Abstract
Soluble sugar is known to improve the sweetness and increase tomato sauce yield. Studies have focused on improving the content of soluble sugar in tomato fruits, usually by promoting functional genes. We studied two genes (SlINVINH1 and SlVPE5) that inhibited the accumulation of soluble sugar in tomato fruits and obtained two genes’ knocked-out lines (CRISPR-invinh1 or CRISPR-vpe5) using CRISPR/Cas9. Aggregated lines with CRISPR-invinh1 and CRISPR-vpe5 were gained by hybridization and self-pollination. Compared to wild-type lines, the glucose, fructose, and total soluble solid (TSS) contents of CRISPR-invinh1 and CRISPR-vpe5 increased significantly. Glucose, fructose, and TSS levels further improved simultaneously with CRISPR-invinh1 and CRISPR-vpe5 than with single gene knock-out lines. This indicates that these genes have a synergistic effect and will increase the soluble sugar content. Thus, the knock-out SlINVINH1 and SlVPE5 may provide a practical basis for improving the sweetness of tomato fruits and their processing quality.
Keywords: Soluble sugars, CRISPR/Cas9, Genome editing, Tomato fruits, Total soluble solids
Introduction
Tomato (Solanum lycopersicum) is a popular and important commercial vegetable crop worldwide. However, modern cultivating practices have been criticized for producing unsatisfactory fruit flavors. Sweetness, an important taste characteristic of tomato fruits, is mainly determined by their soluble sugar content (Mathieu et al., 2009; Tieman et al., 2017). Soluble sugars influence fruit sweetness. They are also the most important component of the total soluble solids (TSS), which affects the production costs of processed tomatoes and tomato sauce. Studies have reported that an increase in processed tomatoes’ TSS content from 4% to 5% could reduce raw material consumption by up to 25% when producing tomato sauce at a concentration of 28% (Gur & Zamir, 2015). Therefore, increasing the soluble sugar content in tomatoes improves fresh tomatoes’ sweetness and increases the efficiency of tomato sauce production.
Several factors directly influence the content of soluble sugars in tomato fruits, including genetic and environmental factors (including temperature, light, moisture, air, fertilizer, plant hormones). Technical factors such as cultivation and management play an indirect role through environmental factors (Beckles, 2012). However, genetic factors are fundamental and the most important. Although some studies have investigated genes related to soluble sugars, the corresponding biological functions of most of these genes are not fully understood (Lupi et al., 2019). This crop’s immense economic value makes it necessary to conduct in-depth research and exploration of these genes. It is also necessary to combine this information with molecular marker-assisted selection or genetic engineering techniques to increase the soluble sugar content in cultivated tomatoes (Gur & Zamir, 2015; Vallarino et al., 2017). Understanding related genes aids the analysis of the molecular mechanisms for the accumulation of soluble sugars in tomato. This knowledge can also help us better understand other fruit types, which has substantial theoretical significance.
Plants have a set of complex metabolic mechanisms for sugars, including the sensing of sugar signals, sugar synthesis, loading, transportation, unloading, transformation, storage, accumulation, and the regulation of several physiological and biochemical processes. Several studies have focused on enzymes and proteins related to the processes of sugar synthesis, transportation, and decomposition. They include invertase, sucrose synthase (SUS), sucrose phosphate synthase (SPS), hexose kinases (Hexokinase, HXK), fructokinases (FRKs), sucrose phosphatase (Sucrose Phosphate Phosphatase, SPP), sugar transporters, and some transcription factors (Beckles et al., 2012; Patrick, Botha & Birch, 2013). These studies have facilitated a deeper understanding of the molecular mechanisms of sugar accumulation.
Invertase has a significant effect on sugar accumulation. Beets (Beta vulgaris) typically accumulate sucrose, while tomatoes largely accumulate hexose. The activity of insoluble acid invertase, also called cell wall invertase, decreases significantly during the early stages of sugar accumulation in beets. However, in tomatoes acid invertase activity increased significantly at the same stage (Patrick, Botha & Birch, 2013). A cloned QTL (Brix9–2–5) is involved the accumulation of soluble sugar is the Lin5 gene. This gene is a cell wall invertase gene and is the key enzyme affecting the unload of sugar in tomato fruits. The soluble sugar content in the introgression line (IL) containing this site is significantly higher than in the IL without this site (Baxter et al., 2005). Silencing Lin5 reduced the concentration of soluble sugar in the fruit (Zanor et al., 2009). Sugar accumulation can also be regulated by controlling the metabolic flow. Sucrose accumulation in sugarcane can generate feedback inhibition. When sucrose is converted to fructose and glucose is promoted by transgenic manipulations, it increases the demand for carbon and reduces the feedback inhibition of sucrose. This increases the leaf photosynthesis rate and elevates the total sugar content (Wu & Birch, 2007). Manipulation of the genes regulating sucrose isomerase (SI) and ADP-glucose pyrophosphorylase (AGPase) promotes the conversion of sucrose to its isomers and starch. It also increases the sugar content of transgenic tomatoes (Petreikov et al., 2009). Some transcription factors are also involved in regulating the metabolism of fruit sugars. For example, the transcription factor RIN, which is related to fruit maturation, can directly bind to the promoter regions of the vacuolar invertase (SlVI) gene and vacuolar invertase inhibitor (SlVIF) gene to regulate their expression. This affects sugar metabolism and maturation (Qin et al., 2016). ABA (abscisic acid) is essential for the accumulation of sugar in tomato fruits (Li et al., 2019). It induces the expression of the NAC transcription factor gene SlNAP2. This in turn can regulate ABA synthesis and influence tomato senescence further influencing sugar metabolism. Transgenic tomatoes with RNA interference exhibited a delayed senescence of leaves, increased yield, and higher content of soluble sugar in fruits (Ma et al., 2018). The overexpression of SlGLK turns the fruit color dark green and significantly increases the starch, sugar, and TSS content. This suggests that there may be a relationship between increased fruit chlorophyll content and enhanced photosynthetic efficiency (Powell et al., 2012).
