Abstract
The peroxisome proliferator-activated receptor γ (PPARγ) is a member of the nuclear receptor superfamily that activates target gene transcription in a ligand-dependent manner. In addition, liganded PPARγ can inhibit transcription of genes induced by gamma interferon (IFN-γ) and/or lipopolysaccharides (LPSs), including the inducible nitric oxide synthase (iNOS) gene. Inhibition of the iNOS promoter is achieved partially through antagonizing the activities of NF-κB, AP-1, and STAT1, which are known to mediate effects of LPS and IFN-γ. Previous studies have suggested that transrepression of these factors by nuclear receptors involves competition for limiting amounts of the general coactivators CREB-binding protein (CBP) and p300. CBP and p300 are thought to be recruited to nuclear receptors through bridging factors that include SRC-1, although CBP also interacts directly with PPARγ through its amino terminus. These observations have raised questions concerning the involvement of SRC-1-like factors in CBP recruitment and transrepression. We here provide evidence that PPARγ's ability to repress iNOS transcription requires the ligand-dependent charge clamp that mediates interactions with CBP and SRC-1. Single amino acid mutations in PPARγ that abolished ligand-dependent interactions with SRC-1 and CBP not only resulted in complete loss of transactivation activity but also abolished transrepression. Conversely, a CBP deletion mutant containing the SRC-1 interaction domain but lacking the N-terminal PPARγ interaction domain was inactive as a PPARγ coactivator and failed to rescue transrepression. Together, these findings are consistent with a model in which transrepression by PPARγ is achieved by targeting CBP through direct interaction with its N-terminal domain and via SRC-1-like bridge factors.
Peroxisome proliferator-activated receptor γ (PPARγ) is a member of the nuclear hormone receptor superfamily that is capable of both positive and negative regulation of gene expression in response to ligand binding. PPARγ has been suggested to be involved in a broad range of cellular functions, including adipocyte differentiation (44, 48, 51), glucose homeostasis (12, 56), inflammatory responses (25, 40), and apoptosis (9). These physiologic actions suggest that synthetic PPARγ ligands may be of use in several disease settings, including type 2 diabetes mellitus, atherosclerosis, and cancer. The thiazolidinedione class of PPARγ ligands have already proven to be effective in the treatment of type 2 diabetes (34), and recent studies suggest that these agents may also be clinically beneficial in inflammatory bowel disease (49). The molecular mechanisms responsible for these activities are not understood.
In addition to the highly conserved DNA binding domain (DBD), PPARγ contains two transactivation domains: an N-terminal ligand-independent activation function 1 (AF1 or A/B) domain and a C-terminal domain that mediates ligand binding, dimerization, and ligand-dependent transactivation (Fig. 1A) (reviewed in reference 32). PPARγ positively regulates gene expression by binding to response elements in target genes as a heterodimer with retinoid X receptors (RXRs) (28). When either the PPARγ or RXR components of the heterodimer are bound by agonists, the respective ligand binding domains (LBDs) undergo a conformational change that leads to the recruitment of coactivators and consequent transcription of target genes (33, 52). Coactivator recruitment and transcriptional activation by nuclear receptors require a highly conserved helical motif located at the extreme C terminus of the LBD, called activation function 2 (AF2) (4, 10, 14). Structural analysis of unliganded and liganded nuclear receptor LBDs suggests that the AF2 domain is randomly oriented or extends away from the ligand-binding pocket in the absence of ligand. In the presence of ligand, the AF2 folds against the LBD, serving as part of the ligand-binding pocket. These ligand-induced conformational changes are thought to regulate transcriptional activity by regulating interaction of coactivator and corepressor complexes.
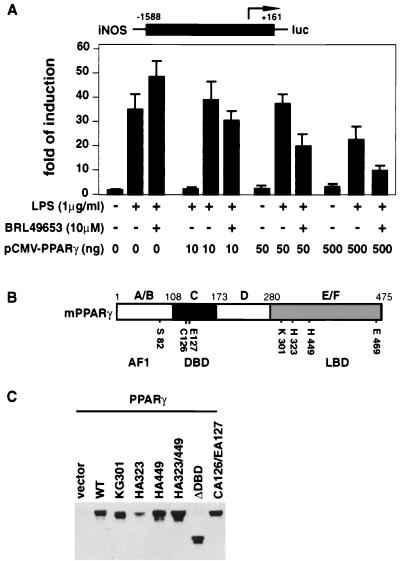
FIG. 1.
(A) Inhibition of LPS-induced iNOS activity by liganded PPARγ. RAW 264.7 cells were cotransfected with 0.5 μg of iNOS-luc reporter construct and different amounts of PPARγ expression plasmid as shown. Cells were treated with BRL49653 (10 μM) for 2 h and then induced with LPS (1 μg/ml) for 24 h before harvesting. (B) Schematic map of the wild-type murine PPARγ1 protein. Functional domains of PPARγ and sites for point mutations are indicated. (C) Whole-cell extracts were made from HeLa cells transfected with empty vector or different PPARγ constructs. Approximately 100 μg of total proteins from each extract was loaded on a 10% polyacrylamide gel, and the level of PPARγ was detected by Western blotting, using a monoclonal antibody generated against the C terminus of PPARγ (Santa Cruz Biotechnology). WT, wild type.
Biochemical and expression screening approaches have led to the identification of a large number of putative coactivator and corepressor proteins that interact with nuclear receptors in a ligand-dependent manner (33, 52). Among the best characterized of these factors are proteins of approximately 160 kDa in molecular mass, including SRC-1, GRIP-1/TIF2, and p/CIP/ACTR/AIB1 (8, 22, 23, 31, 36, 52, 54). Overexpression of SRC-1 potentiates ligand-dependent transcription by many nuclear receptors in cells (37), and microinjection studies suggest that SRC-1 is required for PPARγ-dependent transcription in some contexts (55).
