Abstract
The targeting of proteolytic substrates is accomplished by a family of ubiquitin-conjugating (E2) enzymes and a diverse set of substrate recognition (E3) factors. The ligation of a multiubiquitin chain to a substrate can promote its degradation by the proteasome. However, the mechanism that facilitates the translocation of a substrate to the proteasome in vivo is poorly understood. We have discovered that E2 proteins, including Ubc1, Ubc2, Ubc4, and Ubc5, can interact with the 26S proteasome. Significantly, the interaction between Ubc4 and the proteasome is strongly induced by heat stress, consistent with the requirement for this E2 for efficient stress tolerance. A catalytically inactive derivative of Ubc4 (Ubc4C86A), which causes toxicity in yeast cells, can also bind the proteasome. Purified proteasomes can ligate ubiquitin to a test substrate without the addition of exogenous E2 protein, suggesting that the ubiquitylation of some proteolytic substrates might be directly coupled to degradation by the proteasome.
The degradation of many cellular proteins is mediated by the ubiquitin (Ub)/proteasome pathway (14, 22, 35). Ub is mobilized by a series of trans-esterification reactions prior to its ligation to substrates by Ub-conjugating (E2) enzymes and E3 factors (also known as Ub protein ligases and recognins) (13, 35). Substrates of the proteasome are degraded after their ligation to a multi-Ub chain, which is generally believed to improve targeting to the 26S proteasome (20, 22). Subunits in the proteasome that might play a role in the recognition of multiubiquitylated substrates have been isolated from yeast (Rpn10) (34), plants (Mbp1) (8, 33), and humans (S5a) (7). Significantly, long multi-Ub chains can interact with recombinant Rpn10 and S5a, and multiubiquitylated substrates can be degraded by purified 26S proteasomes.
The composition of the 19S regulatory complex of the 26S proteasome was recently determined, and 17 subunits were identified (12). There is growing evidence, however, that additional factors can also undergo substoichiometric interactions with the proteasome. For instance, the Doa4 Ub-processing protease (Ubp), Ufd5, and Rad23 copurify with the proteasome (10, 21, 25). In addition, Fujimuro et al. reported that the composition of the proteasome is altered in a growth stage-specific manner (9), while Kaiser et al. found that specific regulatory components of the cell cycle machinery can interact with intact proteasomes (18). Other reports have also described interactions between various cellular proteins and components of the 26S proteasome, although it is not clear from these studies if the interactions occurred with intact proteasomes (3, 36, 38). Taken together, these diverse findings indicate that the proteasome is a highly dynamic complex whose composition may be subject to regulation.
We considered the possibility that E2 proteins might be localized at the proteasome: an arrangement that would reduce the likelihood of inadvertent dismantling of multi-Ub chains by cellular Ubp proteins (37), while potentially enhancing the rate of degradation by coupling the assembly of a multi-Ub chain to the proteolytic apparatus. The multi-Ub chain could play an important role in tethering the substrate to the proteasome, consistent with previous studies (20). As a first step toward testing this hypothesis, we examined the subcellular distribution of Ubc4, an abundant E2 protein in yeast (27). We show here that Ubc4 and several other E2 enzymes are associated with the proteasome. Previous studies showed that a yeast mutant lacking Ubc4 and Ubc5 (ubc4Δ ubc5Δ) is exceedingly sensitive to stress-inducing conditions (27). In agreement with these genetic studies, we found that the interaction between Ubc4 and the proteasome was significantly increased following heat stress. Furthermore, purified proteasomes could conjugate Ub to a test protein, confirming the presence of catalytically active E2 proteins and demonstrating that the targeting components of the Ub pathway can interact with the 26S proteasome. The implications of these findings are discussed.
MATERIALS AND METHODS
Fractionation of yeast extracts.
The yeast strains used in this study are listed in Table 1. Yeast cultures were propagated in 100 ml of minimal medium containing 0.1 mM CuSO4. The cells were collected by centrifugation, washed with distilled H2O, and resuspended in lysis buffer (20 mM HEPES [pH 7.5], 100 mM potassium acetate, 5 mM EDTA, 20% glycerol) containing aprotinin, leupeptin, pepstatin A, and Pefabloc-SC. Protease inhibitors were used at concentrations recommended by the manufacturer (Boehringer Mannheim, Inc.). The cells were lysed by vortexing with 0.5-mm-diameter acid-washed glass beads, and the cell debris was removed by centrifugation for 1 h at 17,000 × g. The volume of lysate was adjusted to 10 ml with lysis buffer (containing protease inhibitors), and the proteins were precipitated by the addition of ammonium sulfate to yield 80% saturation. The precipitated proteins were dissolved in 1 ml of column buffer (20 mM Tris-HCl [pH 7.5], 20 mM potassium acetate, 20% glycerol, 1 mM dithiothreitol). Insoluble material was removed by centrifugation, and approximately 3 mg of protein was applied to a precalibrated, 70-ml Sepharose-4B column. Fractions (1 ml) were collected and examined by immunoblotting. The nitrocellulose filter was incubated sequentially with antibodies against Ubc4, FLAG epitope, Ubc2, and Rpt1. In immunoprecipitation experiments we combined 200 μl of buffer A (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) (1) with an equal volume of each column fraction. FLAG-agarose was added, and the reaction mixture was mixed by end-over-end rotation for 4 to 5 h at 4°C. The beads were washed twice with buffer A and then examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. Proteins that were fractionated in Superose-6 were prepared as described above, although only 1.2 mg of protein was applied to the column, and 0.6-ml fractions were collected.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JD47-13C | MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-3,112 | 23 |
| JD126 | MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-3,112 UMP1-HA PRE1-FLAG-His6 | 23 |
| JD138 | MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-3,112 PRE2-HA | 23 |
| JD139 | MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-3,112 PUP1-HA | 23 |
| KMY1003 | MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-3,112 ubc2Δ::URA3 | This study |
| MHY501 | MATα his3-Δ200 leu2-2,112 ura3-52 lys2-801 trp1-1 | 5 |
| MHY513 | MATa his3-Δ200 leu2-2,112 ura3-52 lys2-801 trp1-1 ubc4Δ::HIS3 | 5 |
| MHY499 | MATα his3-Δ200 leu2-2,112 ura3-52 lys2-801 trp1-1 ubc5Δ::LEU2 | 5 |
| MHY508 | MATα his3-Δ200 leu2-2,112 ura3-52 lys2-801 trp1-1 ubc4Δ::HIS3 ubc5Δ::LEU2 | 5 |
| PHY209 | JD47-13C transformed with PCUP1::PRE1-FLAG (LEU2 2μm) | This study |
| PHY210 | PHY209 transformed with PGAL1::UBC4 (URA3 CEN3) | This study |
| PHY211 | PHY209 transformed with PGAL1::UBC2 (URA3 CEN3) | This study |
| PHY212 | PHY209 transformed with PGAL1::UBC1 (URA3 CEN3) | This study |
Coimmunoprecipitation and immunoblotting.
