Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is responsible for the current pandemic coronavirus disease of 2019 (COVID-19). Like other pathogens, SARS-CoV-2 infection can elicit production of the type I and III interferon (IFN) cytokines by the innate immune response. A rapid and robust type I and III IFN response can curb viral replication and improve clinical outcomes of SARS-CoV-2 infection. To effectively replicate in the host, SARS-CoV-2 has evolved mechanisms for evasion of this innate immune response, which could also modulate COVID-19 pathogenesis. In this review, we discuss studies that have reported the identification and characterization of SARS-CoV-2 proteins that inhibit type I IFNs. We focus especially on the mechanisms of nsp1 and ORF6, which are the two most potent and best studied SARS-CoV-2 type I IFN inhibitors. We also discuss naturally occurring mutations in these SARS-CoV-2 IFN antagonists and the impact of these mutations in vitro and on clinical presentation. As SARS-CoV-2 continues to spread and evolve, researchers will have the opportunity to study natural mutations in IFN antagonists and assess their role in disease. Additional studies that look more closely at previously identified antagonists and newly arising mutants may inform future therapeutic interventions for COVID-19.
Keywords: COVID-19, coronavirus disease of 2019; SARS-CoV-2, SARS coronavirus 2; IFN, interferon; IFNAR, interferon alpha/beta receptor; IFNLR, interferon lambda receptor; ISRE, interferon stimulated response element; MAVS, mitochondrial antiviral-signaling protein; MDA-5, melanoma differentiation-associated protein 5; RIG-I, retinoic acid-inducible gene I; TLR, toll-like receptor; nsp, non-structural protein; ORF, open reading frame; PLpro, papain-like protease; UTR, untranslated region; IRF, interferon response factor; STAT, signal transducer and regulator of transcription; 3CLpro, 3-chymotrypsin like protease; SRP, signal recognition particle; TBK1, TANK-binding kinase 1; TAB1, TGF-beta activated kinase 1 binding protein 1; SeV, Sendai virus; TAK1, TGF-beta activated kinase 1; eIF, eukaryotic initiation factor; TRIF, TIR domain-containing adapter-inducing interferon beta
Keywords: SARS-CoV-2, Type I interferon, Immune evasion, nsp1, ORF6
Graphical abstract
1. Introduction
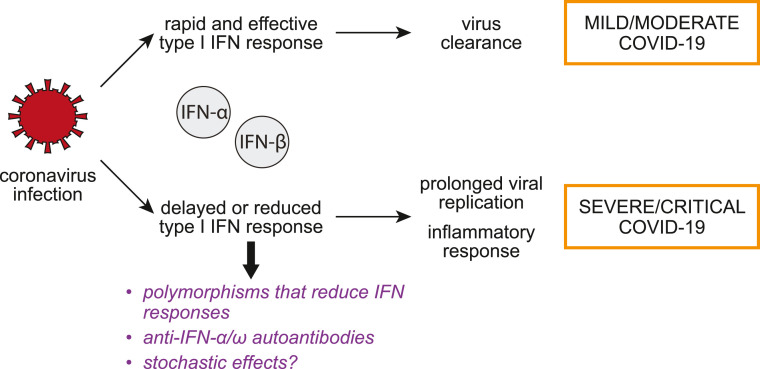
As of November 15th, 2021, SARS coronavirus 2 (SARS-CoV-2) has infected at least 253 million people and killed more than 5 million worldwide. The scientific community is making a continuous effort to understand this virus and to better combat the disease it causes, COVID-19. Like all successful pathogens, SARS-CoV-2 needs to evade host responses to replicate. Indeed, SARS-CoV-2 suppresses the ability of the infected host to turn on the expression and secretion of the type I and possibly type III interferons (IFNs) (Blanco-Melo et al., 2020). Type I and III IFNs are the first line of defense against viral infections and are crucial early anti-viral factors. SARS-CoV-2 may also reduce cellular responses to IFNs, which normally drive the expression of anti-viral interferon stimulated genes (ISGs) (Rebendenne et al., 2021), although not all studies agree on this point (Lokugamage et al., 2020; Shemesh et al., 2021). Here we will review efforts to identify and characterize proteins from SARS-CoV-2 that can inhibit IFN induction and signaling, as well as evidence that this early response has a crucial role in COVID-19 infection (Fig. 1 ).
Fig. 1.
Early response to SARS-CoV-2 infection can influence the course of COVID-19. The diagram represents the model that the efficiency of the early type I IFN response to infection may influence the outcome of disease.
2. Type I and III IFNs have a key role in the protective response to SARS-CoV-2 and in COVID-19 severity
Evidence that the type I IFNs are crucial for COVID-19 pathogenesis comes from studies in COVID-19 patients that had disease of varying severity. Patients with mild and moderate COVID-19 have higher IFN-α levels in the blood compared with patients with more severe disease and their type I IFN response is more sustained (Hadjadj et al., 2020). This reduction may be due to a failure of innate immune cells to mount a type I IFN response to infection, as evidenced by the reduced frequency and activity of plasmacytoid dendritic cells, the main producers of IFN-α during viral infection (Arunachalam et al., 2020). There may also be a transient IFN response in the lungs that is stronger in patients with moderate disease compared to those with severe COVID-19 (Arunachalam et al., 2020). These results suggest that the failure to mount a strong type I IFN response contributes to severe COVID-19, presumably because an effective type I IFN response is needed to rapidly clear SARS-CoV-2 (Fig. 1).
