Abstract
The aim of the present study was to evaluate the resistance-associated mutations in 302 human immunodeficiency virus type 1 (HIV-1)-infected patients receiving combination therapy and monitored in Marseille, France, hospitals from January 1997 to June 1998. In the reverse transcriptase (RT) gene, the most frequent mutations were found at codons 215 (53%), 41 (34%), and 67, 70, 184, and 210 (>20%). One deletion and two insertions in the β3-β4 hairpin loop of the finger subdomain (codon 69) were detected. Interesting associations and/or exclusions of specific mutations were observed. In 96% of RT genes, a mutation at codon 70 (most frequently, K70R) was associated with a wild-type genotype at position 210 (P < 10−5). Similarly, a mutation at codon 210 (most frequently, L210W) was generally associated with mutations at codons 41 (92%) and 215 (96%) but not at codon 219 (16%) or codon 70 (4%) (P < 10−5). In the protease gene, the most prevalent mutations were at codons 63 (84%), followed by codons 10, 36, 71, 77, and 93 (ca. 20%). As for RT, pairwise associations of mutations were observed. Analysis of the mutation patterns for patients with undetectable HIV-1 loads revealed a high proportion (65%) of wild-type RT genotypes but only 18% wild-type protease genotypes. For patients with high viral loads (>100,000 copies/ml), more than 50% of the RT and protease genes displayed three or more mutations. The significant correlation between the level of viremia in plasma and the number of resistance mutations in the protease (P = 0.007) and RT (P = 0.00078) genes strengthens the importance of defining the genotype of the predominant HIV-1 quasispecies before initiating antiretroviral therapy.
Replication of drug-resistant human immunodeficiency virus (HIV) type 1 (HIV-1) during multidrug therapy is considered a major cause of treatment failure (8). The currently available arsenal of drugs for the treatment of HIV infection includes agents that fall into three classes: nucleoside analog reverse transcriptase (RT) inhibitors, nonnucleoside analog RT inhibitors, and HIV-1 protease inhibitors. Drug resistance arises from mutations in the viral genome, specifically, in the genes that encode the molecular targets of therapy (i.e., the HIV-1-encoded enzymes RT and protease). An understanding of the genetic changes that render a particular drug ineffective is important for the development of new drugs, for designing the optimal drug combinations, and, potentially, even for the clinical management of individual patients (21).
The extreme variability of HIV-1 is mainly due to (i) the high replication rate of the virus (5), (ii) the lack of 3′ exonuclease proofreading activity by RT (1), and (iii) the low processivity of RT, which, according to Temin (28), has been evolutionarily conserved to facilitate the strand transfer reaction during retroviral DNA synthesis. Because of this high mutation rate, HIV-1 exists within an individual as a complex mixture of genetically related but distinguishable variants often referred to as quasispecies (18). In this mixture, HIV-1 strains containing many of the possible single amino acid substitutions (naturally occurring mutant strains) are likely to exist even before the administration of antiviral drug therapy (6, 14, 19). However, these mutations may affect viral fitness, i.e., the efficiency of virus replication in a given environment, so that wild-type genotypes predominate in the absence of drug-induced selective pressure (7). During drug therapy, those viruses that carry or develop mutations that confer drug resistance are selected and eventually predominate (2). In addition, during drug therapy, mutations that do not confer resistance at all but that, instead, compensate for the diminished activity (loss of fitness) associated with other drug resistance mutations are selected (8). As recently recommended by the International AIDS Society—USA Panel, “primary” (or major) mutations that confer drug resistance by themselves should be distinguished from “secondary” (or accessory) mutations that could improve the fitness of virus containing primary mutations (8). Therefore, it is of fundamental importance to detect the preexistence and the emergence of resistance-associated mutations in clinical HIV-1 isolates from treated patients. In this respect, the sequences of global isolates are needed to identify naturally occurring polymorphisms of HIV-1 RT and protease genes. Conversely, sequences from patients treated with different antiretroviral drugs and drug combinations are needed to identify the spectrum of genetic changes selected by drug therapy.
In the present study, we collected and analyzed HIV-1 RT and protease gene sequence data for a population of 302 patients undergoing combination therapy between January 1997 and June 1998. We observed 787 sequences and stored them in a specifically designed database that allowed the retrieval of sequences that met specific criteria such as the occurrence and frequency of a particular mutation, the nature and frequency of the amino acid substitution at a given codon, and/or the rate of association of two resistance mutations. In addition, the relationship between the number of resistance mutations and the plasma viral load was analyzed for a subpopulation of 136 patients.
MATERIALS AND METHODS
Patients.
