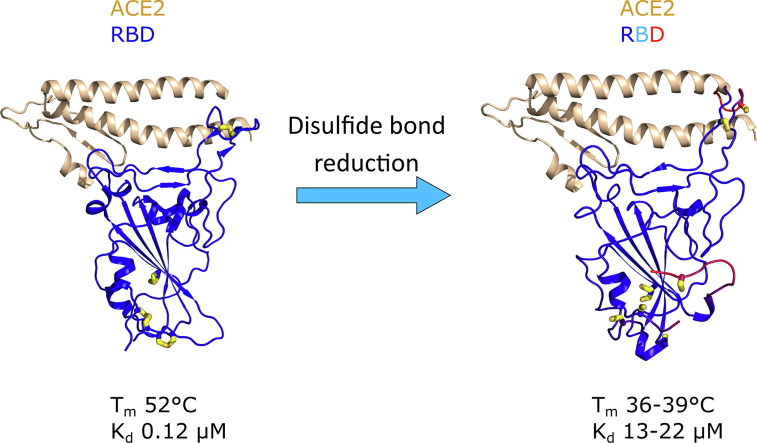
Graphical abstract
Abbreviations: ACE2, Angiotensin-converting Enzyme 2; CD, Circular Dichroism; cryo-EM, Cryo-Electron Microscopy; CV, Column Volume; DMEM, Dulbecco's Modified Eagle Medium; DPP4, Dipeptidyl Peptidase-4; DTT, Dithiothreitol; EC90, Effective Concentration 90%; ELF, Epithelial Lining Fluid; FBS, Fetal Bovine Serum; GSH, Reduced Glutathione; hAPN, Human Aminopeptidase N; HCoV, Human Coronavirus; MERS, Middle East Respiratory Syndrome; MST, Microscale Thermophoresis; MTS, 5-(3-carboxymethoxyphenyl)-2-(4,5-dimethyl-thiazoly)-3-(4-sulfophenyl) tetrazolium; NAC, N-Acetyl-L-cysteine; PCR, Polymerase Chain Reaction; RBD, Receptor-binding Domain; RMSD, Root Mean Square Deviation; RMSF, Root Mean Square Fluctuation; RT, Room Temperature; SARS, Severe Acute Respiratory Syndrome; TCEP, tris(2-carboxyethyl)phosphine; TCID50, Tissue Culture 50% Infectious Dose
Keywords: SARS-CoV-2, disulfide-reducing agent, disulfide bond, receptor-binding domain, RBD
Abstract
The current coronavirus pandemic is exerting a tremendously detrimental impact on global health. The Spike proteins of coronaviruses, responsible for cell receptor binding and viral internalization, possess multiple and frequently conserved disulfide bonds raising the question about their role in these proteins. Here, we present a detailed structural and functional investigation of the disulfide bonds of the SARS-CoV-2 Spike receptor-binding domain (RBD). Molecular dynamics simulations of the RBD predict increased flexibility of the surface loops when the four disulfide bonds of the domain are reduced. This flexibility is particularly prominent for the disulfide bond-containing surface loop (residues 456–490) that participates in the formation of the interaction surface with the Spike cell receptor ACE2. In vitro, disulfide bond reducing agents affect the RBD secondary structure, lower its melting temperature from 52 °C to 36–39 °C and decrease its binding affinity to ACE2 by two orders of magnitude at 37 °C. Consistent with these in vitro findings, the reducing agents tris(2-carboxyethyl)phosphine (TCEP) and dithiothreitol (DTT) were able to inhibit viral replication at low millimolar levels in cell-based assays. Our research demonstrates the mechanism by which the disulfide bonds contribute to the molecular structure of the RBD of the Spike protein, allowing the RBD to execute its viral function.
Introduction
The family Coronaviridae contains four genera: Alpha-, Beta-, Gamma- and Deltacoronaviruses, in which the Alpha- and Betacoronaviruses include the human pathogens: HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, SARS-CoV, SARS-CoV-2 and MERS-CoV.1 The first step in infection of human cells, common to all these viruses, involves the binding of the envelope Spike glycoprotein to a cell surface receptor followed by membrane fusion and virus internalization.2, 3 Different coronaviruses use different surface molecules as receptors: SARS-CoV, SARS-CoV-2 and HCoV-NL63 use angiotensin-converting enzyme 2 (ACE2)4, 5, 6; MERS-CoV binds to dipeptidyl peptidase-4 (DPP4) with high affinity7 and to some sialosides with low affinity8; HCoV-229E uses aminopeptidase N (hAPN)2; HCoV-OC43 binds 9-O-acetylated sialic acids9; while HCoV-HKU1 is internalized through an unknown receptor.10
The structures of the Spike protein and its domains have been determined for all these human pathogens. The functional Spike protein is a homotrimer, in which each protomer is over 1000 amino acids in length.11 The Spike protomers consist of two subunits, connected by a linker, which in many coronaviruses is cleaved during maturation. The S1 subunit is responsible for receptor binding12 and contains the receptor-binding domain (RBD), while the S2 subunit is responsible for membrane fusion.3 The structures of the Spike proteins and their RBDs from different coronaviruses show that these proteins possess multiple disulfide (S—S) bonds. For example, the Spike protein of SARS-CoV-2 contains 14 disulfide bonds in well-defined regions,13, 14 while that of MERS-CoV contains 1115 and the Spike of HCoV-229E contains 13 S—S bonds.16 Such an abundance of S—S bridges implies their important structural roles in the formation and stabilization of the proper Spike protein architecture. This is even more prominent for the relatively small RBD, which has four S—S bonds in SARS-CoV-2,17, 18, 19 four in MERS-CoV20, 21 and three in HCoV-229E.22 Mutations of these cysteine residues (Cys) resulted in either a significant decrease in expression levels or the loss of function.
Specifically, individual mutations of seven Cys residues to alanines (Ala) in the SARS-CoV RBD resulted in two out of seven mutants losing expression and three other mutants losing the ability to bind to ACE2. While only two mutants retained both expression and binding ability, only one of these mutants contained the Cys residue that was involved in the formation of a disulfide bond.23 The knockout of a single S—S bond in the surface loop of the HCoV-229E RBD that interacts with hAPN resulted in the loss of binding.22 Similarly, individual mutations of two conserved Cys residues, forming a disulfide bridge in the S2 domain of SARS-CoV, resulted in a drop of membrane fusion capability to 10% of the wild type, with an even greater loss for the double mutant.24 These data suggest the critical importance of the disulfide bridges for the Spike protein structure and function although our understanding of the precise mechanisms is limited to only general terms.
Respiratory pathways are the main point of entry for SARS-CoV-2 with the epithelial lining fluid (ELF) of the lungs playing a paramount role in protecting airways from a wide range of pathogens, allergens and harmful chemicals. In contrast to most extracellular environments (e.g. blood serum) possessing oxidizing conditions, ELF has a high reducing potential (−200 mV), similar to the intracellular environment.25 Experimental evidence of the sensitivity of coronaviruses to reducing conditions is very limited. For the murine leukemia virus, pseudotyped with the Spike protein from SARS-CoV, 3 mM dithiothreitol (DTT) (but not 0.5 mM) could inhibit the entry of the pseudovirus. This result led to the conclusion that SARS-CoV is quite insensitive towards disulfide-reducing agents.26 A brief in silico molecular modeling study suggested that reduction of only ACE2 disulfide bonds or ACE2 disulfide bonds and SARS-CoV-2 RBD disulfide bonds has a large negative impact on the binding energy, while the reduction of only RBD disulfide bonds does not.27 A recent experimental publication showed that thiol-reducing agents can inhibit infection with vesicular stomatitis virus infection, pseudotyped with the SARS-CoV-2 Spike protein.28
Although it is well known that disulfide bonds stabilize protein architecture, the actual contribution to protein stability and the sensitivity to thiol-reducing agents depends on many factors and is a priori difficult to predict.29 Here, we provide the mechanistic explanation for the critical role of the disulfide bonds in the structure and function of the SARS-CoV-2 Spike RBD by extensive biophysical investigation of RBD and SARS-CoV-2 viral assays. As predicted by our molecular dynamics simulations, the structure of the RBD becomes more flexible when it is not constrained by four S—S bonds. In particular, the surface loop (residues 456–490) participating in binding to ACE2 undergoes a very fast conformational opening after the S—S bond between Cys480 and Cys488 is reduced. Experimentally, this flexibility is evident by the ability of DTT and tris(2-carboxyethyl)phosphine (TCEP) to affect the secondary structure of the RBD and the ability of DTT, TCEP, N-Acetyl-L-cysteine (NAC) and reduced glutathione (GSH) to decrease the RBD melting temperature (Tm). DTT and TCEP could decrease the Tm to as low as 36–39 °C. As a consequence of increased flexibility and partial unfolding in the presence of DTT or TCEP at human body temperature, the binding constant of the RBD to ACE2 of 120 nM increases by 100–200 times. Finally, viral propagation in cell-based assays can be inhibited by TCEP and DTT, consistent with the ability to weaken or completely abrogate the Spike RBD – ACE2 interaction.