Genetic studies of patients with severe disease also support the idea that an efficient type I IFN response is crucial for COVID-19 recovery (Fig. 1). Zhang et al. found that at least 3.5% of patients with life-threatening COVID-19 had polymorphisms in one of eight genes involved in type I IFN responses and previously associated with severe influenza or other viral infections, including the pathogen sensor TLR3 and the type I IFN receptor subunits IFNAR1 and IFNAR2 (Zhang et al., 2020). At least some of the polymorphisms analyzed were loss-of-function or hypomorphic mutations, although they had had no noticeable effect on the patients’ health previously (Zhang et al., 2020). This may be because SARS-CoV-2 is more virulent than other more common infections that these patients had previously experienced. Other studies found mutations in the pathogen sensor TLR7 in otherwise healthy men with severe COVID-19 (Asano et al., 2021; Solanich et al., 2021; van der Made et al., 2020). Similarly, some (Pairo-Castineira et al., 2021) but not all (Shelton et al., 2021; The Severe Covid-19, 2020) genome-wide association studies found significant association for severe disease with the IFNAR2 receptor locus, as well as some other genes involved in IFN signaling. The IFNAR2 mutations were linked to lower expression of this protein, which in turn was associated with more severe COVID-19 (Pairo-Castineira et al., 2021). Lastly, ∼5–20% of severe and critical COVID-19 patients have autoantibodies against type I IFNs in their blood, which likely prevented an appropriate IFN response in these patients (Bastard et al., 2020; Wijst et al., 2021) (Fig. 1).
Type III IFNs (IFN-λs) are also considered important in host defense against SARS-CoV-2. Because the type III IFN receptor IFNLR1 is preferentially expressed in epithelial cells, type III IFN responses are more localized and induce inflammatory cytokines less robustly than type I IFNs (Park and Iwasaki, 2020). IFN-λ1 and IFN-λ3 in the upper airways induce production of protective ISGs in mild COVID-19 cases (Sposito et al., 2021). Type III IFN response may be particularly important in the upper airways. Patients with high type III IFN and low type I IFN in the upper airways tend to develop mild disease, while patients with a type I IFN-dominated response in the upper airways are 10 times more likely to have a severe illness resulting in hospitalization or admission to the intensive care unit (Sposito et al., 2021).
All of these studies reinforce the notion that a faulty type I/III IFN response can predispose individuals to severe and life-threatening COVID-19. Ultimately, these responses start in the infected cells in the lungs that sense SARS-CoV-2 infection, and are then amplified by cells of the immune system, some of which may also be infected by SARS-CoV-2 (Yang et al., 2020). As SARS-CoV-2 viral replication can be suppressed by IFN treatment, the efficient inhibition of type I and III IFN response is necessary for successful SARS-CoV-2 infection (Lokugamage et al., 2020; Mantlo et al., 2020). Moreover, the ability of SARS-CoV-2 to block these responses in the infected cells may influence the course of systemic IFN response and contribute to disease progression.
3. MDA-5 is the main sensor for SARS-CoV-2
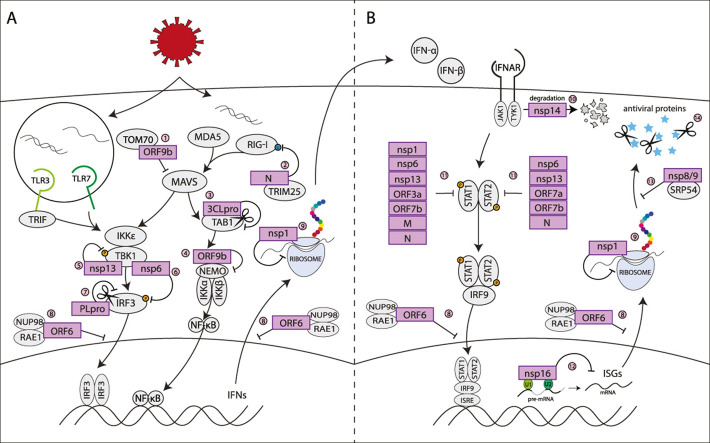
Cells are equipped with pathogen sensors that can alert them of infection and trigger a signaling pathway that culminates in type I (and in some cells III) IFN secretion (Fig. 2 , left). Type I IFN signaling can then recruit innate immune cells to the site of infection and drive expression of ISGs that can directly limit viral infection (Fig. 2, right). In particular, several pathogen sensors are designed to detect viral RNAs, including RIG-I and MDA-5 in the cytoplasm and TLR3 and TLR7 in endosomes. These sensors can distinguish viral nucleic acid from cellular ones because of location, in the case of TLR3/7, or structural elements that are not usually found in cellular RNAs (double-stranded regions, a 5’ triphosphate etc.). All these receptors converge on the same downstream pathway, which drives transcription of type I IFNs (Fig. 2, left). Several studies using lung adenocarcinoma Calu-3 cells, CRISPR knockouts and siRNA knockdowns have concluded that MDA-5 is the main pathogen receptor that senses SARS-CoV-2 (Rebendenne et al., 2021; Sampaio et al., 2021; Thorne et al., 2021), although one study also reported that RIG-I had an effect (Thorne et al., 2021). However, multiple studies noted that IFN induction occurs later than expected and cannot effectively limit SARS-CoV-2 replication (Rebendenne et al., 2021; Thorne et al., 2021). Interestingly, Yamada et al. reported that RIG-I may have a separate role in defenses against SARS-CoV-2, in an IFN-independent manner (Yamada et al., 2021). RIG-I binds the viral RNA and sequesters it, preventing viral replication (Yamada et al., 2021). This activity may serve as a first layer of defenses, with IFN induction via MDA-5 as a backup. These studies confirm that the canonical RNA sensing pathway can detect SARS-CoV-2, but also suggest the virus can impede this response early in infection to reduce clearance.