We monitored 302 patients receiving combination therapy, including various associations of zidovudine, lamivudine, didanosine, stavudine (d4T), nevirapine, indinavir, ritonavir, nelfinavir, or saquinavir, in Marseille, France, hospitals (from January 1997 to June 1998). Most patients received different combinations of drugs during the course of their antiretroviral treatment. Therefore, it was not possible to subdivide the patients on the basis of a common treatment history. At the time of sequence analysis, zidovudine was included in the combination regimen of 40% of the patients. The distribution of the other drugs was as follows: lamivudine, 77%; didanosine, 23%; d4T, 49%; nevirapine, 1%; indinavir, 33%; ritonavir, 36%; nelfinavir, 5%; and saquinavir, 20%. A total of 787 sequences corresponding to the RT gene (287 patients), the protease gene (285 patients), or both genes (270 patients) were analyzed. For 136 patients, the quantitation and the genetic analysis of the viral RT and protease genes were performed with the same sample. When indicated, the genetic analysis was performed at several time points (160 sequences of the RT gene for 52 patients and 158 sequences of the protease gene for 51 patients).
Plasma HIV-1 load.
Viral load was quantitated by the Roche Amplicor method. The limit of detection of the assay is 400 copies/ml.
HIV-1 DNA extraction and sequencing.
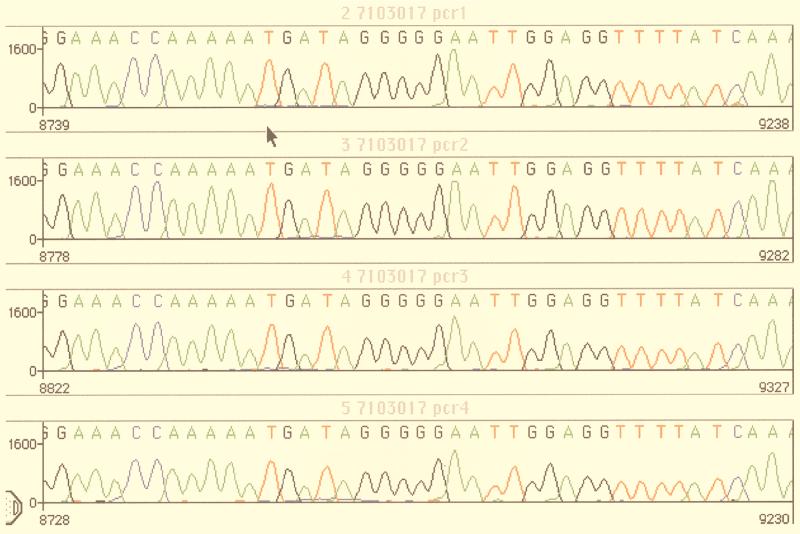
Proviral DNA was isolated ex vivo from patients' peripheral blood mononuclear cells with the QIAamp kit (Qiagen) without a coculture step in order to minimize viral selection. To avoid potential contamination, laboratory HIV-1 strains (e.g., strain HXB2, LAV, or IIIB) were not handled in the rooms used for extraction, PCR, and sequencing (16). Genomic DNA could be sequenced from about 70% of the samples with less than 400 HIV-1 RNA copies/ml. Starting with 1 μg of total DNA, accurate sequences could be obtained for most samples with a plasma HIV-1 load of >1,000 copies/ml. The protease-RT gene region of HIV-1 was amplified by nested PCR, as follows. The first round was performed with primers 5′eprb and MJ4, and 1 μl of the product that was obtained was amplified in a second round of PCR with primers 5′prb and NE1. The resulting PCR-amplified DNA fragments (about 700 bp containing the RT region from codon 20 to codon 230 and 300 bp corresponding to the protease gene) were purified over a PCR purification spin column. The PCR product was used as the DNA template for nucleotide sequencing analysis of the HIV-1 RT and protease gene region with the following primer pairs: primers 5′prb and 5′eprb (protease gene) and primers A and NE1 (RT gene). Cycle sequencing of both strands was performed on the GeneAMP PCR system 9600 instrument (Perkin-Elmer) with the PRISM Ready Dye Terminator Cycle sequencing kit with AmpliTaq DNA polymerase FS (Taq-FS; Perkin-Elmer, Applied Biosystems Division). Excess dye-labeled terminators were removed from the extension products by ethanol-magnesium precipitation. Once separated, the extension products were evaporated to dryness. Each sample was resuspended in loading buffer, heated for 2 min at 90°C, and loaded onto an Applied Biosystems 377 sequencer (Perkin-Elmer, Applied Biosystems Division). The nucleotide sequences of the RT-protease gene were translated and aligned with Sequence Navigator software by using the sequence of HIV-1 HXB2 as the reference sequence. The accuracies of the sequences obtained directly from the PCR products were assessed by performing several amplifications and sequencing of the same sample. As shown in Fig. 1, the sequences obtained from four distinct PCR products of the same DNA sample were generally similar, suggesting that the major HIV-1 quasispecies was preferentially amplified and subsequently sequenced. However, it is important to note that the use of fluorescent dye terminator cycle sequencing was effective in the detection of mixed viral populations representing at least 10 to 20% of the total genomes, as reported previously (17) (see also Fig. 3).
FIG. 1.
