Abstract
Acyl CoA Dehydrogenase 9 (ACAD9) is a member of the family of flavoenzymes that catalyze the dehydrogenation of acyl-CoAs to 2,3 enoyl-CoAs in mitochondrial fatty acid oxidation (FAO). Inborn errors of metabolism of all family members, including ACAD9, have been described in humans, and represent significant causes of morbidity and mortality particularly in children. ACAD9 deficiency leads to a combined defect in fatty acid oxidation and oxidative phosphorylation (OXPHOS) due to a dual role in the pathways. In addition to its function in mitochondrial FAO, ACAD9 has a second function as one of 14 factors responsible for assembly of complex I of the electron transport chain (ETC). Considerable controversy remains over the relative role of these two functions in normal physiology and the disparate clinical findings described in patients with ACAD9 deficiency. To better understand the normal function of ACAD9 and the pathophysiology of its deficiency, several knock out mouse models were developed. Homozygous total body knock out appeared to be lethal as no ACAD9 animals were obtained. Cre-lox technology was then used to generate tissue-specific deletion of the gene. Cardiac-specific ACAD9 deficient animals had severe neonatal cardiomyopathy and died by 17 days of age. They had severe mitochondrial dysfunction in vitro. Muscle-specific mutants were viable but exhibited muscle weakness. Additional studies of muscle from the muscle specific deficient animals were used to examine the evolutionarily conserved signaling Intermediate in toll pathway (ECSIT) protein, a known binding partner of ACAD9 in the electron chain complex I assembly pathway. As expected, ECSIT levels were significantly reduced in the absence of ACAD9 protein, consistent with the demonstrated impairment of the complex I assembly. The various ACAD9 deficient animals should serve as useful models for development of novel therapeutics for this disorder.
Keywords: Acyl-CoA dehydrogenase 9, Mouse models, Fatty acid oxidation, Respiratory chain, Supercomplexes, Cardiomyopathy, Myopathy
Introduction
Acyl-CoA dehydrogenase 9 (ACAD9) is a member of acyl-CoA dehydrogenase (ACAD) gene family, encoding enzymes catalyzing the α, β dehydrogenation of acyl-CoA esters during fatty acid β-oxidation and branched chain amino acid catabolism in mitochondria [1]. ACAD9 exhibits maximum activity with unsaturated long-chain fatty acids (C16), but is also active with saturated substrates, mirroring that of very long chain acyl-CoA dehydrogenase (VLCAD) [2]. Despite this substrate overlap, these two proteins have separate and distinct physiologic functions and deficiency of one is not rescued by the presence of the other, originally thought to be due to differences in tissue expression [3–5]. While low levels of ACAD9 can be seen in all tissues, it is present in quantities significant enough to contribute to fatty acid oxidation (FAO) only in liver [2, 3].
The ACAD9 gene is located on chromosome 3q21.3 and is comprised of 18 exons encoding a precursor protein of 621 amino acids with a molecular mass of 68 kDa [6]. The precursor protein is imported into mitochondria and processed to its mature form through the action of two mitochondrial peptidases that cleave a 37 amino acid sequence from the N-terminus end before the mature protein assembles as a homodimer associated with the inner mitochondrial membrane. ACAD9 shares the greatest sequence homology (47%) with VLCAD, also a homodimer, while the other ACADs are tetramers [1, 2].
ACAD9 is unique among the ACADs in that it has a second function (often denoted as “moonlighting”) in the assembly of complex I of the mitochondrial electron transfer chain (ETC) [6, 7]. Complex I (NADH (nicotinamide adenine dinucleotide) ubiquinone oxidoreductase) is the largest complex of the ETC, located primarily in the inner mitochondrial membrane, but with an NADH+ binding domain that extends into the mitochondrial matrix [8]. The latter domain accepts reducing equivalents from a variety of mitochondrial dehydrogenases as the first step in OXPHOS, which generates a proton gradient across the inner membrane that is ultimately used by complex V to synthesize adenosine triphosphate (ATP). Complex I is composed of 45 structural subunits, and its assembly is a stepwise process requiring at least 14 known assembly factors [9]. Following its description as an enzyme, ACAD9 was subsequently shown to interact with the proteins NDUFAF1 (NADH:Ubiquinone Oxidoreductase Complex Assembly Factor 1), ECSIT (Evolutionarily Conserved Signaling Intermediate in Toll pathway) and TMEM126B (Transmembrane Protein 126B) to add a domain to the maturing complex I molecule that bridges its intramitochondrial membrane component with the matrix NADH binding domain [4, 6, 7]. However, neither ACAD9 nor ECSIT play a structural role in the final complex I molecule [6, 7].
Deficiency of ACAD9 has been described with variable symptoms including cardiomyopathy, liver dysfunction, episodic metabolic decompensation, and hypotonia [3–5, 10–13]. Deficits of both functions of ACAD9 contribute to the phenotype in deficient patients. Specifically, mutations in ACAD9 protein that affect both the assembly and enzymatic functions lead to more severe clinical symptoms than those that affect only the complex I assembly function [4]. Mutations in ACAD9 are now recognized as one of the most common causes of complex I deficiency, typically identified by exome sequencing rather than specific metabolic studies [5, 14].
In this study, we show that ACAD9 deficiency in mice is lethal. In contrast, tissue specific deficiencies are viable and demonstrate symptoms based on the affected tissue. Western blotting and immunostaining confirm a loss of the ACAD9 binding partner ECSIT in affected tissue, consistent with its known function in complex I assembly. These models provide options for testing potential therapies for ACAD9 deficiency.