The SRC-1 class of coactivators interacts with nuclear receptors through a conserved region that contains three helical motifs (HD1, HD2, and HD3) with the consensus sequence LXXLL (13, 20, 52, 53). Cocrystal studies indicated that two highly conserved amino acids, Glu469 in the AF2 helix and Lys301 in H3 of the LBD, form a charge clamp that places the HD motif into a hydrophobic pocket in the receptor (11, 18, 35, 47). Glu469 and Lys301 make contacts with the peptide backbone of the LXXLL helix and form the two ends of the charge clamp. These structural findings are consistent with biochemical studies indicating that these two amino acids play a key role in transcriptional activation and coactivator interaction (14, 15, 21).
SRC-1 and other p160 factors have been suggested to function as coactivators, at least in part, by recruiting CREB-binding protein (CBP) and/or p300. SRC-1, TIF2, and p/CIP contain a conserved C-terminal domain that mediates direct interactions with the C terminus of CBP and p300. CBP and p300 also contain an N-terminal LXXLL domain that can interact directly with many nuclear receptors and has a particularly high affinity for PPARγ (43). The presence of LXXLL motifs in both the CBP-p300 and SRC-1 classes of coactivators has raised questions concerning mechanisms of coactivator assembly. In the case of DNA-bound retinoic acid receptor (RAR)-RXR heterodimers, effective recruitment of CBP requires SRC-1, and the N-terminal LXXLL motif of CBP is dispensable for coactivator function (29, 55). In addition to the ligand-dependent transactivation function of AF2, the ligand-independent transactivation domain AF1 also affects receptor functions. A serine residue (Ser82 in PPARγ1 or Ser112 in PPARγ2) located in the AF1 domain was shown to be phosphorylated in vitro by mitogen-activated protein kinase (1, 6, 24, 59) and could be phosphorylated in vivo in response to mitogenic stimulation by epidermal growth factor, 12-O-tetradecanoylphorbol-13-acetate, insulin, and serum (6, 24, 59). Mutation of the Ser to Ala to prevent receptor phosphorylation increased transcriptional activity of the receptor in transient-transfection experiments (1, 24). These findings suggest that phosphorylation of this residue negatively regulates receptor function (24, 39, 45). Studies by Shao and coworkers suggested that Ser phosphorylation lowers ligand-binding affinity, resulting in decreased efficiency of coactivator (SRC-1) recruitment (45).
Liganded PPARγ was also able to inhibit upregulation of monocyte/macrophage-specific gene expression, such as activation of the inducible nitric oxide synthase (iNOS) gene in response to gamma interferon (IFN-γ) and/or lipopolysaccharide (LPS) (40). This inhibition is considered transrepression because it does not appear to involve direct binding to the iNOS promoter. Transrepression of iNOS by PPARγ is achieved at least partially by antagonizing the activities of STAT1, NF-κB, and AP-1, which are known to mediate the effects of IFN-γ and LPS, respectively (25, 40). However, the molecular mechanism of transrepression by PPARγ remains unclear. Recent studies suggest that transcriptional activation by AP-1, STAT1, and NF-γB (along with many other transcription factors) requires the coactivators CBP and/or p300. Since CBP and p300 are structurally and functionally conserved proteins that have been shown to be critical for multiple cellular functions, it has been suggested that competition for limiting amounts of these proteins represents a mechanism for transrepression by PPARγ and other nuclear receptors (27). Potential roles of p160 proteins in PPARγ-mediated transrepression have not been established, although it has been suggested that SRC-1 plays a role in transrepression of NF-κB by the glucocorticoid receptor (GR) (46).
In the present studies, we have examined effects of mutations in the AF1, LBD, and DBD of PPARγ on its transactivation and transrepression activities and the mechanisms of assembly of coactivator (SRC-1 and CBP) complexes on RXR-PPARγ heterodimers in vitro. Our results indicate a strong correlation between the transactivation and transrepression activities of PPARγ. Mutations that resulted in significantly weakened interactions between PPARγ and the SRC-1 and CBP coactivators resulted in coordinate loss of transactivation and transrepression activities. Together, these data are consistent with a model in which PPARγ-dependent transrepression of the iNOS promoter involves the targeting of CBP–SRC-1 coactivator complexes by a mechanism involving the LXXLL interaction domains of both CBP and SRC-1.
MATERIALS AND METHODS
DNA constructs.
Murine PPARγ1 expression plasmid pCMX PPARγ and reporter constructs for the iNOS and AOx-TK promoters have been previously described (40). Point mutations in PPARγ were made using the QuickChange site-directed mutagenesis kit (Stratagene). A fragment containing the mutation was sequenced and subcloned into an unmutagenized plasmid vector to ensure the absence of undesired mutations. PPARγ ΔDBD was made by deleting the StuI-EcoRV fragment that contains the DBD and most of the hinge region of PPARγ. PPARγ ΔN93 was constructed by first introducing an HpaI site at the third residue of the PPARγ coding sequence and subsequently deleting the HpaI-StuI fragment that contains Asp3 to Arg95. PPARγ ΔAF2 lacks the last 17 amino acid residues of the full-length protein. PPARγ N198 was made by deleting the EcoRV-NheI fragment that contains part of the hinge region and the entire LBD of PPARγ (Ile199 to COOH terminus). Expression plasmids for wild-type CBP, CBP ΔN450, glutathione S-transferase (GST)–CBP1–450, and GST–SRC-1633–715 were described previously (29, 55).
Transient transfections.