Assessment of accuracy of sequence data. The data show the electropherograms resulting from a PCR amplification experiment performed in quadruplicate. Four aliquots (1 μg each) of the same DNA extract were amplified, and the protease gene was sequenced as described in Materials and Methods. The arrow shows the position of codon 46 (ATG) of the protease gene. The sequences were aligned with Sequence Navigator software.
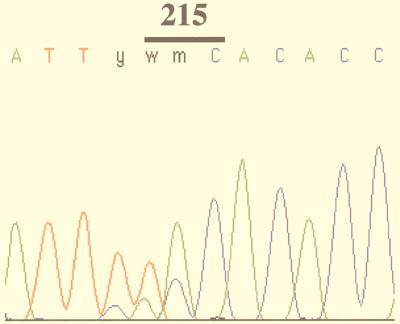
FIG. 3.
Electropherogram of an HIV-1 RT sequence in the region of codon 215. Mixed populations are detected at codon 214 (TTy, i.e., TTC and TTT) and codon 215 (wmC, i.e., AAC, ACC, TAC, or TCC).
Database.
Sequence data were stored in a specifically designed database that allowed the retrieval of genotypes on the basis of the absence or presence of any combinations of resistance-associated mutations. Microsoft Access software was used to create the database. When indicated, specific queries were done with the HIV RT and Protease Database (8a), which is referred to as the “Stanford database” throughout this report (25).
Statistical analysis.
A chi-square test was applied to assess the significance of specific associations (or reciprocal exclusions) of resistance-associated mutations in the RT and protease genes. When the sample size was not sufficient for the chi-square test, a Kendall test was used. Fisher's two-tailed test was used to assess the significance of the number of mutations in the RT and protease genes in different subgroups of plasma viral loads. In all cases, the data were analyzed with Statgraphics software.
RESULTS
Mutational pattern of RT gene.
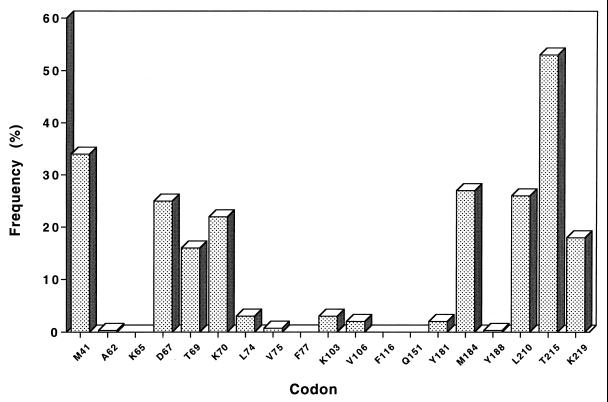
The pattern of resistance-associated mutations detected in the RT gene sequenced from 287 patients is shown in Fig. 2. The most prevalent mutations were found at codons 215 (53%), 41 (34%), and 67, 69, 70, 184, 210, and 219 (>15%). The amino acid substitutions found at these codons are summarized in Table 1. The main substitutions were M41L, D67N, T69D, K70R, M184V, L210W, T215Y, and K219Q. A significant variability was observed at codons 67 and 69, which belong to a random coil loop between two β strands (β3 and β4) in the finger subdomain of RT (12). A major polymorphism associated with resistance to zidovudine (15) was also observed at codon 215. Interestingly, a double mutation in the nucleotide sequence of this codon was detected for 26% of patients, leading to an ambiguity in the assignment of the amino acid substitution. In the example documented in Fig. 3, the wmC codon can be interpreted as AAC, ACC, TAC, or TCC, which would correspond to amino acid N, T, Y, or S, respectively.
FIG. 2.
Frequency of resistance-associated mutations in the RT gene: cross-sectional analysis of 287 sequences.
TABLE 1.
Nature and frequency of amino acid substitutions at resistance-associated codons in the RT gene: cross-sectional analysis of 287 sequences
| Mutation at the following codona:
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41 (M) | 62 (A) | 65 (K) | 67 (D) | 69 (T) | 70 (K) | 74 (L) | 75 (V) | 103 (K) | 106 (V) | 151 (Q) | 181 (Y) | 184 (M) | 188 (Y) | 210 (L) | 215 (T) | 219 (K) |
| L (98) | V (1) | —b | N (66) | D (20) | R (59) | I (4) | L (1) | R (5) | I (5) | — | N (1) | V (75) | L (1) | W (73) | Y (82) | Q (36) |
| I (1) | G (1) | N (15) | A (2) | V (2) | A (1) | E (1) | C (1) | I (1) | S (2) | F (12) | N (8) | |||||
| E (1) | A (7) | G (2) | A (1) | N (1) | D (1) | F (1) | D (5) | E (4) | ||||||||
| L (1) | S (6) | S (1) | G (1) | I (5) | R (4) | |||||||||||
| S (1) | E (2) | N (5) | D (2) | |||||||||||||
| C (1) | C (3) | |||||||||||||||
| I (1) | c (26) | |||||||||||||||
| K (1) | ||||||||||||||||
| L (1) | ||||||||||||||||
| Y (1) | ||||||||||||||||
A mutation was considered when it was unambiguously detected in the electropherogram of the sequenced sample. The letters in parentheses are the wild-type amino acid. The numbers in parentheses refer to the number of sequences with the indicated mutation.