Results
Disulfide bonds are predicted to rigidify the structure of the RBD domain
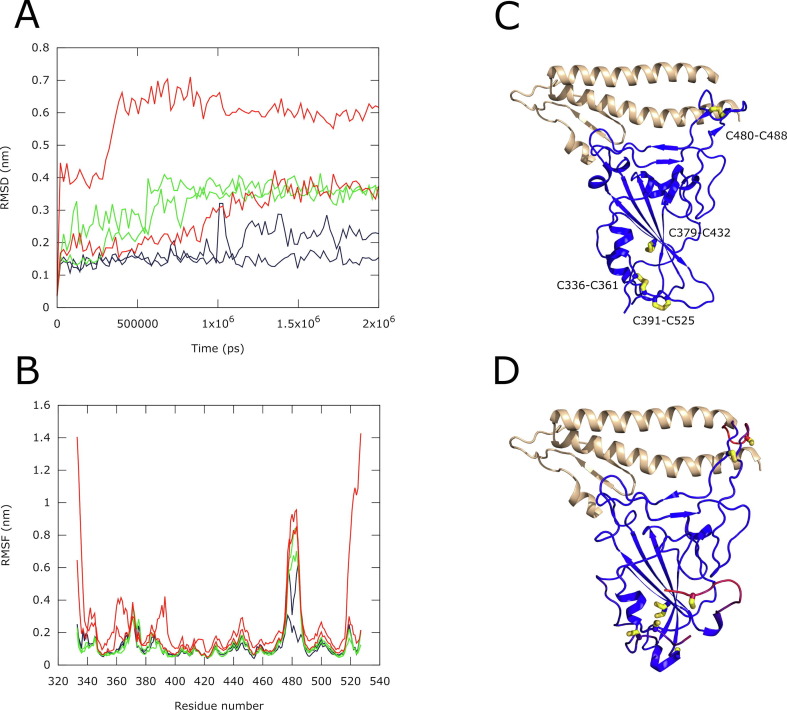
Many simulations of the Spike protein were conducted to date including an impressive worldwide effort for a 1.2-millisecond simulation of the full-length protein.30 Nevertheless, the majority of these simulations start from experimental structures possessing intact S—S bonds. A recent modeling study, exploring the role of the disulfide bonds in the RBD domain, performed by Hati and colleagues, uses relatively short trajectories of only 20 ns.27 To understand whether disulfide bonds are important in the stabilization of the structure of RBD, we performed six independent molecular dynamics simulations of the RBD domain in water of 2 µs each at 37 °C using GROMACS software.31 The RBD domain (residues 319–541) has four disulfide bonds: C480-C488 is located in the loop interacting with the ACE2 receptor, while the three other S—S bonds, C379-C432, C336-C361 and C391-C525, are located on the opposite side of the RBD domain and do not participate in ACE2 binding. We started with three RBD models, first with all four disulfide bonds intact (“RBD”), second with the single disulfide bond between C480-C488 reduced “RBD (−1 SS)” and third with all S—S bonds reduced “RBD (−4 SS)”. Two simulations of 2 µs were performed for each model.
Throughout all simulations, the RBD domain retained its overall fold, although structural changes, measured with the root mean square deviation (RMSD) of Cα atoms, behaved differently in each of the three models (Figure 1 (A)). While the RBD with four intact S—S bonds remained similar to the starting structure (RMSD of 1.0–2.5 Å for two independent simulations), the structures of RBD (−1 SS) and RBD (−4 SS) underwent much larger changes with RMSD increasing to 3.5–6.0 Å. The analysis of the root mean square fluctuation (RMSF) values for the residues of the domain revealed the regions of increased flexibility (Figure 1(B)). These are the regions immediately adjacent to the S—S bonds and include primarily the surface loop, comprising residues 454–492, which directly interacts with ACE2, the N- and the C-termini of the domain as well as protein elements around residues 370 and 390 (Figure 1(B–D)).
Figure 1.
Results of six molecular dynamics simulations of the Spike RBD domain at 37 °C. (A) Root mean square deviation (RMSD) of the Cα atomic coordinates (nm) vs. time (ps). The data for the RBD with 4 S—S bonds intact is colored blue; the RBD without C480-C488 bond – RBD (−1 SS) – green; the RBD with all four bonds reduced – RBD (−4 SS) – red. Two curves are shown for each model, corresponding to two independent simulations. The results show higher RMSD values for simulations with S—S bonds reduced; (B) Root mean square fluctuation (RMSF, nm) of Cα atoms of the RBD residues. Coloring identical to A. The high RMSF values show the parts of the RBD structure that undergo increased molecular flexibility upon reduction of the S—S bonds; (C) Ribbon representation of the experimental structure of Spike RBD – ACE2. RBD is colored blue with the four S—S bonds shown as wires and colored in shades of yellow. A part of ACE2, contacting RBD, is shown as a wheat-colored ribbon. (D) A snapshot from the RBD (−4 SS) simulation, superimposed on the experimental structure of RBD – ACE2 complex and shown instead of the experimental RBD structure. The model was colored by RMSD with a blue-red gradient. Blue corresponds to small structural deviations while red indicates the highest levels of structural change. The cysteine side chains are shown as wires, colored in shades of yellow.
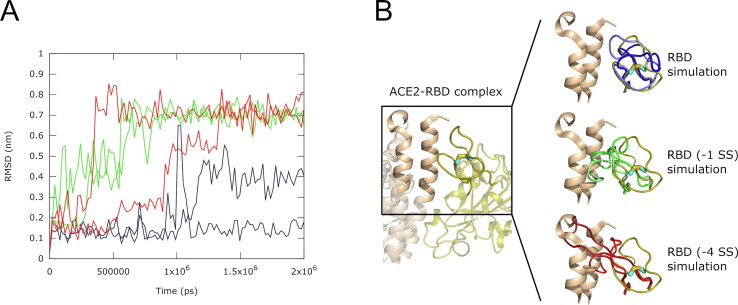
The most prominent difference is that the ACE2-binding loop remains relatively stable in its “closed” conformation, as seen in the crystal structure, when the disulfide bond C480-C488 is present, but undergoes a structural transition into a series of “open” conformations, not compatible with ACE2 binding, when the disulfide is absent (Figure 2 ).
Figure 2.