The macrophage cell line RAW 264.7 was maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 20 μg of l-glutamine per ml, and antibiotics. Typically, 2 × 105 cells were transfected with 1 to 1.5 μg of DNA (vector DNA was used to balance the total amount of DNA if necessary) in six-well plates, using Lipofectamine according to the manufacturer's instructions (Gibco-BRL). After incubation in OptiMEM (Gibco) at 37°C for 5 h, the medium was removed and cells were fed with fresh Dulbecco's modified Eagle medium containing 0.5% fetal bovine serum, in the presence or absence of PPARγ ligand BRL49653. For transactivation assays, cells were harvested 24 h later. For transrepression assays, inducing reagents (IFN-γ, 30 U/ml; LPS, 1 μg/ml) were added 2 h later and cells were further incubated for 24 h before harvest. CV1 cells were transfected by calcium phosphate precipitation as previously described (40).
GST fusion protein binding assays.
GST fusion proteins were produced as crude bacterial lysates and immobilized on glutathione agarose beads. After being preincubated with appropriate ligands for 30 min at room temperature in CHAPS buffer [8 mM Tris-phosphate buffer (pH 7.4), 0.12 M KCl, 8% (vol/vol) glycerol, 4 mM dithiothreitol, 0.1 mg of poly(dI/dC) per ml, and 0.5% (wt/vol) CHAPS detergent], 35S-labeled, in vitro-translated PPARγ (including wild type and mutants) was mixed with immobilized GST-CBP1–450 or GST–SRC-1633–715 in NET-N buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40) and incubated for 1 h at room temperature. The beads were then washed three times with H buffer (20 mM HEPES [pH 8.0], 50 mM KCl, 20% glycerol, 0.1% NP-40), immediately boiled in 2× sodium dodecyl sulfate (SDS) sample buffer, and loaded on an SDS–10% polyacrylamide gel, unless otherwise specified.
DNA-dependent protein-protein interaction assays.
Biotinylated DNA oligonucleotides containing the PPARγ response element (sense strand, 5′-AAGGGGATCCGTACAGGTCACAGGTCACTCGAGATCT-3′) were synthesized, gel purified, and annealed. One microgram of double-stranded DNA fragment was incubated with purified PPARγ and RXR (produced as GST fusion proteins in Escherichia coli, purified on glutathione agarose beads, and eluted by thrombin cleavage) in CHAPS buffer to allow PPARγ-RXR heterodimer formation on DNA. The DNA-receptor complex was captured on streptavidin agarose beads at 4°C for 30 min. After washing of the beads twice with H buffer and once with CHAPS buffer, appropriate ligands were added to allow ligand binding in CHAPS buffer at room temperature for 30 min. Baculovirus-produced, FLAG-tagged full-length CBP lysate was then added to the mixture and incubated for 1 h at room temperature. The beads were washed three times with H buffer, boiled in 2× SDS sample buffer, and immediately loaded on an SDS–6% polyacrylamide gel. The bound CBP was detected by Western blotting, using anti-FLAG antibody (Kodak).
RESULTS
Transient-transfection experiments using increasing amounts of PPARγ expression plasmid showed that inhibition of LPS-induced iNOS promoter activity is both receptor dependent and ligand dependent (Fig. 1A). The hypothesis that transrepression can be caused by competition among different transcription factors and/or pathways for limiting amounts of essential coactivators predicts that mutations in PPARγ affecting coactivator binding and/or transactivation should also affect transrepression. To test this hypothesis, we examined the effects of point mutations and deletions in different regions of PPARγ (Fig. 1B) on ligand-dependent activation of a PPARγ-activated promoter [(AOx)3-TK-luc] and ligand-dependent repression of the iNOS promoter in transient-transfection assays. Western blot analysis indicated approximately equivalent expression levels for each PPARγ mutant except PPARγ HA323 (Fig. 1C and data not shown).
AF1 domain and N-terminal phosphorylation are minor factors in transrepression.
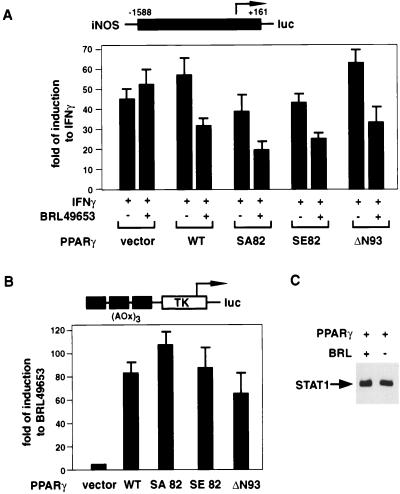
Deletion of the ligand-independent transcriptional activation domain AF1 (PPARγ ΔN93) had little effect on transrepression (Fig. 2A) or transactivation (Fig. 2B). Phosphorylation of Ser82 by activated mitogen-activated protein kinases has been shown to decrease ligand-binding affinity and ligand-dependent functions of PPARγ. Ser82 was changed to Glu (SE82) to mimic the constitutively phosphorylated PPARγ and to Ala (SA82) to mimic the unphosphorylated PPARγ. Both mutants behaved similarly to wild-type protein in a transactivation assay (Fig. 2B), possibly because the ligand concentration (10 μM) under which the experiments were performed was at a saturating level that masked any difference that the two mutants might have in ligand-binding affinity. They also behaved similarly to wild-type PPARγ in transrepression assays (Fig. 2A). Phosphorylation of Ser82, therefore, does not appear to account for why higher concentrations of PPARγ ligands are required for half-maximal transrepression than for transactivation. Inhibition of iNOS transcription by liganded PPARγ was not due to changes in STAT1 levels, as documented by Western blot assay (Fig. 2C).
FIG. 2.