—, none.
In the case of codon 215, mutations could be detected at both the first and the second positions of the codon (Fig. 2).
Associations and/or exclusions of mutations in RT gene.
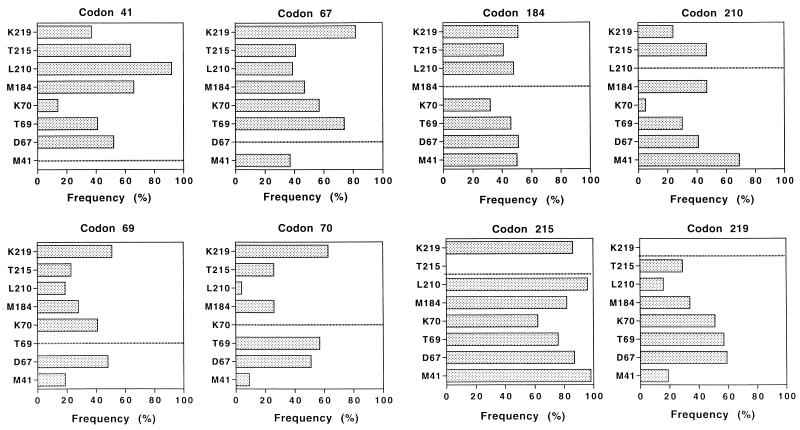
Our database was also used to study the potential association (or exclusion) of specific resistance-associated mutations in the RT gene (Fig. 4). The statistical significance of these data was assessed by the chi-square test. A mutation at codon 41 was generally (>90%) associated with a mutation at codon 210 (P < 10−5). In contrast, this mutation was rarely (<20%) associated with a mutation at codon 70, and this lack of association was statistically significant (P < 10−5). The frequency of the other mutations associated with a mutated codon 41 varied from 40% (codon 219; P < 10−5) to 65% (codons 184 and 215; P < 10−5 for each codon). In the same way, a mutation at codon 70 was generally associated with a wild-type genotype at codons 210 (P < 10−5) and 41 (P = 0.0002) and with a mutation at codons 67, 69, and/or 219 (>50%). Mutations at codons 184 and 215 could be associated with any of the other mutations without a marked preference for a given codon. A mutation at codon 210 was preferentially associated with mutations at codons 41, 184, and 215 (P < 10−5) but was less preferentially associated with a mutation at codon 219 and very rarely with a mutation at codon 70 (P < 10−5). Finally, a mutation at codon 219 was generally associated with mutations at codons 67, 69, and/or 70 (P < 10−5) but was less frequently associated with mutations at the other codons.
FIG. 4.
Pairwise associations between amino acid sites in HIV-1 RT. The results are expressed as the percentage of genomes expressing two resistance mutations. For instance, in the graph for codon 41, one can read that a mutation at codon 70 is found in 14% of the genomes with a mutation at codon 41. The statistical significance of these data (chi-square test) is indicated in the text.
Insertions and deletions in the RT gene.
In addition to point mutations, genetic rearrangements associated with resistance to multiple nucleoside analogs have recently been detected in the β3-β4 hairpin loop of HIV-1 RT (26, 29). For the HXB2 reference strain RT, the amino acid sequence of the loop connecting the two β strands is KKKDSTKWR (i.e., codons 64 to 72). In this study, a deletion of codon 69 was noted in the DNA extracted from 1 of the 302 patients. Insertions in this region were observed for two of these patients (with amino acid sequences NKKGSTTRWR and KKKDSSSTKWR [the insertions are underlined]).
Mutational pattern of protease gene.
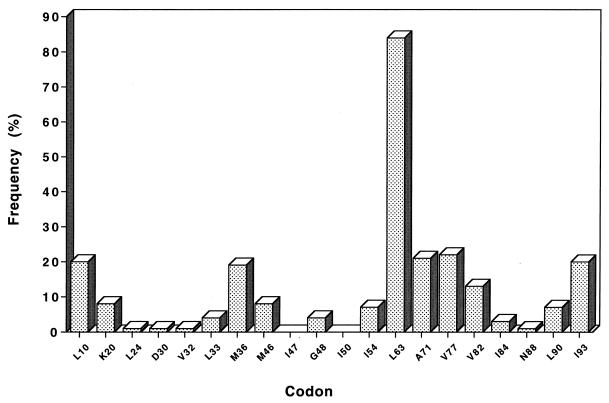
The pattern of resistance-associated mutations detected in the protease gene sequenced from 285 patients is shown in Fig. 5. The most prevalent mutations were found at codons 63 (84%), 77 (22%), 71 (21%), 10 (20%), 93 (20%), 36 (19%), 82 (13%), 46 (8%), 20 (8%), 90 (7%), and 54 (7%). The amino acid substitutions found at these codons are summarized in Table 2. The main substitutions were L10I, K20R, M36I, M46I, I54V, L63P, A71V, V77I, V82A, L90M, and I93L.