The gene INVINH1 is an inhibitor of the acid invertase gene Lin5, which specifically inhibits cell wall invertase activity. Studies have reported that silencing the invertase inhibitor corresponding to the Lin5 gene can increase invertase activity. This also significantly increases soluble sugar and TSS content in the fruit (Jin, Ni & Ruan, 2009; Ruan, Jin & Huang, 2009). The gene SlVPE5 belongs to the VPE (vacuolar processing enzyme) family of genes. In this family, 15 VPE encoding genes are present in the tomato genome (Matarasso, Schuster & Avni, 2005; Aoki et al., 2010; Ariizumi et al., 2011; Wang et al., 2016, 2017; Teper-Bamnolker et al., 2017). SlVPE5 negatively regulates sugar accumulation (Ariizumi et al., 2011; Wang et al., 2016). However, the mechanism by which INVINH1 and SlVPE5 negatively regulate sugar accumulation has not been determined, and the interactive relationship between INVINH1 and SlVPE5 has not been studied.
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 9 (Cas9) genome editing system is a powerful tool for targeted gene modifications in plants. The first publication of the five CRISPR/Cas9 gene editing literatures in plants (Feng et al., 2013; Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013; Xie & Yang, 2013) led to explosive growth in this field and enabled the creation of genetic variations. Variations had previously been made randomly by physical radiation and chemical mutagens. However, this method also introduced precise genomic modifications such as deletion, insertion, inversion, and base edits of specific DNA positions in plant genomes. Scientists and plant breeders have the potential to revolutionize crop improvement by developing new plant breeding techniques (NPBTs) (Shao, Punt & Wesseler, 2018; Shan-e-Ali Zaidi et al., 2019) and usher in a new era of precise and efficient crop improvement.
We analyzed the structural types and genetic relationship of two genes (INVINH1 and SlVPE5) that could inhibit the accumulation of soluble sugar with the goal of increasing soluble sugar content in tomato fruits. Using CRISPR/Cas9 technology, both genes were knocked out separately to obtain mutants (the mutants were named CRISPR-invinh1 or CRISPR-vpe5) with loss of function in each gene. We then analyzed the influence of each gene on glucose, fructose, and TSS during fruit ripening. Through hybridization and self-pollination, we obtained lines with loss of function in both genes. We further explained the regulatory relationship between these genes and the accumulation of soluble sugar and provided significant theoretical guidance for improving tomato fruit sweetness and trait processing. Our research will aid in the accelerated introgression of a single gene, introduce some elite genes into the breeding lines, and provide a high-efficiency and promising strategy for plant breeding.
Materials & methods
Plant material and growth conditions
We use cultivated tomato M82 as the experimental material. The tomatoes were grown in a greenhouse at the seedling stage (environmental conditions: 14 h light/10 h dark, daytime temperature of 25 °C, night temperature of 18 °C, and daily automatic irrigation by sprinkler equipment). The seedlings were transplanted to the field before entering the flowering stage. The management of the test field was the same as that of the general field with drip irrigation once a week, nitrogen as the main fertilizer at the seedling stage, and P and K as the main fertilizer at the reproductive stage. The test field was located at the comprehensive testing field (87°47′E, 43°95′N) of Xinjiang Academy of Agricultural Sciences in Urumqi, China.
Sample preparation
The genotypes of the transgenic seedlings and wild-type seedlings were identified when the plants grew stronger, with at least four leaves, and genomic DNA from the leaves was extracted for the detection of mutations. We tested for soluble sugar and TSS content in the fruit when the fruit was red and ripe (about 50 days post anthesis). At least six uniform fruits (two from each replicate with three replications) were collected from the second and third spikes of each plant, crushed, and homogenized into a sauce. The sauce was then analyzed for soluble sugar and TSS content.
Phylogenetic analysis
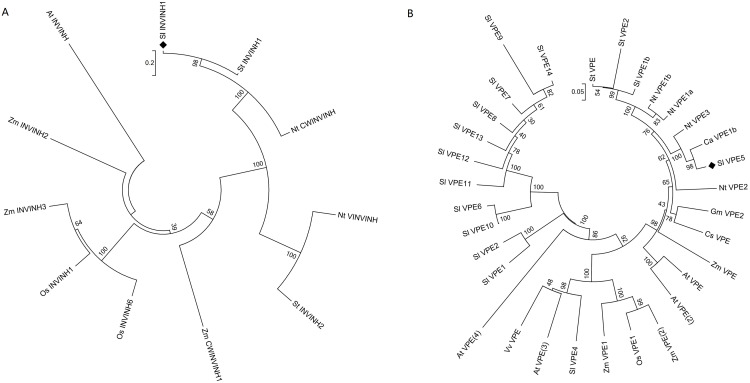
The protein sequences of INVINH and VPE from tomato (Solanum lycopersicum), Arabidopsis thaliana, pepper (Capsicum annuum), citrus (Citrus sinensis), soybean (Glycine max), tobacco (Nicotiana tabacum), rice (Oryza sativa), potato (Solanum tuberosum), grape (Vitis vinifera), and maize (Zea mays) were collected using the BLAST database search tool at NCBI (Fig. 1). The following protein sequences were downloaded from GenBank or SGN: Sl INVINH1, accession number AJ010943; Os INVINH1, AK288558; Os INVINH6, AK110798; Zm CWINVINH1, XP_008668976; Zm INVINH2, AX214357; ZM INVINH3, AX214336; At INVINH, AT1G48010; NT VINVINH, AY594179; NT CWINVINH, Y12805; St INVINH1, JQ043180; St INVINH2, GU980594; St VPE, XP_006355320; St VPE2, ACC68681; Nt VPE1b, BAC54828; NtVPE1a,BAC54827; NtVPE3, BAC54830; Nt VPE2, BAC54829; Gm VPE2, NP_001236564; Cs VPE, NP_001275777; Zm VPE, ACG34144; Zm VPE1, AAL58571; Zm VPE2, ACG47915; At VPE, NP_195020; At VPE2, NP_180165; At VPE3, NP_176458; At VPE4, NP_001189938; Vv VPE,AGC54786; Os VPE1, XP_015636414; Ca VPE1b, CA12g18500; Sl VPE1b, Solyc08g065610; Sl VPE5, Solyc12g095910; Sl VPE4, Solyc08g079160; Sl VPE1, Solyc08g065790; Sl VPE2, Solyc08g065780; Sl VPE10, Solyc08g065690; Sl VPE6, Solyc08g065530; Sl VPE11, Solyc08g065710; Sl VPE12, Solyc08g065720; Sl VPE8, Solyc08g065590; Sl VPE13, Solyc08g065740; Sl VPE7, Solyc08g065570; Sl VPE9, Solyc08g065600; Sl VPE14, Solyc08g065750. Multiple alignments of the INVINH and VPE protein sequences were processed using MEGA7 (Kumar, Stecher & Tamura, 2016). A phylogenetic tree was constructed with the neighbor-joining statistical method using 1,000 bootstrap replicates (Kumar, Stecher & Tamura, 2016).