Yeast cultures were grown in minimal medium containing 0.1 mM CuSO4 and harvested by centrifugation. Cell extracts were prepared as described above, and equal amounts of protein (∼1 mg) were adjusted to the same volume with buffer A and incubated with appropriate antibodies and affinity beads. Following incubation at 4°C for 4 to 5 h, the beads were washed twice with buffer A and examined by SDS-PAGE and immunoblotting. Plasmids that expressed V5-tagged proteasome subunits were purchased from Invitrogen, Inc. (San Diego, Calif.) and transformed into JD47-13C (23) and PHY209. Protein A-Sepharose was purchased from Repligen, and FLAG-agarose was purchased from Sigma Chemical Co. Antibodies against Rpt1, Ubc1, Ubc2, and Ubc4 were generated against full-length proteins at Pocono Rabbits, Inc. The immunoblots were developed by enhanced chemiluminescence (New England Nuclear).
Release of immunopurified proteasome with FLAG peptide.
Yeast strain JD47-13C was transformed with a plasmid encoding Pre1-FLAG expressed from the copper-inducible PCUP1 promoter. The resulting strain, PHY209, was grown in minimal medium plus 0.1 mM CuSO4 and suspended in lysis buffer containing protease inhibitors. Extracts were adjusted to ∼5 mg/ml, and 1.5 mg was incubated with 60 μl of FLAG-agarose for 4 h at 4°C. The beads were washed twice with buffer A and incubated with 60 μl of FLAG elution buffer (178 mM Tris-borate, 0.5% Triton X-100, 1 mM ATP, 200 μg of FLAG peptide per ml) at 30°C for 15 min with occasional mixing. The reaction mixture was centrifuged, and the supernatant was removed. The elution step was repeated, and the supernatants were combined. The eluates were concentrated by ultracentrifugation in Centricon-10 and examined by SDS-PAGE and immunoblotting. FLAG peptide was purchased from Sigma Chemical Co.
Ubiquitylation assays with purified proteasomes.
The proteasome was purified by immunoprecipitating Pre1-FLAG from PHY211. The FLAG-agarose beads were washed twice with Ub reaction buffer (50 mM Tris-HCl [pH 7.5], 40 mM KCl, 4 mM MgCl2) and then resuspended in 25 μl of Ub reaction buffer containing either 5 μl of histone H2B (1 mg/ml) or buffer. Wheat E1 (0.5 μg) and 5 μl of 32P-Ub were added to the reaction mixture, which was then adjusted to 5 mM ATP and incubated at 30°C for 45 min. (Detailed experimental details were described recently [32].) The reactions were terminated by adding loading dye containing SDS, and the products were resolved in an SDS–11% polyacrylamide gel and exposed to X-ray film.
Purification of the proteasome.
In addition to purifying the proteasome by immunoprecipitation, we used conventional chromatography as described previously (11, 21). Yeast strain JD126 (23) was grown in YEPD, pelleted, suspended in buffer D (50 mM Tris-HCl [pH 7.4], 10% glycerol, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP), and lysed using glass beads. The extracts were centrifuged at 17,000 × g for 1 h to remove cell debris. The extract was adjusted to a final volume of 10 ml at ∼10 mg/ml and applied to Blue-Sepharose that was equilibrated in the same buffer containing ATP. The column was then washed with 2 volumes of buffer D, and the bound proteins were eluted with a linear NaCl gradient (0 to 250 mM) at a flow rate of 1 ml/min. An aliquot from each 3-ml fraction was examined by immunoblotting with antibodies against Ubc4 and Rpt1. Aliquots (0.5 ml) were also tested for post-glutamyl peptide hydrolysis (PGPH) activity, and fractions that contained peak levels of activity were combined and further fractionated as described by Glickman et al. (11).
RESULTS
Ubc4 cosediments with components of the proteasome.
Ubc4 is a small, evolutionarily conserved Ub-conjugating (E2) enzyme whose counterparts in yeast, rats, plants, and humans have been isolated (15, 27). Ubc4 contains a conserved catalytic domain that is present in all E2 proteins. However, most other E2 proteins also contain highly divergent amino acid sequences that may contribute to E3 binding and substrate selectivity, and their absence in Ubc4 has suggested that it might lack substrate specificity. Although Ubc4 is required for the general elimination of damaged proteins, it is also evident that it can play a more specific role in recognizing proteolytic substrates in association with other targeting factors (16, 17).
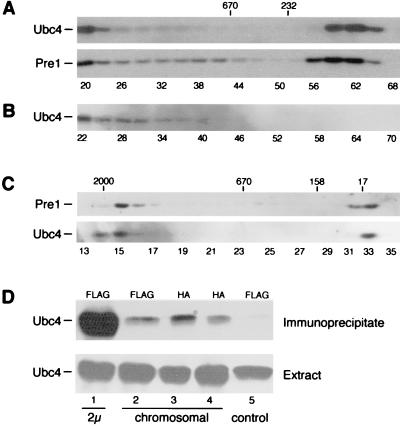
We examined the distribution of Ubc4 in a wild-type yeast strain by gel exclusion chromatography in Sepharose-4B and discovered that a significant fraction was present in a complex with a relative molecular mass of >106 kDa (Fig. 1A), consistent with the size of the 26S proteasome (6). Monomeric Ubc4 (14 kDa) was also present in column fractions that contained low-molecular-mass proteins (Fig. 1A, fractions 58 to 64). We examined the distribution of Pre1-FLAG, an epitope-tagged proteasome subunit, and found that it was also present in fractions that contained Ubc4 (Fig. 1A). To avoid autoubiquitylation of Ubc4, the chromatography in Sepharose-4B was performed in the absence of ATP. However, these conditions can promote the dissociation of the 26S proteasome into the 19S regulatory and 20S catalytic particles (11) and may have contributed to the distribution of Pre1-FLAG in a large number of column fractions. Nonetheless, we showed previously that intact, catalytically active 26S proteasomes can be efficiently precipitated with Pre1-FLAG (25). We incubated aliquots from the Sepharose-4B column fractions with FLAG-agarose and separated the precipitated proteins in an SDS-polyacrylamide gel. The resolved proteins were transferred to nitrocellulose and incubated with antibodies against Ubc4. We found that Ubc4 could be recovered with Pre1-FLAG only from fractions that contained high-molecular-mass complexes (Fig. 1B). Significantly, Ubc4 was not recovered with Pre1-FLAG from fractions that contained the monomeric forms of these proteins, despite the presence of high levels of both proteins (fractions 56 to 64). We conclude that Ubc4 and Pre1-FLAG can be copurified only when they are both present in the proteasome, because the monomeric proteins do not interact nonspecifically.