FIG. 5.
Frequency of resistance-associated mutations in the protease gene: cross-sectional analysis of 285 sequences.
TABLE 2.
Nature and frequency of amino acid substitutions at resistance-associated codons in the protease gene: cross-sectional analysis of 285 sequences
| Mutation at the following codona:
| |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 (L) | 20 (K) | 24 (L) | 30 (D) | 32 (V) | 33 (L) | 36 (M) | 46 (M) | 48 (G) | 54 (I) | 63 (L) | 71 (A) | 77 (V) | 82 (V) | 84 (I) | 88 (N) | 90 (L) | 93 (I) |
| I (44) | R (11) | I (3) | N (3) | I (3) | V (9) | I (48) | I (12) | V (9) | V (18) | P (203) | V (36) | I (58) | A (30) | V (8) | S (2) | M (21) | L (54) |
| V (10) | I (6) | I (2) | L (4) | L (11) | Q (1) | I (9) | S (19) | T (21) | A (1) | I (5) | L (2) | D (1) | F (1) | ||||
| F (3) | M (6) | R (1) | R (1) | T (2) | A (17) | I (2) | L (1) | F (3) | G (1) | N (1) | |||||||
| L (1) | V (1) | A (1) | H (10) | P (1) | T (1) | T (2) | |||||||||||
| Q (1) | S (1) | V (5) | M (1) | C (1) | |||||||||||||
| T (1) | F (2) | D (1) | |||||||||||||||
| C (1) | G (1) | ||||||||||||||||
| D (1) | |||||||||||||||||
| I (1) | |||||||||||||||||
| N (1) | |||||||||||||||||
| Q (1) | |||||||||||||||||
See footnote a of Table 1.
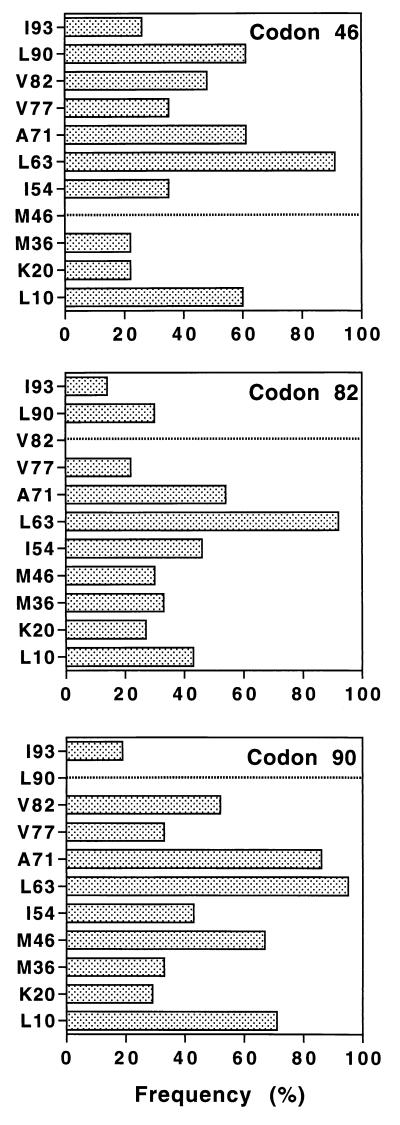
The L63P mutation is found in most genomes and appears to be randomly associated with any of the other resistance codons (data not shown). Indeed, this mutation is frequently detected in HIV-1-infected patients naive for protease inhibitors (14), and proline may thus represent the most common amino acid at this position. In contrast to HIV-1 RT, the sequences of the protease gene had a relatively low frequency of primary resistance mutations, with M46, V82, and L90 being the most common. As shown in Fig. 6, a mutation at codon 46 was frequently associated with mutations at codons 10, 71, and 90 (each >60%), while the percentage of association of the other codons (except L63P) was between 20% (codons 20 and 36) and 47% (codon 82). Mutation codon 82 was preferentially associated with mutation codon 71 and, to a lesser extent, mutations codon 10, codon 54, and codon 90. Finally, mutation codon 90 was generally associated with mutation codon 71 and was frequently associated with mutations codon 10, codon 46, codon 54, codon 77, and codon 82. The statistical significance of these results was assessed by using the Kendall test (P < 10−5 for all pairs).
FIG. 6.
Frequency of mutations found in association with mutations at codons 46, 82, or 90 in the protease genome.
Mutational pattern and viral load.