Materials and Methods
Mouse strains
The mouse strain used for this research project, KOMP ES cell line Acad9tm1a(KOMP)Wtsi, RRID:MMRRC_051676-UCD, was obtained from the Mutant Mouse Resource and Research Center (MMRRC) at University of California at Davis, an NIH-funded strain repository, and was donated to the MMRRC by The KOMP Repository, University of California, Davis; Originating from Pieter de Jong, Kent Lloyd, William Skarnes, Allan Bradley, Wellcome Sanger Institute [15]. The knockout-first allele produces a constitutive whole-body knockout unless Flp recombinase is used to convert the allele to a conditional allele with a “floxed” exon [15]. We first created the whole-body constitutive knockout at the Genome Editing, Transgenic, and Virus Core Facility at the Magee Women’s Research Institute (Pittsburgh, PA). Then, after preliminary characterization of the whole-body knockout, the constitutive ACAD9 knockout mice were bred to a transgenic strain expressing Flp recombinase (Jackson Laboratories, Bar Harbor, ME) to convert the ACAD9 knockout-first allele to a conditional allele in which exon four of the ACAD9 gene is flanked by LoxP sites. Floxed ACAD9 mice were then interbred with cardiac and muscle-specific Cre-expressing strains obtained from the Jackson Laboratories (Bar Harbor, ME). The FVB-Tg(Myh6-cre)2182Mds/J strain, which utilizes the myosin heavy chain-6 (Myh6) promoter, was used to provide cardiac specificity, while the FVB.Cg-Tg(ACTA1-cre)79Jme/J was used to provide muscle specificity via the human alpha-skeletal actin-1 (ACTA1) promoter. Genotyping of all experimental animals was verified by PCR using genomic tail tip DNA. Cre-negative, homozygous floxed littermate controls (hereafter designated as Acad9+/+) were used for all experiments. All animal protocols were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC), and all experiments were conducted in accordance with the guidelines and regulations set forth in the Animal Welfare Act (AWA) and PHS Policy on Humane Care and Use of Laboratory Animals.
Extracting mouse tissue for processing and protein analysis
Mice were euthanized by CO2 inhalation followed by cervical dislocation. Mouse heart, liver, and skeletal muscle were excised, and each tissue was washed in ice-cold phosphate buffered saline PBS until clear of blood. Tissues were cut into two (ratio of approximately 25:75), and the smaller piece of tissue was treated with 4% paraformaldehyde, which was then submitted for histologic analysis to the UPMC Children’s Hospital Mouse Histology Core for immunofluorescent, hemotoxylin and eosin staining (H&E), and trichrome staining. The larger piece of tissue was placed in a cryofreeze-safe tube and immediately submerged in liquid nitrogen to flash freeze. The sample was stored at −80 °C.
Blue native gel electrophoresis
A 1:5 master mix of digitonin:HEPES (Millipore. Burlington, MA) was made and incubated at 95 °C for 5 min to dissolve into solution. The digitonin mixture was placed on ice for at least 20 min then added to mouse mitochondria samples to give a 1:5 ratio of protein:digitonin and a 3 mg/mL protein concentration. The samples were incubated on ice for an additional 30 min, then Coomassie blue stain (0.1% R250, BioRad) in 7% acetic acid, 40% methanol in water was added at a dilution between 1:20 – 1:30 and resuspended by pipette. Samples were centrifuged at 17,000 rpm for 1 hr at 4 °C. The supernatants were transferred to new vials then loaded onto a 3–12%, 10-well bis-tris gel (Native-PAGE; Thermo Fisher Scientific, Waltham, MA). The gel was run at 80V for 4–5 hours at 4°C, then stained with Coomassie R250 stain for 15 min, and de-stained for 30 minutes to 1 hour or until the stain is sufficiently removed in 10% acetic acid, 40% methanol mixture in water.
Western blotting
Twenty-five micrograms of protein treated with SDS sample buffer (S3401–10VL, Sigma-Aldrich, St. Louis, MO) were loaded onto a 4–15 % criterion™ TGX™ precast midi-protein gels (5671084, Bio-Rad, Hercules, CA) along with a Precision plus protein™ dual color molecular weight maker (1610374, Bio-Rad, Hercules, CA). Following electrophoresis, the gels were transferred onto a nitrocellulose membrane (1620097, Bio-Rad, Hercules, CA), the membranes were blocked for 1 hour at room temperature in blocking buffer [10% nonfat dry milk in phosphate buffered saline, pH 7.4 containing 0.05% tween 20 (PBST)], and then washed with PBST three times for 5 min each at room temperature. The membranes were then incubated with an in-house produced (Cocalico Biologicals, Inc) primary rabbit anti-ACAD9, ECSIT, and VLCAD or MCAD (used for heart tissue only) polyclonal antibody (1:1000 dilution) or HRP-conjugated Beta Actin Monoclonal antibody (HRP-66009, Proteintech Group Inc, Rosemont, IL) in 1% non-fat dry milk in PBST for 1 hour at room temperature, and washed three times with PBST for 5 min each at room temperature. Anti-ACAD9, ECSIT, and VLCAD were incubated with a dilution of 1:3000 goat anti-rabbit, horseradish peroxidase (HRP) secondary antibody (1706515, Bio-Rad, Hercules, CA) in 1% non-fat dry milk in PBST for 1 hour at room temperature. Anti-GAPDH was incubated with a dilution of 1:3000 goat anti-mouse, horseradish peroxidase (HRP) secondary antibody (1706516, Bio-Rad, Hercules, CA) in 1% non-fat dry milk in PBST for 1 hour at room temperature. The membrane was washed three times with PBST for 5 min each, and then left in 1-Step™ NBT/BCIP substrate solution (PI34042, Thermo Fisher Scientific, Waltham, MA) for 4 minutes until bands became visible.
In-vivo Cardiac MRI
Animal preparation
Anesthesia induction was first achieved with 3–4% isoflurane mixed with oxygen in an induction box. The depth of anesthesia was monitored by toe reflex, extension of limbs, and spine positioning. Once the plane of anesthesia was established, the mouse was placed on a designated animal bed for imaging and the anesthesia was maintained by 1.0 to 1.5 % isoflurane with 100% oxygen via a nose cone. Body temperature was monitored by an optical rectal temperature probe used for feedback control of a warm air blower around the animal to maintain the core temperature 37 °C + 0.5 °C. Respiration was continuously monitored by placing a small pneumatic pillow under the animal’s diaphragm connected to a magnet-comparable pressure transducer feeding to a physiological monitoring computer equipped with respiration-waveform measuring software (SA Instruments, Stony Brook, NY). The respiration waveform was automatically processed to detect inspiration, expiration and respiration rate.
In vivo CMR acquisition
In-vivo cardiac MRI (CMR) was performed on a Bruker Biospec 7T/30 system (Bruker Biospin MRI, Billerica, MA) with the 35-mm quadrature coil for both transmission and reception. Free-breathing-no-gating cine MRI with retrospective navigators was acquired with the Bruker Intragate module. White-blood multi-planar cine movies with 20 cardiac phases were acquired for short-axis and long-axis 4-chamber views with the following parameters: Field of view (FOV) = 2 cm × 2 cm, slice thickness = 1 mm, inter-slice gap = 0, acquisition matrix = 256 × 256, in-plane resolution = 78 μm, flip angle (FA) = 10 degrees, echo time (TE) = 3.059 msec, repetition time (TR) = 5.653 msec, number of repetition = 250, total scan time = 3 min 2 sec.