FIG. 1.
Ubc4 interacts with the proteasome. (A) Protein extracts were prepared from JD47-13C that expressed Pre1-FLAG and separated on a Sepharose-4B column. The fractions were examined by immunoblotting using antibodies against Ubc4 and FLAG. The fraction numbers and positions of gel filtration standards (in kilodaltons) are shown. Fractions 14 to 18 correspond to the column void volume. (B) An equal volume from the indicated Sepharose-4B fractions was incubated with FLAG-agarose, and the precipitated proteins were examined in an immunoblot with antibodies against Ubc4. (C) Protein extracts were separated on a Superose-6 column, and the migration of Pre1-FLAG and Ubc4 was determined as in panel A. (D) The Ubc4-proteasome interaction was examined by immunoprecipitating the proteasome from a wild-type strain (PHY209). Immunoprecipitation of 2μm plasmid-based Pre1-FLAG yielded high levels of Ubc4 (lane 1, Immunoprecipitate). However, the recovery of Pre1-FLAG-6His (lane 2), Pre2-HA (lane 3), and Pup1-HA (lane 4) expressed at single-copy levels yielded much lower levels of the proteasome as well as Ubc4, due to inefficient antibody reactions. Ubc4 was not precipitated from a control extract that did not express an epitope-tagged protein (lane 5). The expression of Ubc4 was similar in all the strains (Extract).
A significant amount of Ubc4 eluted in fractions close to the void volume in the Sepharose-4B column. We therefore resolved yeast extracts by Superose-6 fast protein liquid chromatography to exclude the possibility that Ubc4 was present in a large nonphysiological aggregate. Ubc4 and Pre1-FLAG were both detected in fractions that contained high-molecular-mass complexes that were well separated from the void volume (Fig. 1C). The reduced dissociation of the 26S proteasome (into 19S and 20S particles) in Superose-6 is due in part to the shorter duration of this chromatography procedure. Taken together, the results in Fig. 1A and C demonstrate that Ubc4 and Pre1-FLAG, as well as other proteasome subunits (see below), are present in the same high-molecular-mass complex.
Although Ubc4 was coprecipitated with Pre1-FLAG only from fractions that contained high-molecular-mass complexes (Fig. 1B), we were concerned that the high level expression of Pre1-FLAG may have contributed to a nonphysiological interaction between Ubc4 and the proteasome. We therefore examined the Ubc4-proteasome interaction in yeast strains that expressed derivatives of 20S proteasome subunits (Pre1-FLAG-6His, Pre2-HA, and Pup1-HA) at physiological levels from their chromosomal loci. Equal amounts of yeast extract were incubated with antibodies against the FLAG or HA epitope, and the precipitated proteins were recovered and examined by immunoblotting. We found that Ubc4 copurified with Pre1-FLAG-6His, Pre2-HA, and Pup2-HA, which were expressed at single-copy levels (Fig. 1D, lanes 2 to 4). In contrast, Ubc4 was not recovered from mock-treated extracts that did not contain an epitope-tagged proteasome subunit (lane 5). Much higher levels of Ubc4 were recovered using plasmid-borne Pre1-FLAG (2μm; lane 1), in contrast to the integrated derivative of Pre1, which contained carboxy-terminal FLAG and His6 epitopes. We found that the immunoprecipitation of the proteasome using Pre1-FLAG-6His was much less efficient than a similar reaction using the same anti-FLAG antibodies against Pre1-FLAG, possibly due to the carboxy-terminal His6 epitope. Similarly, immunoprecipitating the proteasome using HA-tagged proteasome subunits was inefficient (data not shown). We therefore used 2μm-based Pre1-FLAG in subsequent experiments because of the efficient recovery of catalytically active proteasomes.
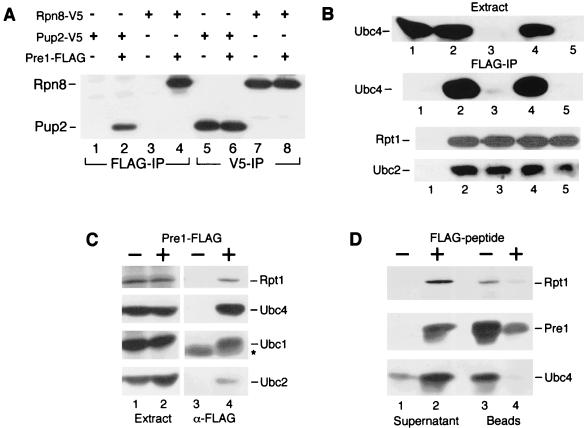
We expressed Rpn8-V5 (a 19S subunit [12]) and Pup2-V5 (a 20S subunit [6]) in a wild-type strain that also contained Pre1-FLAG (PHY209). Equal amounts of cell extract were incubated with anti-FLAG or anti-V5 antibodies, and the precipitated proteins were analyzed by immunoblotting. We found that both V5-tagged proteins were specifically purified on FLAG-agarose from strains that expressed Pre1-FLAG (Fig. 2A, lanes 2 and 4) but not from strains lacking Pre1-FLAG (lanes 1 and 3). The V5-tagged proteins were expressed at equivalent levels (lanes 5 to 8). The ability to rapidly immunoprecipitate intact, catalytically active proteasomes using Pre1-FLAG (25) permitted a rapid assessment of E2-proteasome interactions under different environmental conditions.