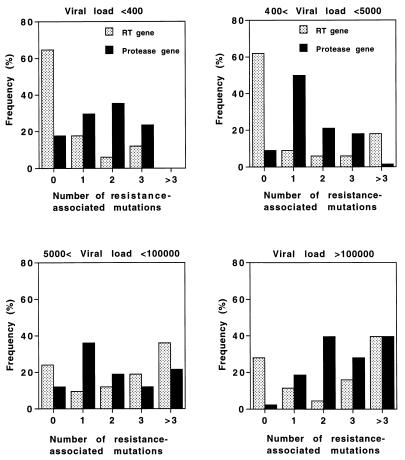
The relationship between the genotype and the viral load was investigated for a subgroup of 136 patients for whom sequence and viral quantitation were done with the same blood sample (Fig. 7). Four viral load groups were considered: <400 HIV-1 copies/ml (17 patients), between 400 and 5,000 HIV-1 copies/ml (34 patients), between 5,000 and 100,000 HIV-1 copies/ml (42 patients), and >100,000 HIV-1 copies/ml (43 patients). For the first group of patients with an undetectable plasma HIV-1 load, 65% of genomes did not contain any resistance-associated mutations in the RT gene (Fig. 7). In contrast, the ratio of genomes without resistance-associated mutations in the protease gene was only 18% for this group of patients with undetectable viral loads. However, it is noteworthy that the number of resistance-associated mutations in both the RT and protease genes was between zero and three for all these patients. When the viral load increased, genomes with more than three resistance-associated mutations were detected and the ratio of genomes with fewer than three such mutations decreased. Indeed, among the patients with the highest viral load, 56% of the patients had at least three resistance-associated mutations in the RT gene, while 68% had three or more resistance-associated mutations in the protease gene. The statistical significance of these data was assessed by Fisher's two-tailed exact test. In particular, the number of mutations (zero or greater than three) was found to correlate closely with the most extreme groups of viral load (i.e., <400 and >100,000 HIV-1 copies/ml) (P = 0.00078 for the RT gene and P = 0.007 for the protease gene).
FIG. 7.
Number of resistance-associated mutations in HIV-1 RT and protease genes: correlation with viremia. The patients were classified into four groups according to their levels of plasma viremia. The number of resistance-associated mutations in the RT and protease genes is indicated. Statistical analysis was performed by Fisher's two-tailed exact test (P < 0.007).
Evolution of HIV-1 RT and protease genotypes.
For 52 patients, the sequencing of the HIV-1 RT gene was performed at several time points over a maximal period of 18 months. Accordingly, 160 sequences were analyzed and entered in a specific file of the database. Overall, the sequences at the resistance-associated codons appeared to be remarkedly stable, with a mean variation of +0.1 resistance mutation per patient (range, −1 to +4), and 82 sequences had no change at the resistance-associated codons. Only two cases of reversion at codon 184 were observed.
Similarly, 158 sequences of the protease gene were analyzed at different time points for 51 patients. The mean variation was +0.15 resistance mutation per patient (range, −2 to +6), and 73 sequences had no change at the resistance-associated codons. For one patient, a double reversion of the mutations at codons 30 and 36 was observed 9 months after the first sequence analysis. For another patient, the resistance-associated pattern of mutations evolved from a single mutation at codon 63 to a polymutated genotype at codons 10, 24, 33, 46, 54, 63, and 88 after 10 months.
DISCUSSION
Scope of study.
In the present study, we have analyzed the mutational pattern of the HIV-1 pol gene directly sequenced from cellular DNA from a large population of patients under combination therapy. The data provide a cross-sectional snapshot of the HIV-1 pol gene diversity in treated patients from southern France. The DNA sequences of the RT and protease genes, which were derived from one PCR product encompassing both the RT and protease genes, were stored in a specifically designed database. The sequences obtained in this study from a homogeneous population of patients were compared with the sequences stored in the Stanford database, an on-line relational database containing a compilation of nearly all published HIV RT and protease sequences obtained in various laboratories worldwide (25).
Resistance mutations in RT gene.
The nucleoside analog 3′-azido-3′-deoxythymidine (zidovudine) is the most widely used clinical therapy for HIV-1 infection. However, zidovudine monotherapy invariably results in the appearance of HIV-1-resistant strains with multiple mutations in the RT gene, especially M41L, D67N, K70R, L210W, T215Y/F, and K219Q (2, 4, 9, 15). Thus, the high prevalence of these mutations (Fig. 2) reflects the systematic use of zidovudine in HIV-infected patients (40% of the patients were still receiving zidovudine at the time of inclusion in the present study). Interestingly, similar frequencies of zidovudine resistance mutations can be obtained from the Stanford database. Moreover, one major observation from our study was the low frequency of genotypes with both K70R and L210W mutations (Fig. 4). This apparent exclusion of the mutations at codons 70 and 210 is consistent with previously published data on the evolution of zidovudine-resistant genotypes in treated patients (4, 9). Taken together with the data obtained with the sequences stored in the Stanford database, this suggests that the concomitant detection of the L210W and K70R mutations in the same genome is only transient. A detailed discussion of these results on a biochemical basis will be published elsewhere.