Fig. 2.
Viral proteins disrupt the interferon response at many different points
A) SARS-CoV-2 proteins antagonize interferon induction 1. ORF9b inhibits IFN induction via MDA5 and RIG-I by binding with TOM70 and inhibiting MAVS activation (Gao et al., 2021; Jiang et al., 2020). 2. The N protein binds to TRIM25, inhibiting RIG-I activation (Gori Savellini et al., 2021). 3. Nsp5 viral protease 3CLpro cleaves TAB1, inhibiting the activation of NFkB (Moustaqil et al., 2021) 4. ORF9b binds to NEMO and blocks NFkB signaling (Wu et al., 2021) 5. Nsp13 binds TBK1 to prevent its phosphorylation (Vazquez et al., 2021; Xia et al., 2020). 6. Nsp6 binds to TBK1 preventing the phosphorylation of transcription factor IRF3 (Xia et al., 2020). 7. Nsp3 viral protease PLpro cleaves IRF3 prior to phosphorylation (Moustaqil et al., 2021). 8. ORF6 binds to the Nup98-Rae1 complex in the nuclear pore complex and prevents nuclear translocation of transcription factors and nuclear export of mRNAs and mRNA transporters (Kimura et al., 2021; Addetia et al., 2021; Kato et al., 2021; Miorin et al., 2020) 9. SARS-CoV-2 nsp1 prevents host translation by binding to the 40s ribosomal unit and blocking the mRNA entry channel (Banerjee et al., 2020; Schubert et al., 2020; Thoms et al., 2020; Yuan et al., 2020). B) SARS-CoV-2 proteins inhibit interferon response and ISG production 10. Nsp14 marks the interferon receptor IFNAR1 for lysosomal degradation (Hayn et al., 2021). 11. STAT1 and/or STAT2 phosphorylation after type I IFN stimulation is inhibited by nsp1, nsp6, nsp13, ORF3a, ORF7b, M, and N (Xia et al., 2020). 12. Nsp16 binds to U1 and U2 small nuclear RNAs and blocks splicing of pre-mRNA (Banerjee et al., 2020) 13. Nsp8 and 9 bind 7SL in the signal recognition particle (SRP) complex, disrupting protein trafficking and resulting in the degradation of newly translated proteins (Banerjee et al., 2020) 14. Viral proteases have been found to cleave interferon-stimulated antiviral proteins after they are formed. PLpro cleaves ISG15 (Shin et al., 2020), and 3CLpro cleaves RNF20 (Zhang et al., 2021).
4. Multiple SARS-CoV-2 proteins contribute to evasion of type I IFN responses
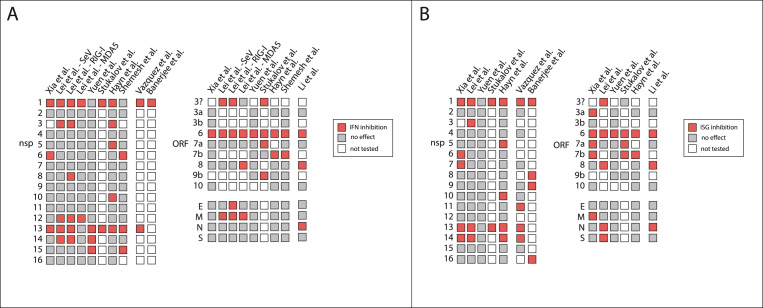
Several studies have sought to identify SARS-CoV-2 encoded proteins that directly contribute to inhibition of type I IFN responses (Fig. 3 ). Multiple studies have used luciferase reporters driven by the IFN-β promoter and/or the IFN-stimulated response element (ISRE) to screen SARS-CoV-2 viral proteins for inhibition of the IFN response. Most groups used HEK293T cells for their luciferase reporter assays, although Yuen et al. used the fast growing HEK293FTs, and Vazquez et al. used HeLa cells (Shemesh et al., 2021; Hayn et al., 2021; Lei et al., 2020; Li et al., 2020; Vazquez et al., 2021; Xia et al., 2020; Yuen et al., 2020). While none of these are lung cells, they are all commonly used to study the type I IFN response in response to RNA stimuli. Depending on the study, IFN induction was triggered by Sendai Virus (SeV) infection, or overexpression of the RNA sensors MDA5 and RIG-I or their adaptor MAVS (Shemesh et al., 2021; Hayn et al., 2021; Lei et al., 2020; Li et al., 2020; Vazquez et al., 2021; Xia et al., 2020; Yuen et al., 2020). ISRE-driven luciferase expression was triggered by recombinant IFN-α treatment (Hayn et al., 2021; Lei et al., 2020; Li et al., 2020; Vazquez et al., 2021; Xia et al., 2020; Yuen et al., 2020). In addition to these studies, Stukalov et al. identified potential IFN antagonists based on the analysis of SARS-CoV-2/host protein interactions through proteomics and statistical modelling in