AF1 domain and N-terminal phosphorylation of PPARγ play a minor role in transactivation and transrepression. (A) For the transrepression assay, RAW 264.7 cells were cotransfected with 0.5 μg of iNOS-luc reporter construct and 0.5 μg of PPARγ expression plasmid. Cells were treated with BRL49653 (10 μM) for 2 h and then induced with IFN-γ (30 U/ml) for 24 h before harvesting. (B) For the transactivation assay, RAW 264.7 cells were cotransfected with 1 μg of (AOx)3-TK-luc reporter construct and 0.1 μg of PPARγ expression plasmid. Cells were treated with BRL49653 (10 μM) for 24 h before harvesting. Transfections were done in triplicate and repeated at least two times. Data shown are representative of one typical experiment. (C) Assessment of STAT1 levels. Whole-cell extracts were prepared from RAW 264.7 cells transfected with wild-type PPARγ expression plasmid, with or without subsequent treatment with 10 μM BRL49653. Approximately 100 μg of total proteins from each extract was loaded on a 10% polyacrylamide gel, and the level of STAT1 was detected by Western blotting, using an antibody from Santa Cruz Biotechnology. WT, wild type.
The ligand-dependent charge clamp is required for transactivation and transrepression.
Previous studies of selected nuclear receptors, including RAR, RXR, GR, thyroid hormone receptor (TR), and vitamin D receptor, indicated that the ligand-dependent activation domain AF2 is important for recruiting coactivators to the activation complex (3, 7, 19, 27, 30). Not surprisingly, AF2 domain deletion and point mutations affecting the charge clamp (PPARγ EA469 and KG301) rendered PPARγ completely inactive for transactivation (Fig. 3A). In addition, the same mutations also abolished ligand-dependent transrepression (Fig. 3B). Protease sensitivity experiments indicated ligand-dependent changes in protected fragments, indicating that these proteins retain the ability to bind ligands (data not shown). Western blotting experiments indicated that both KG301 and EA469 were expressed at levels similar to those of the wild-type receptor, while ΔAF2 was expressed at a slightly lower level (Fig. 2C and data not shown). These observations suggest that transactivation and transrepression are mechanistically linked.
FIG. 3.
The charge clamp and AF2 domain are essential for transactivation, transrepression, and coactivator interactions. Transactivation (A) and transrepression (B) assays were performed as described in the Fig. 2 legend. Charge clamp and AF2 mutants are unable to interact with GST-CBP1–450 (C) or GST–SRC-1633–715 (D) efficiently. GST–SRC-1633–715 or CBP1–450 was incubated with in vitro-translated PPARγ, wild-type or mutant protein, in the presence or absence of ligand (15d-PGJ2, 0.1 μM; BRL49653, 2 μM) as described in Materials and Methods. GST in panel C indicates the amount of wild-type 35S-PPARγ pulled down by GST alone. WT, wild type.
To further explore whether the ability of PPARγ to repress gene expression was directly linked to its ability to interact with coactivators, GST fusion protein binding assays were performed, using 35S-labeled, in vitro-translated PPARγ and GST–SRC-1633–715, which contains LXXLL helical domains 1 and 2, or GST-CBP1–450, which contains the N-terminal LXXLL motif previously demonstrated to interact with PPARγ. Interaction experiments using GST-CBP1–450 demonstrated a significant ligand-independent interaction with PPARγ that was modestly increased by addition of BRL49653 and abolished by the AF2 deletion and the charge clamp mutations (Fig. 3C). Wild-type PPARγ exhibited a nearly exclusive ligand-dependent interaction with the SRC-1 fragment. These interactions were abolished by the charge clamp and AF2 mutations (Fig. 3D).
The cocrystal structure of the PPARγ LBD and its ligand BRL49653 indicates that His323 and His449 of PPARγ form multiple hydrogen bonds with the ligand (35), predicting that these residues are important for ligand binding and ligand-dependent receptor functions. We changed these two His residues to Ala, both separately and together, and tested their effects on transactivation and transrepression. Surprisingly, the HA449 mutation had no significant effect on PPARγ's ability to activate or repress transcription (Fig. 4). PPARγ HA323 was inactive in both transactivation and transrepression assays. However, as this may have been due in part to poor expression (Fig. 1C), further experiments were not performed with this mutant. Additional evidence that H323 is critical for ligand binding was provided by the HA323-HA449 double mutant. This mutant was expressed equivalently to the wild-type receptor but exhibited less than 10% of wild-type activity in both transactivation and transrepression assays, consistent with a mechanistic link between these two processes.
FIG. 4.
His323 and His449 in the PPARγ ligand-binding pocket are critical for both transactivation (A) and transrepression (B). Transfections were performed as described in the Fig. 2 legend using the following concentrations of BRL49653: 1 nM (lanes 1), 10 nM (lanes 2), 100 nM (lanes 3), 1 μM (lanes 4), 10 μM (lanes 5), and 100 μM (lanes 6). In panel B, BRL49653 was used at 10 μM. WT, wild type.
Differential effects of DBD mutations on transactivation and transrepression.
A large deletion of the DBD and part of the hinge region (PPARγ ΔDBD) resulted in complete loss of not only transactivation (data not shown) but also transrepression function (Fig. 5A). To examine the effects of this mutation on coactivator interaction, GST fusion protein binding assays were performed using GST–SRC-1633–715 and GST-CBP1–450. Intriguingly, the PPARγ ΔDBD mutation abolished interaction with the nuclear receptor interaction domain of SRC-1 but did not abolish interaction with the N terminus of CBP (Fig. 5B). These experiments suggest that, in addition to the charge clamp, the hinge and/or DBD of PPARγ plays a role in stabilizing interactions with SRC-1. Studies using a series of N-terminal fragments of PPARγ revealed that there is a strong ligand-independent interaction between the N terminus of CBP and the N-terminal A/B domain of PPARγ (Fig. 5C), consistent with previous reports (16). To more selectively disrupt DNA binding, we made point mutations of two critical amino acids in the first zinc finger (CA126-EA127) involved in base-specific interaction. As expected, this mutant was completely inactive for transactivation (data not shown). In contrast to the ΔDBD mutant, the PPARγ CA126-EA127 mutant retained partial transrepression activity, exhibiting 40 to 60% of the activity of wild-type PPARγ in four experiments, with a representative experiment illustrated in Fig. 5A. Unlike the PPARγ ΔDBD deletion mutant, PPARγ CA126-EA127 interacted with GST–SRC-1633–715 in a ligand-dependent manner (Fig. 5B).