RMSD and snapshots of the conformation of the RBD ACE2-binding surface loop, residues 454–492, along six molecular dynamics trajectories at 37 °C. (A) Root mean square deviation (RMSD) of the Cα atomic coordinates (nm) vs. time (ps). The data for the RBD with 4 S—S bonds is colored blue; the RBD without C480-C488 bond – RBD (−1 SS) – green; the RBD with all four bonds reduced – RBD (−4 SS) – red. Two curves are shown for each model, corresponding to two independent simulations. The RMSD values show that models with the reduced C480-C488 bond have higher levels of structural deviation in the ACE2-binding surface loop 454–492; (B) (left) The loop-focused view of the crystal structure of the Spike RBD – ACE2 complex in ribbon representation. Only a part of the structure is shown. The Spike RBD domain is colored olive, while ACE2 is colored wheat. The C480-C488 S—S bond is shown as a wire with cyan carbon and yellow sulfur atoms. (right) Snapshots of molecular dynamics simulations, showing the conformations of the ACE2 – binding loop. The conformations from the RBD simulations are shown in shades of blue, RBD (−1 SS) – shades of green and RBD (−4 SS) – shades of red and are superimposed on the experimental structure.
Additional differences are seen for the second trajectory of RBD (−4 SS), where the last 10 C-terminal residues and the first 6 N-terminal residues of the domain adopt a completely random conformation, while the regions around the residues 370 and 390 change their position by up to 7.5 Å. Because of these motions, the second simulation of RBD (−4 SS) has elevated RMSD values (Figure 1(A and D)).
To increase the sampling of conformational space, we have also conducted one 2 µs simulation for each of the three RBD models at elevated temperature (77 °C), which provided additional insight into the ACE2-binding loop conformational equilibrium (Figure S1). Increasing the temperature of simulations is a well-known approach towards efficient conformational sampling used for example in replica exchange molecular dynamics.32, 33 In these simulations, we see an opening of the ACE2-binding loop in all three trajectories. However, while for RBD (−1 SS) and RBD (−4 SS) the opening happens right at the beginning of the simulations, for RBD with the C480-C488 bond present the loop holds its conformation for at least 1 µs. This result provides additional proof that the disulfide bond plays an important role in stabilizing the loop in ACE2-binding conformation.
Our simulation results agree that without ACE2 the loop may be present in its open conformation, as seen in the cryo-EM structures of the unbound Spike trimer13, 14 where this loop is disordered. However, the presence of the S—S bond may help in loop stabilization upon binding to the receptor, as seen in the crystal structure of RBD complexed to ACE2.17, 18, 19 Reciprocally, C480-C488 bond reduction causes increased loop fluctuations, as suggested by molecular dynamics, which may render the adoption of the correct bound conformation less energetically favorable.
Disulfide-reducing agents destabilize the Spike RBD structure
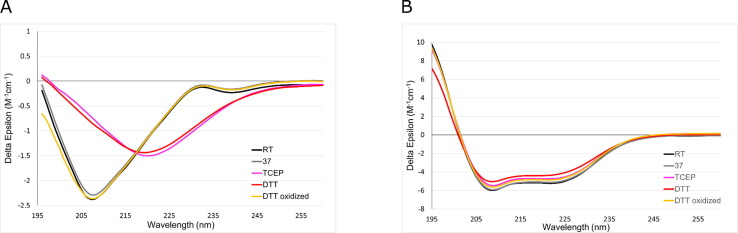
Since our molecular dynamics results suggest increased flexibility in the RBD domain upon the reduction of disulfide bonds, we investigated whether reagents capable of reducing S—S bonds would introduce structural changes in RBD and ACE2. To this end, we incubated the RBD and ACE2 in the presence of DTT, trans-4,5-dihydroxy-1,2-dithiane (oxidized DTT) or TCEP for 1 hour at 37 °C and measured their circular dichroism (CD) spectra at 37 °C. Oxidized DTT served as a control compound, being identical to DTT, except for the two thiol groups forming an intramolecular S—S bond. The compounds were added at a final concentration of 2.5 mM, the highest concentration possible without causing a significant increase in the UV light absorption.34
Incubation of the RBD at 37 °C did not change its CD spectrum as compared to an RBD sample kept at room temperature (RT) (Figure 3 (A)), suggesting that the RBD is stable at human body temperature. However, the addition of DTT and TCEP but not oxidized DTT caused a significant change in the CD spectrum of the RBD (Figure 3(A)), indicating changes in the secondary structure composition, induced by disulfide bond reduction. These changes persisted when the samples were cooled and analyzed at RT following the preincubation at 37 °C (Figure S2). The same experiments conducted for ACE2 (Figure 3(B)) showed that the addition of DTT or TCEP followed by preincubation and data acquisition at 37 °C caused no significant spectral changes, indicating that the ACE2 structure is insensitive to exogenous reducing agents in our experimental conditions. Indeed, ACE2 has only three S—S bonds for a domain of ∼600 residues long and none of these bonds are located in the proximity to the RBD binding site.
Figure 3.
Comparison of circular dichroism spectra of (A) RBD and (B) ACE2. Both the RBD and ACE2 were preincubated for 1 hour at 37 °C in the presence of disulfide-reducing agents at 2.5 mM concentration. Spectra were acquired at 37 °C immediately after preincubation. The spectrum of the RBD and ACE2, never exposed to elevated temperatures and acquired at room temperature (RT), is provided for reference.
Having demonstrated these spectral changes in the presence of DTT or TCEP, we next investigated whether reducing compounds affect the melting temperature (Tm) of the RBD. Indeed, the presence of four S—S bonds in the relatively small RBD domain of 320 residues should provide a significant contribution to its stability. We conducted a melting experiment of the RBD in the presence of 2.5 mM DTT, oxidized DTT and TCEP in which CD spectra were acquired as a function of increasing temperature. The RBD exhibited a Tm of 52.1 °C (Table 1 ), which did not change in the presence of 2.5 mM oxidized DTT (53.3 °C). However, a significantly lower Tm was measured in the presence of 2.5 mM DTT (39.6 °C) and 2.5 mM TCEP (36.3 °C).
Table 1.
Melting temperatures (Tm) of the RBD in the presence of disulfide-reducing agents at different concentrations.
Temperatures are expressed in Celsius (C) degrees.
|
Method |
Thermal shift assay |
Circular dichroism |
||||
|---|---|---|---|---|---|---|
| No compound |
47.0 |
52.1 |
||||
| Concentration | 5 mM | 2.5 mM | 1.25 mM | 0.62 mM | 0.31 mM | 2.5 mM |
| TCEP | ND1 | ND1 | 35.0 | 36.7 | 37.9 | 36.3 |
| DTT | 37.0 | 38.5 | 39.2 | 39.5 | 40.5 | 39.6 |
| Oxidized DTT | 46.6 | 46.4 | 46.5 | 46.6 | 46.3 | 53.3 |
| Concentration | 10 mM | 5 mM | 2.5 mM | 1.25 mM | 0.62 mM | |
| NAC | 41.5 | 42.7 | 44.0 | 44.9 | 45.5 | |
| GSH | 41.5 | 42.4 | 43.4 | 44.1 | 44.8 | |
1 ND, the melting curve profile did not allow for value determination.
The standard deviation is less than 5% for all values.
To broaden our concentration ranges and to explore more disulfide bond reducers, we used a thermal shift assay in which protein unfolding causes a significant increase in the fluorescence of Sypro® orange dye.35 We determined the RBD Tm values in the presence of DTT, oxidized DTT and TCEP in the concentration range of 0.31–5 mM and monothiols NAC and GSH in the range of 0.62–10 mM (Table 1). While all compounds, except oxidized DTT, caused an observable decrease in the Tm value of RBD, DTT and TCEP were most potent. NAC and GSH had a lesser impact than 0.31 mM DTT or TCEP even when added in 10 mM concentrations. High concentrations of TCEP (2.5 mM and 5 mM) precluded accurate Tm determination as at these concentrations melting curves lost their typical sigmoidal shape.
