Supplemental Digital Content is available in the text.
Keywords: animals, cardiomyopathies, heart failure, mutation, proteomics
Background:
The p.(Arg14del) pathogenic variant (R14del) of the PLN (phospholamban) gene is a prevalent cause of cardiomyopathy with heart failure. The exact underlying pathophysiology is unknown, and a suitable therapy is unavailable. We aim to identify molecular perturbations underlying this cardiomyopathy in a clinically relevant PLN-R14del mouse model.
Methods:
We investigated the progression of cardiomyopathy in PLN-R14Δ/Δ mice using echocardiography, ECG, and histological tissue analysis. RNA sequencing and mass spectrometry were performed on cardiac tissues at 3 (before the onset of disease), 5 (mild cardiomyopathy), and 8 (end stage) weeks of age. Data were compared with cardiac expression levels of mice that underwent myocardial ischemia-reperfusion or myocardial infarction surgery, in an effort to identify alterations that are specific to PLN-R14del–related cardiomyopathy.
Results:
At 3 weeks of age, PLN-R14Δ/Δ mice had normal cardiac function, but from the age of 4 weeks, we observed increased myocardial fibrosis and impaired global longitudinal strain. From 5 weeks onward, ventricular dilatation, decreased contractility, and diminished ECG voltages were observed. PLN protein aggregation was present before onset of functional deficits. Transcriptomics and proteomics revealed differential regulation of processes involved in remodeling, inflammation, and metabolic dysfunction, in part, similar to ischemic heart disease. Altered protein homeostasis pathways were identified exclusively in PLN-R14Δ/Δ mice, even before disease onset, in concert with aggregate formation.
Conclusions:
We mapped the development of PLN-R14del–related cardiomyopathy and identified alterations in proteostasis and PLN protein aggregation among the first manifestations of this disease, which could possibly be a novel target for therapy.
WHAT IS NEW?
Mice homozygous for the phospholamban p.(Arg14del) pathogenic variant (PLN-R14Δ/Δ) rapidly develop progressive dilated cardiomyopathy with ECG microvoltages, myocardial fibrosis, PLN (phospholamban) protein aggregation, and biventricular failure, which exactly mimics human PLN cardiomyopathy.
Tissue analysis revealed that PLN protein aggregation in hearts of PLN-R14Δ/Δ mice was observed before the onset of functional impairment or other structural abnormalities, which may implicate a causative role in the disease.
An unbiased approach of combined transcriptomics and proteomics identified differential expression of genes and proteins involved in protein homeostasis, which was not observed in the most common etiology of heart failure, that is, myocardial ischemia.
WHAT ARE THE CLINICAL IMPLICATIONS?
Our results reiterate the concept of genotype-specific treatment in genetic cardiomyopathies, which is important as patients with PLN-R14del–related cardiomyopathy do not optimally benefit from standard heart failure therapies.
Especially, our findings suggest that targeting PLN aggregation or boosting protein quality control systems may present a bona fide target for therapy of PLN-R14del–related cardiomyopathy.
Comprehensive omics studies, taking into account multiple stages of heart failure severity and multiple etiologies, are useful tools in the identification of disease-specific pathways.
Cardiomyopathies are heart muscle disorders and an important cause of heart failure (HF).1 Among cardiomyopathies, dilated cardiomyopathy is the most prevalent cause of HF.1 Although reported estimations for a pathogenic variant underlying dilated cardiomyopathy vary up to 40%, a genetic basis is well established.2 The p.(Arg14del) pathogenic variant of the PLN (phospholamban) gene (PLN-R14del) is a prevalent genetic deviation, identified in 14% of Dutch dilated cardiomyopathy and arrhythmogenic right ventricular cardiomyopathy patients.3,4 PLN-R14del–related cardiomyopathy is characterized by high prevalence of malignant ventricular arrhythmias and end-stage HF, with poor prognosis and high mortality from adolescence.3,4
PLN is a key regulator of cardiomyocyte calcium handling.5 For cardiac muscle contraction, calcium is required in the cytoplasm of cardiomyocytes to facilitate sarcomeric shortening. During diastole, most of the cytoplasmic calcium is transported into the sarcoplasmic reticulum by SERCA (sarco/endoplasmic reticulum Ca2+-ATPase) and stored for the next cycle.5 PLN exerts its regulatory function by inhibiting Ca2+-uptake by SERCA, thereby controlling cardiac lusitropy.5 Although aberrant calcium handling and PLN protein aggregation have been implicated in PLN-R14del–related cardiomyopathy,6,7 their contribution to this disease is unknown.