Cardiac function from cine CMR
The left ventricular endocardium and epicardium boundaries of each imaging slice at the end-systole (ES) and the diastole (ED) were manually traced by a blinded operator on the Paravision 5.1 Xtip software (Bruker Biospin MRI, Billerica, MA) to calculate the following functional parameters: left ventricular blood volume (LVV), left ventricular wall volume (LV wall), LV mass, stroke volume (SV), and ejection fraction (EF). LVV is calculated by summation of blood volume of all the short-axis slices at the end-diastole. , where the internal left ventricle area of slice i at end diastole, and hi is the thickness of each short-axis slice. The ejection fraction (EF) was calculated using the following equation: , where is the internal left ventricle area of slice i at end systole, the internal left ventricle area of slice i at the end diastole, and hi is the thickness of each slice. The LV wall volume was calculated by summation of LV wall volume (LVW) of all short-axis slices at the end diastole. hi, where ( is the area encompassed by the epicardial boundary, is the area encompassed by the endocardial boundary at the end-diastole and hi is the thickness of each slice. The left ventricle wall thickening was calculated through equation LV wall thickening = (des – ded)/ded × 100% where des and ded are the wall thickness measured at the end-systole (des) and the end diastole (ded). LV wall mass was calculated from the end-diastolic LV wall volume times the specific myocardial tissue density 1.055 g/ml [16, 17]. Myocardial feature tracking analysis was performed with Circle cvi4 software. The endocardial and epicardial surface was defined in short-axis cine MRI to calculate the peak global circumferential and radial strain and peak global circumferential and radial displacement for both LV and RV.
Hanging wire test.
A custom hanging wire apparatus was constructed from standard household hardware. Before the trial, each mouse started with a fall score defined as 10. The mouse was lowered onto the middle of the wire and allowed to hang by its front paws. The mouse was released, and a timer for 180 seconds was started. If the mouse fell, the timer was immediately stopped, the mouse’s fall score was reduced by one, the mouse was placed back on the center of the wire, and the timer was restarted. If the mouse reached either end of the wire, the timer was stopped, the mouse was placed back on the center of the wire, and the timer was restarted again. This continued for 180 secs or until the mouse’s fall score reached zero. Fall score, and elapsed time were recorded. Each 180-second trial was repeated consecutively, in triplicate for each mouse.
Measurement of blood glucose and L-lactate.
A drop of blood was extracted by making a small incision near the tip of the tail using a pair of surgical scissors. The directions of the glucometer (AimStrip Plus. Thermo Fisher Scientific, Waltham, MA) and strips and L-lactometer (Lactate Scout. EKF Diagnostics, Elkhart, IN) and strips were followed to measure each parameter. Bleeding was halted using Kwik Stop (ARC Laboratories, Dayton, OH). Glucose and lactate were measured before and after the hanging wire test. For all samples, stastical significance was assessed by performing the unpaired and two-tailed Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey multiple range test using GraphPad Prism version 7.04 for Windows (GraphPad Software, La Jolla California USA). Differences were considered significant when P < 0.05. The corresponding p-values are provided in the figure legends.
Immunofluorescent staining of paraffin-embedded tissues.
The formalin fixed paraffin embedded (FFPE) mice tissue sections were deparaffinized with xylene, rehydrated with decreasing concentrations of ethanol (100, 70 and 50%) and subjected to heat-induced epitope retrieval in 10mM citrate buffer (pH6.0) followed by cell permeabilization with 0.1% Triton X-100 in phosphate buffered saline (PBS) followed by brief PBS washes. Using a PAP (Peroxidase Antiperoxidase, Thermofisher Scientific, Fremont, CA) hydrophobic pen, each section was circled to hold staining solutions in place, then incubated with a series of hour-long, room temperature stains beginning with blocking buffer (5% donkey serum in PBS), the primary antibodies (Table 1), then secondary fluorescent antibody, washing with PBS in between each step. After the final wash, the nuclei were stained with DAPI counterstain. Anti-fade fluorescence mounting media permafluor (Thermofisher Scientific, Fremont, CA) was applied to each section, a coverslip was placed over it and sealed with clear nail polish, left to dry for 2 hours before imaging. The slides were analyzed using an Olympus FluoView FV1000 confocal microscope (Olympus) at 60x magnification.
Table 1.
Antibodies used for immunfluoresence and western blotting studies
| Antigen | Host Species | Source | Dilution | Type |
|---|---|---|---|---|
| ACAD9 | Rabbit | In-house (Cocalico Biologicals, Inc) | 1:1000 | Polyclonal |
| VLCAD | Rabbit | In-house (Cocalico Biologicals, Inc) | 1: 1000 | Polyclonal |
| MCAD | Rabbit | In-house (Cocalico Biologicals, Inc) | 1: 1000 | Polyclonal |
| ECSIT | Rabbit | In-house (Cocalico Biologicals, Inc) | 1: 500 | Polyclonal |
| MTCOI | Mouse | Abcam, Cambridge, MA (Catalog # ab14705,) | 1: 1000 | Monoclonal |
Oroboros high-resolution respirometry.
Mitochondria were isolated from fresh P14 mouse hearts as described and resuspended in Mir05 respiration media (110mM sucrose,0.5mM EGTA, 3mM MgCl2, 60mM K-lactobionate,20mM taurine, 10mM KH2PO4, 20mM HEPES, 1g/L fatty acid free BSA) [18]. Mitochondrial respiration was measured in an Oroboros Oxygraph-2K as described [19]. Substrate additions were 2 mM malate and 5 mM pyruvate, followed by 5 mM ADP to stimulate state 3 respiration. Next, 10 mM glutamate was added to further drive Complex I and 10 mM succinate to stimulate respiration through Complex II. Oxygen consumption rates were normalized to protein content. Total Complex I respiration was defined as the rate of oxygen consumption in the presence of malate, pyruvate, ADP, and glutamate. Total Complex I + II was defined as the maximum rate of respiration achieved in the presence of malate, pyruvate, ADP, glutamate, and succinate, while Complex II was defined as the remaining rate of respiration upon addition of rotenone (0.5 μM) to inhibit Complex I.