Disulfide-reducing agents decrease the Spike RBD – ACE2 binding affinity
Since the presence of reducing agents causes the RBD domain to be less stable, we hypothesized that these compounds should negatively affect the binding affinity of the RBD to its receptor ACE2. We used microscale thermophoresis (MST) to determine the extent to which 2.5 mM DTT, oxidized DTT or TCEP, as well as 10 mM NAC and GSH, would affect the binding. The RBD and ACE2 were preincubated for 1 hr at 37 °C in the presence of the reducing agents, after which time the proteins were mixed to yield a constant 20 nM concentration of fluorescently-labeled RBD and a variable concentration of ACE2 in the range of 1 nM–50 µM. MST was conducted at 37 °C.
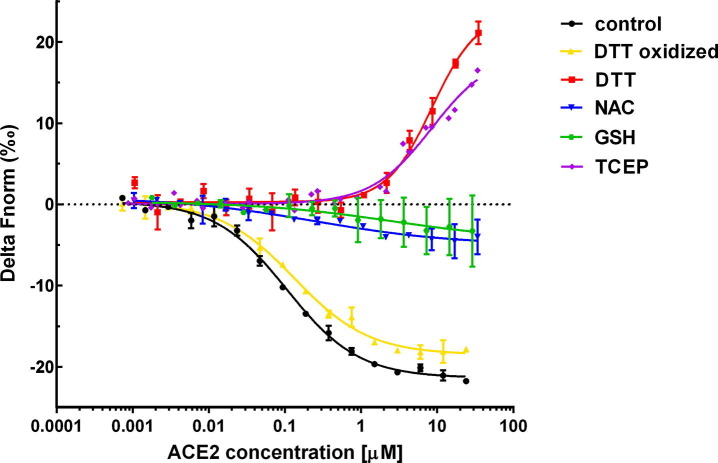
The measured affinity of the RBD for ACE2 was 120 nM (Table 2 , Figure 4 ), which was not significantly affected by oxidized DTT (170 nM). However, the addition of 2.5 mM TCEP or DTT caused a 100–200 times decrease in the affinity to 13.3 µM and 21.9 µM, respectively. NAC and GSH also decreased the affinity of the binding between ACE2 and RBD, although the change was not statistically significant even at 10 mM concentration.
Table 2.
The affinity of the RBD – ACE2 interaction in the presence of disulfide-reducing agents at 37 °C determined from microscale thermophoresis (MST).
| Affinity (µM) | |
|---|---|
| No compound | 0.12 ± 0.05 |
| 2.5 mM TCEP | 13.3 ± 4.0* |
| 2.5 mM DTT | 21.9 ± 13.6* |
| 2.5 mM DTT oxidized | 0.17 ± 0.08 |
| 10 mM NAC | 0.57 ± 0.40 |
| 10 mM GSH | 0.60 ± 0.38 |
Significant at p < 0.05.
Figure 4.
Affinity determination of the RBD – ACE2 interaction in the presence of disulfide-reducing compounds at 37 °C by microscale thermophoresis (MST). The graph shows the change in baseline-corrected normalized fluorescence (delta Fnorm (‰)) of labeled RBD as a function of ACE2 concentration. The graph shows a sigmoidal binding curve with a plateau at high ACE2 concentrations for the negative control, oxidized DTT, NAC and GSH. For DTT and TCEP the plateau at high ACE2 concentrations is absent, indicating weaker binding.
Disulfide-reducing agents prevent coronavirus infectivity in cell-based assays
To evaluate whether the reduction in binding affinity and partial melting of the RBD would impact viral infectivity, we tested disulfide-reducing agents in cell-based assays. We contrasted DTT and TCEP, which caused a prominent change in the RBD architecture to NAC and GSH, which had a milder effect, and additionally included an approved drug carboxymethyl-cysteine lacking an active SH-group as a negative control. Viral infectivity assays were conducted by infecting Vero’76 cells with SARS-CoV-2 and determining the virus titer 48 hr later by the tissue culture 50% infectious dose (TCID50) assay. Both cells and the virus were preincubated separately for 1 hr with the compounds. Following thereafter cellular media was discarded and the virus solution containing compounds was added to the cells for 1 hr for infection. Next, the virus was washed away and the cells were incubated for another 48 hr in fresh media in the absence of the compounds, allowing the virus to replicate, after which the viral titers in cell media were determined. In parallel, cell viability was determined using two assays – MTS and resazurin assays, and confirmed by phase-contrast microscopy.
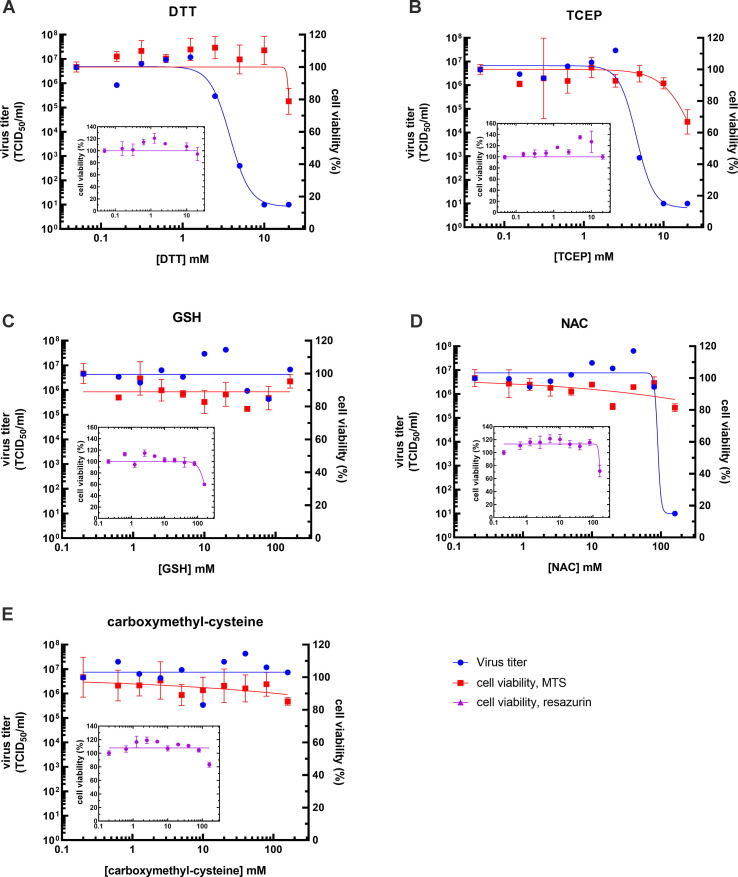
Two independent experiments were performed and the results of the first experiment are presented in Figure 5 , while of the second one – Figure S4. The two experiments showed largely the same results. Cell viability was unaffected by a 2-hour exposure to the compounds and it was only at the highest concentration of TCEP (20 mM), DTT (20 mM), GSH (160 mM) and NAC (160 mM) where a slight decrease in cell viability was noted. The negative control carboxymethyl-cysteine had no effect on cell viability.
Figure 5.
Virus titer and cell viability as a function of the concentration of (A) DTT, (B) TCEP, (C) GSH, (D) NAC and (E) carboxymethyl-cysteine. The figure shows data from a representative experiment. The viral titer, expressed as a TCID50/ml (left Y-axis) of SARS-CoV-2 and the viability of Vero’76 cell (% of the viability of untreated cell control) (right Y-axis), determined by the MTS assay, are plotted as the function of compound concentration (mM) (X-axis). The insets show the viability of Vero’76 cell, determined by the resazurin assay. Virus titers are colored in blue, while the viability data is colored in red for the MTS assay and purple for the resazurin assay. Because X-axis is logarithmic, viral titers and cell viability at 0 mM compound concentration are represented at 0.05 mM concentration for DTT and TCEP and at 0.2 mM concentration for GSH, NAC and carboxymethyl-cysteine.
Viral propagation, however, was affected by the same compounds that most affected the structure of the RBD in vitro. TCEP and DTT inhibited viral replication in the low mM range, while GSH and NAC had a negligible effect on viral titers if any even at 10-times higher concentrations. The negative control carboxymethyl-cysteine had no effect on viral propagation.