On the other hand, the high frequency of genomes with a mutation at codon 184 is indicative of the use of 2′,3′-dideoxyinosine and 2′,3′-dideoxy-3′-thiacytidine in the therapeutic regimen (27). M184 is the X residue in the catalytically important YXDD motif common to all RTs. The mutant with the M184 mutation has been studied extensively (22), and the M→V substitution, which is predominant in vivo (Table 1 and Stanford database), confers greater fidelity to the HIV-1 RT (10) through an increase in processivity (20).
One important result obtained in this study was the very low frequency of genomes with mutations at codons potentially associated with resistance to nonnucleoside analog RT inhibitors (i.e., codons 103, 106, 181, and 188) (27). The low prevalence of mutations at those codons reflects the limited number of patients (1%) treated with nonnucleoside analog RT inhibitors at the time of inclusion. One should also note the absence of genotypes with a mutation at codon Q151, previously shown to confer multidideoxynucleoside resistance (23).
Finally, one deletion and two insertions were detected in the vicinity of codon 69, which belongs to the β3-β4 hairpin loop in the finger subdomain. These genetic rearrangements have been discovered very recently (26) and were therefore not documented in the Stanford database at the time of this analysis.
Resistance mutations in protease gene.
As discussed above, primary mutations that confer drug resistance by themselves should be distinguished from secondary mutations that could improve the fitness of virus containing primary mutations. For the protease gene, the main primary mutations that confer resistance to antiprotease drugs are M46I/L and/or V82A/F/T for indinavir, V82A for ritonavir, G48V and/or L90M for saquinavir, and D30N for nelfinavir (8). The limited use of nelfinavir by the patients in this study (5%) explains the very low frequency of occurrence of the D30N mutation (in only 1 of 285 sequences). One important result of this study is the high prevalence of secondary mutations detected in the protease gene. This result is consistent with the high degree of polymorphism of the protease gene, which has previously been evidenced in nontreated patients by using high-density nucleotide arrays (14). Since primary resistance mutations are selected during the course of treatment with protease inhibitors, their occurrence depends on (at least) two parameters: (i) the presence of protease inhibitors in the combination regimen and (ii) elapsed time since the initiation of treatment. Even though resistance mutations in the protease gene may be more easily detected in plasma HIV-1 RNA than in cellular DNA (13), our data may reflect the more recent use of protease inhibitors as part of anti-HIV combination therapy. Indeed, primary resistance mutations may occur rapidly in the protease gene for those patients who do not respond to treatment. Subsequently, secondary mutations are expected to accumulate in the protease gene on the basis of their compensatory effect on viral fitness (24). In agreement with this concept, we could observe in some patients in our cohort the occurrence of up to five secondary mutations for only one primary mutation after 10 months of treatment. Since the restoration of viral fitness may be due to the selection of virus quasispecies that exhibit various types of secondary mutations, the analysis of the associations and/or exclusions of resistance mutations is particularly complex for the protease gene (3). Nevertheless, some interesting features emerge from this analysis. The L90M substitution is a primary mutation that confers resistance to saquinavir. Subsequently, secondary mutations may be detected at residues 36, 71, and 84 (11). In our database, L90M was preferentially associated with a mutation at codon 71 (P < 10−5 by the Kendall test). Interestingly, the reciprocal was not true since 42 sequences of the protease gene displayed a mutation at codon 71 but had a wild-type sequence at codon 90 (L90). These data, together with previously published studies (14), suggest that (i) secondary mutations may exist before the occurrence of a primary resistance mutation and (ii) once a virus population with a primary mutation has been selected, the loss of fitness of these viruses is important enough to favor the emergence of variants with secondary mutations. Therefore, most genomes with primary mutations also display one or several secondary mutations, whereas secondary mutations can exist in genomes without a primary mutation. According to this concept, one would expect that some secondary mutations may confer a replicative advantage even in the absence of selective pressure from drugs.
Viral load and genotype.
The relationship between the genotype and the viral load is a central issue for the validation of genotype analysis as a means of predicting therapeutic efficiency or failure. The data reported here show that a significant correlation exists between the number of mutations in the RT and protease genes and the viral load. Clearly, most patients with high-level viremia (>100,000 HIV-1 copies/ml of plasma) have viruses with more than three mutations in both genes. Reciprocally, all sequenced viruses from patients with undetectable viral loads displayed a maximal number of three mutations in both genes. However, the natural polymorphism of the protease gene is evident from our data since genomes with a wild-type protease gene (for resistance-associated codons) were detected in only 12 of the 136 (9%) patients in this study. In comparison, wild-type RT genes (for resistance-associated codons) were detected in 54 of these 136 (40%) patients.
Conclusion and perspectives.
In conclusion, this survey of 787 sequences revealed specific patterns of mutations in the RT gene, some of them being mutually exclusive, and confirmed the high degree of polymorphism of the protease gene in treated patients. The data obtained with a homogeneous population of treated patients from southern France were generally consistent with the analysis of a worldwide compilation of RT and protease sequences available through the Stanford database. This cross-sectional snapshot of HIV-1 pol gene diversity in treated patients will serve as a statistically significant baseline characterization for future studies of the evolution of the RT and protease genes.