Results
Mouse strains.
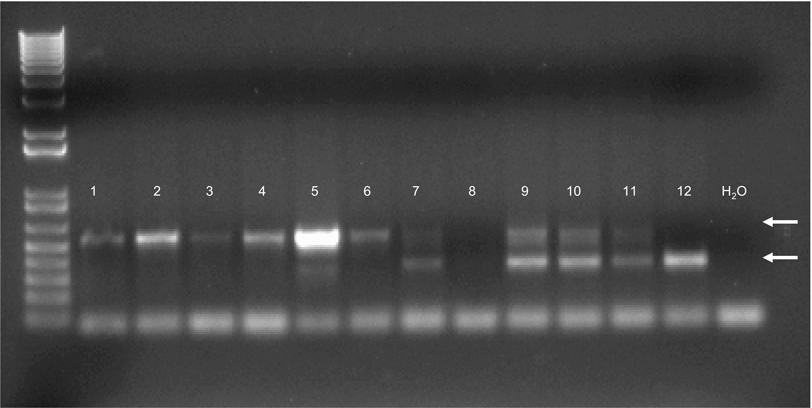
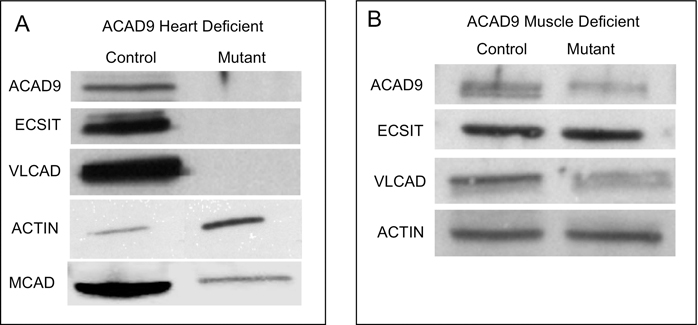
Initial attempts at creating constitutive whole body ACAD9 deficient animals from heterozygote animals carrying a constitutively deleted Acad9 allele were unsuccessful. A total of 55 pups generated from heterozygous mating were genotyped. 40% were wild type and 60% were heterozygous for the mutant allele, indicating that, unlike other ACAD deficiencies, ACAD9 deficiency in mice is embryonic lethal. To circumvent this problem, mice with a floxed Acad9 were generated and bred to Cre-expressing transgenic strains to produce conditional heart and skeletal muscle-specific Acad9 knockout strains. Figure 1 shows a screening gel demonstrating the presence of the floxed Acad9 allele in mice 1–6. Mice with a floxed ACAD9 allele were then bred to a strain expressing the cre recombinase gene driven from tissue specific promotors. Figure 2 shows western blots for heart and skeletal muscle from breeding of the floxed Acad9 mice with mice homozygous for cardiac and skeletal muscle specific promotor driven Cre.
Figure 1. A 2.5% agarose genotyping gel for floxed Acad9 mice.
The floxed gene forms a band that is larger (538 bp; top arrow) than the wildtype allele (349 bp; white arrow). In this example, mice 1 through 6 are homozygous for the floxed allele; 7 and 9–11A are heterozygotes; and mouse 12 (wild type) is homozygous for the wild type allele of Acad9 (bottom arrow). Mouse 8 was inconclusive due to PCR failure. Water was used as a negative control. The 100 bp ladder starts with 100 bp as the lowest band. The left most column is a 1 Kb Plus ladder (Invitrogen, Carlsbad, CA), and the right most is a PCR reaction primed with water without and no DNA sample.
Figure 2. Protein expression of ACAD9, ECSIT and MCAD in cardiac and skeletal muscle tissues of wild type, floxed uninduced and floxed induced mice.
A. Tissue extracts from wild type and cardiac (A) or muscle (B) specific mutant animals were separated on SDS-PAGE gels, transferred to a membrane, then analyzed by western blotting and, following stripping, stained sequentially with antibodies to GAPDH, ACAD9, ECSIT, VLCAD, and actin. Extracts from ACAD9 deficient hearts were also blotted with MCAD antiserum. The images shown were all from a single gel so that the intensity of the bands are a relative measure of protein abundance.
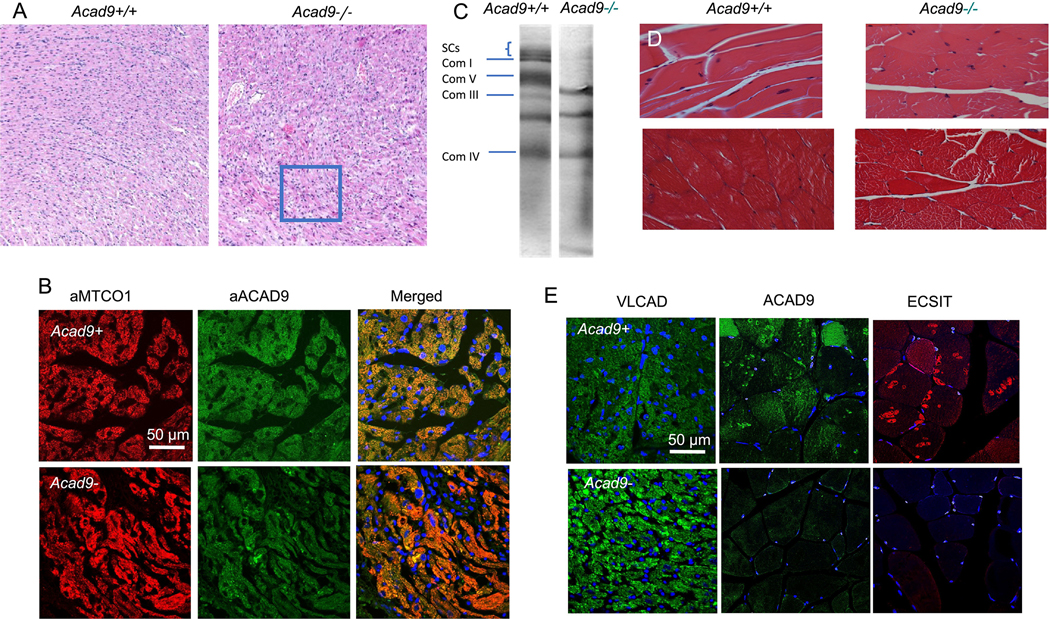
As expected, the cardiac specific Acad9 knockout animals (Acad9−/−) showed lack of ACAD9 protein in heart tissue (Figure 2A), while actin antigen was normal. As expected ECSIT was undetectable, but surprisingly, VLCAD antigen was also not visualized while MCAD was greatly reduced compared to wild type with a strong actin, suggesting a severe global mitochondrial derangement. Next tissue histology was examined in all tissues in the ACAD9 deleted animals. Cardiac cellular morphology was abnormal on hematoxylin and eosin staining (Figure 3A), and ACAD9 protein was reduced or absent as shown by immunofluorescent staining of cardiac tissue (Figure 3B). Note that the ACAD9 staining seen in this figure was likely non-specific as it did not overlap with the MTCO1 signal in the merged image. This conclusion is consistent with the western blotting. The respiratory chain supercomplexes and isolated complex I were absent on blue native gel electrophoresis (Figure 3C), consistent with findings reported in human patients [5]. Muscle specific ACAD9 deficient mice similarly showed reduced ACAD9 antigen (Figure 2B), though less so than in the cardiac mutants, while VLCAD, actin, and ECSIT were normal or