We recently generated a novel PLN-R14del mouse model and demonstrated that this model accurately mimics human disease.8 Mice heterozygous for the PLN-R14del pathogenic variant (PLN-R14Δ/+) developed cardiomyopathy at late age, while homozygotes (PLN-R14Δ/Δ) exerted an accelerated phenotype with early mortality.8 Since PLN-R14Δ/Δ mice develop a translational and fast-forward phenotype, this model provides a great opportunity to study the pathophysiology of this disease.
In this study, we investigate the development of cardiomyopathy in PLN-R14Δ/Δ mice. Combining in vivo functional analyses with tissue and molecular analyses, including RNA sequencing (RNA-Seq) and mass spectrometry, we map the sequelae of this disease over time and identify aberrations in protein homeostasis (proteostasis) as an early hallmark. Moreover, we describe that proteostasis is uniquely affected in PLN-R14Δ/Δ mice and not in models of ischemic HF. These findings may help to identify disease-specific therapies.
Methods
All supporting data are available within the article and its Data Supplement. Detailed methods are available in the Data Supplement.
Experimental Animals
All animal experiments were approved by the Animal Ethical Committee of the University of Groningen (permit numbers AVD10500201583 and IVD1583-02-001) and performed conform the ARRIVE guidelines.9 Generation of PLN-R14del mice has been published elsewhere.8 To investigate cardiomyopathy development in PLN-R14Δ/Δ mice, 4 male PLN-R14Δ/Δ mice and WT (wild-type) littermates were monitored weekly by serial echocardiography and ECG from 4 to 7 weeks of age. Additionally, at the age of 3, 4, 5, 6, 7, and 8 weeks, hearts were collected from PLN-R14Δ/Δ and WT mice (male and female; in total, n=8 per group) for histological and molecular analyses. Furthermore, additional cardiac tissues were isolated from male PLN-R14Δ/Δ, PLN-R14Δ/+, and WT mice at 3, 5, and 8 weeks of age for transcriptomics (n=4 per group) and proteomics (n=6 per group). All in vivo procedures were performed under continuous isoflurane (2%; TEVA Pharmachemie, the Netherlands) anesthesia. To compare to other HF models, we performed RNA-Seq on left ventricular (LV) tissues of male mice 3 days after myocardial ischemia-reperfusion (I/R) or 8 weeks after myocardial infarction (MI) surgery (n=4 per group) and corresponding sham controls (n=2 for I/R and n=2 for MI) from a previous study.10
Echocardiography
Echocardiography was performed using a Vevo 3100 preclinical imaging system (FUJIFILM VisualSonics, Canada) to assess LV morphology and function from 3-dimensional reconstructions and global longitudinal strain (GLS) by speckle tracking. Data acquisition and analysis were executed in line with the recommendations of the European Society of Cardiology Working Group on Myocardial Function.11
Surface ECG
ECG recordings were acquired and analyzed in LabChart Pro using 2-lead subdermal needle electrodes (lead II configuration) connected to a PowerLab 8/30 system (ADInstruments, New Zealand).12
Sacrifice
Euthanasia was performed as reported previously.13 Cardiac tissue weights were indexed by tibia length to the power 3 to normalize for body size.14
Histological Analysis
Masson trichrome stain was performed on formalin-fixed, paraffin-embedded cardiac transverse mid-slices to detect collagen deposition.15 PLN localization was determined using a fluorescently labeled anti-PLN antibody (Invitrogen, CA),7,8,16 costained with fluorescein isothiocyanate–conjugated WGA (wheat germ agglutinin; Sigma-Aldrich, MO) and 4′,6-diamidino-2-phenylindole (Vector Laboratories, CA).17
Quantitative Polymerase Chain Reaction
RNA was isolated from LVs using TRI Reagent (Sigma-Aldrich). cDNA synthesis was performed using the QuantiTect reverse transcription kit (Qiagen, Germany). Gene expressions were determined using iQ SYBR green supermix and a CFX384 Touch real-time PCR system (Bio-Rad, CA) and normalized to reference values of a component of the large 60S ribosomal subunit (Rplp0, 36B4).18
Western Blot
Proteins were isolated from LVs using radioimmunoprecipitation assay lysis buffer.19 Proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride membranes, and incubated with an anti-ANP (atrial natriuretic peptide) antibody (Abcam, United Kingdom). Detection was performed by enhanced chemiluminescence using a horseradish peroxidase–conjugated secondary antibody.