A549 cells (Stukalov et al., 2021). Candidates were then validated using luciferase assays for type I IFN induction and response in HEK293-R1 cells (Stukalov et al., 2021). Also, Banerjee et al. used an RNA crosslinking approach in HEK293T cells to detect RNAs bound by SARS-CoV-2 proteins, and then tested the IFN antagonism of proteins that bind to cellular machinery essential for protein production (Banerjee et al., 2020). As shown in Fig. 3, different proteins were identified as IFN inhibitors in the different studies, as well as within the same study using different stimuli (Lei et al., 2020). Nonetheless, several proteins were identified by multiple studies, suggesting these proteins are most likely bona fide and strong type I IFN antagonists: nsp1, nsp13, and ORF6 as inhibitors of IFN induction, and nsp1, nsp13, nsp14, ORF6, and ORF7b as inhibitors of ISG induction (Shemesh et al., 2021; Hayn et al., 2021; Lei et al., 2020; Li et al., 2020; Vazquez et al., 2021; Xia et al., 2020; Yuen et al., 2020; Stukalov et al., 2021; Banerjee et al., 2020). Also, while most of the screening studies did not test the protease nsp3/PLpro, individual studies revealed that PLpro cleaves the IFN transcription factor IRF3 and interferon-stimulated protein ISG15 (Fig. 2) (Shin et al., 2020; Moustaqil et al., 2021). Most of the screening studies did not find an effect for the protease nsp5/3CLpro on IFN induction, but 3CLpro does inhibit the pro-inflammatory NFkB pathway by cleaving TAB1, a regulator of the TAK1 kinase (Fig. 2) (Moustaqil et al., 2021).
Fig. 3.
Multiple SARS-CoV-2 proteins inhibit type I IFN induction and response. Charts of studies that have tested the role of SARS-CoV-2 proteins on type I IFN (A) or interferon-stimulated gene (ISG) induction (B). Proteins that showed an effect in each study are in red, proteins that did not have an effect in grey and proteins that were not tested in white. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Follow-up studies reveal that SARS-CoV-2 proteins antagonize the type I IFN response at many different points to evade host detection and antiviral response (Fig. 2), although thorough follow up studies are limited for most proteins. We will discuss results for nsp1 and ORF6 in more detail in subsequent sections, as these proteins have been studied more in-depth. They may also be more potent IFN antagonists, as they have been identified in most of the screening studies (Fig. 3). Strikingly, collectively these studies indicate that the majority of SARS-CoV-2 proteins may play some role in the disruption of type I IFN signaling (Fig. 2, Fig. 3). Given that SARS-CoV-2 viral replication can be suppressed by IFN-β treatment (Lokugamage et al., 2020; Mantlo et al., 2020), this redundancy in IFN antagonism may be important to viral replication and survival. While none of the studies specifically looked at type III IFN, the same or similar signaling pathways are thought to induce type III IFNs as well. Therefore, these SARS-CoV-2 proteins are generally expected to also impact type III IFNs.
To note, all of the screening studies (Fig. 3) utilized luciferase reporter assays in some capacity. Luciferase assays are a helpful tool, but it is important to acknowledge the limitations of this method. For example, nsp12 is a potent inhibitor of the IFN-β promoter luciferase reporter, but does not have an effect on endogenous IFN-β transcription (Li et al., 2021). Given this discrepancy and the varying results from the screens, additional studies are needed to determine whether particular viral proteins truly inhibit the IFN response during infection in relevant cell types.