FIG. 2.
Ubc4 and subunits in the 19S and 20S particles can be immunoprecipitated with Pre1-FLAG. (A) The proteasome subunits Rpn8-V5 (19S) and Pup2-V5 (20S) could be precipitated on FLAG-agarose beads, and detected with V5 antibodies, from extracts that expressed Pre1-FLAG (lanes 2 and 4, respectively). Despite equivalent levels of expression (lanes 5 to 8), neither Pup2-V5 nor Rpn8-V5 was precipitated from extracts that lacked Pre1-FLAG (lanes 1 and 3). (B) Ubc4 was detected in extracts prepared from wild-type (lanes 1 and 2) and ubc5Δ strains (lane 4) but not in ubc4Δ (lane 3), or ubc4Δ ubc5Δ strains (lane 5). Ubc4 was coimmunoprecipitated only from strains that contained Pre1-FLAG (lanes 2 and 4, FLAG-IP) and not from a strain that did not express Pre1-FLAG (lane 1, FLAG-IP). The same immunoblot was incubated with antibodies against Rpt1 and Ubc2, and, as expected, both proteins were recovered only from the strains that expressed Pre1-FLAG (lanes 2 to 5). (C) Multiple E2 enzymes associate with the proteasome. Extracts prepared from a wild-type yeast strain containing (+) or lacking (−) Pre1-FLAG were separated by SDS-PAGE, and a single immunoblot was incubated sequentially with antibodies against Rpt1, Ubc4, Ubc1, and Ubc2. The expression of Pre1-FLAG did not alter the level of Rpt1 or the E2 proteins (lane 1 and 2, Extract). Equal amounts of extract were incubated with FLAG-agarose, and Rpt1, Ubc4, Ubc1, and Ubc2 were precipitated from the strain that expressed Pre1-FLAG (lane 4, α-FLAG). (The asterisk indicates a cross-reaction against the immunoglobulin light chain, which has a higher mobility than Ubc1.) (D) Samples were prepared as in panel C, and the beads were incubated for 15 min at 30°C with buffer containing (+) or lacking (−) FLAG peptide. Proteins present in the supernatant and beads were separated by SDS-PAGE and examined by immunoblotting. Pre1-FLAG, Rpt1, and Ubc4 were quantitatively released following exposure to FLAG peptide (lane 2). In contrast, treatment with buffer alone resulted in the retention of all three proteins on the FLAG-agarose beads (lane 3).
Multiple E2 enzymes bind the proteasome.
To verify the interaction between Ubc4 and the 26S proteasome, we immunoprecipitated Pre1-FLAG from wild-type, ubc4Δ, ubc5Δ, and ubc4Δ ubc5Δ strains and also separated equal amounts of extract by SDS-PAGE (Fig. 2B, lanes 2 to 5). We incubated an immunoblot with polyclonal antibodies and detected Ubc4 in wild-type and ubc5Δ strains (lanes 2 and 4) and also detected a weak cross-reaction against Ubc5 in ubc4Δ (lane 3, FLAG-IP). Ubc4 was not precipitated from extracts that lacked Pre1-FLAG (lane 1, FLAG-IP), despite the high physiological levels of endogenous Ubc4 (lane 1, Extract). The same filter was subsequently incubated with antibodies against the proteasome subunit Rpt1 and the E2 protein Ubc2, which were expressed at physiological levels. Rpt1 was precipitated from all extracts that contained Pre1-FLAG (lanes 2 to 5), demonstrating that equivalent levels of the proteasome were recovered from the various strains. We were also able to precipitate Ubc2 with Pre1-FLAG, although an ∼10-fold-longer exposure was required to detect this E2 enzyme. The interaction between Ubc2 and the proteasome was not affected in ubc4Δ, ubc5Δ, or ubc4Δ ubc5Δ mutants. Similar to Ubc4, neither Rpt1 nor Ubc2 was precipitated from an extract that did not contain Pre1-FLAG (lane 1, FLAG-IP).
To further corroborate these results, we immunoprecipitated Pre1-FLAG from a wild-type strain (JD47-13C) and incubated the affinity beads with buffer alone or buffer containing FLAG peptide (23). We then examined the supernatant and affinity beads for the presence of Rpt1, Ubc1, Ubc2, and Ubc4, which were each expressed at physiological levels from the chromosome (Fig. 2C and D). The expression of Pre1-FLAG did not alter the cellular levels of Rpt1 or the E2 enzymes (Fig. 2C). Consistent with the results in Fig. 2B, Rpt1 and all three E2 proteins were specifically immunoprecipitated with Pre1-FLAG (Fig. 2C, lane 4). None of these proteins was recovered on FLAG-agarose beads from extracts prepared form JD47-13C that did not express Pre1-FLAG (lane 3). Significantly, the addition of FLAG peptide to the FLAG-agarose beads released Pre1-FLAG as well as Rpt1 and Ubc4 into the supernatant (Fig. 2D, lane 2), in agreement with our hypothesis that E2 proteins can interact with the 26S proteasome. In contrast, the addition of buffer lacking FLAG peptide failed to release these proteins into the supernatant (lane 1), and they were retained on the FLAG-agarose beads (lane 3).
E2 enzymes do not compete for proteasome binding.