DTT caused a sharp decline in viral titers of one order of magnitude or more between 1.25–2.5 mM in the first and between 0.62–1.25 mM in the second experiment. Averaging the results from both experiments yielded the EC90 of 1.2 mM (0.58–2.2 mM confidence interval). Similarly, TCEP inhibited viral replication between 2.5–5 mM in the first and 1.25–2.5 mM in the second experiment, yielding EC90 of 2.7 mM (1.24–5.4 mM confidence interval). At these concentrations of DTT and TCEP, cell viability remained at 100% as confirmed by two viability assays (Figure 5, S4) and phase-contrast microscopy (Figure S5).
To determine to which extent the observed antiviral effect may be related to disulfide bond reduction in other SARS-CoV-2 proteins apart from Spike, we inspected available structures of SARS-CoV-2 proteins in the Protein Data Bank, presently covering a large percentage of the viral proteome. Structures of many protein products of the Orf1ab polyprotein are currently available and all of them have their Cys residues reduced.36, 37, 38, 39 Apart from Spike, only one additional protein – ORF8 – is rich in disulfide bonds,40 however, a natural mutation leading to the loss of its expression does not abolish viral infectivity neither in cellular assays nor in the human population.41 Thus, we considered that Spike is the most likely target of the disulfide-reducing agents.
Discussion
In this paper we show that disulfide bonds play a critical role in maintaining the proper structure of the RBD, allowing for a high-affinity interaction with its cellular receptor ACE2. As expected, DTT and TCEP were much more potent in comparison to NAC and GSH, as the redox potential of monothiols is generally weaker.42, 43, 44, 45, 46 It is well-known that both the dithiol DTT and the phosphine TCEP are capable of two-electron reduction of disulfide bonds without the involvement of any other species. DTT and TCEP have similar redox potentials (−330 mV and −290 mV, respectively) and form relatively unreactive oxidized products – a six-membered dithiane ring and a phosphine oxide, respectively.47, 48 The monothiols NAC and GSH, on the other hand, require the presence of two such thiol-containing molecules to reduce disulfide bonds and thus are expected to be inherently less effective.
Many toxins and pathogens rely on the proper redox state of their S—S bonds or sulfhydryl groups for host cell entry. Similar to the SARS-CoV-2 Spike protein, the entry of hepatitis C virus depends on the S—S bond-rich E2 envelope protein and can be blocked by 1 mM DTT.49 Additionally, the oxidized status of E2 thiol groups is important for evading the production of neutralizing antibodies.49 Botulinum neurotoxin B is a complex of 2 chains, connected by a single S—S bond. The reduction of this bond by TCEP prior to the neurotoxin internalization prevents the penetration of the active catalytic chain and saves the cells from intoxication.50
The opposite is also true for other toxins and pathogens – disulfide bond reduction and exposure of sulfhydryl groups promotes the internalization of HIV-1,51 rotavirus,52 Newcastle disease virus53 and diphtheria toxin.54
Knowing the sensitivity of the Spike protein towards reducing environments, it would be expected that the ELF of lungs, which possesses reductive properties25, 55, 56 would antagonize coronaviral entry. The levels of glutathione in ELF are approximately 50–100 times higher than those in blood serum (200–400 μM in ELF versus 2–8 μM in serum)55, 56, 57 and approach those in the intracellular environment.58 Additionally, ELF contains high levels of ascorbic acid and redox-active enzymes, such as superoxide dismutase, catalase, glutathione reductase, glutathione peroxidase, etc.56 to strengthen its reductive potential.
The first work in this direction used SARS-CoV pseudovirus and demonstrated its relative insensitivity towards DTT,26 suggesting molecular adaptations of the Spike protein towards tolerating reducing conditions of ELF. Our results for SARS-CoV-2 also show that it is in the low mM range that the potent disulfide reducers show their effect. As we improve our knowledge regarding the adaptation of pathogenic strategies towards the redox state of the environment, pathogen sensitivities could be uncovered and potentially used for medicinal purposes. Such work is gaining momentum. Recently, it was shown that N-acetyl cysteine amide (NACA) could inhibit a SARS-CoV-2 pseudovirus infection and the formation of multinucleated cells.28 A recent manuscript submitted to BioRxiv revealed a promising impact of some approved thiol-containing drugs on SARS-CoV-2 infectivity and cell entry.59 The effect of inhibition of viral entry by cysteamine was confirmed in yet another manuscript.60 One more BioRxiv manuscript explored the idea of an SH-group warhead attached to a delivery polar moiety, which guides the drug molecule to a cryptic pocket in the RBD architecture and positions the SH group in the proximity to a disulfide bond.61
Materials and Methods
Molecular dynamics
Molecular dynamics simulations of the Spike RBD domain were conducted with Gromacs 2019.31 For the force field, AMBER99S-ILDN62 was chosen with TIP3P water.63, 64 The cut-off for Van der Waals and short-range electrostatic interactions was 1.0 nm. Long-range electrostatics were calculated using the Particle Mesh Ewald (PME) approach65, 66 with a grid spacing of 0.12 nm × 0.12 nm × 0.085 nm. Temperature coupling was done by the v-rescale algorithm67 to maintain the system temperature of 310°K or 350°K, while the pressure coupling was isotropic and maintained by the Parinello-Rahman algorithm68, 69 at 1 atm. Rigid bonds between hydrogens and heavy atoms allowed for a 2-fs leap-frog integrator.
The RBD domain was taken from the ACE2 – Spike RBD protein complex structure PDB ID 6LZG.17 The RBD structure is comprised of residues 333–527. The sugar moieties were erased from the PDB files. Standard protonation states at pH 7.0 were used, while the only His519 in the RBD structure was protonated at Nε. Three independent simulations were made: (1) RBD with all 4 S—S bonds present as in the X-ray structure (control); (2) RBD with 3 S—S bonds present, while the S—S bond between Cys480 and Cys488 was treated as reduced; and (3) RBD with all 4 S—S bonds reduced. The modeling was conducted in a rhombic dodecahedral box in which the proteins were surrounded by a 10 nm thick layer of water. The system consisted of around 44,000 atoms total, 13,700 water molecules and 2 Cl- atoms. The systems were energy-minimized by 1000 steps of the steepest descent method. Minimized systems were subjected to further equilibrium simulations, first for 1 ns with all protein atoms restrained, followed by 10 ns with only backbone atoms restrained. The production simulations were conducted for 2 µs, saving the coordinates every 1 ns. Each of the three models was simulated twice for 2 µs per trajectory at 310°K and once at 350°K.
The starting models, topology files, molecular dynamics parameter files and trajectories, compressed to show 10 ns steps, were uploaded to Mendeley Data (https://doi.org/10.17632/v6mxmp9t3t.1)
Preparation of compounds
The stock solutions of DL-dithiothreitol (DTT) (Bioshop, DTT002), tris(2-carboxyethyl)phosphine (TCEP) (Bioshop; TCE101), glutathione reduced (GSH) (Bioshop GTH001), N-acetyl-cysteine (NAC) (Sigma-Aldrich; A7250) and S-carboxymethyl-L-cysteine (Sigma-Aldrich; C7757) were titrated to pH 7.5 and filter sterilized through a 0.2 µM filter. trans-4,5-Dihydroxy-1,2-dithiane (oxidized DTT) (Sigma-Aldrich; D3511) was dissolved in common buffers (10 mM Phosphate pH 7.5, PBS) in concentrations up to 100 mM.