Figure 1. Phylogenetic analysis of INVINH and VPE proteins in plants.
(A) Phylogenetic tree of INVINH. (B) Phylogenetic tree of VPE. At, Arabidopsis thaliana; Ca, Capsicum annuum; Cs, Citrus sinensis; Gm, Glycine max; Nt, Nicotiana tabacum; Os, Oryza sativa; Sl, Solanum lycopersicum; St, Solanum tuberosum; Vv, Vitis vinifera; Zm, Zea mays. The size bar shows the estimated evolutionary distance.
Design of sgRNA and construction of CRISPR/Cas9 vector
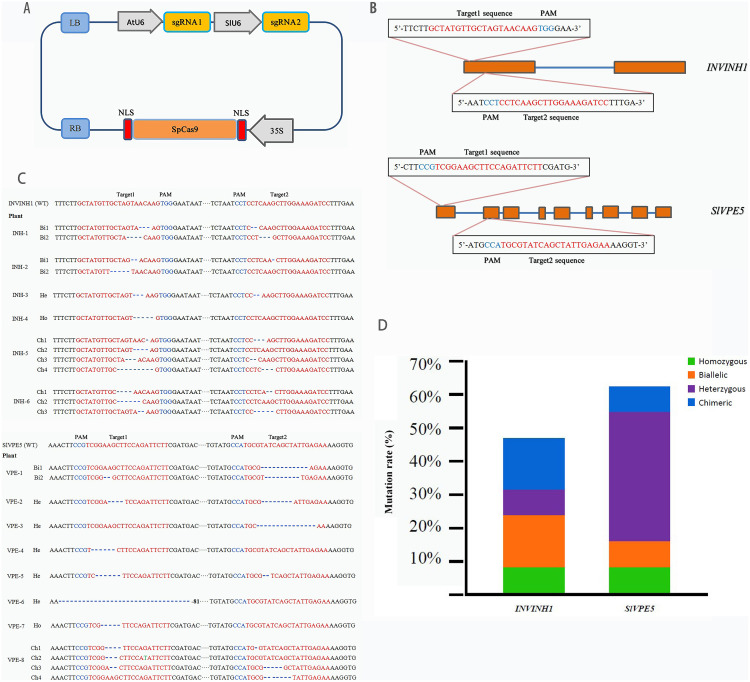
CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) was utilized to design the target sites of the target genes SlINVINH1 (Solyc12g099200.1) and SlVPE5 (Solyc12g095910.1). Each gene was provided with two target sites for a more efficient gene knockout. SlINVINH1 target sites were selected in the first exon, and SlVPE5 target sites were selected in the first and second exon (Fig. 2B). The CRISPR/Cas9 original vector was obtained from Prof. Cai-xia Gao (Chinese Academy of Science, Beijing, China). The optimization of CRISPR/Cas9 gene codon and the synthesis of sgRNA sequence was provided by the Biological Company of Hong Xun, Suzhou, China. We used the construction method described by Yu et al. (2017), adding the 35S promoter driving CRISPR/Cas9 gene, the U6 promoter of Aribidopsis thaliana, and tomato control the sgRNA1 and sgRNA2, separately. The cloned fragments were assembled using the Circular Polymerase Extension Cloning (CPEC) method (Quan & Tian, 2009). The pCAMBIA1301 binary vector (AtU6-sgRNA1-SlU6-sgRNA2-35S-Cas9) was constructed and used for gene knockout (Fig. 2A).
Figure 2. Target genes editing by CRISPR/Cas9.
(A) The schematic description of CRISPR/Cas9-sgRNA expression cassette. SpCas9 is controlled by a CaMV 35S promoter, the first sgRNA is expressed by the AtU6 promoter, the second sgRNA is expressed by the SlU6 promoter. (B) Four target sites were designed in two target genes. Straight blue lines and rectangular orange boxes are the introns and exons of the target genes, respectively. (C) The sequenced targeted InDel mutation of two target gene. A total of 26 clones of the each PCR amplicon were picked and sequenced. “WT” means Wild-type, “Bi” means Biallele, “He” means Heterozygote, “Ho” means Homozygote, “Ch” means Chimera. (D) Specific types of each target gene in T0 lines. Green, orange, purple, and blue represent homozygous, biallelic, heterozygous, and chimeric mutations, respectively.
Plant transformation
We used the Agrobacterium-mediated transformation method as described previously (Van Eck, Kirk & Walmsley, 2006) to transform pCAMBIA1301 vectors containing the CRISPR/Cas9 and sgRNA cassette into M82. Tomato seeds were germinated after sterilization with 10% NaClO on ½ MS medium. The apical segments of hypocotyls were punctured with Agrobacterium suspension after 9 to 12 days of culturing (OD600 = 0.5–0.6). The explants were then inoculated on a selective medium containing three mg/L hygromycin until the transgenic plants were regenerated from the callus. After the plants were grown in vitro, they were transplanted in a pot with soil and placed in light growth chamber.
DNA extraction and mutation detection
Genomic DNA from fresh frozen leaves was extracted using a high-efficiency plant genome DNA extraction kit (Tiangen, Beijing, China). Specific primers were used to amplify the flanks of genomes containing the target site (Table S1) and the PCR products were isolated by 1% agarose. The PCR products were linked to the pZERO-T vector after cutting and purifying (Transgen, Beijing, China), and 26 positive clones were randomly selected for mutagenesis from each plant. The Sanger method was used for sequencing with M13 primers. We obtains T1 and T2 lines for genotyping analysis by strict self-pollination of the T0 plants. The target fragments were directly sequenced. The genotyping of F2 plants was obtained from the F1 lines using strict self-pollination. The F1 lines were obtained from the two cross-pollinated T0 lines. Their target fragments were also directly sequenced.
Determinations of soluble sugars, TSS and lycopene
Soluble sugars were determined as described previously by Freschi et al. (2010). Glucose, fructose, and sucrose levels were determined using high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD; Dionex, Sunnyvale, CA, USA). We used a Carbopac PA1 column (250 × 4 mm, five μm particle size, Dionex, Sunnyvale, CA, USA) as an isocratic run with 18 mM NaOH as the mobile phase. A standard curve made with pure glucose, fructose, and sucrose was used to calculate sugar content. TSS were measured with a refractometer DR201-95 (Kruess, Germany).