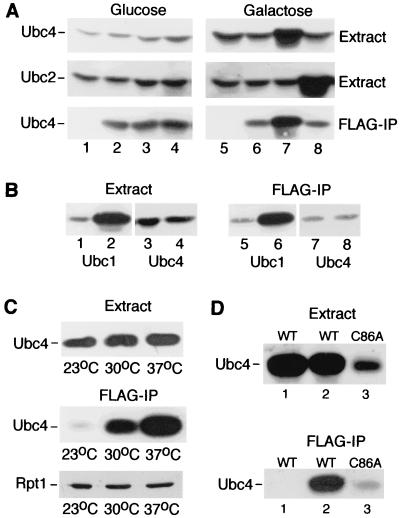
We surmised that if E2 enzymes could bind the proteasome directly, in the absence of E3 or substrate, their overexpression might lead to higher levels in the proteasome. We therefore overexpressed specific E2 enzymes, in the absence of stress-inducing conditions, and examined their interaction with the proteasome. Plasmids containing galactose-inducible UBC2 and UBC4 were expressed in JD47-13C that contained Pre1-FLAG. Yeast cells were grown in glucose- or galactose-containing medium, and protein extracts were prepared (Fig. 3A). The expression of endogenous Ubc4 and Ubc2 was readily detected in extracts prepared from uninduced (glucose) cultures (lanes 1 to 4). Ubc4 was efficiently precipitated on FLAG-agarose from strains that expressed Pre1-FLAG (lanes 2 to 4) but not from a strain lacking this epitope-tagged proteasome subunit (lanes 1 and 5). Growth in inducing (galactose) medium led to significantly higher intracellular levels of Ubc4 (lane 7, Extract) and Ubc2 (lane 8, Extract). Equal amounts of extract, which were isolated from galactose-grown cells, were incubated with FLAG-agarose, and the precipitated proteins were examined (lanes 5 to 8, FLAG-IP). Higher levels of Ubc4 were associated with the proteasome that was purified from galactose-grown cells that expressed PGAL1::UBC4 (lane 7, FLAG-IP). Significantly, the overexpression of Ubc2 did not affect Ubc4 levels (lane 8, Extract) or proteasome interaction (lane 8, FLAG-IP). Similarly, overexpression of Ubc1 in galactose-containing medium (Fig. 3B, lane 2) led to increased association with the proteasome (lane 6) but did not affect Ubc4 abundance (lane 4) or proteasome binding (lane 8). We also note that the Ubc2-proteasome interaction was unaffected in the ubc4Δ ubc5Δ mutant (Fig. 2B). The failure to detect competition for proteasome interaction among the various E2 enzymes suggests that only a small fraction of proteasomes are bound to E2 enzymes, similar to other proteins, including Ufd5 (10) and Rad23 (25). Alternatively, it is possible that E2 proteins interact with distinct proteasome subunits. Although E2 proteins might interact with the proteasome without binding physiological substrates, our data do not exclude the possibility that high-level expression of Ubc1, Ubc2, and Ubc4 results in proteasome binding through interactions with substrates that are not normally targeted by these E2 enzymes.
FIG. 3.
High-copy expression and heat stress result in an increased E2-proteasome interaction. (A) UBC4 and UBC2 were expressed from the galactose-inducible GAL1 promoter in wild-type yeast (JD47-13C). With the exception of lanes 1 and 5, all the strains also expressed Pre1-FLAG (PHY209). The effect of overexpressing Ubc4 was examined in lanes 3 and 7, while the effect of overexpressing Ubc2 was tested in lanes 4 and 8. The expression of the endogenous genes was observed in extracts prepared from cells grown in glucose medium (Glucose panel, lanes 1 to 4). Equal amounts of extract were incubated with FLAG-agarose, and the precipitated proteins were analyzed by immunoblotting (FLAG-IP). Ubc4 was recovered from extracts that contained Pre1-FLAG (lanes 2 to 4) but not from a strain that did not express this epitope-tagged proteasome subunit (lane 1). A similar analysis of extracts prepared from galactose-grown cultures showed that high-level expression of Ubc4 (lane 7, Extract) resulted in an increased proteasome interaction (lane 7, FLAG-IP). Interestingly, overexpression of Ubc2 (lane 8, Extract) did not affect the Ubc4-proteasome interaction noticeably (lane 8, FLAG-IP). Overexpression of Ubc2 also led to its increased interaction with the proteasome (see Fig. 4A). (B) We also examined the effect of PGAL1::UBC1 on the Ubc4-proteasome interaction. In agreement with the results in panel A, growth of PHY212 in galactose medium led to higher expression of Ubc1 (lane 2) and an increased proteasome interaction (lane 6). However, the expression of endogenous Ubc4 (lanes 3 and 4) and its interaction with the proteasome (lanes 7 and 8) were unaffected. Even-numbered lanes represent extracts prepared from PHY212 that expressed PGAL1::UBC1, while the extracts examined in the odd-numbered lanes represent a control that contained vector. Note that endogenous Ubc1 was also detected in the proteasome (lane 5). (C) Heat stress increased the Ubc4-proteasome interaction. Yeast cells expressing Pre1-FLAG were grown at 23°C for 24 h and then incubated at 23, 30, and 37°C for 2 h. We found that the cellular levels of Ubc4 increased ∼twofold at 37°C (Extract). However, the level of Ubc4 that was bound to the proteasome increased ∼25-fold when the temperature was increased from 23 to 37°C. The relative increase in the levels of Ubc4 in the proteasome was >10-fold. In contrast, the recovery of Rpt1 with Pre1-FLAG was unaffected at the different temperatures. (D) A catalytically inactive derivative of Ubc4 (Ubc4C86A) can interact with the proteasome. Extracts prepared from the ubc4Δ ubc5Δ mutant that expressed both Pre1-FLAG and Ubc4C86A (lane 3, Extract) were incubated with FLAG-agarose. Ubc4C86A was precipitated with the proteasome (lane 3, FLAG-IP), although its intracellular levels were much lower than those of wild-type (WT) Ubc4 (lane 2). Lane 1 represents a control strain that did not express Pre1-FLAG.
Ubc4-proteasome interaction increases following heat stress.
The Ubc4 class of E2 enzymes is required for the stress response and is implicated in the degradation of a broad range of proteolytic substrates (27). Ubc4 and Ubc5 are 93% identical, and highly conserved counterparts have been isolated from diverse species. A strain lacking both E2 enzymes (ubc4Δ ubc5Δ) displays growth and proteolytic defects. Since the double mutant is acutely sensitivity to heat stress (27), we examined the Ubc4-proteasome interaction at elevated temperature. Yeast cells (ubc5Δ) expressing Pre1-FLAG were grown at 23°C for 24 h, and the culture was diluted into fresh medium that was equilibrated to 23, 30, or 37°C. The cells were allowed to propagate for 2 h and then pelleted and frozen in liquid nitrogen. Protein extracts were incubated with FLAG-agarose, and the precipitated proteins were resolved by SDS-PAGE (Fig. 3C, Extract). We found that Ubc4 levels in the proteasome increased ∼25-fold compared to those in an untreated cell (Fig. 3C, FLAG-IP), demonstrating that the interaction is responsive to stress-inducing conditions. Since the cellular levels of Ubc4 also increased slightly (∼2-fold) as the temperature was raised from 23 to 37°C, we estimate that the relative amount of Ubc4 associated with the proteasome increased >10-fold at high temperature. The efficiency of immunoprecipitation was not altered under these conditions, because the recovery of Rpt1 with Pre1-FLAG was similar at 23, 30, and 37°C (Fig. 3C, FLAG-IP). These results show that the fraction of proteasomes that contain Ubc4 at low temperature can be rapidly increased at high temperature, consistent with the requirement for this E2 in stress resistance. Increased interaction with the proteasome at 37°C is unlikely to be caused by heat-induced denaturation, because the melting temperature (Tm) of Ubc4 is 58.5°C (31). Furthermore, Ubc4 is not ubiquitylated or degraded at high temperature. In addition, we have shown previously that Ubc4 retains catalytic activity following incubation at 42°C (31).
