%). The increased sensitivity of fixed-tissue PCR was attributed to the increased accuracy of sample selection inherent in histologic specimens. Fixed-tissue sections contained known tissue lesions, while the lesion status of fresh-tissue specimens was unknown. In summary, both fixed-tissue and fresh-tissue pNC-5 PCR using the Np4-Np7 primer pair provided adequate methods to confirm N. caninum infection in aborted fetuses.
Fixed-tissue PCR detected N. caninum infection in one true-negative fetus. It is well known that congenital infection is the primary mode of parasite transmission and that most congenitally infected calves are born clinically normal (30, 37). It is highly likely some fetuses that aborted for other causes or were diagnosed as resulting from idiopathic abortions in the present study may have had a mild, subclinical N. caninum infection that was below the detection limits of the gold standard methods (HP and IHC). The one PCR-positive fetus detected in the fixed-tissue true-negative group may be a good example of the new test (PCR) being more sensitive than the gold standard and possibly skewing the specificity data.
The pNC-5 PCR analyses showed that brain was the most reliable tissue overall for PCR analysis and that a nested PCR procedure (to increase sensitivity and specificity) was not necessary to detect N. caninum-infected fetuses. Reliable detection of N. caninum DNA in brain tissue by PCR is not surprising and is consistent with previous studies showing tissue parasites detected most frequently in brain by IHC or PCR (7, 14, 20, 39). Demonstrating the reliability of a standard PCR procedure for detecting N. caninum infection is important because one of the main disadvantages of PCR for routine diagnosis of infectious diseases is amplicon contamination, which may lead to false-positive tests. The chances of amplicon contamination increase significantly with a nested procedure, in which there is increased handling of amplicons from the first-round PCR and up to 1,000 times increased efficiency at generating second-round amplicons (22).
Another goal of the present study was to determine whether pNC-5 PCR was more sensitive than IHC in detecting N. caninum infection in fetal tissues. The finding of fetuses with N. caninum-compatible tissue lesions but no demonstrable parasites by immunohistochemistry (HP+ IHC−) is a common occurrence in routine diagnostic examinations of aborted bovine fetuses. The classification of these fetuses is unclear because Neospora-like lesions can also occur with other infectious agents (1, 17). This finding may ironically overrepresent N. caninum infections in fetuses because the cause of the abortion is not definitively confirmed. In the present study, PCR analysis of formalin-fixed tissues from HP+ IHC− fetuses showed that both standard and seminested pNC-5 PCR detected N. caninum DNA at similar rates in six of eight fetuses that were IHC negative, a significant difference by chi-square analysis. This data supports other studies suggesting that the routine practice of screening aborted fetal tissues by HP and confirming infection by using IHC is not a particularly sensitive or consistent way to identify true N. caninum infections (14, 18). Diagnosis of N. caninum abortion would be more accurate if a strategy of screening fetal tissues with histopathology followed by confirmation of N. caninum infection with standard pNC-5 PCR on serial sections from the same paraffin block was used.
In conclusion, the present study demonstrates the utility of PCR-based assay to identify N. caninum infection in spontaneously aborted bovine fetuses. PCR detection of N. caninum DNA worked well on formalin-fixed paraffin-embedded tissue, which increases the practical application of a PCR-based assay. When interpreted in conjunction with significant histopathologic changes in aborted fetal tissues, PCR should provide a valuable confirmatory tool to diagnose N. caninum abortion in cattle and makes possible retrospective analyses of archived clinical samples. In addition, the ability to amplify parasite-specific DNA from formalin-fixed or fresh clinical samples provides a new method to obtain N. caninum genetic material for analysis of strain genotype or variation of specific parasite genes.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of the Washington Animal Disease Diagnostic Laboratory, in particular, Victor Tobias and Nancy Weber of the histology laboratory and Pam Dilbeck and Ruth Brown of the IHC laboratory.