just slightly reduced. Surprisingly, the ECSIT band was nearly the same intensity in WT and mutant animals. Overall, these findings suggest a less efficient excision of the floxed ACAD9 allele in muscle and would predict a milder phenotype, an often reported phenomenon with Cre excision strains [20]. On histologic examination, there was some disruption of normal muscle architecture with presence of abnormal myofibril bundles and centralized nuclei, consistent with persistent muscle damage (Figure 3D). Finally, the immunofluorescence signal for ACAD9 and ECSIT in situ was reduced but detectable, consistent with residual retention of functional ACAD9 gene. A robust signal was obtained with VLCAD antiserum, confirming its presence in the ACAD9 deleted muscle (Figure 3E). In total, our findings confirm the efficient deletion of ACAD9 in cardiac muscle, but less so in skeletal muscle, depending on the promotor used to drive the Cre recombinase gene.
Figure 3. Histology studies of cardiac and skeletal muscle tissues from floxed induced heart and skeletal muscle mice.
A. H&E staining of mouse cardiac tissue. Acad9 deficient cardiac tissue (right panel) show disarray of myocardial fibers consistent with cardiomyopathy as compared to the striated and organized orientation of the cardiac cells in a wild type littermate (left panel). This is seen prominently in the swirled patterns and clustered nuclei of the cells (highlighted in the square in the right panel). The photos were taken with a 10x microscope objective. B. Immunofluorescent stains of cardiac tissue-specific Acad9 mouse heart tissue. Cardiac tissue from wild type (top) and mutant (bottom) animals were stained with anti-MTCOI antibody (cytochrome c oxidase subunit 1 of respiratory complex IV; left panel) and visualized with a red fluorescently labeled secondary antibody, and an anti-ACAD9 antibody (center panel) visualized with a green fluorescently labeled secondary antibody. The two images are shown merged (right panel) with yellow representing overlapping signals. Nuclei were stained with DAPI (blue) in the right panel. The image was captures at 60 x magnification. C. Blue-native gel electrophoresis of cardiac tissue mitochondria from cardiac specific Acad9 deficient and wild type animals. Extracts from wild type (left panel) and mutant (right panel) animal hearts were separated by BNGE and stained with Coomasie Blue. The known migration positions of ETC complexes are indicated on the left. Supercomplexes (SC) consist of various combinations of complex I, III, and IV, and are absent in mutant animals, as is isolated complex I. D. Hematoxalyn and eosin (H&E) (top) and trichrome (bottom) stains of skeletal muscle specific Acad9 deficient mouse muscle tissue. Normal muscle morphology is present in wild type animals (left panel). Muscle specific ACAD9 deficient animals show disruption of the normal myofibril bundles and centralized nuclei suggestive of chronic muscle damage. The trichrome staining highlights the more disorganized morphology of the ACAD9 deficient muscle as compared to the more striated pattern of the normal littermate. The photos were taken with a 40x microscope objective. E. Immunofluorescent stain of cardiac specific Acad9 deficient mouse heart tissue. Acad9+/+ animals show normal immunofluorescence of heart when stained with VLCAD, ACAD9, and ECSIT antibodies (top panel). While cardiac specific Acad9−/− animals show little or no signal with ACAD9 and ECSIT antibodies. All panels have nuclei stained blue with DAPI. Images were captures at 60 x magnification.
Clinical and biochemical phenotype of heart ACAD9 deficient animals.