A catalytically inactive derivative of Ubc4 (Ubc4C86A) can bind the proteasome.
To characterize the mechanism of the E2-proteasome interaction, we determined if a catalytically inactive derivative of Ubc4 could bind the proteasome. We converted the catalytic cysteine residue in Ubc4 to alanine and confirmed that Ubc4C86A was unable to form a thioester-linked intermediate with Ub. Ubc4C86A accumulated at very low levels in wild-type yeast cells (Fig. 3D, lane 3, Extract) and caused moderate growth inhibition. However, high-level expression of Ubc4C86A (from the copper-inducible PCUP1 promoter) inhibited the growth of the ubc4Δ ubc5Δ mutant, suggesting that it might form a deleterious interaction with an important physiological effector, in the absence of native Ubc4. We expressed Ubc4C86A and Pre1-FLAG in ubc4Δ and found that the catalytically inactive mutant protein could interact with the proteasome (Fig. 3D, lane 3, FLAG-IP). The reduced level of Ubc4C86A in the Pre1-FLAG immunoprecipitates reflects the low in vivo abundance of this mutant protein. In contrast to Ubc4C86A, overexpression of a similar mutant derivative of Ubc2 (Ubc2C88A) did not cause growth inhibition (data not shown), which may reflect its significantly weaker interaction with the proteasome. Because the 26S proteasome is essential for viability, it is conceivable that a nonproductive interaction with Ubc4C86A can lead to severe growth inhibition. However, it is also possible that Ubc4C86A interacts with some other important regulator to cause toxicity. Ubc4C86A might interfere with other components of the UFD pathway, such as Ufd1, which is essential for cell viability. Although Ubc4C86A could interfere with a component of the UFD pathway, neither Ubc4 nor Ubc5 is required for the toxicity, because high levels of Ubc4C86A inhibited the growth of the ubc4Δ ubc5Δ mutant.
Proteasome-associated E2 can ligate Ub to a test protein.
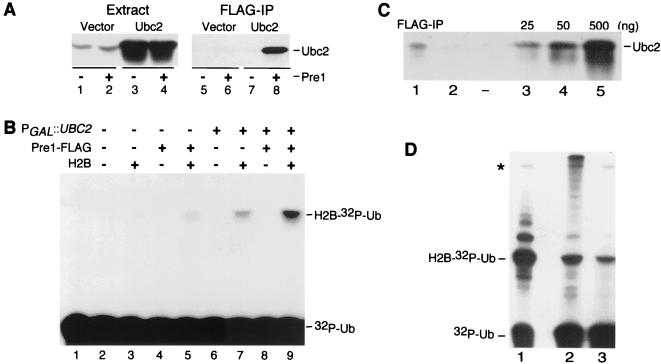
We investigated if proteasome-associated E2 enzymes could ligate Ub to a test protein. Since the ubiquitylation of substrates by Ubc4 requires several additional targeting factors (17), we addressed this question by using a simpler system. Histone H2B is a physiological substrate of Ubc2 (24) that can be efficiently ubiquitylated in the absence of other targeting factors (28–30, 32). Based on our finding that Ubc2 can bind the proteasome, we examined the ubiquitylation of H2B by proteasome-associated Ubc2. The lower abundance of cellular Ubc2, and its correspondingly reduced levels in the proteasome, required high expression from a galactose-inducible promoter. Growth in galactose led to ∼25-fold-increased levels of Ubc2 (Fig. 4A, lanes 3 and 4). Protein extracts were incubated with FLAG-agarose, and, in agreement with previous results, Ubc2 was recovered in the FLAG-agarose beads only if the strain also expressed Pre1-FLAG (lane 8) and not from extracts lacking Pre1-FLAG (lane 7). We added histone H2B, 32P-Ub, and E1 to FLAG-agarose beads that contained immunopurified proteasomes and detected specific ubiquitylation of H2B (Fig. 4B, lane 9). The ubiquitylation of H2B was strictly dependent on the addition of E1 and was significantly increased when Ubc2 was overexpressed. A low level of H2B ubiquitylation was detected in extracts prepared from cells lacking PGAL1::UBC2 and is due to the presence of endogenous Ubc2 expressed from the chromosome (lane 5). Western blot experiments confirmed that Ubc2 could be purified with the proteasome even when it was expressed at physiological levels (Fig. 2B). Longer exposures revealed high-molecular-mass derivatives that are a characteristic of multiubiquitylated proteins, an activity that has been previously ascribed to Ubc2 (30). We also compared the ubiquitylation of H2B by recombinant and proteasome-associated Ubc2. We first determined the amount of Ubc2 that could be recovered in association with the proteasome. Proteasomes were immunoprecipitated from ∼3 mg of cell extract from a strain that expressed PGAL1::UBC2 and Pre1-FLAG. We used antibodies against Ubc2 and estimated that ∼10 ng of Ubc2 was precipitated with Pre1-FLAG (Fig. 4C, lane 1), based on a reaction against different amounts of recombinant Ubc2 present on the same filter (lanes 3 to 5). We then examined the ubiquitylation of H2B with proteasome-associated Ubc2 that was isolated from 25 mg of cell extract (Fig. 4D). A comparable reaction with 250 ng of recombinant Ubc2 was also performed (Fig. 4D, lane 1). We found that both forms of Ubc2 conjugated Ub to H2B, although Ubc2 associated with the proteasome appeared to generate higher-molecular-mass multi-Ub conjugates (lane 2). The 32P-ubiquitylation of H2B by proteasome-associated Ubc2 was abolished in the absence of E1 (data not shown) and significantly reduced if excess unlabeled Ub was added to the reaction mixture (lane 3).