REFERENCES
- 1.Anderson M L, Barr B C, Conrad P A. Protozoal causes of reproductive failure in domestic ruminants. Vet Clin N Am Food Anim Pract. 1994;10:439–461. doi: 10.1016/s0749-0720(15)30531-4. [DOI] [PubMed] [Google Scholar]
- 2.Anderson M L, Blanchard P C, Barr B C, Dubey J P, Hoffman R L, Conrad P A. Neospora-like protozoan infection as a major cause of abortion in California dairy cattle. J Am Vet Med Assoc. 1991;198:241–244. [PubMed] [Google Scholar]
- 3.Anderson M L, Palmer C W, Thurmond M C, Picanso J P, Blanchard P C, Breitmeyer R E, Layton A W, McAllister M, Daft B, Kinde H, et al. Evaluation of abortions in cattle attributable to neosporosis in selected dairy herds in California. J Am Vet Med Assoc. 1995;207:1206–1210. [PubMed] [Google Scholar]
- 4.Barr B C, Anderson M L, Blanchard P C, Daft B M, Kinde H, Conrad P A. Bovine fetal encephalitis and myocarditis associated with protozoal infections. Vet Pathol. 1990;27:354–361. doi: 10.1177/030098589002700508. [DOI] [PubMed] [Google Scholar]
- 5.Barr B C, Anderson M L, Dubey J P, Conrad P A. Neospora-like protozoal infections associated with bovine abortions. Vet Pathol. 1991;28:110–116. doi: 10.1177/030098589102800202. [DOI] [PubMed] [Google Scholar]
- 6.Barr B C, Anderson M L, Sverlow K W, Conrad P A. Diagnosis of bovine fetal Neospora infection with an indirect fluorescent antibody test. Vet Rec. 1995;137:611–613. [PubMed] [Google Scholar]
- 7.Barr B C, Anderson M L, Woods L W, Dubey J P, Conrad P A. Neospora-like protozoal infections associated with abortion in goats. J Vet Diagn Investig. 1992;4:365–367. doi: 10.1177/104063879200400331. [DOI] [PubMed] [Google Scholar]
- 8.Barr B C, Conrad P A, Dubey J P, Anderson M L. Neospora-like encephalomyelitis in a calf: pathology, ultrastructure, and immunoreactivity. J Vet Diagn Investig. 1991;3:39–46. doi: 10.1177/104063879100300109. [DOI] [PubMed] [Google Scholar]
- 9.Barr B C, Rowe J D, Sverlow K W, BonDurant R H, Ardans A A, Oliver M N, Conrad P A. Experimental reproduction of bovine fetal Neospora infection and death with a bovine Neospora isolate. J Vet Diagn Investig. 1994;6:207–215. doi: 10.1177/104063879400600212. [DOI] [PubMed] [Google Scholar]
- 10.Baszler T V, Knowles D P, Dubey J P, Gay J M, Mathison B A, McElwain T F. Serological diagnosis of bovine neosporosis by Neospora caninum monoclonal antibody-based competitive inhibition enzyme-linked immunosorbent assay. J Clin Microbiol. 1996;34:1423–1428. doi: 10.1128/jcm.34.6.1423-1428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns W C, Liu Y S, Dow C, Thomas R J S, Phillips W A. Direct PCR from paraffin-embedded tissue. Biotechniques. 1997;22:638–640. doi: 10.2144/97224bm13. [DOI] [PubMed] [Google Scholar]
- 12.Conrad P A, Sverlow K, Anderson M, Rowe J, BonDurant R, Tuter G, Breitmeyer R, Palmer C, Thurmond M, Ardans A, et al. Detection of serum antibody responses in cattle with natural or experimental Neospora infections. J Vet Diagn Investig. 1993;5:572–578. doi: 10.1177/104063879300500412. [DOI] [PubMed] [Google Scholar]
- 13.Dubey J P, Lindsay D S. Neosporosis. Parasitol Today. 1993;9:452–458. doi: 10.1016/0169-4758(93)90099-2. [DOI] [PubMed] [Google Scholar]