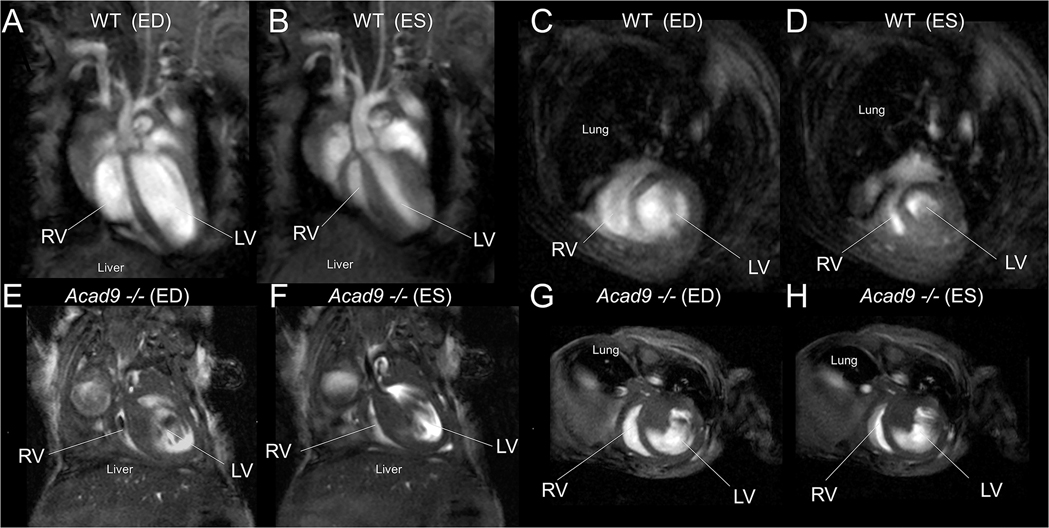
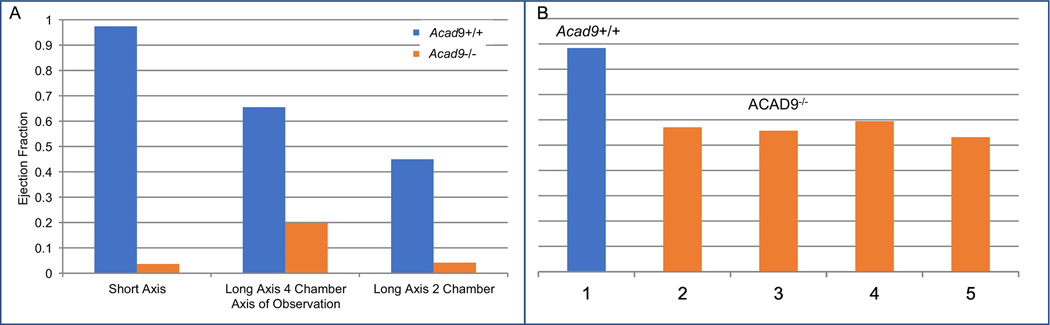
Cardiac MRIs were performed on cardiac specific deficient animals at age p14 as they began to die shortly thereafter (the earliest death was p13). Mutant animals exhibited dramatic cardiomyopathy with noticeable thickening of the atrial and ventricular walls, and severe enlargement of all chambers showed (Figure 4). The ejection fraction of affected hearts (a measure of dynamic blood flow) was also dramatically reduced in the mutant mice by as much as 27-fold (Figure 5A). MRIs of animals at day p3 already showed significant cardiomyopathy and reduced ejection fraction (Figure 5B). These findings suggest that the cardiomyopathy began in utero rather than after birth, though prenatal imaging was not performed.
Figure 4.
Cardiac MRI for an Acad9−/− and a wild-type (WT) littermate control acquired on post-natal day p14. (A-D) WT control, (E-H) Acad9−/−, (A, B, E, F) long-axis 4-chamber view, (C, D, G, H) short-axis view, (A, C, E, G) end-diastole, (B, D, F, H) end-systole. LV: left ventricle, RV: right ventricle, ED: end-diastole, ES: end-systole. ACAD9 deficient hearts show severe dysfunction. Video can be found at https://pitt-my.sharepoint.com/:v:/g/personal/gev1_pitt_edu/EXZb9_UgPydHuApINpZuMeQBA8dwWvotqrcXFfEp3ecZxA?e=GAcbAn.
Figure 5. Ejection fraction (EF) in Acad9 cardiac specific deficient compared to wild type mouse.
A. Comparison of ejection fraction between a single Acad9 deficient mouse and a wild type littermate at day P14. EFs were calculated by determining the difference in surface area of the visible blood in the left ventricle when it is at its largest and smallest (i.e. an image where the left ventricle completely closes would have an EF of 1. Cardiac EFs were calculated using all three orientations. B. EF in day P3 hearts of Acad9−/− and wild type animals. EF measured at day P3 was significantly decreased in mutant animals (2–5) compared to wild type (1) suggesting that the pathologic process started in utero. EF was measured from the short axis.
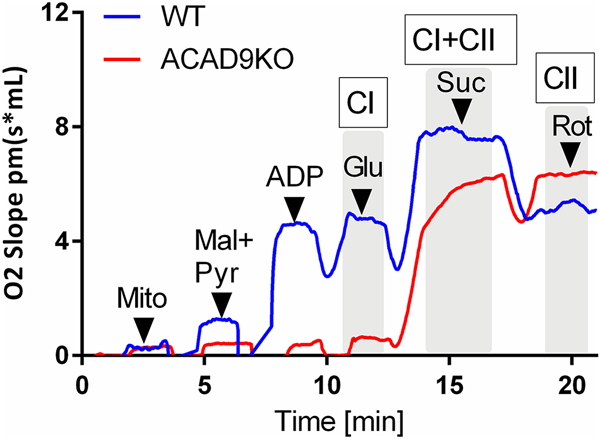
Since the primary function of ACAD9 in most tissues including heart is as a respiratory chain complex I assembly factor, we examined mitochondria from cardiac-specific ACAD9 deficient animals for respiratory chain driven oxygen consumption. These mitochondria did not respond to substrates that drive complex I (malate, pyruvate, and glutamate), but have a robust response to the complex II substrate succinate (Figure 6). Additional proof of an isolated complex I deficiency comes from the injection of rotenone, which leads to a loss of respiration upon addition in wild type mitochondria, but no response in ACAD9 deficient mitochondria.
Figure 6. Oroboros high-resolution respirometry of isolated heart mitochondria from wild type and heart specific Acad9−/− mice.
Isolated mitochondria were added to the assay media and oxygen consumption was measured after the addition of malate and pyruvate (mal+pyr), adenosine diphosphate (ADP), glutamate (Glu), succinate (Suc), and rotenone (Rot). Robust oxygen consumption due to complex I (CI) and complex II (CII) was evident in wild type mitochondria, while only complex II activity was present in ACAD9−/− mitochondria.
Clinical phenotype of skeletal muscle ACAD9 deficient animals.