RNA Sequencing
mRNA libraries were generated using a TruSeq Stranded mRNA Library Prep kit and sequenced using a NextSeq 500 Sequencing System (Illumina, CA).20 Principal component (PC) analysis and differential expression analysis were performed using the DESeq2,21 and gene set enrichment analysis was performed using the fgsea.22 Genes with Benjamini-Hochberg–adjusted P<0.05 were considered significant. For a subset of mice (3- and 8-week-old PLN-R14Δ/Δ, PLN-R14Δ/+, and WT mice), raw data from a previous run were reanalyzed.8 RNA-Seq data have been deposited in NCBI Gene Expression Omnibus23 and are accessible through GEO Series accession number GSE168610.
Mass Spectrometry
Proteins were isolated from LVs, followed by denaturation, reduction, alkylation, and tryptic digestion. Peptides were labeled with tandem mass tag isobaric labels (Thermo Scientific, MA), fractionated by high-pH reversed-phase liquid chromatography, and analyzed by liquid chromatography-mass spectrometry using an UltiMate 3000 chromatography system and an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific). Proteome Discoverer (Thermo Scientific) was used to search raw data against the mouse UniProtKB/Swiss-Prot database. The limma R package, eBayes method, and 2-way ANOVA were used to compare between phenotypes.24 Enriched gene sets were identified using the Database for Annotation, Visualization, and Integrated Discovery.25 Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE26 partner repository with data set identifier PXD024594.
Statistical Analyses
Data are presented as mean±SD. Since group sizes (n≤8) were insufficient to test for parametric distribution, comparisons between 2 groups were performed using Mann-Whitney U tests and Kruskal-Wallis tests for multigroup comparisons using the SPSS Statistics software (IBM, NY). P<0.05 was considered significant. Statistical analyses of -omics data are described in the Data Supplement.
Results
Ventricular Contractility Is Impaired in PLN-R14Δ/Δ Mice Starting at 4 Weeks of Age
To investigate the development of PLN-R14del–related cardiomyopathy, we performed serial echocardiography and ECG in PLN-R14Δ/Δ and WT mice. We have previously reported that PLN-R14Δ/Δ mice exhibited accelerated cardiomyopathy, resulting in severe HF and premature mortality within 2 months.8 Therefore, in vivo analyses started at the age of 4 weeks and were repeated every week until 7 weeks of age (due to severe HF in PLN-R14Δ/Δ mice, later time points could not be investigated).
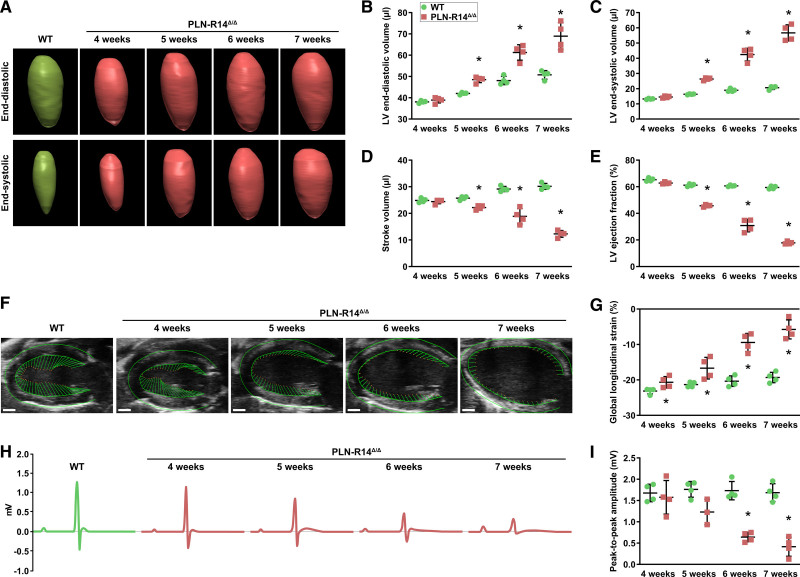
To accurately determine cardiac morphology and contractility, we generated 3-dimensional reconstructions of the LVs using echocardiography (Figure 1A).27 We observed that LV end-diastolic volume, end-systolic volume, stroke volume, and ejection fraction (LVEF) of PLN-R14Δ/Δ mice were not significantly different from control at 4 weeks of age (Figure 1B through 1E), but GLS was significantly impaired (Figure 1F and 1G). At the age of 5 weeks, GLS was further impaired, LV volumes were significantly increased, and stroke volume and LVEF were significantly decreased in PLN-R14Δ/Δ mice as compared with 5-week-old WT littermates. At 6 and 7 weeks of age, ventricular dilatation and dysfunction progressively worsened. WT mice also showed gradually increasing LV volumes as a result of physiological growth associated with aging (Figure I in the Data Supplement). Conventional 2-dimensional short-axis M-mode echocardiographic analysis demonstrated similar outcomes (Figure II in the Data Supplement).