ADDENDUM IN PROOF
Through September 1999, 2,200 DNA and RNA HIV-1 RT and protease gene sequences had been analyzed in our database. Since the initial submission of this paper, multidrug resistance mutations (especially Q151M) have been detected in 10 patients. The frequency of mutations associated with resistance to nonnucleoside RT inhibitors is increasing dramatically, consistent with the more frequent use of these drugs in combination regimens.
REFERENCES
- 1.Beard W A, Wilson S H. 8. Reverse transcriptase. In: Karn J, editor. HIV: a practical approach, vol. 2. Biochemistry, molecular biology and drug discovery. Oxford, United Kingdom: IRL Press; 1995. pp. 15–36. [Google Scholar]
- 2.Boucher C A, O'Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M, Goudsmit J, Larder B A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Brown A J, Korber B T, Condra J H. Associations between amino acids in the evolution of HIV type 1 protease sequences under indinavir therapy. AIDS Res Hum Retroviruses. 1999;15:247–253. doi: 10.1089/088922299311420. [DOI] [PubMed] [Google Scholar]
- 4.Cleland A, Watson H G, Robertson P, Ludlam C A, Brown A J. Evolution of zidovudine resistance-associated genotypes in human immunodeficiency virus type 1-infected patients. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12:6–18. doi: 10.1097/00042560-199605010-00002. [DOI] [PubMed] [Google Scholar]
- 5.Coffin J. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 6.Cornelissen M, van den Burg R, Zorgdrager F, Lukashov V, Goudsmit J. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol. 1997;71:6348–6358. doi: 10.1128/jvi.71.9.6348-6358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo E. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vézinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 8a.HIV RT and protease database. [Online.] http://hivdb.stanford.edu/hiv.
- 9.Hooker D J, Tachedjian G, Solomon A E, Gurusinghe A D, Land S, Birch C, Anderson J L, Roy B M, Arnold E, Deacon N J. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad V R, Wainberg M A. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1997;25:4532–4536. doi: 10.1093/nar/25.22.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ives K J, Jacobsen H, Galpin S A, Garaev M M, Dorrell L, Mous J, Bragman K, Weber J N. Emergence of resistant variants of HIV in vivo during monotherapy with the protease inhibitor saquinavir. J Antimicrob Chemother. 1997;39:771–779. doi: 10.1093/jac/39.6.771. [DOI] [PubMed] [Google Scholar]
- 12.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kramer G, Ferris A L, Clark P, Hizi A, Hugues S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch N, Yahi N, Ariasi F, Fantini J, Tamalet C. Comparison of human immunodeficiency virus type 1 (HIV-1) mutations in HIV-1 genomes detected in plasma and in peripheral blood mononuclear cells from patients receiving combination drug therapy. J Clin Microbiol. 1999;37:1595–1597. doi: 10.1128/jcm.37.5.1595-1597.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merrigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 15.Larder B A, Kellam P, Kemp S. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Learn G H, Jr, Korber B T, Foley B, Hahn B H, Wolinsky S M, Mullins J I. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyö E M, Uhlen M. Analysis of heterogeneous viral populations by direct DNA sequencing. BioTechniques. 1993;15:120–127. [PubMed] [Google Scholar]
- 18.Meyerhans A E, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson J, Asjö B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 19.Najera I, Holguin A, Quinones-Mateu M E, Munoz-Fernandez M A, Najera R, Lopez-Galindez C, Domingo E. pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no therapy. J Virol. 1995;69:25–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P M, Modak M J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 21.Pazzani M J, See D, Schroeder E, Tilles J. Application of an expert system in the management of HIV-infected patients. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:356–362. doi: 10.1097/00042560-199708150-00005. [DOI] [PubMed] [Google Scholar]
- 22.Quan Y, Gu Z, Li X, Liang C, Parniak M A, Wainberg M A. Endogenous reverse transcriptase assays reveal synergy between combinations of the M184V and other drug resistance-conferring mutations in interactions with nucleoside analog triphosphates. J Mol Biol. 1998;277:237–247. doi: 10.1006/jmbi.1997.1592. [DOI] [PubMed] [Google Scholar]
- 23.Rezende L F, Curr K, Ueno K, Mitsuya H, Prasad V R. The impact of multideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J Virol. 1998;72:2890–2895. doi: 10.1128/jvi.72.4.2890-2895.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts N A, Craig J C, Sheldon J. Resistance and cross-resistance with saquinavir and other HIV protease inhibitors: theory and practice. AIDS. 1998;12:453–460. doi: 10.1097/00002030-199805000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Shafer R W, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamalet C, Izopet J, Koch N, Fantini J, Yahi N. Stable rearrangements of the β3-β4 hairpin loop of HIV-1 reverse transcriptase in plasma viruses from patients receiving combination therapy. AIDS. 1998;12:F161–F166. doi: 10.1097/00002030-199814000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hugues S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 28.Temin H. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winters M A, Cooley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merrigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Invest. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]