4.1. Nsp1
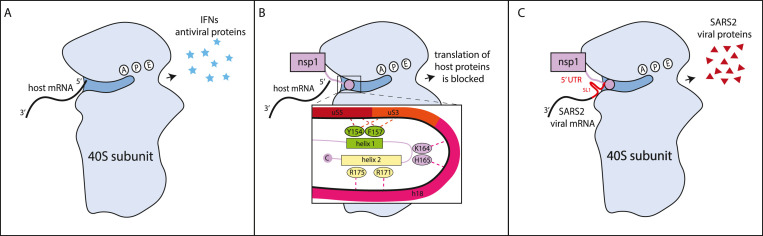
One of the best studied IFN antagonists is non-structural protein 1 (nsp1). Nsp1 is the most N-terminal peptide released from the SARS-CoV-2 polyprotein. Interest in this protein stems from the fact that its homologs in SARS-CoV and murine hepatitis virus (MHV) are virulence factors (Nakagawa and Makino, 2021). Indeed, mutations in nsp1 have been proposed as a way to make attenuated coronavirus strains for vaccine development (Züst et al., 2007). As in other coronaviruses, SARS-CoV-2 nsp1 is a host shutoff protein that controls anti-viral responses by globally reducing host gene expression (Nakagawa and Makino, 2021). In multiple studies, nsp1 expression reduces translation, measured through metabolic labeling methods in cells, reporter constructs in cells, or in vitro translation assays (Vazquez et al., 2021; Banerjee et al., 2020; Rao et al., 2021; Schubert et al., 2020; Shen et al., 2021; Thoms et al., 2020; Yuan et al., 2020). This inhibition also results in reduced type I IFN induction and/or responses (Vazquez et al., 2021; Banerjee et al., 2020; Rao et al., 2021; Schubert et al., 2020; Thoms et al., 2020; Yuan et al., 2020). Interactions between nsp1 and the 40S subunit of the ribosome underlie the translational inhibition, as shown by cryogenic electron microscopy structures of the nsp1-40S complex and RNA crosslinking data analyzing nsp1-RNA interactions (Banerjee et al., 2020; Schubert et al., 2020; Thoms et al., 2020; Yuan et al., 2020). These studies have uncovered specific interactions between residues in the C terminus of nsp1 and proteins and RNAs of the 40S subunit that are required for activity (Fig. 4 ). Nsp1 likely binds 40S in the context of the 43S pre-initiation complex, which includes translation initiation factors and the initiator transfer RNA, but can also bind the full 80S ribosome (Schubert et al., 2020; Thoms et al., 2020; Yuan et al., 2020). However, since many of the structural studies have reconstituted complexes in vitro, mixing lysates or ribosomal preparations with recombinant nsp1, not all these complexes may form in cells. In addition to direct blocking of the mRNA channel, nsp1 binding may also competitively prevent binding of the initiation factor eIF3j, and thus the eIF3 complex, blocking further steps in translation initiation (Yuan et al., 2020; Chen and Chen, 2021; Lapointe et al., 2021).
Fig. 4.
Nsp1 binds to ribosomal subunit 40s and blocks the mRNA entry channel. Structural and protein-RNA interactions study show that two helices at the C terminus of nsp1 bind to the mRNA channel on the 40S subunit blocking mRNA access to it and inhibiting translation (Banerjee et al., 2020; Schubert et al., 2020; Thoms et al., 2020; Yuan et al., 2020). The C-terminal nsp1 helices and the hairpin between them make contact with the uS3, uS5 and uS30 ribosomal proteins, and with the helix h18 of the 18S rRNA (Banerjee et al., 2020; Schubert et al., 2020; Thoms et al., 2020; Yuan et al., 2020). The residues highlighted in the figure mediate key interactions and are required for translation inhibition by nsp1 (Vazquez et al., 2021; Banerjee et al., 2020; Schubert et al., 2020; Shen et al., 2021; Thoms et al., 2020; Yuan et al., 2020). As shown in the panel on the right, the 5′ UTR sequence of SARS-CoV-2 can restore translation of transcripts. This is mediated by the SL1 stem loop. SL1 needs to be located at the very 5′ end of the transcript to promote translation, suggesting that it does not simply function to recruit ribosomes, like an IRES (Banerjee et al., 2020).
In SARS-CoV, nsp1 also drives degradation of host RNAs (Nakagawa and Makino, 2021). While SARS-CoV-2 infection does reduce host mRNA levels (Yuan et al., 2020; Burke et al., 2021; Finkel et al., 2021), only a subset of studies report that nsp1 expression reduces mRNA levels (Yuan et al., 2020; Burke et al., 2021; Finkel et al., 2021; Mendez et al., 2021). Others do not see a difference (Shen et al., 2021; Thoms et al., 2020). To note, the studies that do not report an effect used specific reporter constructs, whereas some of the others used more unbiased approaches. Therefore, it is possible that SARS-CoV-2 nsp1 does trigger RNA degradation but does not affect all transcripts equally.
Viral mRNA are protected from the activity of nsp1, similar to what was described for SARS-CoV nsp1 (Fig. 4). All viral mRNA bear the same 5′ untranslated region (UTR), and the stem loop SL1 at the beginning of the UTR is responsible for the protection (Banerjee et al., 2020; Rao et al., 2021; Schubert et al., 2020; Mendez et al., 2021; Tidu et al., 2021). Some studies have proposed that this sequence generally increases translation, thus partially overcoming nsp1 inhibition (Schubert et al., 2020; Lapointe et al., 2021), while others suggest that it directly affects nsp1 activity (Mendez et al., 2021, Tidu et al., 2021). Tidu et al. suggest that the formation of translation complexes on mRNAs with the SARS-CoV-2 5′ UTR is different, although it is unclear what this means and how this occurs (Tidu et al., 2021). Mendez et al. found that specific mutations in nsp1 make it able to target RNAs with the SARS-CoV-2 5′ UTR, suggesting a direct mechanism of regulation (Mendez et al., 2021). In any case, nsp1 inhibits host mRNA translation, which prevents IFN and ISG production, but does not prevent translation of SARS-CoV-2 viral mRNAs. In addition, some sequences in the 5′ UTR of host transcripts, including the 5’ terminal oligopyrimidine motif, may also promote translation during nsp1-mediated inhibition, which could lead to specific translation of some key proteins (Rao et al., 2021).