Figure 1.
PLN-R14Δ/Δ mice develop progressive dilated cardiomyopathy with heart failure between 4 and 7 wk of age. A through E, Representative 3-dimensional echocardiographic reconstructions of left ventricular (LV) end-diastolic (top) and end-systolic (bottom) volumes of male WT (wild-type; 5 wk old) and PLN-R14Δ/Δ mice (A) with quantification of end-diastolic volume (B), end-systolic volume (C), stroke volume (D), and ejection fraction (E) in WT and PLN-R14Δ/Δ mice at 4, 5, 6, and 7 wk of age (n=4 per group). F and G, Representative long-axis B-mode echocardiographic images of male WT (5 wk old) and PLN-R14Δ/Δ mice with delineation of LV epicardial and endocardial borders showing vectors that indicate direction and magnitude of wall motion (scale bars, 1 mm; F) with quantification of global longitudinal strain in WT and PLN-R14Δ/Δ mice at 4, 5, 6, and 7 wk of age (n=4 per group; G). H and I, Representative average tracings of 1-min ECG recordings of male WT (5 wk old) and PLN-R14Δ/Δ mice (H) with quantification of peak-to-peak amplitudes in WT and PLN-R14Δ/Δ mice at 4, 5, 6, and 7 wk of age (n=4 per group, except n=3 for 5-wk-old PLN-R14Δ/Δ mice; I). Data are presented as mean±SD. *P<0.05 vs age-matched WT mice (Mann-Whitney U test).
Additionally, ECG voltages of PLN-R14Δ/Δ mice were not significantly different from control at 4 weeks of age and thereafter gradually decreased over time, which is a typical hallmark of PLN-R14del–related cardiomyopathy,28 while peak-to-peak amplitude remained unchanged over time in WT mice (Figure 1H and 1I). Diminished ECG potentials were seen in 5-week-old PLN-R14Δ/Δ mice and reached statistical significance compared with age-matched control mice starting at the age of 6 weeks. Other ECG parameters are shown in Figure III in the Data Supplement.
Intracardiomyocyte PLN Protein Aggregation Precedes Functional Deficits
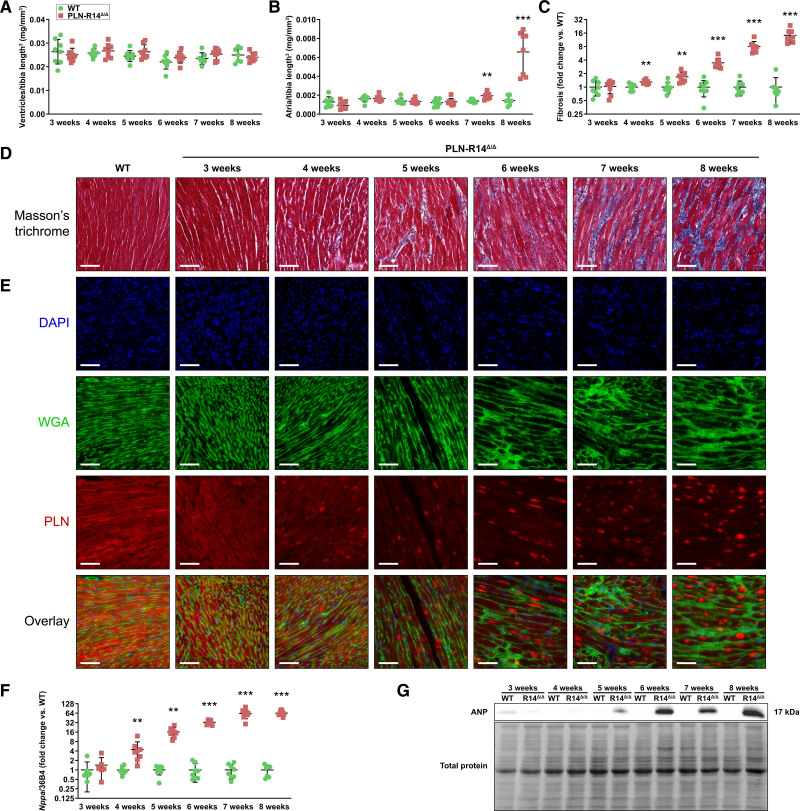
In addition to functional analyses, we collected heart tissue from 3- to 8-week-old PLN-R14Δ/Δ and WT mice with weekly intervals. At sacrifice, biventricular weights, normalized to tibia length to the power 3,14 were not significantly different between groups, indicating the absence of ventricular hypertrophy (Figure 2A). Atrial weights were not significantly different between 3- to 6-week-old PLN-R14Δ/Δ and WT mice but were significantly and progressively increased in PLN-R14Δ/Δ mice as compared with age-matched controls at 7 and 8 weeks of age as a result of volume overload due to impaired contractility (Figure 2B).