For determination of lycopene, pigments were extracted from two g of fresh ripe fruits using acetone/petroleum ether (1:1, v/v), then dried under a stream of N2 and dissolved in 100% dichloromethane. The HPLC analysis was performed with 10 μL dichloromethane-dissolved pigments on ACQUITY UPLC (Waters, Milford, MA, USA). Lycopene was identified by the characteristic absorption spectra (472 nm) and distinctive retention times, compared with the standard substance (Sigma, St. Louis, MO, USA). Each lycopene content was calculated through the linear regression equation generated from the corresponding calibration curve, which was made using the corresponding standard substance.
Each sample contained three replicates with two fruits per replicate.
Results
Characteristics and phylogenetic analysis of genes
The invertase inhibitor (INVINH1) protein consists of 171 amino acid residues, with a signal peptide of 19 amino acid residues at the N-terminus. The differences between INVINH1 and another type of invertase inhibitor in tomato, SolyCIF, include distinctions in protein sequences and protein structures (Reca et al., 2008). Compared with the sequence of the INVINH1 protein in other crops, four cysteine residues were conserved, which was a significant feature of all invertase inhibitors (Rausch & Greiner, 2004). Cluster analysis revealed that the INVINH1 protein of tomato had the closest relationship with that of potato and tobacco, which are also solanaceous crops, but was distantly related to Arabidopsis, corn, and rice (Fig. 1A).
The vacuolar processing enzyme (VPE) has been classified in the cysteine protease family and is mainly involved in the regulation of post-translational levels of proteins in vacuoles. The VPE proteins are highly conserved in most plants and animals (Hara-Nishimura, Takeuchi & Nishimura, 1993). Among the numerous VPE proteins in tomatoes, SlVPE3 and SlVPE5 have been reported to regulate the accumulation of soluble sugars. The soluble sugar content was higher in RNA interference lines (RNAi-SlVPE3 and RNAi-SlVPE5) compared with the control plants. However, the soluble sugar content of RNAi-SlVPE5 line was significantly higher than that of RNAi-SlVPE3 line. Therefore, only SlVPE5 was chosen as the target gene for genome editing in this study (Ariizumi et al., 2011). Cluster analysis revealed that SlVPE3 and SlVPE5, which are involved in sugar metabolism, did not cluster with other tomato VPE proteases but were rather the most closely related to tobacco (Nicotiana tabacum), the NtVPE3 protein, and the sweet pepper CaVPE1b protein (Fig. 1B). This suggests that there may be a class of proteases among the VPE family proteins that specifically regulate the accumulation of sugars.
Characterization of targeted editing in transgenic plants
The transformation system of the SlINVINH1 gene knock-out finally yielded 14 T0 generation lines positive for the Cas9 gene. Genomic DNA was extracted from the leaves of each line. PCR using primers designed for the target region was then conducted to amplify the mutant region fragments, and each clone of the amplified products was sequenced and analyzed to obtain information on mutations in the target region of the INVINH1 gene. The sequencing results demonstrated that among the 14 lines that were positive for Cas9, only six had a mutation in the target sequence of the SlINVINH1 gene (probability of mutation = 42.86%). Among the mutants, one line was a homozygous mutant (both alleles had the same type of mutation), two were biallelic mutants (both alleles were mutated, but with different mutations), one was a heterozygous mutant (only one allele was mutated), and two were chimeric mutants (only some of the cells are mutated) (Figs. 2C and 2D; Table S2). Detailed mutations in the target sequence were mainly the deletions of single or multiple nucleotides (Fig. 2C).
The transformation system of the SlVPE5 gene knock-out yielded 13 T0 generation lines that were positive for the Cas9 gene. The results of mutation detection revealed that among the 13 Cas9-positive lines, eight lines had mutations in the target sequence of the SlVPE5 gene (probability of mutation = 61.54%). Among the mutants, one line was a homozygous mutant, another line was a biallelic mutant, five lines were heterozygous mutants, and one line was a chimeric mutant (Figs. 2C and 2D; Table S2). Detailed mutations in the target region were mainly the deletion and replacement of single or multiple nucleotides (Fig. 2C). Further analysis revealed that the mutation types of the SlINVINH1 gene were mainly biallelic mutations and chimeras, while the mutation type of the SlVPE5 gene was mainly heterozygotic (Figs. 2C and 2D; Table S2).
CRISPR/Cas9-mediated mutants exhibited soluble sugar and TSS content increase
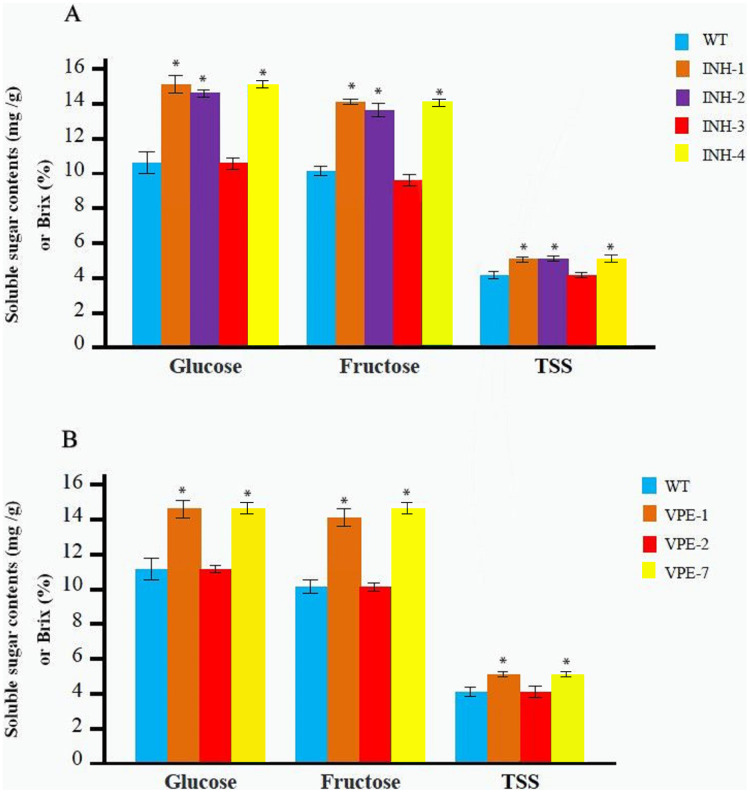
The T0 mutant line was obtained as described above. Soluble sugar and TSS contents were determined after maturation with three replicates for each line. Among the T0 mutant lines of the SlINVINH1 gene four lines (INH-1, INH-2, INH-3, and INH-4) (Fig. 2C) were selected. The INH-5 and INH-6 lines were not selected as they were chimeras and displayed an extremely low probability of passing the mutations on to the next generation. We observed that the contents of both glucose and fructose in the fruits from lines INH-1, INH-2, and INH-4 were significantly increased compared to the wild-type. INH-1 showed a 40.19% increase in glucose and a 42.42% increase in fructose; INH-2 showed a 36.39% increase in glucose and a 35.69% increase in fructose; INH-4 showed a 40.82% increase in glucose and a 42.76% increase in fructose (Fig. 3A; Table S3). In addition, the TSS content also showed a significant increase (31.90% in INH-1, 30.17% in INH-2, 32.76% in INH-4) (Fig. 3A; Table S3), indicating that the SlINVINH1 gene mainly regulated the accumulation of glucose and fructose in the fruits. However, the content of soluble sugars in the fruit of heterozygous INH-3 did not increase significantly compared to the wild-type (Fig. 3A; Table S3), which could be due to the recessive function of the mutated slinvinh1 gene.