Protein expression and purification
ACE2 19-615 untagged and Spike RBD 320-541, bearing the C-terminal TEV-His6 tag, were cloned into pCEP4 vector, with the N-terminal hemagglutinin signal sequence (KTIIALSYIFCLVFA). The constructs of ACE2 and Spike protein of SARS-CoV-2 were PCR amplified from pCEP4-myc-ACE2, which was a gift from Erik Procko (Addgene plasmid #141185; http://n2t.net/addgene:141185; RRID:Addgene_141185) and pcDNA3.1-SARS2-Spike, which was a gift from Fang Li (Addgene plasmid #145032; http://n2t.net/addgene:145032; RRID:Addgene_145032).
The proteins were expressed in Expi293F cells (Thermo Fisher Scientific) according to the manufacturer’s protocol. The cells, grown in Gibco Expi293 Expression Medium at 37 °C at 125 rpm, humidified atmosphere and 8% CO2 to a density of 3 × 106 cells/ml, were transfected with 1 µg of plasmid DNA per 1 ml of cell culture and Expifectamine® 293 or FectoPro® transfection reagents. Transfection enhancers were added the next day. The media was harvested 4 days after transfection, cleared by centrifugation at 20,000xg for 1 hour at +4 °C, filtered through a 0.22 µm filter and applied for further purification.
For the purification of ACE2 19-615, the media was applied on HiTrap® Q HP anion exchanger, 5 ml volume (GE Healthcare), preequilibrated with 10 column volumes (CV) of 20 mM Tris–HCl, pH 7.5. After the sample application, the column was washed with 10 CV of 20 mM Tris-HCl, pH 7.5. ACE2 was eluted with 20 mM Tris–HCl, pH 7.5 and NaCl gradient 0–0.5 M over 10 CV.
For the purification of RBD, the media was first dialyzed against 20 mM MES, pH 6.1, then applied on HiTrap® SP HP cation exchanger, 5 ml volume (GE Healthcare), preequilibrated with 10 CV of 20 mM MES, pH 6.1. After sample application, the column was washed with 10 CV of 20 mM MES, pH 6.1. The RBD was eluted with 20 mM MES, pH 6.1 and NaCl gradient 0–0.5 M over 10 CV.
ACE2 and RBD fractions, eluted from ion exchange, were concentrated and applied on gel-filtration, conducted on a Superdex 75 16/600 column in 10 mM phosphate buffer, pH 7.5, 50 mM NaCl. The final protein purity was >95% (Figure S3).
Circular Dichroism (CD)
Proteins were dialyzed overnight against 10 mM phosphate, pH 7.5 and diluted to 0.2 mg/ml. The CD spectra were collected by Chirascan CD spectrophotometer (Applied Photophysics, UK) in the range of 195–260 nm in a quartz glass cuvette at either room temperature (RT) or 37 °C, using 1-nm step size and an acquisition time of 3 s/nm. Data were collected over two accumulations, averaged, smoothed, the background was subtracted. The raw data in millidegrees were converted into molar circular dichroism (Δε).
For spectra acquisition at RT, the proteins were first preincubated with compounds at 2.5 mM final concentration for 1 hr at 37 °C, then cooled down to RT. For spectra acquisition at 37 °C, the proteins were preincubated with the compounds at 2.5 mM final concentration for 1 hr at 37 °C and spectra were acquired at 37 °C. RBD with no added compound was also measured at RT without preincubation at 37 °C for reference.
For the thermal denaturation experiments, the proteins were melted in the temperature range of 20–70 °C at a rate of 1 °C/min in the presence or absence of the compounds at 2.5 mM final concentration. The CD spectra were acquired every 1 °C between 195 and 250 nm with an acquisition time of 1 s/nm. The changes in a CD spectrum as a function of temperature were analyzed with Global3 software (Applied Photophysics) to yield the Tm of transitions between protein species observed by CD. The results represent an average of two independent experiments.
Thermal shift assays
The RBD was assayed at a final concentration of 0.5 mg/ml in 10 mM phosphate, 50 mM NaCl, pH 7.5. Sypro® orange protein fluorescent stain (Thermo Fisher; S6650) was added at a 1:1000 v/v ratio. DTT, oxidized DTT and TCEP were added to yield final concentrations in the range of 0.312–5.0 mM, while NAC and GSH were assayed in the concentration range of 0.625–10.0 mM. Melting was performed in an Applied Biosystems StepOne Plus Real-Time PCR amplifier in the temperature range of 20.0–60.0 °C in steps of 1 °C/min. Fluorescence measurements were taken at every step. The inflection point of a sigmoidal melting curve, calculated in StepOne software, was used as a measure of the melting temperature (Tm). The results represent an average of two independent experiments.
Binding analysis
The affinity constants of ACE2 and RBD were analyzed by microscale thermophoresis, using a Monolith instrument (NanoTemper Technologies GmbH). First, the RBD was labeled with the RED-NHS 2nd generation Monolith Protein Labeling Kit according to the manufacturer’s instructions (Nanotemper; MO-L011). The fluorescent group was conjugated to the RBD through primary amines, while ACE2 was unlabeled. The RBD and ACE2 solutions were separately preincubated for 1 hr at 37 °C with or without compounds added at the following final concentrations: 2.5 mM DTT, oxidized DTT and TCEP; 10 mM NAC and GSH. Then, the RBD was mixed with ACE2 in 20 nM RBD: 1 nM−50 µM ACE2 ratios in binding buffer 10 mM phosphate, pH 7.5, 500 mM NaCl, 0.1% pluronic acid. The binding experiments were conducted at 37 °C according to the Monolith’s instruction manual. The results were analyzed in MO.Control and MO.Affinity Analysis software (NanoTemper Technologies GmbH). The results represent an average of at least two independent experiments.
Coronavirus infectivity
The ability of SARS-CoV-2 to infect and propagate in eukaryotic cells was determined in the containment level 3 (CL3) facility available at the Vaccine and Infectious Disease Organization, (VIDO, Saskatoon, SK, Canada). Overall, four disulfide-reducing compounds were tested with 2-fold serial-dilutions at the following final concentrations for DTT and TCEP: 0.15 mM, 0.31 mM, 0.625 mM, 1.25 mM, 2.5 mM, 5 mM, 10 mM and 20 mM; for GSH and NAC: 0.625 mM, 1.25 mM, 2.5 mM, 5 mM, 10 mM, 20 mM, 40 mM, 80 mM and 160 mM. One control compound – S-carboxymethyl-L-cysteine – was included in the series at 0.625 mM, 1.25 mM, 2.5 mM, 5 mM, 10 mM, 20 mM, 40 mM, 80 mM and 160 mM final concentrations. Each compound was tested in two independent experiments for its effect on viral infectivity and cell viability.
Vero’76 cells were purchased from the American Type Culture Collection (ATCC; #CRL-1587) and grown to a confluence of 80–90% in Dulbecco's Modified Eagle Medium (DMEM) (Sigma-Aldrich; D5796) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher; 16000-044) and 1X Penicillin-Streptomycin (Pen-Strep) (Gibco; 15140148). To evaluate the antiviral potency of these compounds, Vero’76 cells were seeded in DMEM with 10% FBS and 1X Pen-Strep in 96-well flat-bottom tissue culture plates (Millipore Sigma; CLS3595). The SARS-CoV-2/BetaCoV/Canada/ON/VIDO-01/2020 virus (Sequence available at GISAID: EPI_ISL_425177) was diluted in DMEM supplemented with 2% FBS and 1X Pen-Strep to obtain a multiplicity of infection (m.o.i.) of 0.1 (2000 TCID50/well). The compounds were serially-diluted in DMEM supplemented with 2% FBS and 1X Pen-Strep, then added individually to both the cells (cells-compound) and the virus (virus-compound) to reach the above-mentioned final concentrations and preincubated for 1 hour at 37 °C. After 1 hour, the compounds were removed from the Vero’76 cells (cells-compound) and the pre-incubated virus-compound mixture was added to the cells and incubated for 1 hour at 37 °C to allow for viral infection in the presence of the serially-diluted compounds. After 1 hour, the virus-compound mixture was removed and replaced with fresh media containing no compounds. The cells were incubated for 48 hours, after which the viral supernatants were harvested and titrated by the tissue culture 50% infectious dose (TCID50) assay, in quadruplicate.