- 14.Dubey J P, Lindsay D S. A review of Neospora caninum and neosporosis. Vet Parasitol. 1996;67:1–59. doi: 10.1016/s0304-4017(96)01035-7. [DOI] [PubMed] [Google Scholar]
- 15.Ellis J T. Polymerase chain reaction approaches for the detection of Neospora caninum and Toxoplasma gondii. Int J Parasitol. 1998;28:1053–1060. doi: 10.1016/s0020-7519(98)00096-4. [DOI] [PubMed] [Google Scholar]
- 16.Ellis J T, Amoyal G, Ryce C, Harper P A, Clough K A, Homan W L, Brindley P J. Comparison of the large subunit ribosomal DNA of Neospora and toxoplasma and development of a new genetic marker for their differentiation based on the D2 domain. Mol Cell Probes. 1998;12:1–13. doi: 10.1006/mcpr.1997.0143. [DOI] [PubMed] [Google Scholar]
- 17.Fayer R, Dubey J P. Bovine sarcocystosis. Compendium for Continuing Education of Practicing Veterinarians. 1986;8:F130. [Google Scholar]
- 18.Gottstein B, Hentrich B, Wyss R, Thur B, Busato A, Stark K D, Muller N. Molecular and immunodiagnostic investigations on bovine neosporosis in Switzerland. Int J Parasitol. 1998;28:679–691. doi: 10.1016/S0020-7519(98)00006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho M S, Barr B C, Marsh A E, Anderson M L, Rowe J D, Tarantal A F, Hendrickx A G, Sverlow K, Dubey J P, Conrad P A. Identification of bovine Neospora parasites by PCR amplification and specific small-subunit rRNA sequence probe hybridization. J Clin Microbiol. 1996;34:1203–1208. doi: 10.1128/jcm.34.5.1203-1208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho M S Y, Barr B C, Tarantal A F, Lai L T Y, Hendrickx A G, Marsh A E, Sverlow K W, Packham A E, Conrad P A. Detection of Neospora from tissues of experimentally infected rhesus macaques by PCR and specific DNA probe hybridization. J Clin Microbiol. 1997;35:1740–1745. doi: 10.1128/jcm.35.7.1740-1745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmdahl O J, Mattsson J G. Rapid and sensitive identification of Neospora caninum by in vitro amplification of the internal transcribed spacer 1. Parasitology. 1996;112(part 2):177–182. doi: 10.1017/s0031182000084742. [DOI] [PubMed] [Google Scholar]
- 22.Jackson D P, Hayden J D, Quirke P. Extraction of nucleic acid from fresh and archival material. In: McPherson J J, Quirke P, Taylor G R, editors. PCR: a practical approach. New York, N.Y: Oxford University Press; 1992. pp. 29–50. [Google Scholar]
- 23.Lally N C, Jenkins M C, Dubey J P. Development of a polymerase chain reaction assay for the diagnosis of neosporosis using the Neospora caninum 14-3-3 gene. Mol Biochem Parasitol. 1996;75:169–178. doi: 10.1016/0166-6851(95)02530-8. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay D S, Dubey J P. Immunohistochemical diagnosis of Neospora caninum in tissue sections. Am J Vet Res. 1989;50:1981–1983. [PubMed] [Google Scholar]
- 25.Lindsay D S, Lenz S D, Cole R A, Dubey J P, Blagburn B L. Mouse model for central nervous system Neospora caninum infections. J Parasitol. 1995;81:313–315. [PubMed] [Google Scholar]
- 26.Long M T, Baszler T V, Mathison B A. Comparison of intracerebral parasite load, lesion development, and systemic cytokines in mouse strains infected with Neospora caninum. J Parasitol. 1998;84:316–320. [PubMed] [Google Scholar]