FIG. 5.
Contributions of PPARγ domains to coactivator interaction and transrepression. (A) Point mutation, but not deletion, of the PPARγ DBD partially retains transrepression activity. Transfections were performed as described in the Fig. 2 legend, except that cells were induced with LPS (1 μg/ml) instead of IFN-γ. (B) DBD deletion and point mutation mutants show different patterns of interaction with GST–SRC-1633–715 and GST-CBP1–450. (C) N-terminal A/B and C domains of PPARγ strongly interact with GST-CBP1–450 in a ligand-independent manner. GST pulldown experiments were performed as described in the Fig. 3 legend. Ligand concentrations were 2 μM BRL49653 (for PPARγ) and 2 μM TTNPB (for RARα). In panels B and C, incubation with GST alone resulted in no detectable background binding (data not shown). WT, wild type.
Coactivator complex assembly and CBP domain requirements.
To better assess the interaction between PPARγ and CBP, we performed a protein-protein interaction experiment using full-length PPARγ fused to GST and full-length CBP produced in baculovirus. This experiment demonstrated a strong ligand-independent interaction between the two proteins that was further increased upon ligand binding (Fig. 6A). Since GST fusion protein binding assays dealt only with interactions between the coactivators and PPARγ in solution, we performed DNA-based protein-protein interaction assays. Recruitment of full-length CBP to DNA-bound PPARγ-RXR heterodimers was evaluated in the absence or presence of recombinant SRC-1621–1207, which contains both the nuclear receptor interaction domain and the CBP interaction domain. As seen in Fig. 6B, in the absence of SRC-1, CBP binding was extremely weak and barely detectable only when both ligands for PPARγ and RXR were present. These results are in marked contrast to the interaction of soluble full-length PPARγ with GST-CBP1–450 (Fig. 5) or the interaction of soluble full-length CBP and full-length GST-PPARγ (Fig. 6A). However, when recombinant SRC-1621–1207 containing the nuclear receptor and CBP interaction domains was added to the reaction, CBP was recruited much more efficiently in a ligand-dependent manner (Fig. 6B). These results suggest that, like RAR-RXR heterodimers, DNA-bound PPARγ-RXR is likely to need SRC-1 or a related p160 protein as a bridge factor to recruit CBP efficiently.
FIG. 6.
Coactivator assembly and CBP domain requirements in transactivation and transrepression. (A) Interaction between full-length CBP and full-length PPARγ. Immobilized GST-PPARγ (full length) was incubated with full length CBP-WT in the presence or absence of BRL49653 (2 μM). The specifically bound fraction was resolved on a 6% polyacrylamide gel, transferred to a nitrocellulose membrane, and visualized by probing with anti-FLAG antibody (Kodak). (B) SRC-1-dependent interactions of CBP with PPARγ-RXR heterodimers bound to DNA. PPARγ-RXR heterodimers were assembled on a biotinylated PPARγ response element and incubated with full-length CBP (CBP-WT) expressed in baculovirus in the presence of the indicated ligands (2 μM) and recombinant SRC-1 fragment (SRC-1621–1207). (C) The N-terminal 450 amino acid residues of CBP are required for potentiation of PPARγ's transactivation function. RAW 264.7 cells were cotransfected with 0.2 μg of (AOx)3-TK-luc reporter construct, 0.02 μg of PPARγ expression plasmid, and 0.8 μg of CBP (wild-type or ΔN450) expression plasmid. Cells were treated with BRL49653 (0.1 μM) for 24 h before harvesting. (D) Overexpression of wild-type CBP but not CBP Δ450 or wild-type SRC-1 partially relieves transrepression mediated by PPARγ. RAW 264.7 cells were cotransfected with 0.5 μg of iNOS-luc reporter construct, 0.5 μg of PPARγ expression plasmid, and 0.5 μg of coactivator (CBP WT, CBP ΔN450, or SRC-1 WT) expression plasmid. Cells were pretreated with BRL49653 (10 μM) for 2 h and then induced with LPS (1 μg/ml) for 24 h before harvesting. WT, wild type.
To examine the functional role of the N terminus of CBP in transactivation and transrepression by PPARγ, transient-transfection assays were performed to assess the activities of wild-type CBP and CBP ΔN450. As illustrated in Fig. 6C, wild-type CBP strongly potentiated PPARγ-dependent transcription of the (AOx)3-TK-luc reporter plasmid, while CBP ΔN450 was inactive. These results are consistent with an essential role of the N-terminal 450 amino acids in mediating the activities of PPARγ. Wild-type CBP, but not CBP ΔN450, potentiated LPS-dependent activation of the iNOS promoter, indicating an unexpected role of the N terminus of CBP in mediating transcriptional effects of LPS. Overexpression of wild-type CBP reversed PPARγ-dependent transrepression of iNOS promoter activity relative to control values (i.e., LPS-induced activity in the absence of BRL49653) but did not abolish inhibitory effects of BRL49653 (Fig. 6C). Overexpression of SRC-1 had a relatively modest effect on activation of the iNOS promoter and was relatively ineffective in relieving PPARγ-dependent repression (Fig. 6D), suggesting that it is not a limiting target for the transrepressive effects of PPARγ.