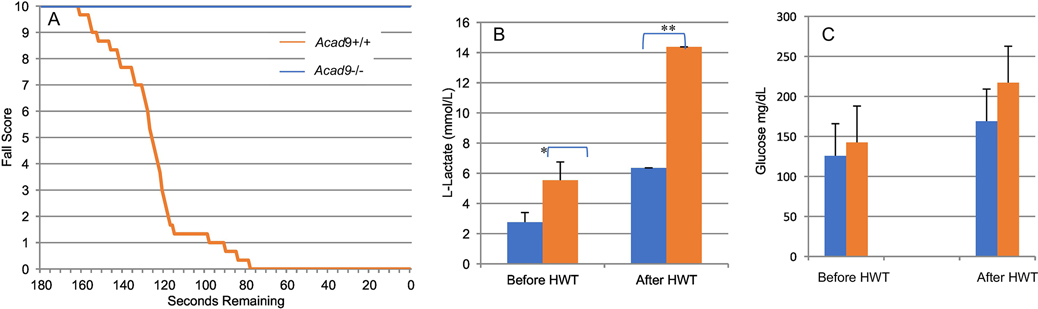
Skeletal muscle deficient ACAD9 deficiency animals were viable and showed no obvious clinical phenotype. At 2–6 months of age mutant mice and their normal littermates were subjected to a hanging wire test and the time to fall was recorded to create a Kaplan-Meier-like survival curve (Figure 7A). Unaffected animals were easily able to hang on the apparatus for 3 min. However, no mutant animals successfully completed the test with half the animals falling just after 60 secs and the longest animal hanging for just over 100 sec. Additionally, wildtype animals returned to normal activity levels immediately after the test, while the ACAD9 deficient animals were inactive for 15–30 minutes after they fell. Together, these findings indicate impaired exercise tolerance. Since lactic acidosis and hypoglycemia are reported in human patients with ACAD9 deficiency, these parameters were measured at the start of the hanging wire test and at the time of fall. Mutant animals had resting L-lactic acid levels of 6.4 mmol/L, greater than double wild type animals prior to the start of the test 2.8 mmol/lL, and increased to 14.4 mmol/L, again over double the wild type animals’ level of 5.5 mmol/L(Figure 7B). Glucose levels were similar in both animals before and after the test (Figure 7C).
Figure 7. Functional testing of skeletal muscle Acad9−/− mice.
A. Kaplan-Meier-like curve measuring fall score of Acad9 deficient skeletal muscle-specific mice. Average of fall scores over a period of 180 seconds. While all normal mice were able to last the full three minutes, All mutants had received a final fall score of zero by 78 seconds. B. Blood L-lactate levels in skeletal-muscle specific Acad9 deficient mice measured before and after a hanging wire test. Muscle specific ACAD9 deficient mice have an increased resting L-Lactate levels compared to wild type littermates (blue), which increases more than wild type following exercise (orange; the hanging wire test). *p-value = 0.002, **p-value = 0.004. C. Blood glucose Levels in skeletal-muscle specific Acad9 deficient mice measured before and after a hanging wire test. Muscle specific Acad9 deficient mice do not have a significant difference of resting glucose levels or after exercise (orange) compared to wild type littermates (blue).
Discussion
ACAD9 Deficiency is recognized as one of the most common causes of ETC deficiency and can present in infancy with severe systemic lactic acidosis, cardiomyopathy, and early death [4, 5, 10, 14]. However, later presentations are common, predominantly with cardiomyopathy and milder symptoms including exercise intolerance that are sometimes responsive to riboflavin supplementation. Our first attempt to create a constitutive whole body deficient mouse model failed to yield any affected animals, suggesting embryonic lethality. To circumvent this problem, we constructed several tissue models using Cre-lox technology, which allows deletion in the earliest stages of embryogenesis without subsequent lethality. Heart-specific ACAD9 deficient mice showed a severe phenotype consistent with human cardiac pathology, developing severe cardiomyopathy present at the earliest time point examined (p3), suggesting the development of cardiac dysfunction in utero, with ultimate death before three weeks of age. Cardiac tissue from deficient animals demonstrated significant and broad molecular and biochemical changes, with complete loss of mitochondrial ETC supercomplexes, as well as other unrelated mitochondrial proteins, likely reflecting “end stage” mitochondria near the end of life when tissue was harvested for study. While the severity of the cardiomyopathy limited the physiologic testing that could be performed, this model will be valuable for testing therapeutic agents, though in utero treatment may be necessary given early onset symptoms.
To study the myopathy associated with ACAD9 deficiency, a skeletal muscle specific ACAD9 deficient mouse was created. Unlike the cardiac tissue-specific deficient mice, the skeletal muscle deficient animals were viable and survived to adulthood, allowing direct study of the effect of the ACAD9 deficiency on muscle tissue. Notably, although the mice appeared grossly normal, they demonstrated systemic lactic acidosis and exercise intolerance, mimicking late-onset human phenotype, making them excellent candidates for therapeutic testing. The viability of these animals suggests that therapies should target cardiac symptoms in the most severely affected patients in order to allow survival to an older age.
Ultimately, these models provide novel tools to study the interaction of ACAD9 protein with the complex I assembly machinery, the dynamics of ETC supercomplex formation and recycling following loss of ACAD9 function, and therapeutic options to abrogate the effects of ACAD9 deficiency. While the proteins involved with complex I assembly have been identified, the dynamics of their interactions have not. Inducible ACAD9 deficiency in animals or cells would allow characterization of prescribed loss of this protein, while expression experiments adding it back will demonstrate the timing of interaction of the complex I assembly proteins. Similarly, prescribed loss and expression of ACAD9 would provide the opportunity to examine sequential assembly of ETC super complexes, not possible in other current systems. Tissue specific or whole-body inducible models of ACAD9 deficiency provide the rigorous pre-clinical testing of novel therapeutic agents that is necessary to move them to clinical trials in humans.
The role of ACAD9 in fatty acid oxidation was not directly addressed in these studies, but the lack of significant hypoglycemia and no evidence of abnormal acylcarnitines (data not shown) in blood as is characteristic in other mouse models of fatty acid oxidation disorders suggests that its role is at least limited in the heart and muscle. Additional studies on this topic will likely require use of the conditional knock out model to allow for evaluation of fatty acid oxidation in all tissues.
Acknowledgements.