Figure 2.
Hearts of PLN-R14Δ/Δ mice undergo extensive remodeling between 3 and 8 wk of age. A and B, Ratios of biventricular (A) and atrial (B) weights to tibia length to the power 3 of WT (wild-type) and PLN-R14Δ/Δ mice at 3, 4, 5, 6, 7, and 8 wk of age (n=8 per group, mixed males and females). C and D, Quantification of myocardial fibrosis in WT and PLN-R14Δ/Δ hearts at 3, 4, 5, 6, 7, and 8 wk of age, shown as fold change compared with age-matched WT mice (n=8 per group, mixed males and females; C) with representative images of Masson trichrome–stained cardiac tissue sections (scale bars, 70 μm; D). E, Representative fluorescent staining images of WT (5 wk old) and PLN-R14Δ/Δ hearts at 3, 4, 5, 6, 7, and 8 wk of age stained for nuclei (4′,6-diamidino-2-phenylindole [DAPI]) in blue (top), extracellular matrix (WGA [wheat germ agglutinin]) in green (second), PLN (phospholamban) in red (third), and an overlay of all channels (bottom; scale bars, 70 μm). F, Left ventricular gene expression levels of Nppa normalized to housekeeping gene Rplp0 (36B4) of WT and PLN-R14Δ/Δ mice at 3, 4, 5, 6, 7, and 8 wk of age as measured by quantitative polymerase chain reaction (qPCR), shown as fold change compared with age-matched WT mice (n=8 per group, mixed males and females). G, Western blot images of relative protein expression levels of ANP (natriuretic peptide type A; top) and total protein levels (bottom) of WT and PLN-R14Δ/Δ mice at 3, 4, 5, 6, 7, and 8 wk of age. Full blot images are included in Figure IV in the Data Supplement. Data are presented as mean±SD. **P<0.01 and ***P<0.001 vs age-matched WT mice (Mann-Whitney U test).
Masson trichrome stain of formalin-fixed, paraffin-embedded midventricular sections demonstrated equal levels of myocardial collagen in PLN-R14Δ/Δ and WT mice at the age of 3 weeks (Figure 2C and 2D). At 4 weeks of age, a mild (1.3-fold) but significant (P=0.002) increase of fibrotic tissue was found in PLN-R14Δ/Δ hearts, which rapidly increased thereafter to ≈15-fold increased fibrosis at the age of 8 weeks.
Immunofluorescence revealed uniform distribution of PLN proteins in cardiomyocytes of WT mice (Figure 2E). In contrast, intracardiomyocyte PLN protein aggregates were observed in hearts of PLN-R14Δ/Δ mice, which, although less prominently, were already present when mice were 3 weeks old, before any other abnormalities were seen. In the following weeks, localization of PLN proteins increasingly progressed from a diffuse toward an aggregated distribution.
In line with in vivo functional observations, gene and protein expression levels of Nppa/ANP, a well-established HF marker,29 confirmed the absence of cardiomyocyte distress at the age of 3 weeks, followed by gradual increases in the subsequent weeks with significantly elevated ANP levels compared with control starting at 4 weeks of age, which ultimately reached a ≈64-fold increase at end stage (Figure 2F and 2G).