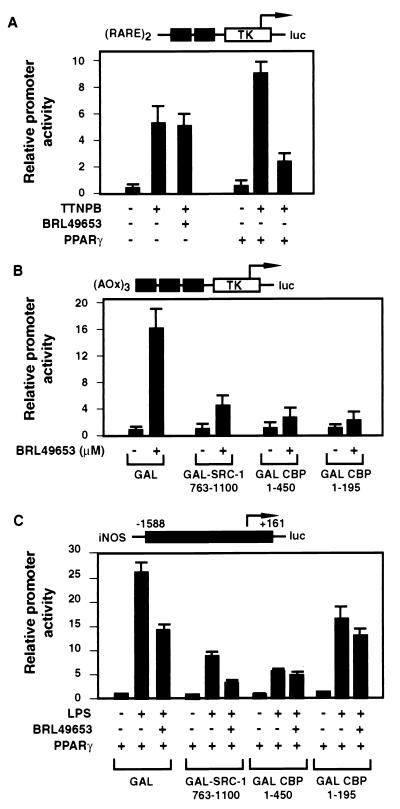
To further examine the mechanism of transrepression, we tested whether PPARγ can mediate repression of another CBP-dependent promoter that is independent of the factors involved in iNOS response (such as NF-κB and STAT1). We used a reporter construct that has two copies of the DR5 element, which mediates RAR response, fused to the luciferase gene. Transient transfection of CV1 cells with this reporter construct and the PPARγ expression plasmid demonstrated a PPARγ ligand-dependent repression of the (DR5)2-luc activity in response to the RAR ligand TTNPB (Fig. 7A), compatible with the hypothesis that competitive sequestration or inhibition of CBP coactivator complexes accounts for transrepression.
FIG. 7.
CBP is the target of competitive recruitment by different transcription factors. (A) RAR signaling is repressed by ligand-activated PPARγ. CV1 cells were cotransfected with 0.2 μg of (DR5)2-luc reporter construct and 1 μg of PPARγ expression plasmid (or empty vector). Cells were treated with 10 μM BRL49653 for 2 h and then induced with 0.1 μM TTNPB for 24 h before harvesting. (B and C) Transactivation of (AOx)3-TK-luc (B) and transrepression of iNOS-luc (C) were differentially affected by overexpression of GAL-CBP and GAL–SRC-1 fusions. (B) RAW 264.7 cells were cotransfected with 0.2 μg of (AOx)3-TK-luc reporter construct, 0.02 μg of PPARγ expression plasmid, and 0.8 μg of GAL fusion construct. Cells were treated with BRL49653 (0.1 μM) for 24 h before harvesting. (C) RAW 264.7 cells were cotransfected with 0.5 μg of iNOS-luc reporter construct, 0.5 μg of PPARγ expression plasmid, and 0.5 μg of GAL fusion construct. Cells were pretreated with BRL49653 (10 μM) for 2 h and then induced with LPS (1 μg/ml) for 24 h before harvesting.
To further evaluate the functional role of the N terminus of CBP in transactivation and transrepression, GAL fusion proteins were overexpressed and tested for dominant-negative effects on PPARγ-dependent transactivation and transrepression. As illustrated in Fig. 7B, GAL-CBP1–450 strongly inhibited ligand-dependent activation of the (AOx)3-TK-luc reporter gene. GAL-CBP1–450 also inhibited the transcriptional response of the iNOS promoter to LPS and blocked ligand-dependent transrepression by PPARγ (Fig. 7C). In contrast, a GAL fusion protein containing the CBP interaction domains of SRC-1 (GAL–SRC-1763–1100), which inhibited ligand-dependent activation of (AOx)3-TK-luc and LPS-dependent induction of iNOS, did not prevent ligand-dependent transrepression of the residual iNOS activity (Fig. 7B and C). A further truncation of GAL-CBP to amino acid 195, which retains the LXXLL motif, did not affect dominant-negative effects on transactivation of (AOx)3-TK-luc but largely relieved dominant-negative effects on LPS-dependent induction of iNOS. GAL-CBP1–195 also retained the ability to inhibit PPARγ-dependent transrepression of iNOS (Fig. 7C).
DISCUSSION
Previous studies have demonstrated that both CBP and SRC-1 can interact directly with PPARγ via LXXLL motifs and that the interaction of the N-terminal LXXLL motif of CBP is particularly strong when assessed by GST pulldown assays. These observations have raised a number of questions regarding the mechanism of CBP recruitment to RXR heterodimers. In the case of RAR-RXR heterodimers, the amino terminus of CBP could be deleted without significantly altering its coactivator activity (29). Further, two LXXLL helices within SRC-1 were shown to make contacts with each component of the RXR-RAR heterodimer (55). In contrast, the N-terminal region of CBP appears to be necessary for PPARγ function, as its deletion abolished coactivator activity. However, the LXXLL motif in CBP does not appear to be sufficient to mediate effective recruitment to DNA-bound PPARγ-RXR heterodimers. This presumably reflects differences in available interaction surfaces of PPARγ when it is bound to DNA as a heterodimer with RXR compared to when it is in solution. The interaction of full-length CBP with DNA-bound PPARγ-RXR heterodimers was strongly enhanced by addition of SRC-1 (Fig. 6B). These findings suggest that PPARγ-RXR heterodimers may recruit CBP-p160 complexes by a mechanism in which an LXXLL motif from the p160 factor interacts with RXR and the N-terminal LXXLL motif from CBP interacts with PPARγ. Thus, the potential exists for a high degree of plasticity in coactivator assembly on different members of the nuclear receptor superfamily.