JV was funded in part by NIH grant R01 DK78755. LGG is funded in part by the National Human Genome Research Institute (NHGRI) grant #1K08 HG010490. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the NIH. Special thanks to Dr. Jung-Ja Kim, Medical College of Wisconsin for providing purified ECSIT for antibody production.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Zhang J, Zhang W, Zou D, Chen G, Wan T, Zhang M, Cao X, Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family Biochemical and Biophysical Research Communications 297 (2002) 1033–1042. [DOI] [PubMed] [Google Scholar]
- [2].Ensenauer R, He M, Willard JM, Goetzman ES, Corydon TJ, Vandahl BB, Mohsen AW, Isaya G, Vockley J, Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty acids Journal of Biological Chemistry 280 (2005) 32309–32316. [DOI] [PubMed] [Google Scholar]
- [3].He M, Rutledge SL, Kelly DR, Palmer CA, Murdoch G, Majumder N, Nicholls RD, Pei Z, Watkins PA, Vockley J, A new genetic disorder in mitochondrial fatty acid beta-oxidation: ACAD9 deficiency American Journal of Human Genetics 81 (2007) 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schiff M, Haberberger B, Xia C, Mohsen AW, Goetzman ES, Wang Y, Uppala R, Zhang Y, Karunanidhi A, Prabhu D, Alharbi H, Prochownik EV, Haack T, Haberle J, Munnich A, Rotig A, Taylor RW, Nicholls RD, Kim JJ, Prokisch H, Vockley J, Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency Human Molecular Genetics 24 (2015) 3238–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Repp BM, Mastantuono E, Alston CL, Schiff M, Haack TB, Rotig A, Ardissone A, Lombes A, Catarino CB, Diodato D, Schottmann G, Poulton J, Burlina A, Jonckheere A, Munnich A, Rolinski B, Ghezzi D, Rokicki D, Wellesley D, Martinelli D, Wenhong D, Lamantea E, Ostergaard E, Pronicka E, Pierre G, Smeets HJM, Wittig I, Scurr I, de Coo IFM, Moroni I, Smet J, Mayr JA, Dai L, de Meirleir L, Schuelke M, Zeviani M, Morscher RJ, McFarland R, Seneca S, Klopstock T, Meitinger T, Wieland T, Strom TM, Herberg U, Ahting U, Sperl W, Nassogne MC, Ling H, Fang F, Freisinger P, Van Coster R, Strecker V, Taylor RW, Haberle J, Vockley J, Prokisch H, Wortmann S, Clinical, biochemical and genetic spectrum of 70 patients with ACAD9 deficiency: is riboflavin supplementation effective? Orphanet J Rare Dis 13 (2018) 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nouws J, Te Brinke H, Nijtmans LG, Houten SM, ACAD9, a complex I assembly factor with a moonlighting function in fatty acid oxidation deficiencies Human Molecular Genetics 23 (2014) 1311–1319. [DOI] [PubMed] [Google Scholar]
- [7].Nouws J, Nijtmans L, Houten SM, van den Brand M, Huynen M, Venselaar H, Hoefs S, Gloerich J, Kronick J, Hutchin T, Willems P, Rodenburg R, Wanders R, van den Heuvel L, Smeitink J, Vogel RO, Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I Cell Metab 12 (2010) 283–294. [DOI] [PubMed] [Google Scholar]
- [8].Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA, Mitochondrial complex I: structure, function and pathology J Inherit Metab Dis 29 (2006) 499–515. [DOI] [PubMed] [Google Scholar]
- [9].Smith PM, Fox JL, Winge DR, Biogenesis of the cytochrome bc(1) complex and role of assembly factors Biochimica et Biophysica Acta 1817 (2012) 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D, Uziel G, Lamantea E, Invernizzi F, Poulton J, Rolinski B, Iuso A, Biskup S, Schmidt T, Mewes HW, Wittig I, Meitinger T, Zeviani M, Prokisch H, Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency Nature Genetics 42 (2010) 1131–1134. [DOI] [PubMed] [Google Scholar]
- [11].Gerards M, van den Bosch BJ, Danhauser K, Serre V, van Weeghel M, Wanders RJ, Nicolaes GA, Sluiter W, Schoonderwoerd K, Scholte HR, Prokisch H, Rotig A, de Coo IF, Smeets HJ, Riboflavin-responsive oxidative phosphorylation complex I deficiency caused by defective ACAD9: new function for an old gene Brain 134 (2011) 210–219. [DOI] [PubMed] [Google Scholar]
- [12].Haack TB, Haberberger B, Frisch EM, Wieland T, Iuso A, Gorza M, Strecker V, Graf E, Mayr JA, Herberg U, Hennermann JB, Klopstock T, Kuhn KA, Ahting U, Sperl W, Wilichowski E, Hoffmann GF, Tesarova M, Hansikova H, Zeman J, Plecko B, Zeviani M, Wittig I, Strom TM, Schuelke M, Freisinger P, Meitinger T, Prokisch H, Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing Journal of Medical Genetics 49 (2012) 277–283. [DOI] [PubMed] [Google Scholar]
- [13].Leslie N, Wang X, Peng Y, Valencia CA, Khuchua Z, Hata J, Witte D, Huang T, Bove KE, Neonatal multiorgan failure due to ACAD9 mutation and complex I deficiency with mitochondrial hyperplasia in liver, cardiac myocytes, skeletal muscle, and renal tubules Hum Pathol 49 (2016) 27–32. [DOI] [PubMed] [Google Scholar]
- [14].Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, Karkucinska-Wieckowska A, Pajdowska M, Jurkiewicz E, Halat P, Kosinska J, Pollak A, Rydzanicz M, Stawinski P, Pronicki M, Krajewska-Walasek M, Ploski R, New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre J Transl Med 14 (2016) 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A, A conditional knockout resource for the genome-wide study of mouse gene function Nature 474 (2011) 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nahrendorf M, Streif J, Hiller K, Hu K, Nordbeck P, Ritter O, Sosnovik D, Bauer L, Neubauer S, Jakob P, Ertl G, Spindler M, Bauer W, Multimodal functional cardiac MRI in creatine kinase-deficient mice reveals subtle abnormalities in myocardial perfusion and mechanics Am J Physiol Heart Circ Physiol 2006 (2006) H2516–2521. [DOI] [PubMed] [Google Scholar]
- [17].Vinnakota K, Bassingthwaighte J, Myocardial density and composition: a basis for calculating intracellular metabolite concentrations Am J Physiol Heart Circ Physiol 286 (2004) H1742–1749. [DOI] [PubMed] [Google Scholar]
- [18].Krumschnabel G, Fontana-Ayoub M, Sumbalova Z, Heidler J, Gauper K, Fasching M, Gnaiger E, Simultaneous high-resolution measurement of mitochondrial respiration and hydrogen peroxide production Methods Mol Biol 1264 (2015) 245–261. [DOI] [PubMed] [Google Scholar]
- [19].Zhang YBSSBMEGES, The fatty acid oxidation enzyme long-chain acyl-CoA dehydrogenase can be a source of mitochondrial hydrogen peroxide Redox Biology 26 (2019) 101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Murray SA, Eppig JT, Smedley D, Simpson EM, Rosenthal N, Beyond knockouts: cre resources for conditional mutagenesis Mamm Genome 23 (2012) 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]