To titrate the viral supernatants and determine when viral entry and replication was not blocked by disulfide-reducing compounds, evident by cytopathic effects (CPE), the TCID50 assay was performed. To this end, Vero’76 cells were infected with viral supernatants that were serially diluted 10-fold for 1 hour at 37 °C. After 1 hour, the virus inoculum was removed and the maintenance medium was added composed of DMEM supplemented with 2% FBS, 1X Pen-Strep and 1 μg/mL L-[(toluene-4-sulphonamido)-2-phenyl] ethyl chloromethyl ketone (TPCK)-trypsin (Sigma-Aldrich). The CPE was observed by visual microscopy at 3 and 5 days post-infection (d.p.i.) for the SARS-CoV-2-induced phenotype, evident by cell death, clustering and rounding of cells. The TCID50 titers were determined by the Reed and Muench algorithm.
In parallel, the serially-diluted compounds were added to uninfected control cells, allowed to remain for two hours, after which the compounds were carefully washed away and replaced by fresh media, as described above. Cell viability was determined 48-hours post-exposure. Two cell viability assays were used – CellTiter 96 Aqueous One Solution Assay (Promega; G3580), which yields colored formazan product when metabolically active cells reduce the MTS tetrazolium (Owen’s reagent), and resazurin (Alamar Blue, Sigma-Aldrich; R7017) assay, which yields fluorescent resorufin upon reaction with dehydrogenase enzymes of metabolically active cells. Both assays were conducted according to manufacturers’ protocols.70 Briefly, for the CellTiter Aqueous assay, the cells were incubated with the MTS tetrazolium reagent for 2 hours at 37 °C to allow the development of the color and the absorbance values were determined at 490 nm. For resazurin, the growth media was removed and substituted by 100 µl of fresh media, containing 88 µM resazurin. Cells were incubated for 2 hours and the fluorescence was measured at an excitation of 530 nm and emission of 590 nm. The assays were conducted in triplicates in each independent experiment.
Statistical analysis
TCID50 viral titers were plotted against the concentration of compounds. The effective concentration EC90 was determined as the concentration of compound that reduces the viral titer by 90% when compared to untreated infected control. Both cell viability and viral titer results were plotted and EC90 values were calculated in GraphPad Prism 9 software, using the non-linear four-parameter regression analysis, along with a 95% confidence interval (95% CI).
Acknowledgments
Acknowledgments
We are grateful to two reviewers of our manuscript, whose suggestions and constructive criticism helped us to substantially improve present research. We gratefully acknowledge the use of instruments at the Protein Characterization and Crystallization Facility, College of Medicine, the University of Saskatchewan funded by the Canada Foundation for Innovation. Operational support of VIDO is provided in part by the Canadian Foundation for Innovation through the Major Science Initiative and Innovation Saskatchewan.
We thank Information and Communications Technology, the University of Saskatchewan for access to the Research Computing cluster, and Compute Canada for providing additional computational time. We feel indebted to the members of the Health Sciences Supply Centre (HSSC) and the laboratory managers of the Health Sciences Building of the University of Saskatchewan, specifically Mss. Angela Seto, Gail Lasiuk, Kazia Anderson, Elaine Carlson and Heather Neufeld, whose work and quality of service allowed for streamlined execution of experiments and peace of mind. We thank Dr. Oleg Dmitriev, Department of Biochemistry, Microbiology and Immunology, College of Medicine, the University of Saskatchewan and members of his laboratory for support and help with equipment. We thank Drs. Ivy Chung, Michal Boniecki and Thirumalai Selvi Ulaganathan from Dr. Miroslaw Cygler’s laboratory for their help and support.
This work was supported by the Canadian Institutes of Health Research grant GSP-48370 (M.C.) and OV5-170349 (D.F.), the Natural Sciences and Engineering Research Council RGPIN-05280-2016 (D.F.), RGPIN-2016-05810 (I.J.P.) and RGPIN-2019-05351 (G.N.G.), the University of Saskatchewan and the Canada Research Chairs Programs (I.J.P., G.N.G., M.C.).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Edited by M.F. Summers
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2021.167357.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
We have shared the link to molecular dynamics data in the text of the main manuscript and revision notes. The DOI link will become active upon acceptance of the manuscript.
References
- 1.Zhao X., Ding Y., Du J., Fan Y. 2020 update on human coronaviruses: One health, one world. Med. Nov. Technol. Devices. 2020;8 doi: 10.1016/j.medntd.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoo D., Parker M.D., Babiuk L.A. The S2 subunit of the spike glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology. 1991;180:395–399. doi: 10.1016/0042-6822(91)90045-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W., Moore M.J., Vasllieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofmann H., Pyrc K., Van Der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., Marasco W.A., Baric R.S., et al. Human Coronavirus HKU1 Spike Protein Uses O -Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/jvi.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou X., Guan H., Qin B., Mu Z., Wojdyla J.A., Wang M., Dominguez S.R., Qian Z., Cui S. Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1. Nature Commun. 2017;8:1–10. doi: 10.1038/ncomms15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogue B.G., Brian D.A. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986;5:131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68 doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-.) 2020 doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Commun. 2017;8:1–9. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Tomlinson A.C.A., Wong A.H.M., Zhou D., Desforges M., Talbot P.J., Benlekbir S., Rubinstein J.L., et al. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife. 2019;8 doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 19.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong A.H.M., Tomlinson A.C.A., Zhou D., Satkunarajah M., Chen K., Sharon C., Desforges M., Talbot P.J., et al. Receptor-binding loops in alphacoronavirus adaptation and evolution. Nature Commun. 2017;8:1–10. doi: 10.1038/s41467-017-01706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-Amino Acid Fragment of the SARS Coronavirus S Protein Efficiently Binds Angiotensin-converting Enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madu I.G., Belouzard S., Whittaker G.R. SARS-coronavirus spike S2 domain flanked by cysteine residues C822 and C833 is important for activation of membrane fusion. Virology. 2009;393:265–271. doi: 10.1016/j.virol.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzpatrick A.M., Jones D.P., Brown L.A.S. Glutathione redox control of asthma: From molecular mechanisms to therapeutic opportunities. Antioxidants Redox Signal. 2012;17:375–408. doi: 10.1089/ars.2011.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavillette D., Barbouche R., Yao Y., Boson B., Cosset F.L., Jones I.M., Fenouillet E. Significant redox insensitivity of the functions of the SARS-CoV spike glycoprotein: Comparison with HIV envelope. J. Biol. Chem. 2006;281:9200–9204. doi: 10.1074/jbc.M512529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hati S., Bhattacharyya S. Impact of Thiol-Disulfide Balance on the Binding of Covid-19 Spike Protein with Angiotensin-Converting Enzyme 2 Receptor. ACS Omega. 2020;5:16292–16298. doi: 10.1021/acsomega.0c02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manček-Keber M., Hafner-Bratkovič I., Lainšček D., Benčina M., Govednik T., Orehek S., Plaper T., Jazbec V., et al. Disruption of disulfides within RBD of SARS-CoV-2 spike protein prevents fusion and represents a target for viral entry inhibition by registered drugs. FASEB J. 2021;35 doi: 10.1096/FJ.202100560R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betz S.F. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2:1551–1558. doi: 10.1002/PRO.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman M.I., Porter J.R., Ward M.D., Singh S., Vithani N., Meller A., Mallimadugula U.L., Kuhn C.E., et al. SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome. Nature Chem. 2021;137(13):651–659. doi: 10.1038/s41557-021-00707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.AM G., KR B., RX G., DP T., GS S., M C. Regulation of Shigella Effector Kinase OspG through Modulation of Its Dynamic Properties. J. Mol. Biol. 2018;430:2096–2112. doi: 10.1016/J.JMB.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Bernardi R.C., Melo M.C.R., Schulten K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim. Biophys. Acta – Gen. Subj. 1850;2015:872–877. doi: 10.1016/J.BBAGEN.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Micsonai A., Bulyáki É., Kardos J. Methods Mol. Biol. Humana Press Inc.; 2021. BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy; pp. 175–189. [DOI] [PubMed] [Google Scholar]