Several mechanisms have been described for negative regulation of gene expression by nuclear receptors. A subset of nuclear receptors, including TR and RAR, harbor ligand-independent repressor function and actively repress transcription upon binding to cognate sites within the promoter region of target genes. These active repressive functions require the recruitment of corepressor complexes that are dismissed upon ligand binding and replaced by coactivator complexes (reviewed in reference 17). Alternatively, many nuclear receptors can exert inhibitory effects through DNA-binding independent mechanisms. One established mechanism involves direct interactions between nuclear receptors and negatively regulated transcription factors, resulting in the inhibition of DNA-binding and/or transactivating activity of one or both factors. For example, when activated by ligand, GR has been suggested to inhibit NF-κB-mediated gene expression at least in part by physically interacting with NF-κB (termed “cross coupling”) and blocking its ability to bind DNA (41). A second mechanism for transrepression involves the inhibition of signal transduction pathways necessary for activation of specific transcription factors. Activation of c-Jun is greatly enhanced by phosphorylation of Ser63 and Ser73 by members of the Jun amino-terminal kinase superfamily. GR, RAR, and TR have been suggested to inhibit the Jun amino-terminal kinase induction pathway, hence preventing c-Jun from being activated by Ser63-Ser73 phosphorylation and inhibiting AP-1-dependent gene expression (5).
A third proposed mechanism for transrepression involves coactivator competition, which attributes transrepression to competition for limiting amounts of essential coactivators by different transcription factors and/or pathways. This mechanism potentially accounts for mutual antagonism between transcription factors, such as the mutual antagonism observed between nuclear hormone receptors and AP-1 pathways (26, 27, 42, 57). CBP and p300 are essential coactivators for a large family of signal-dependent transcription factors and have been proposed as critical targets for the transrepressive actions of nuclear receptors. Genetic studies indicate that CBP and p300 are functionally limiting in cells, and overexpression of CBP and p300 has been shown to rescue transrepressive effects of nuclear receptors in several contexts (2, 38, 50, 58). If the sequestering of CBP by activated PPARγ accounts for this mechanism of transrepression, there should be a direct correlation between structural determinants required for transactivation and transrepression.
In the present studies, we have demonstrated a strong correlation among PPARγ's abilities to interact with coactivators, to activate target genes, and to repress the iNOS promoter in response to ligand binding. A deletion of the ligand-dependent activation domain AF2 (PPARγ ΔAF2), as well as point mutations of the critical charge clamp residues (PPARγ EA469 and KG301), led to complete loss of both transactivation and transrepression functions, as well as significantly weakened interaction with GST–SRC-1633–715 and GST-CBP1–450 (Fig. 3). An independent assay based on fluorescence resonance energy transfer also showed that PPARγ EA469 was unable to interact with SRC-1568–780 and CBP1–453 (60). In addition to mutations in the charge clamp, mutations in the ligand-binding pocket of PPARγ were made that were predicted to influence the ability of ligand to activate transcription based on X-ray crystal structures. While one of these mutations (HA449) had little effect on transactivation and transrepression, the double mutation (HA323-HA449) abolished both activities. Together, these results indicate that PPARγ-mediated transrepression is tightly correlated with transactivation and support a model in which interactions with LXXLL-containing coactivators are critical components of the transrepression mechanism.
In addition to the ligand-activated charge clamp, the DBD and/or part of the hinge region of PPARγ was required for transrepression (Fig. 5A) but not for interaction with CBP. Furthermore, point mutations (PPARγ CA126-EA127) targeted to selectively alter DNA recognition retained the ability to interact with SRC-1 and were only partially active for transrepression (Fig. 5A and B). The role of the DBD suggests two alternative models for transrepression. On the one hand, DNA binding activity may be required for sequestration of coactivator complexes. In this model, transrepression should be relieved by overexpression of the limiting coactivator complex, and this was achieved by overexpression of CBP in the present studies. However, the observation that iNOS activity could still be inhibited by BRL49653 even when CBP was overexpressed suggests either that CBP levels were still limiting or that additional mechanisms were involved. An alternative model that is consistent with these observations is that PPARγ in solution binds to CBP coactivator complexes through a charge clamp-dependent mechanism and inhibits their ability to mediate transcriptional effects of other CBP-dependent transcription factors. In this model, inhibitory effects would presumably require protein-protein interaction surfaces provided by the DBD. As this model represents a coactivator inactivation mechanism, rather than a coactivator sequestration mechanism, it is consistent with the ability of overexpressed CBP to partially reverse transrepressive effects of BRL49653. To distinguish between these models, it will be necessary to determine whether loss of transrepression by mutations in the DBD of PPARγ results from alterations in DNA binding or alterations in specific protein-protein interactions.