While the role of the C-terminus of nsp1 is clear (Fig. 4), whether and how the N terminal globular domain of nsp1 (aa 1–125) contributes to nsp1 function is not. The N terminal domain can be substituted with an unrelated globular protein of similar size with minimal loss of function (Schubert et al., 2020; Zhao et al., 2021). Nonetheless, the N terminus is very conserved at a sequence level to that of SARS-CoV nsp1 and at a structural level to those of multiple other coronaviruses, which suggests it may also have a specific function (Zhao et al., 2021; Clark et al., 2020). Moreover, Mendez et al. showed that deletion of the N terminus or of the linker between the N and C terminal domains impaired nsp1 activity (Mendez et al., 2021). It is also possible that the N terminal domain could contribute to other functions of nsp1, such as the proposed direct inhibition of the type I IFN pathway and/or of inflammasome activation (Kim et al., 2021; Kumar et al., 2021).
The potential importance of nsp1 for SARS-CoV-2 virulence is underscored by the isolation of circulating variants with 1–11 aa deletions in nsp1, which in most cases should disrupt proper folding of the protein (Benedetti et al., 2020; Islam et al., 2020; Lin et al., 2021). Indeed, some of the mutations appear to inactivate the protein, although replication of mutants bearing those deletions is not impaired in Calu-3 cells and the mutations do not prevent ribosome association (Lin et al., 2021). It should be noted that this study, to our knowledge, is the only one that has tested SARS-CoV-2 bearing nsp1 mutations (Lin et al., 2021). Therefore, much of the current model of nsp1 function remains to be confirmed in the context of viral replication, including the translation inhibition vs. RNA degradation functions.
4.2. ORF6
All ORF6 proteins in human and non-human sarbecoviruses inhibit type I IFN induction and response, which prompted investigation of the ORF6 homolog in SARS-CoV-2 (Kimura et al., 2021). ORF6 in SARS-CoV blocks IFN induction and response by sequestering the importin proteins KPNA2 and KPNB1 at the endoplasmic reticulum and preventing nuclear translocation of the transcription factors IRF3 and STAT1/2 (Frieman et al., 2007). SARS-CoV-2 ORF6 also blocks IRF3 and STAT1/2 translocation, as well as potentially a range of other nuclear proteins, but through a different mechanism of action (Fig. 5 ) (Kimura et al., 2021; Addetia et al., 2021; Kato et al., 2021; Miorin et al., 2020). While SARS-CoV-2 ORF6 also interacts with KPNA1 and KPNA2, overexpression of KPNA1 and KPNA2 does not rescue inhibition of STAT1 translocation (Miorin et al., 2020). Instead, the effect of ORF6 on nuclear/cytoplasmic trafficking is mediated by interactions with the mRNA export factors Rae1 and Nup98 (Kimura et al., 2021; Kato et al., 2021; Miorin et al., 2020; Gordon et al., 2020). Rae1 and its binding partner Nup98 are normally located in the nuclear membrane, but ORF6 binding causes cytoplasmic localization of Rae1 and Nup98 (Kato et al., 2021). Mutations of residues in ORF6 that mediate these interactions, as well as overexpression of Nup98, can rescue the nuclear translocation defect, confirming the functional importance of these interactions (Miorin et al., 2020). Shemesh et al. reported that ORF6 blocks MAVS-induced - but not TRIF-induced - production of IFN-β (Shemesh et al., 2021) This is a curious finding, as MAVS- and TRIF-induced interferon responses are both IRF3-dependent. Further investigation is required to determine how ORF6 can block IRF3 translocation induced by MAVS, but not by TRIF.
Fig. 5.
ORF6 binds to the Nup98-Rae1 complex and blocks bidirectional nucleocytoplasmic transport.
ORF6 obstructs nuclear pore trafficking by binding to mRNA export proteins Nup98 and Rae1 (Kimura et al., 2021; Kato et al., 2021; Miorin et al., 2020; Gordon et al., 2020). ORF6 binding to the Nup98-Rae1 complex prevents nuclear translocation of transcription factors including IRF3 and STATs (Kimura et al., 2021; Addetia et al., 2021; Kato et al., 2021; Miorin et al., 2020). Mutation of methionine 58 to an arginine (M58R) prevents ORF6 binding to Nup98-Rae1, but does not affect interactions with importins (KPNA1 and KPNA2) (Miorin et al., 2020). The M85R mutation rescues the effect of ORF6 on STAT1/2 translocation to the nucleus, demonstrating that SARS-CoV-2 ORF6 activity does not depend on interactions with importins, unlike that of its SARS-CoV ortholog (Frieman et al., 2007; Miorin et al., 2020). In addition to preventing nuclear entry of proteins, ORF6/Nup98-Rae1 binding inhibits nuclear export of mRNAs and results in an accumulation of polyA + mRNAs in the nucleus (Addetia et al., 2021).
In addition to blocking immune signaling proteins from entering the nucleus, interactions between ORF6 and Nup98-Rae1 also prevent nuclear export of mRNAs (Addetia et al., 2021). Indeed, SARS-CoV-2 infected cells have an accumulation of polyA + mRNA in the nucleus compared to mock-infected cells (Addetia et al., 2021). Moreover, the mRNA transporter hnRNPA1, which is thought to chaperone mRNA through the nuclear pore complex, also accumulates in the nucleus in cells overexpressing ORF6 (Kato et al., 2021). This disruption of nucleocytoplasmic RNA transport may also contribute to reducing the immune response from infected cells, and could globally alter host gene expression during infection.