Figure 3. Determination of the soluble sugar and TSS content in red fruit from different T0 lines and WT.
(A) The contents of the soluble sugar and TSS of red fruit in four different T0 lines of editing SlINVINH1 gene and WT. (B) The contents of the soluble sugar and TSS of red fruit in three different T0 lines of editing SlVPE5 gene and WT. The contents of glucose and fructose is measured in mg/g in fruit weigh, and the TSS is measured in Brix (%). *(P < 0.05, Student’s t-test, n = 3) indicate statistically significant differences between T0 mutant lines and wild-type.
Among the T0 lines that carried the mutations of the SlVPE5 gene, three lines, VPE-1, VPE-2, and VPE-7, were selected. The VPE-8 line was not selected as it was chimera and showed an extremely low probability of passing the mutations to the next generation. Among the five heterozygous, only one line, namely VPE-2, was selected as a representative for testing. The test revealed that the glucose and fructose content in the lines fruits of VPE-1 and VPE-7 increased significantly (glucose increased 35.20% in VPE-1, and increased 35.83% in VPE-7; fructose increased 37.33% in VPE-1, and increased 43.00% in VPE-7). The TSS content also increased significantly (30.63% in VPE-1, 32.43% in VPE-7) (Fig. 3B; Table S3), indicating that the SlVPE5 gene also regulated the accumulation of glucose and fructose in fruits. However, the soluble sugar content in the fruits of heterozygous INH-2 did not increase significantly compared to the wild-type (Fig. 3B; Table S3), which might also be caused by the recessive gene characteristics of the mutant slvpe5 gene.
The double homozygous mutants further increased the soluble sugar and TSS content
The T0 generation homozygous mutant lines INH-4 and VPE-7 were cross-pollinated to generate the F1 generation. The F2 generation was obtained by self-pollination of the F1 generation, and 515 well-grown lines were obtained after sowing the F2 generation seeds. We screened for the gene segregation and obtained 32 lines with homozygous mutated SlINVINH1 and SlVPE5 sites (genotype: inh/inh-vpe/vpe) from 515 lines. We identified two lines without the exogenous Cas9 genes by further screening for the exogenous Cas9 genes among the 32 lines. The two lines were numbered F2–1 and F2–2.
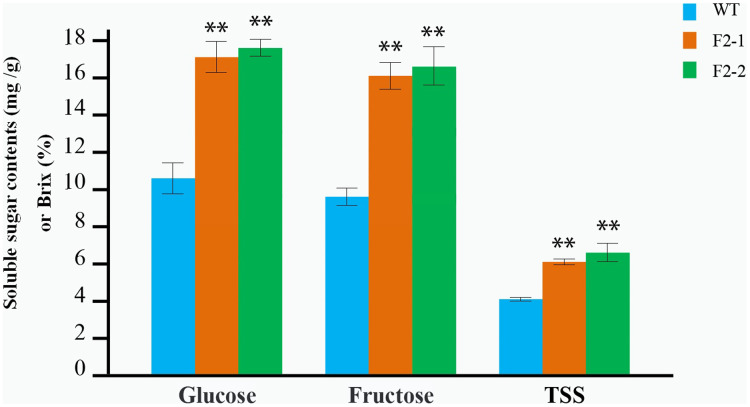
The soluble sugar and TSS in fruit from the F2–1 and F2–2 lines were determined after maturation, each line with three replicates. Glucose and fructose content in F2–1 and F2–2 fruits significantly increased compared to the wild-type. In F2–1, glucose increased by 64.86%, and fructose increased by 68.40%, while in F2–2, glucose increased by 67.41%, and fructose increased by 69.44% (Fig. 4; Table S4). The TSS content also showed a significant increase (55.17% in F2–1 and 62.07% in F2–2) (Fig. 4; Table S4).
Figure 4. Determination of the soluble sugar and TSS content in red fruit from the WT and the two F2 lines of double homozygous mutants of edited SlINVINH1 and SlVPE5 genes.
The contents of glucose and fructose is measured in mg/g in fruit weigh, and the TSS is measured in Brix (%). **(P < 0.01, Student’s t-test, n = 3) indicate statistically highly significant differences between F2 lines and wild-type.
We observed that both the SlINVINH1 and SlVPE5 gene mainly regulated the accumulation of glucose and fructose in the fruit. In addition, both glucose and fructose content was significantly synergistically increased in the F2–1 and F2–2 fruit (compared to the average of INH-4 and VPE-7 lines, glucose increased by an average of 18.12% in two F2 lines; and fructose increased by 14.06%) (Table S5). The TSS content was also significantly higher than the single locus-mutated homozygote (which increased by 22.31%) (Table S5). This indicated that the SlINVINH1 and SlVPE5 genes displayed a synergistic effect in regulating the content of soluble sugar.