- 27.Mirsky M L, Da Y, Lewin H A. Detection of bovine leukemia virus proviral DNA in individual cells. PCR Methods Appl. 1993;2:333–340. doi: 10.1101/gr.2.4.333. . [Erratum, 3:81, 1993.] [DOI] [PubMed] [Google Scholar]
- 28.Moen A R, Wouda W. Proceedings of the Symposium Neospora abortus Bij Het Rund 8 November 1995, Morra 2. 1995. Field experiences with bovine Neospora abortion in Dutch dairy herds; pp. 11–17. Drachten. [Google Scholar]
- 29.Otter A, Jeffrey M, Griffiths I B, Dubey J P. A survey of the incidence of Neospora caninum infection in aborted and stillborn bovine fetuses in England and Wales. Vet Rec. 1995;136:602–606. doi: 10.1136/vr.136.24.602. [DOI] [PubMed] [Google Scholar]
- 30.Pare J, Thurmond M C, Hietala S K. Congenital Neospora caninum infection in dairy cattle and associated calfhood mortality. Can J Vet Res. 1996;60:133–139. [PMC free article] [PubMed] [Google Scholar]
- 31.Pare J, Thurmond M C, Hietala S K. Neospora caninum antibodies in cows during pregnancy as a predictor of congenital infection and abortion. J Parasitol. 1997;83:82–87. [PubMed] [Google Scholar]
- 32.Payne S, Ellis J. Detection of Neospora caninum DNA by the polymerase chain reaction. Int J Parasitol. 1996;26:347–351. doi: 10.1016/0020-7519(96)00030-6. [DOI] [PubMed] [Google Scholar]
- 33.Reichel M P, Drake J M. The diagnosis of Neospora abortions in cattle. N Z Vet J. 1996;44:151–154. doi: 10.1080/00480169.1996.35960. [DOI] [PubMed] [Google Scholar]
- 34.Sasavage N L, Nilson J H, Horowitz S, Rottman F M. Nucleotide sequence of bovine prolactin messenger RNA. Evidence for sequence polymorphism. J Biol Chem. 1982;257:678–681. [PubMed] [Google Scholar]
- 35.Thilsted J P, Dubey J P. Neosporosis-like abortions in a herd of dairy cattle. J Vet Diagn Investig. 1989;1:205–209. doi: 10.1177/104063878900100301. [DOI] [PubMed] [Google Scholar]
- 36.Thornton R N, Gajadhar A, Evans J. Neospora abortion epidemic in a dairy herd. N Z Vet J. 1994;42:190–191. doi: 10.1080/00480169.1994.35819. [DOI] [PubMed] [Google Scholar]
- 37.Thurmond M C, Hietala S K. Effect of congenitally acquired Neospora caninum infection on risk of abortion and subsequent abortions in dairy cattle. Am J Vet Res. 1997;58:1381–1385. [PubMed] [Google Scholar]
- 38.Wouda W, Dubey J P, Jenkins M C. Serological diagnosis of bovine fetal neosporosis. J Parasitol. 1997;83:545–547. [PubMed] [Google Scholar]
- 39.Wouda W, Moen A R, Visser I J, van Knapen F. Bovine fetal neosporosis: a comparison of epizootic and sporadic abortion cases and different age classes with regard to lesion severity and immunohistochemical identification of organisms in brain, heart, and liver. J Vet Diagn Investig. 1997;9:180–185. doi: 10.1177/104063879700900212. [DOI] [PubMed] [Google Scholar]
- 40.Wouda W, van den Ingh T S, van Knapen F, Sluyter F J, Koeman J P, Dubey J P. Neospora abortion in cattle in The Netherlands. Tijdschr Diergeneeskd. 1992;117:599–602. [PubMed] [Google Scholar]
- 41.Yamage M, Flechtner O, Gottstein B. Neospora caninum: specific oligonucleotide primers for the detection of brain “cyst” DNA of experimentally infected nude mice by the polymerase chain reaction (PCR) J Parasitol. 1996;82:272–279. [PubMed] [Google Scholar]