SARS-CoV-2 variants with ORF6 deletions have been detected during the pandemic, although there were no observable differences in clinical severity between ORF6 deletion cases compared to WT SARS-CoV-2 (Quéromès et al., 2021). However, viral infection of cells with SARS-CoV-2 carrying these ORF6 deletions resulted in higher levels of inflammatory cytokines including CCL2/MCP1, PTX3, and TNFα which correlate with severe COVID-19 infections (Quéromès et al., 2021). Kimura et al. also analyzed the ORF6 region of SARS-CoV-2 sequences from a global database and found that 0.2% of cases have mutations that inactivate ORF6 (Kimura et al., 2021). While they proposed that loss of the potent IFN inhibitor ORF6 could result in attenuation of pathogenicity, they also noted that the viral sequences with inactivated ORF6 came from a database of symptomatic cases (Kimura et al., 2021). Given the strong anti-IFN activity of ORF6 in cells, it is interesting that infections with mutant viruses that have an inactivated ORF6 do not seem to alter the severity of disease. It is possible that the activity of other viral proteins can compensate for the loss of ORF6 function, resulting in minimal changes to clinical disease severity. Close monitoring of clinical cases with ORF6 inactivation mutations, as well as animal models using viral mutants will be helpful in understanding the full effect of this protein on type I IFN response.
5. Conclusions
The type I IFN response is important to combat SARS-CoV-2 infection, and robust IFN responses correlate with improved clinical outcomes (Hadjadj et al., 2020). In light of these early observations, recombinant IFN therapies for COVID-19 are currently in clinical trials, with early IFN treatments resulting in reduced mortality (Lee and Shin, 2020). Another approach to improving responses could be to reduce inhibition of type I IFN responses by SARS-CoV-2. Here, we discussed studies that have begun to uncover a multitude of type I IFN inhibitors in SARS-CoV-2. One of the major limitations at this time is that very few of these proteins have been studied in the context of the virus. It will be important to evaluate the anti-IFN activity of these proteins in the context of the virus to understand what role they play in infection. While many of these proteins are strong type I IFN inhibitors individually, there may be a temporal element that contributes to their activity. For example, the non-structural proteins (nsps) are expressed prior to the accessory proteins (ORFs). Interactions with other viral proteins may also alter a protein's role in infection. Variants in some of these genes have arisen as SARS-CoV-2 spreads, as discussed in the context of nsp1 and ORF6, but their contribution to changes in strain transmission and/or virulence remains unclear. Naturally occurring mutations that alter the function of these IFN antagonists will present a valuable opportunity to learn more about the impact of these proteins in SARS-CoV-2 infection.
Funding
Work on SARS-CoV-2 in the Gaglia laboratory is supported by an American Lung Association COVID-19 & Respiratory Virus Research Award and a donation by Dr. Marie Rozan to MMG. ACS is supported by the Lalor-Rozan fellowship.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Marta M. Gaglia reports financial support was provided by American Lung Association.
Footnotes
Marta M. Gaglia reports financial support was provided by American Lung Association.
References
- Addetia A., Lieberman N.A.P., Phung Q., Hsiang T.-Y., Xie H., Roychoudhury P., et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio. 2021;12 doi: 10.1128/mBio.00065-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Renkilaraj M.R.L.M., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339. doi: 10.1016/j.cell.2020.10.004. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F., Snyder G.A., Giovanetti M., Angeletti S., Gallo R.C., Ciccozzi M., et al. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020;18:329. doi: 10.1186/s12967-020-02507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J.M., St Clair L.A., Perera R., Parker R. SARS-CoV-2 infection triggers widespread host mRNA decay leading to an mRNA export block. RNA. 2021;121 doi: 10.1261/rna.078923.121. rna.078923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Chen J. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J. 2021 doi: 10.15252/embj.2021107776. n/a:e107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.K., Green T.J., Petit C.M. Structure of nonstructural protein 1 from SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.02019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Gluck A., Nachshon A., Winkler R., Fisher T., Rozman B., et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021:1–9. doi: 10.1038/s41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Zhu K., Qin B., Olieric V., Wang M., Cui S. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nat. Commun. 2021;12:2843. doi: 10.1038/s41467-021-23118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;1–13 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori Savellini G., Anichini G., Gandolfo C., Cusi M.G. SARS-CoV-2 N protein targets TRIM25-mediated RIG-I activation to suppress innate immunity. Viruses. 2021;13:1439. doi: 10.3390/v13081439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020 doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayn M., Hirschenberger M., Koepke L., Nchioua R., Straub J.H., Klute S., et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021;35:109126. doi: 10.1016/j.celrep.2021.109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A.S.M.R.U., Akther M., Puspo J.A., et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10:14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Zhang H., Meng Q., Xie J., Li Y., Chen H., et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Ikliptikawati D.K., Kobayashi A., Kondo H., Lim K., Hazawa M., et al. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem. Biophys. Res. Commun. 2021;536:59–66. doi: 10.1016/j.bbrc.2020.11.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.-E., Kim D.-K., Song Y.-J. SARS-CoV-2 nonstructural proteins 1 and 13 suppress caspase-1 and the NLRP3 inflammasome activation. Microorganisms. 