Dynamic variation patterns of fruit development and coloration
SlINVINH1 is an invertase inhibitor gene and the loss of invertase function may delay fruit ripening. However, the loss of SlVPE5 gene function may also cause changes in other commercial traits like the size and color of fruits. We followed the development of two lines (F2–1 and F2–2), starting from flowering to fruit ripening, in real time. The results indicated that tomatoes of the F2–1 and F2–2 lines were similar to the wild-type tomato fruits, starting from flowering to the physiological fruit expansion stage, and including both shape and size of the fruits. However, during the breaker stage and the beginning of the ripping stage, fruits of the F2–1 and F2–2 lines were significantly delayed the break stage compared to the wild-type fruits, although they were fully ripened at the same time as wild-type fruits (Fig. 5A). This indicated that although the knock-out of the SlINVINH1 and SlVPE5 genes did delay the break stage and the color change of tomato fruits, it neither affected the ripening and harvesting time of the fruits nor the color of fruits after ripening nor did it affect fruit weight (Fig. 5B; Table 1).
Figure 5. Phenotypic detection of the WT fruits and the two F2 lines fruits.
(A) Developmental series of WT fruits (up) and F2 fruits (down), “dpa” is days post anthesis. (B) Comparison of bisected fruit at the ripening stage. Bars = one cm.
Table 1. Comparison of lycopene content and weight of red fruit between WT and F2 lines.
| Lines | Lycopene mg/100 g fw | Weight g | ||||
|---|---|---|---|---|---|---|
| Rep | Avg | P-value | Rep | Avg | P-value | |
| WT | 12.8 | 13.37 ± 0.54 | 79.8 | 80.40 ± 2.24 | ||
| 13.2 | 83.4 | |||||
| 14.1 | 78.0 | |||||
| F2-1 | 13.0 | 13.07 ± 0.33 | 0.741 | 80.2 | 78.73 ± 1.04 | 0.751 |
| 13.5 | 77.9 | |||||
| 12.7 | 78.1 | |||||
| F2-2 | 14.2 | 13.63 ± 0.45 | 0.786 | 83.8 | 81.03 ± 2.98 | 0.956 |
| 13.1 | 76.9 | |||||
| 13.6 | 82.4 | |||||
Note:
WT, Wild-type; fw, fruit weight; Rep, Three test replicates; Avg, Average.
Discussion
The effects of SlINVINH1 and SlVPE5 in increasing the soluble sugar and the TSS content of tomato
SlINVINH1 is an invertase inhibitor gene, which specifically inhibits the activity of cell wall invertase. Invertase hydrolyzes sucrose into glucose and fructose and plays a major role in fruit development. The suppression of SlINVINH1 gene expression increases the glucose and fructose content in fruits (Sturm, 1999; Jin, Ni & Ruan, 2009). SlVPE5 belongs to the family of VPE genes, which targets various types of vacuolar hydrolases (such as β-glycosidase, α-mannnosidases, and α-galactosidases) for degradation and the negative regulatory mechanism for soluble sugar accumulation (Rojo et al., 2003; Ariizumi et al., 2011). We observed that the both genes SlINVINH1 and SlVPE5 mainly regulated the accumulation of glucose and fructose in fruits (Figs. 3A, 3B, and 4), confirming the findings of previous research (Jin, Ni & Ruan, 2009; Ariizumi et al., 2011; Xu et al., 2017). Previous research also found that the SlVPE5 gene also regulates the accumulation of sucrose in fruits (Wang et al., 2016). However, in this study no sucrose content was detected in mature fruits (detection threshold was ≥0.2 mg/g fw) in the T0 or F2 lines. This may be due to the low sucrose content in mature fruits and because the sucrose contribution to the TSS of mature fruits is very small (Tieman et al., 2017). The knock-out of both loci in homozygotes resulted in a significant increase in both tested sugars in the fruit, and the TSS content was also significantly higher than that in single locus-mutated homozygotes, indicating that there was a synergistic effect of SlINVINH1 and SlVPE5 genes in regulating the content of soluble sugar. This proves once again that the increase of soluble sugar content in fruits is a complex process and not regulated by a single gene (Beckles et al., 2012; Patrick, Botha & Birch, 2013). This finding is crucial for the improvement of quantitative genetic traits of soluble sugar.
The knockout efficiency of two genes
The knock-out efficiency of both genes were different. In the knock-out experiment on SlINVINH1, we obtained six lines with the successful knock-out of the target gene from 14 positive lines. Its knock-out efficiency was 42.86%. In the knock-out experiment on SlVPE5, eight lines with the successful knock-out of the target gene were obtained from 13 positive lines, with a knock-out efficiency of 61.54%. Furthermore, the mutation types of the SlINVINH1 gene were mainly biallelic mutations and chimeras, while the mutation types of the SlVPE5 gene were mainly heterozygous (Figs. 2C and 2D; Table S2). This may be linked to the different types and structures of the genes (Pan et al., 2016; Li et al., 2018). According to previous research vector compontents, such as codon optimization of CRISPR/Cas9 gene, promoter, and structure of gRNA, may affect gene knockout efficiency (Dang et al., 2015; Ma et al., 2015; Čermák et al., 2017; Feike et al., 2019; Shao et al., 2020; Grützner et al., 2021).
Effective improvements to the commercial traits of tomato
We obtained two homozygous lines (F2–1 and F2–2) with mutations in both the functional gene loci, excluding exogenous Cas9, and found that both the soluble sugar and TSS content in tomato fruits were significantly increased (Fig. 4), facilitating the rapid improvement of the commercial traits of tomatoes. Our study confirmed the feasibility of target gene editing by the transfer of Cas9 and sgRNA, and the subsequent application of conventional methods such as hybridization and inbreeding. Two mutation sites may be brought together, however, the mutation site could be made homozygous and the exogenous gene Cas9 could be eradicated. This method greatly shortened the breeding cycle to improve tomato varieties. It also completely eliminated various risk factors associated with the exogenous genes. This system will improve technological innovation and progress in tomato breeding, and has tremendous potential in a variety of applications.
Conclusions
The purpose of this study was to further elucidate two tomato genes (SlINVINH1 and SlVPE5) that inhibited the accumulation of soluble sugars in tomato fruits. We also aimed to obtain the knock-out lines of two genes using CRISPR/Cas9. We determined the aggregated lines with both CRISPR-invinh1 and CRISPR-vpe5 using hybridization and self-pollination. The glucose, fructose, and TSS content of CRISPR-invinh1 and CRISPR-vpe5 were significantly increased compared to wild-type tomato lines. In addition, the levels of glucose, fructose, and TSS further improved in the lines with CRISPR-invinh1 and CRISPR-vpe5 simultaneously versus that of the single gene knock-out lines. This result indicated that the two genes had a synergistic effect in increasing the content of these soluble sugars. Thus, the knock-outs, SlINVINH1 and SlVPE5, may effectively increase the content of soluble sugars in tomato and provide an important theoretical and practical basis for improving the sweetness of tomato fruits and their processing quality.