- 35.Senisterra G., Chau I., Vedadi M. Thermal Denaturation Assays in Chemical Biology. Assay Drug Dev. Technol. 2012;10:128–136. doi: 10.1089/adt.2011.0390. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y., Wower J., Maltseva N., Chang C., Jedrzejczak R., Wilamowski M., Kang S., Nicolaescu V., et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 2021;41(4):1–11. doi: 10.1038/s42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nature Commun. 2021;121(12):1–7. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuller M., Correy G.J., Gahbauer S., Fearon D., Wu T., Díaz R.E., Young I.D., Martins L.C., et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 2021;7:eabf8711. doi: 10.1126/SCIADV.ABF8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nature Commun. 2020;111(11):1–11. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X., Hurley J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/PNAS.2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su Y.C.F., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7B and orf8 during the early evolution of SARS-CoV-2. MBio. 2020;11:1–9. doi: 10.1128/MBIO.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cline D.J., Redding S.E., Brohawn S.G., Psathas J.N., Schneider J.P., Thorpe C. New water-soluble phosphines as reductants of peptide and protein disulfide bonds: Reactivity and membrane permeability. Biochemistry. 2004;43:15195–15203. doi: 10.1021/bi048329a. [DOI] [PubMed] [Google Scholar]
- 43.Rothwarf D.M., Scheraga H.A. Equilibrium and kinetic constants for the thiol-disulfide interchange reaction between glutathione and dithiothreitol. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7944–7948. doi: 10.1073/pnas.89.17.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chau M.-H., Nelson J.W. Direct measurement of the equilibrium between glutathione and dithiothreitol by high performance liquid chromatography. FEBS Lett. 1991;291:296–298. doi: 10.1016/0014-5793(91)81305-R. [DOI] [PubMed] [Google Scholar]
- 45.Gilbert H.F. Thiol/disulfide exchange equilibria and disulfidebond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 46.Noszál B., Visky D., Kraszni M. Population, acid-base, and redox properties of N-acetylcysteine conformers. J. Med. Chem. 2000;43:2176–2182. doi: 10.1021/jm9909600. [DOI] [PubMed] [Google Scholar]
- 47.Cleland W.W. Dithiothreitol, a New Protective Reagent for SH Groups. Biochemistry. 1964 doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 48.Pullela P.K., Chiku T., Carvan M.J., Sem D.S. Fluorescence-based detection of thiols in vitro and in vivo using dithiol probes. Anal. Biochem. 2006;352:265–273. doi: 10.1016/j.ab.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 49.Fenouillet E., Lavillette D., Loureiro S., Krashias G., Maurin G., Cosset F.L., Jones I.M., Barbouche R. Contribution of redox status to hepatitis C virus E2 envelope protein function and antigenicity. J. Biol. Chem. 2008;283:26340–26348. doi: 10.1074/jbc.M805221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi X., Garcia G.E., Neill R.J., Gordon R.K. TCEP treatment reduces proteolytic activity of BoNT/B in human neuronal SHSY-5Y cells. J. Cell. Biochem. 2009;107:1021–1030. doi: 10.1002/jcb.22205. [DOI] [PubMed] [Google Scholar]
- 51.Stantchev T.S., Paciga M., Lankford C.R., Schwartzkopff F., Broder C.C., Clouse K.A. Cell-type specific requirements for thiol/disulfide exchange during HIV-1 entry and infection. Retrovirology. 2012;9:97. doi: 10.1186/1742-4690-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calderon M.N., Guerrero C.A., Acosta O., Lopez S., Arias C.F. Inhibiting Rotavirus Infection by Membrane-Impermeant Thiol/Disulfide Exchange Blockers and Antibodies against Protein Disulfide Isomerase. Intervirology. 2012;55:451–464. doi: 10.1159/000335262. [DOI] [PubMed] [Google Scholar]
- 53.Jain S., McGinnes L.W., Morrison T.G. Role of Thiol/Disulfide Exchange in Newcastle Disease Virus Entry. J. Virol. 2009;83:241–249. doi: 10.1128/jvi.01407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryser H.J., Mandel R., Ghani F. Cell surface sulfhydryls are required for the cytotoxicity of diphtheria toxin but not of ricin in Chinese hamster ovary cells. J. Biol. Chem. 1991;266:18439–18442. https://www.jbc.org/content/266/28/18439 (accessed January 1, 2021) [PubMed] [Google Scholar]
- 55.Cantin A.M., North S.L., Hubbard R.C., Crystal R.G. Normal alveolar epithelial lining fluid contains high levels of glutathione. J. Appl. Physiol. 1987;63:152–157. doi: 10.1152/jappl.1987.63.1.152. [DOI] [PubMed] [Google Scholar]
- 56.Cross C.E., Van der Vliet A., O’Neill C.A., Louie S., Halliwell B. Environ. Health Perspect. Public Health Services, US Dept of Health and Human Services; 1994. Oxidants, antioxidants, and respiratory tract lining fluids; pp. 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahman I., MacNee W. Lung glutathione and oxidative stress: Implications in cigarette smoke-induced airway disease. Am. J. Physiol. – Lung Cell. Mol. Physiol. 1999;277:1067–1088. doi: 10.1152/ajplung.1999.277.6.l1067. [DOI] [PubMed] [Google Scholar]
- 58.Kosower N.S., Kosower E.M. The glutathione status of cells. Int. Rev. Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 59.Khanna K., Raymond W., Charbit A.R., Jin J., Gitlin I., Tang M., Sperber H.S., Franz S., et al. Binding of SARS-CoV-2 spike protein to ACE2 is disabled by thiol-based drugs; evidence from in vitro SARS-CoV-2 infection studies. One Sentence Summary: Thiol-based drugs decrease binding of SARS-CoV-2 spike protein to its receptor and inhibit SARS-CoV-2 cell entry. BioRxiv. 2020 doi: 10.1101/2020.12.08.415505. 2020.12.08.415505. [DOI] [Google Scholar]
- 60.Thoene J., Gavin R.F., Towne A., Wattay L., Ferrari M.G., Pal R. In Vitro Activity of Cysteamine Against SARS-CoV-2 Variants Alpha Beta, Gamma and Delta. BioRxiv. 2021 doi: 10.1101/2021.10.02.462862. 2021.10.02.462862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y., Zeida A., Edwards C.E., Mallory M.L., Sastre S., Machado M.R., Pickles R.J., Fu L., et al. Thiol-based mucolytics exhibit antiviral activity against SARS-CoV-2 through allosteric disulfide disruption in the spike glycoprotein. BioRxiv. 2021 doi: 10.1101/2021.07.01.450701. 2021.07.01.450701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinforma. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 64.Neria E., Karplus M. A position dependent friction model for solution reactions in the high friction regime: Proton transfer in triosephosphate isomerase (TIM) J. Chem. Phys. 1996;105:10812–10818. doi: 10.1063/1.472889. [DOI] [Google Scholar]
- 65.Darden T., York D., Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 66.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 67.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 68.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. doi: 10.1080/00268978400101201. [DOI] [Google Scholar]
- 69.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 70.Riss, T.L., Moravec, R.A., Niles, A.L., Duellman, S., Benink, H.A., Worzella, T.J., Minor, L., Cell Viability Assays, Assay Guid. Man. (2016). http://europepmc.org/books/NBK144065 (accessed August 2, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have shared the link to molecular dynamics data in the text of the main manuscript and revision notes. The DOI link will become active upon acceptance of the manuscript.