ACKNOWLEDGMENTS
This work was supported by an institutional fellowship from the National Heart, Lung, and Blood Institute to M.L. and NIH grants to C.K.G. C.K.G. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Adams M, Reginato M J, Shao D S, Lazar M A, Chatterjee V K. Transcriptional activation by peroxisome proliferator-activated receptor γ is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J Biol Chem. 1997;272:5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- 2.Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehop signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 3.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M-J, O'Malley B W. The τ4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barettino D, Vivanco Ruiz M M, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caelles C, Gonzales-Sancho J M, Munoz A. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev. 1997;11:3351–3364. doi: 10.1101/gad.11.24.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camp H S, Tafuri S R. Regulation of peroxisome proliferator-activated receptor γ activity by mitogen-activated protein kinase. J Biol Chem. 1997;272:10811–10816. doi: 10.1074/jbc.272.16.10811. [DOI] [PubMed] [Google Scholar]
- 7.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Chinetti G, Griglio S, Antonucci M, Torra I P, Delerive P, Majd Z, Fruchart J-C, Chapman J, Najib J, Staels B. Activation of proliferator-activated receptors α and γ induces apoptosis of human monocyte-derived macrophages. J Biol Chem. 1998;273:25573–25580. doi: 10.1074/jbc.273.40.25573. [DOI] [PubMed] [Google Scholar]
- 10.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone-dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeb S S, Fajas L, Nemoto M, Pihlajamaki J, Mykkanen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 13.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 14.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng X, Peng Z H, Di W, Li X Y, Rochette-Egly C, Chambon P, Voorhees J J, Xiao J H. Suprabasal expression of a dominant-negative RXR alpha mutant in transgenic mouse epidermis impairs regulation of gene transcription and basal keratinocyte proliferation by RAR-selective retinoids. Genes Dev. 1997;11:59–71. doi: 10.1101/gad.11.1.59. [DOI] [PubMed] [Google Scholar]
- 16.Gelman L, Zhou G, Fajas L, Raspe E, Fruchart J-C, Auwerx J. p300 interacts with the N- and C-terminal part of PPARγ2 in a ligand-independent and -dependent manner, respectively. J Biol Chem. 1999;274:7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- 17.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 18.Greene M E, Blumberg B, McBride O W, Yi H F, Kronquist K, Kwan K, Hsieh L, Greene G, Nimer S D. Isolation of the human peroxisome proliferator activated receptor gamma cDNA: expression in hematopoietic cells and chromosomal mapping. Gene Expr. 1995;4:281–299. [PMC free article] [PubMed] [Google Scholar]
- 19.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 20.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 21.Henttu P M, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu E, Kim J B, Sarraf P, Spiegelman B M. Inhibition of adipogenesis through Map kinase-mediated phosphorylation of PPARγ. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 25.Jiang C, Ting A T, Seed B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 26.Jonat C, Rahmsdorf H J, Park K-K, Ponta H, Herrlich P. Anti-tumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 27.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R, Rose D, Glass C, Rosenfeld M. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 28.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa R, Söderström M, Hörlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 34.Nolan J J, Ludvik B, Beerdsen P, Joyce M, Olefsky J. Improvement in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N Engl J Med. 1994;331:1188–1193. doi: 10.1056/NEJM199411033311803. [DOI] [PubMed] [Google Scholar]
- 35.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 36.Oñate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M J, Edwards D P, O'Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 37.Oñate S A, Tsai S Y, Tsai M-J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 38.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C M, Masuno M, Tommerup N, van Ommen G-J, Goodman R H, Peters D J M, Breuning M H. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 39.Reginato M J, Krakow S L, Bailey S T, Lazar M A. Prostaglandins promote and block adipogenesis through opposing effects on peroxisome proliferator-activated receptor gamma. J Biol Chem. 1998;273:1855–1858. doi: 10.1074/jbc.273.4.1855. [DOI] [PubMed] [Google Scholar]
- 40.Ricote M, Li A C, Willson T M, Kelly C J, Glass C K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 41.Scheinman R I, Gualberto A, Jewell C M, Cidlowski J A, Baldwin A S., Jr Characterization of mechanisms involved in transrepression of NF-κB activated glucocorticoid receptors. Mol Cell Biol. 1995;15:242–244. [Google Scholar]
- 42.Schüle R, Rangarajan P, Kliewer S, Ransone L J, Bolado J, Yang N, Verma I M, Evans R M. Functional antagonism between a coprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 43.Schulman I G, Shao G, Heyman R A. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor γ (PPARγ) heterodimers: intermolecular synergy requires only the PPARγ hormone-dependent activation function. Mol Cell Biol. 1998;18:3483–3494. doi: 10.1128/mcb.18.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sears I B, MacGinnitie M A, Kovacs L G, Graves R A. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 1996;16:3410–3419. doi: 10.1128/mcb.16.7.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao D, Rangwala S M, Bailey S T, Krakow S L, Reginato M J, Lazar M A. Interdomain communication regulating ligand binding by PPAR-γ. Nature. 1998;396:377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 46.Sheppard K-A, Phelps K M, Williams A J, Tbano D, Rosenfeld M G, Glass C K, Gerritsen M E, Collins T. Nuclear integration of glucocorticoid receptor and nuclear factor-κB signaling by CREB-binding protein and steroid receptor coactivator-1. J Biol Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- 47.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 48.Spiegelman B M, Flier J S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 49.Su C G, Wen X, Bailey S T, Jiang W, Rangwala S M, Keilbaugh S A, Flanigan A, Murthy S, Lazar M A, Wu G D. A novel therapy for colitis utilizing PPAR-γ ligands to inhibit the epithelial inflammatory response. J Clin Investig. 1999;104:383–389. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tontonoz P, Hu E, Graves R A, Budavari A I, Spiegelman B M. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 52.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 53.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 55.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 56.Willson T M, Cobb J E, Cowan D J, Wiethe R W, Correa I D, Prakash S R, Beck K D, Moore L B, Kliewer S A, Lehmann J M. The structure-activity relationship between peroxisome proliferator-activated receptor gamma agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 57.Yang-Yen H-F, Chambard J-C, Sun Y-L, Smeal T, Schmidt T J, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 58.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 59.Zhang B, Berfer J, Zhou G, Elbrech A, Biswas S, White-Carrington S, Szalkowski D, Moller D E. Insulin- and mitogen-activated protein kinase-mediated phosphorylation and activation of peroxisome proliferator-activated receptor γ. J Biol Chem. 1996;271:31771–31774. doi: 10.1074/jbc.271.50.31771. [DOI] [PubMed] [Google Scholar]
- 60.Zhou G, Cummings R, Li Y, Mitra S, Wilkinson H A, Elbrecht A, Hermes J D, Schaeffer J M, Smith R G, Moller D E. Nuclear receptors have distinct affinities for coactivators: characterization by fluorescence resonance energy transfer. Mol Endocrinol. 1998;12:1594–1604. doi: 10.1210/mend.12.10.0176. [DOI] [PubMed] [Google Scholar]