Supplemental Information
Funding Statement
The work was funded by the National Natural Science Foundation of China (31860555, 31760581), the Special Incubation Project of Science & Technology Renovation of Xinjiang Academy of Agricultural Sciences (xjkcpy-2021001), and the National Key R&D Program of China (2017YFD0101906). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Juan Wang, Email: 278089648@qq.com.
Qinghui Yu, Email: yuqinghui98@sina.com.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Baike Wang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Ning Li performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Shaoyong Huang performed the experiments, prepared figures and/or tables, and approved the final draft.
Jiahui Hu performed the experiments, prepared figures and/or tables, and approved the final draft.
Qiang Wang performed the experiments, prepared figures and/or tables, and approved the final draft.
Yaping Tang performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Tao Yang performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Patiguli Asmutola performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Juan Wang conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Qinghui Yu conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.
References
- Aoki et al. (2010).Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Suda K, Kurabayashi A, Suzuki T, Tsugane T, Watanabe M, Ooga K, Torii M, Narita T, Shin-i T, Kohara Y, Yamamoto N, Takahashi H, Watanabe Y, Egusa M, Kodama M, Ichinose Y, Kikuchi M, Fukushima S, Okabe A, Arie T, Sato Y, Yazawa K, Satoh S, Omura T, Ezura H, Shibata D. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics. 2010;11(1):210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi et al. (2011).Ariizumi T, Higuchi K, Arakaki S, Sano T, Asamizu E, Ezura H. Genetic suppression analysis in novel vacuolar processing enzymes reveals their roles in controlling sugar accumulation in tomato fruits. Journal of Experimental Botany. 2011;62(8):2773–2786. doi: 10.1093/jxb/erq451. [DOI] [PubMed] [Google Scholar]
- Baxter et al. (2005).Baxter CJ, Carrari F, Bauke A, Overy S, Hill SA, Quick PW, Fernie AR, Sweetlove LJ. Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant and Cell Physiology. 2005;46(3):425–437. doi: 10.1093/pcp/pci040. [DOI] [PubMed] [Google Scholar]
- Beckles (2012).Beckles DM. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biology and Technology. 2012;63(1):129–140. doi: 10.1016/j.postharvbio.2011.05.016. [DOI] [Google Scholar]
- Beckles et al. (2012).Beckles DM, Hong N, Stamova L, Luengwilai K. Biochemical factors contributing to tomato fruit sugar content: a review. Fruits. 2012;67(1):49–64. doi: 10.1051/fruits/2011066. [DOI] [Google Scholar]
- Dang et al. (2015).Dang Y, Jia G, Choi J, Ma H, Anaya E, Ye C, Shankar P, Wu H. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biology. 2015;16(1):823. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feike et al. (2019).Feike D, Korolev AV, Soumpourou E, Murakami E, Reid D, Breakspear A, Rogers C, Radutoiu S, Stougaard J, Harwood WA, Oldroyd GED, Miller JB. Characterizing standard genetic parts and establishing common principles for engineering legume and cereal roots. Plant Biotechnology Journal. 2019;17(12):2234–2245. doi: 10.1111/pbi.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng et al. (2013).Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK. Efficient genome editing in plants using a CRISPR/Cas system. Cell Research. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freschi et al. (2010).Freschi L, Rodrigues MA, Tiné MAS, Mercier H. Correlation between citric acid and nitrate metabolisms during CAM cycle in the atmospheric bromeliad Tillandsia pohliana. Journal of Plant Physiology. 2010;167(18):1577–1583. doi: 10.1016/j.jplph.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Grützner et al. (2021).Grützner R, Martin P, Horn C, Mortensen S, Cram EJ, Lee-Parsons CWT, Stuttmann J, Marillonnet S. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Communications. 2021;2(2):100135. doi: 10.1016/j.xplc.2020.100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur & Zamir (2015).Gur A, Zamir D. Mendelizing all components of a pyramid of three yield QTL in Tomato. Frontiers in Plant Science. 2015;6(64358):344. doi: 10.3389/fpls.2015.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura, Takeuchi & Nishimura (1993).Hara-Nishimura I, Takeuchi Y, Nishimura M. Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell. 1993;5(11):1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Ni & Ruan (2009).Jin Y, Ni DA, Ruan YL. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell. 2009;21(7):2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2019).Li Y, Lu Y, Li L, Chu Z, Zhang H, Li H, Fernie AR, Ouyang B. Impairment of hormone pathways results in a general disturbance of fruit primary metabolism in tomato. Food Chemistry. 2019;274:170–179. doi: 10.1016/j.foodchem.2018.08.026. [DOI] [PubMed] [Google Scholar]
- Li et al. (2013).Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nature Biotechnology. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2018).Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y, Zhu H. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Frontiers in Plant Science. 2018;9:559. doi: 10.3389/fpls.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupi et al. (2019).Lupi ACD, Lira BS, Gramegna G, Trench B, Alves FRR, Demarco D, Peres LEP, Purgatto E, Freschi L, Rossi M. Solanum lycopersicum GOLDEN 2-LIKE 2 transcription factor affects fruit quality in a light- and auxin-dependent manner. PLOS ONE. 2019;14(2):e0212224. doi: 10.1371/journal.pone.0212224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2018).Ma X, Zhang Y, Turečková V, Xue GP, Fernie AR, Mueller-Roeber B, Balazadeh S. The NAC transcription factor SLNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiology. 2018;177(3):1286–1302. doi: 10.1104/pp.18.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2015).Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Matarasso, Schuster & Avni (2005).Matarasso N, Schuster S, Avni A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell. 2005;17(4):1205–1216. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu et al. (2009).Mathieu S, Cin VD, Fei Z, Li H, Bliss P, Taylor MG, Klee HJ, Tieman DM. Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. Journal of Experimental Botany. 2009;60(1):325–337. doi: 10.1093/jxb/ern294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov et al. (2013).Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nature Biotechnology. 2013;31(8):691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- Pan et al. (2016).Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Scientific Reports. 2016;6(1):24765. doi: 10.1038/srep24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, Botha & Birch (2013).Patrick JW, Botha FC, Birch RG. Metabolic engineering of sugars and simple sugar derivatives in plants. Plant Biotechnology Journal. 2013;11(2):142–156. doi: 10.1111/pbi.12002. [DOI] [PubMed] [Google Scholar]