FIG. 4.
Ub-conjugating activity is detected in immunopurified proteasomes. (A) Protein extracts were prepared from galactose-grown cells that contained PGAL1::UBC2 (lanes 3, 4, 7, and 8) or a vector (lanes 1, 2, 5, and 6). The samples examined in the even-numbered lanes also contained Pre1-FLAG. Equal amounts of extract were incubated with FLAG-agarose, and the level of Ubc2 in the proteasome was determined with specific antibodies. Ubc2 was detected in the proteasome only in the strains that expressed Pre1-FLAG (lanes 6 and 8), although only the sample that contained overexpressed Ubc2 is visible in this reproduction (lane 8). (B) 32P-Ub was prepared as described previously (32), and reaction mixtures containing purified E1, 32P-Ub, and histone H2B were incubated with immunopurified proteasomes. The ubiquitylation reactions were terminated by the addition of SDS-containing gel-loading buffer, and the products were separated in an SDS-polyacrylamide gel and exposed to X-ray film. Lane 1 contains only 32P-Ub, and the contents of the various reaction mixtures are indicated. Significant ubiquitylation of H2B was detected in extracts that contained high levels of Ubc2 precipitated with Pre1-FLAG (lane 9). (C) We estimated the amount of Ubc2 that could be recovered with Pre1-FLAG from PHY211. Protein extracts (3 mg) were prepared from galactose-grown PHY211 and incubated with FLAG-agarose. The beads were washed, suspended in SDS sample buffer, and resolved by SDS-PAGE. We also separated various amounts of recombinant Ubc2 in the same gel (lanes 3 to 5). The resolved proteins were transferred to nitrocellulose and examined with antibodies against Ubc2. We recovered ∼10 ng of Ubc2 with Pre1-FLAG (lane 1). A low level of Ubc2 could also be detected in extracts prepared from noninduced cells (lane 2). (D) We examined the ubiquitylation of H2B by recombinant Ubc2 and proteasome-associated Ubc2. We prepared a standard reaction mixture that contained recombinant Ubc2 (250 ng), E1, 32P-Ub and histone H2B (lane 1). A similar reaction mixture containing proteasome-associated Ubc2 was prepared from ∼25 mg of cell extract that was derived from galactose-grown PHY211 (lane 2). An equivalent reaction mixture (containing proteasome-associated Ubc2) was supplemented with excess unlabeled Ub (lane 3). Proteasome-associated Ubc2 appeared to generate more high-molecular-mass 32P-Ub–H2B conjugates (lane 2) than did recombinant Ubc2 (lane 1). Significantly, the generation of multiply 32P-ubiquitylated H2B by proteasome-associated Ubc2 was strongly inhibited in the presence of excess unlabeled Ub.
DISCUSSION
We show in this report that Ub-conjugating (E2) enzymes can interact with the 26S proteasome. We also report that proteasome-associated Ubc2 can conjugate Ub to histone H2B. A recent study reported that H2B is a physiological substrate of Ubc2 in yeast (24) and has raised the interesting question whether H2B ubiquitylation in vivo is accomplished by proteasome-associated Ubc2. The presence of an E2 enzyme in the proteasome could allow the formation of a multi-Ub chain to be directly coupled to degradation. The assembly of multi-Ub chains at the proteasome could promote high-affinity substrate-proteasome interaction, consistent with the current model.
We found that the interaction between Ubc4 and the proteasome increased >10-fold under heat stress conditions. The extreme sensitivity of ubc4Δ ubc5Δ mutants to high temperature (25) suggests that the stress-induced Ubc4-proteasome interaction is biologically relevant. Overexpression of Ubc1, Ubc2, and Ubc4 resulted in higher levels in the proteasome, consistent with a direct interaction with the proteasome rather than with an association that is mediated by other factors. Furthermore, the interaction between Ubc4C86A and the proteasome suggests that catalytic activity may not be required for proteasome interaction and also raises the possibility that the ubiquitylation of some substrates can occur after interaction with the proteasome. Consequently, high levels of Ubc4C86A might inhibit proteasome function through a nonproductive interaction. However, it is equally plausible that Ubc4C86A binds the proteasome after its dimerization with a different E2 enzyme or another proteolytic factor. In addition, the toxicity caused by Ubc4C86A could be the result of an interaction with an important cellular factor and not with the proteasome.
To examine Ubc2- and Ubc4-proteasome interactions in greater detail, we determined if accessory factors were required. We found that the Ubc2-proteasome interaction was unaffected in rad18Δ and ubr1Δ mutants, which lack Ubc2-specific E3 factors (2, 4). This result is not surprising, because Rad18 and Ubr1 mediate only a subset of Ubc2-specific functions. It is possible that the interaction between Ubc2 and the proteasome might be regulated by DNA damage, similar to the heat-induced binding of Ubc4 with the proteasome (Fig. 3C). We also examined the Ubc4-proteasome interaction in strains with mutations of the UFD pathway, which is required for the degradation of a Ubc4-specific substrate (17). Ufd4 encodes an E3 factor, while Ufd2 (E4) promotes the formation of multi-Ub chains that are initiated by Ubc4 (19). However, the Ubc4-proteasome interaction was unaffected in ufd2, ufd4, and ufd5 mutants compared to the isogenic wild-type parental strain (data not shown). Although further study is required to demonstrate if E2 enzymes interact with the proteasome directly, an important conclusion of our results is that the proteasome can bind ubiquitin-conjugating enzymes. Whether an E2 enzyme binds the proteasome directly or as a component of a larger targeting ensemble does not affect this central finding.