2021;9:494. doi: 10.3390/microorganisms9030494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Konno Y., Uriu K., Hopfensperger K., Sauter D., Nakagawa S., et al. Sarbecovirus ORF6 proteins hamper induction of interferon signaling. Cell Rep. 2021;34:108916. doi: 10.1016/j.celrep.2021.108916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Ishida R., Strilets T., Cole J., Lopez-Orozco J., Fayad N., et al. SARS-CoV-2 nonstructural protein 1 inhibits the interferon response by causing depletion of key host signaling factors. J. Virol. 2021;95 doi: 10.1128/JVI.00266-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe C.P., Grosely R., Johnson A.G., Wang J., Fernández I.S., Puglisi J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl. Acad. Sci. Unit. States Am. 2021:118. doi: 10.1073/pnas.2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Shin E.-C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020 doi: 10.1016/j.virusres.2020.198074. 198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Zhao K., Zhang B., Hua R., Fang Y., Jiang W., et al. SARS-CoV-2 NSP12 protein is not an IFN-β antagonist. J. Virol. 2021 doi: 10.1128/JVI.00747-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Tang C., Wei H., Du B., Chen C., Wang M., et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. 2021;29:489–502. doi: 10.1016/j.chom.2021.01.015. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020;94:e01410–e01420. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez A.S., Ly M., González-Sánchez A.M., Hartenian E., Ingolia N.T., Cate J.H., et al. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep. 2021 doi: 10.1016/j.celrep.2021.109841. 109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M., Ollivier E., Chiu H.-P., Van Tol S., Rudolffi-Soto P., Stevens C., et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microb. Infect. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Makino S. Mechanisms of coronavirus nsp1-mediated control of host and viral gene expression. Cells. 2021;10:300. doi: 10.3390/cells10020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Park A., Iwasaki A. Type I and type III interferons – induction signaling evasion and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéromès G., Destras G., Bal A., Regue H., Burfin G., Brun S., et al. Characterization of SARS-CoV-2 ORF6 deletion variants detected in a nosocomial cluster during routine genomic surveillance, Lyon, France. Emerg. Microb. Infect. 2021;10:167–177. doi: 10.1080/22221751.2021.1872351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Hoskins I., Tonn T., Garcia P.D., Ozadam H., Cenik E.S., et al. Genes with 5′ terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein. RNA. 2021;27:1025–1045. doi: 10.1261/rna.078661.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebendenne A., Valadão A.L.C., Tauziet M., Maarifi G., Bonaventure B., McKellar J., et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 2021 doi: 10.1128/JVI.02415-20. JVI.02415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio N.G., Chauveau L., Hertzog J., Bridgeman A., Fowler G., Moonen J.P., et al. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021;11:13638. doi: 10.1038/s41598-021-92940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Sonmez T., Coker D., et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- Shemesh M., Aktepe T.E., Deerain J.M., McAuley J.L., Audsley M.D., David C.T., et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zhang G., Yang Y., Li M., Yang S., Peng G. Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J. Gen. Virol. 2021:102. doi: 10.1099/jgv.0.001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanich X., Vargas-Parra G., van der Made C.I., Simons A., Schuurs-Hoeijmakers J., Antolí A., et al. Genetic screening for TLR7 variants in young and previously healthy men with severe COVID-19. Front. Immunol. 2021;12:2965. doi: 10.3389/fimmu.2021.719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposito B., Broggi A., Pandolfi L., Crotta S., Clementi N., Ferrarese R., et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021;184:4953–4968. doi: 10.1016/j.cell.2021.08.016. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- The Severe Covid-19 GWAS Group Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L.G., Reuschl A.-K., Zuliani-Alvarez L., Whelan M.V.X., Turner J., Noursadeghi M., et al. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021;40 doi: 10.15252/embj.2021107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P., et al. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2021;27:253–264. doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., et al. Presence of genetic variants among young men with severe COVID-19. J. Am. Med. Assoc. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez C., Swanson S.E., Negatu S.G., Dittmar M., Miller J., Ramage H.R., et al. SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijst MGP van der, Vazquez SE, Hartoularos GC, Bastard P, Grant T, Bueno R, et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. 2021:eabh2624. [DOI] [PMC free article] [PubMed]
- Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- Yang D., Chu H., Hou Y., Chai Y., Shuai H., Lee A.C.-Y., et al. Attenuated interferon and proinflammatory response in SARS-CoV-2–infected human dendritic cells is associated with viral antagonism of STAT1 phosphorylation. J. Infect. Dis. 2020;222:734–745. doi: 10.1093/infdis/jiaa356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., et al. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020;80:1055–1066. doi: 10.1016/j.molcel.2020.10.034. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Pen J.L., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang J., Cheng G. Protease cleavage of RNF20 facilitates coronavirus replication via stabilization of SREBP1. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2107108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Ke Z., Hu H., Liu Y., Li A., Hua R., et al. Structural basis and function of the N terminus of SARS-CoV-2 nonstructural protein 1. Microbiol. Spectr. 2021 doi: 10.1128/Spectrum.00169-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragán L., Kuri T., Blakqori G., Weber F., Ludewig B., et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]