- Petreikov et al. (2009).Petreikov M, Yeselson L, Shen S, Levin I, Schaffer AA, Efrati A, Bar M. Carbohydrate balance and accumulation during development of near-isogenic tomato lines differing in the AGPase-L1 allele. Journal of the American Society for Horticultural Science. 2009;134(1):134–140. doi: 10.21273/jashs.134.1.134. [DOI] [Google Scholar]
- Powell et al. (2012).Powell ALT, Nguyen CV, Hill T, Cheng KLL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, Lopez-Baltazar J, Barry CS, Liu Y, Chetelat R, Granell A, Van Deynze A, Giovannoni JJ, Bennett AB. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science. 2012;336(6089):1711–1715. doi: 10.1126/science.1222218. [DOI] [PubMed] [Google Scholar]
- Qin et al. (2016).Qin G, Zhu Z, Wang W, Cai J, Chen Y, Li L, Tian S. A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening. Plant Physiology. 2016;172(3):1596–1611. doi: 10.1104/pp.16.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan & Tian (2009).Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLOS ONE. 2009;4(7):e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch & Greiner (2004).Rausch T, Greiner S. Plant protein inhibitors of invertases. Biochimica et Biophysica Acta-Proteins and Proteomics. 2004;1696(2):253–261. doi: 10.1016/j.bbapap.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Reca et al. (2008).Reca IB, Brutus A, D’Avino R, Villard C, Bellincampi D, Giardina T. Molecular cloning, expression and characterization of a novel apoplastic invertase inhibitor from tomato (Solanum lycopersicum) and its use to purify a vacuolar invertase. Biochimie. 2008;90(11–12):1611–1623. doi: 10.1016/j.biochi.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Rojo et al. (2003).Rojo E, Zouhar J, Carter C, Kovaleva V, Raikhel NV. A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(12):7389–7394. doi: 10.1073/pnas.1230987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Jin & Huang (2009).Ruan YL, Jin Y, Huang J. Capping invertase activity by its inhibitor: roles and implications in sugar signaling, carbon allocation, senescence and evolution. Plant Signaling and Behavior. 2009;4(10):983–985. doi: 10.4161/psb.4.10.9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan et al. (2013).Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nature Biotechnology. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shan-e-Ali Zaidi et al. (2019).Shan-e-Ali Zaidi S, Vanderschuren H, Qaim M, Mahfouz MM, Kohli A, Mansoor S, Tester M. New plant breeding technologies for food security. Science. 2019;363(6434):1390–1391. doi: 10.1126/science.aav6316. [DOI] [PubMed] [Google Scholar]
- Shao, Punt & Wesseler (2018).Shao Q, Punt M, Wesseler J. New plant breeding techniques under food security pressure and lobbying. Frontiers in Plant Science. 2018;9:1324. doi: 10.3389/fpls.2018.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao et al. (2020).Shao X, Wu S, Dou T, Zhu H, Hu C, Huo H, He W, Deng G, Sheng O, Bi F, Gao H, Dong T, Li C, Yang Q, Yi G. Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnology Journal. 2020;18(1):17–19. doi: 10.1111/pbi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm (1999).Sturm A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiology. 1999;121(1):1–8. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper-Bamnolker et al. (2017).Teper-Bamnolker P, Buskila Y, Belausov E, Wolf D, Doron-Faigenboim A, Ben-Dor S, Van der Hoorn RAL, Lers A, Eshel D. Vacuolar processing enzyme activates programmed cell death in the apical meristem inducing loss of apical dominance. Plant Cell and Environment. 2017;40(10):2381–2392. doi: 10.1111/pce.13044. [DOI] [PubMed] [Google Scholar]
- Tieman et al. (2017).Tieman D, Zhu G, Resende MFR, Lin T, Nguyen C, Bies D, Rambla JL, Beltran KSO, Taylor M, Zhang B, Ikeda H, Liu Z, Fisher J, Zemach I, Monforte A, Zamir D, Granell A, Kirst M, Huang S, Klee H. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355(6323):391–394. doi: 10.1126/science.aal1556. [DOI] [PubMed] [Google Scholar]
- Vallarino et al. (2017).Vallarino JG, Yeats TH, Maximova E, Rose JK, Fernie AR, Osorio S. Postharvest changes in LIN5-down-regulated plants suggest a role for sugar deficiency in cuticle metabolism during ripening. Phytochemistry. 2017;142:11–20. doi: 10.1016/j.phytochem.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Van Eck, Kirk & Walmsley (2006).Van Eck J, Kirk DD, Walmsley AM. Methods in Molecular Biology. Clifton: Humana Press; 2006. Tomato (Lycopersicum esculentum) [DOI] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang W, Cai J, Wang P, Tian S, Qin G. Post-transcriptional regulation of fruit ripening and disease resistance in tomato by the vacuolar protease SlVPE3. Genome Biology. 2017;18(1):219. doi: 10.1186/s13059-017-1178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2016).Wang N, Duhita N, Ariizumi T, Ezura H. Involvement of vacuolar processing enzyme SlVPE5 in post-transcriptional process of invertase in sucrose accumulation in tomato. Plant Physiology and Biochemistry. 2016;108:71–78. doi: 10.1016/j.plaphy.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Wu & Birch (2007).Wu L, Birch RG. Doubled sugar content in sugarcane plants modified to produce a sucrose isomer. Plant Biotechnology Journal. 2007;5(1):109–117. doi: 10.1111/j.1467-7652.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- Xie & Yang (2013).Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Molecular Plant. 2013;6(6):1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- Xu et al. (2017).Xu XX, Hu Q, Yang WN, Jin Y. The roles of call wall invertase inhibitor in regulating chilling tolerance in tomato. BMC Plant Biology. 2017;17:195. doi: 10.1186/s12870-017-1145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2017).Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, Asmutola P. CRISPR/Cas9-induced targeted Mutagenesis and gene replacement to generate long-shelf life tomato lines. Scientific Reports. 2017;7:11874. doi: 10.1038/s41598-017-12262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanor et al. (2009).Zanor MI, Osorio S, Nunes-Nesi A, Carrari F, Lohse M, Usadel B, Kühn C, Bleiss W, Giavalisco P, Willmitzer L, Sulpice R, Zhou YH, Fernie AR. RNA interference of LIN5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiology. 2009;150(3):1204–1218. doi: 10.1104/pp.109.136598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák et al. (2017).Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29(6):1196–1217. doi: 10.1105/tpc.16.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.