The composition of the 19S regulatory particle of the yeast proteasome was recently defined, and E2 proteins were not detected (12). We used the purification method described by Glickman et al. (11) and found that Ubc4 copurified with the proteasome following chromatography in Blue-Sepharose but not after subsequent purification steps (11). Maximal peptidase (PGPH) activity (25) was detected in fractions that eluted between 70 and 160 mM NaCl and contained Rpt1 and Ubc4. However, when we combined and resolved these fractions by Q-Sepharose Fast Flow chromatography, both PGPH activity and Rpt1 separated from E2. Similarly, we examined the Ubc4-proteasome interaction by using a purification strategy described by Papa et al., who found that the Doa4 Ub-processing protease was present in the proteasome (21). We separated yeast extracts in fast protein liquid chromatography Mono-Q columns and detected a significant amount of Ubc4 in fractions that contained Rpt1 and peptidase activity, consistent with proteasome function. However, in subsequent chromatography steps, Ubc4 again dissociated from Pre1-FLAG and Rpt1, indicating a lower-affinity interaction. Thus, the failure to detect E2 proteins in the previous study (12) was probably due to substoichiometric levels of these enzymes in the proteasome. Furthermore, proteins smaller than ∼30 kDa (which includes most E2 enzymes) were not analyzed. The presence of Ubc4 in the proteasome complements the finding of Fujimuro et al. that Ufd5 copurified with the proteasome (10). Ufd5 is a component of the UFD targeting system that is required for the degradation of a Ubc4-specific substrate (17). Ufd5 was also not detected in purified preparations of the 19S regulatory particle (12), and it is not known if the other members of the UFD targeting system (Ufd1 to Ufd4) associate with the proteasome.
We speculate that proteasome-associated E2 enzymes might enhance proteolysis by preventing the disassembly of nascent multi-Ub chains by cellular Ubp proteins and by coupling multi-Ub chain assembly to proteolysis. These findings complement the existing model of substrate targeting (13, 14, 22, 26, 35) and offer a novel insight into the poorly understood mechanism of substrate translocation to the proteasome in vivo. An important implication of this study is that the targeting potential of the proteasome could be altered by controlling the repertoire of associated E2 enzymes. This idea is supported by the observation that the level of Ubc4 in the proteasome is influenced by heat stress, in agreement with its role in conferring stress resistance.
ACKNOWLEDGMENTS
This work was supported by grants to K.M. from the National Institutes of Health (GM52058), and the American Heart Association (9850170T). D.L. was supported by a Predoctoral Fellowship from the American Heart Association.
We thank members of the laboratory for helpful discussions and suggestions. M. Colon-Berlingeri and D. Rodriguez are thanked for their contributions during laboratory rotations. We thank J. Dohmen, M. Ellison, and M. Hochstrasser for strains, plasmids, and reagents.
ADDENDUM IN PROOF
While this paper was in review, Y. Xie and A. Varshavsky reported that E3 proteins could interact with the proteasome (Proc. Natl. Acad. Sci. USA 97:2497–2502, 2000).
REFERENCES
- 1.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 2.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 3.Barhite S, Thibault C, Miles M F. Phosducin-like protein (PhLP), a regulator of Gβγ function, interacts with the proteasomal protein SUG1. Biochim Biophys Acta. 1998;1402:95–101. doi: 10.1016/s0167-4889(97)00141-9. [DOI] [PubMed] [Google Scholar]
- 4.Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 6.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 7.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 8.Deveraux Q, van Nocker S, Mahaffey D, Vierstra R, Rechsteiner M. Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit S5a. J Biol Chem. 1995;270:29660–29663. doi: 10.1074/jbc.270.50.29660. [DOI] [PubMed] [Google Scholar]
- 9.Fujimuro M, Takada H, Saeki Y, Toh-e A, Tanaka K, Yokosawa H. Growth-dependent change of the 26S proteasome in budding yeast. Biochem Biophys Res Commun. 1998;251:818–823. doi: 10.1006/bbrc.1998.9560. [DOI] [PubMed] [Google Scholar]
- 10.Fujimuro M, Tanaka K, Yokosawa H, Toh-e A. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1998;423:149–154. doi: 10.1016/s0014-5793(98)00084-2. [DOI] [PubMed] [Google Scholar]
- 11.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–624. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 12.Glickman M H, Rubin D M, Fried V A, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershko A. The ubiquitin pathway for protein degradation. Trends Biochem Sci. 1991;16:265–268. doi: 10.1016/0968-0004(91)90101-z. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 15.Jentsch S, Seufert W, Sommer T, Reiss H. Ubiquitin-conjugating enzymes: Novel regulators of eukaryotic cells. Trends Biochem Sci. 1990;15:195. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- 16.Johnson E S, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson E S, Ma P C M, Ota I M, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser P, Moncollin V, Clarke D J, Watson M H, Bertolaer B L, Reed S I, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 20.Lam Y A, Xu W, DeMartino G N, Cohen R E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 21.Papa F R, Amerik A Y, Hochstrasser M. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 23.Ramos P C, Hockendorff J, Johnson E S, Varshavsky A, Dohmen R J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 24.Robzyk K, Recht J, Osley M A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 25.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 26.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 27.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 1991;10:2187–2193. doi: 10.1002/j.1460-2075.1991.tb07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung P, Prakash S, Prakash L. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc Natl Acad Sci USA. 1990;87:2695–2699. doi: 10.1073/pnas.87.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung P, Prakash S, Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988;2:1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- 31.Tongaonkar P, Madura K. Characterization of a temperature-sensitive mutant of a ubiquitin-conjugating enzyme and its use as a heat-inducible degradation signal. Anal Biochem. 1999;272:263–269. doi: 10.1006/abio.1999.4190. [DOI] [PubMed] [Google Scholar]
- 32.Tongaonkar P, Madura K. Reconstituting ubiquitination reactions with affinity-purified components and 32P-ubiquitin. Anal Biochem. 1998;260:307–319. doi: 10.1006/abio.1998.2697. [DOI] [PubMed] [Google Scholar]
- 33.van Nocker S, Deveraux Q, Rechsteiner M, Vierstra R D. Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc Natl Acad Sci USA. 1996;93:856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Nocker S, Sadis S, Rubin D M, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra R D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Chevray P M, Nathans D. Mammalian Sug1 and c-Fos in the nuclear 26S proteasome. Proc Natl Acad Sci USA. 1996;93:8236–8240. doi: 10.1073/pnas.93.16.8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson K. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 38.Zhu X, Craft C M. Interaction of Phosducin and Phosducin isoforms with a 26S proteasomal subunit, SUG1. Mol Vision. 1998;4:13. [PubMed] [Google Scholar]