Protein Homeostasis Is Differentially Regulated in PLN-R14Δ/Δ Hearts But Not in Mouse Models of Myocardial Ischemia
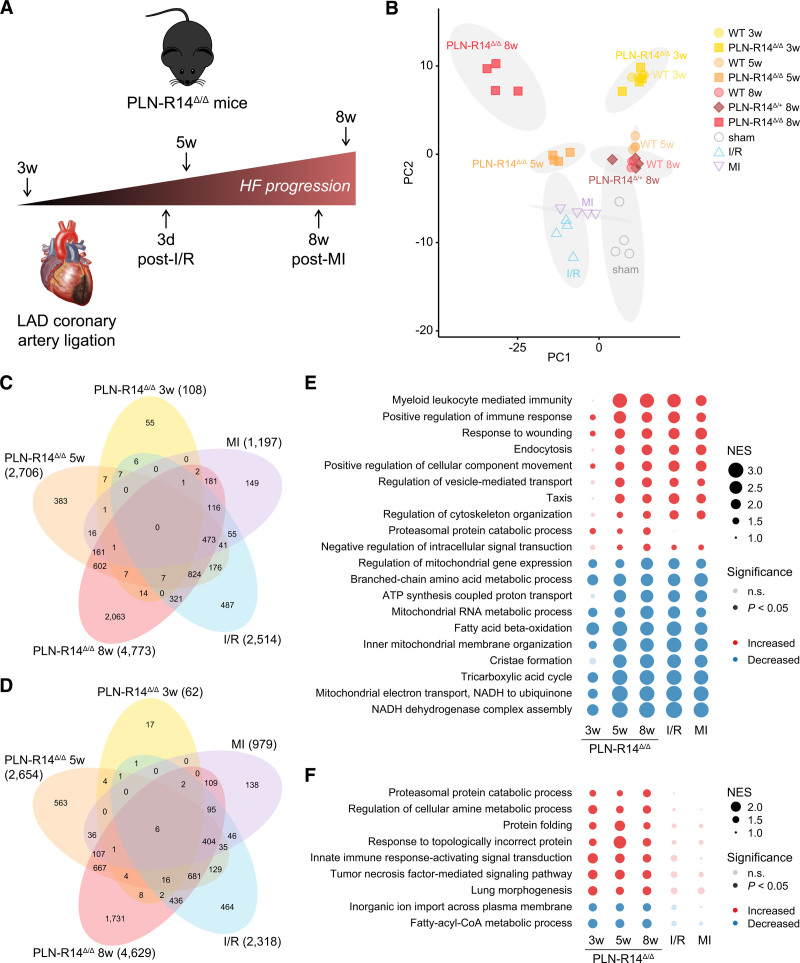
To investigate the molecular changes that underlie PLN-R14del–related cardiomyopathy, we performed RNA-Seq on LV tissue of PLN-R14Δ/Δ, PLN-R14Δ/+, and WT mice. We included hearts at multiple stages of disease: (1) before onset of HF or cardiac remodeling (3 weeks of age)8; (2) when the first signs of LV dysfunction became apparent with mild fibrosis (5 weeks of age); and (3) end-stage HF, indicated by severe LV dilatation and dysfunction, atrial hypertrophy, and extensive fibrosis (8 weeks of age; Figure 3A). Additionally, we aimed to distinguish between PLN-R14del–related processes and generic HF-related gene expression changes by including LVs of mice undergoing cardiac ischemia by temporary (I/R) or permanent (MI) ligation of the left anterior descending coronary artery.10 Myocardial I/R injury led to a rapid decline of LVEF within 1 week with an acute induction of genes involved in cardiac remodeling as early as 3 days after injury (Figure V in the Data Supplement).10 MI surgery resulted in more severe cardiac dilatation and functional impairment, together with aggravated fibrotic and inflammatory responses after 8 weeks, implying chronic stress (Figure VI in the Data Supplement).10 Based on these findings, we included samples of acute (3 days after I/R), chronic pathological cardiac remodeling (8 weeks post-MI), and corresponding sham controls (Figure 3A).
Figure 3.
RNA sequencing identified differential regulation of protein homeostasis systems in PLN-R14Δ/Δ hearts but not in models of myocardial ischemia. A, Schematic overview of the left ventricular tissues that were included for RNA sequencing and the corresponding stage of heart failure development. B, Principal component (PC) analysis plot of RNA sequencing results of 3-, 5-, and 8-wk-old WT (wild-type; circles) and PLN-R14Δ/Δ (squares) mouse hearts, 8-wk-old PLN-R14Δ/+ (diamonds) mice, mice 3 d post-ischemia/reperfusion (I/R; triangles, up) or 8 wk post–myocardial infarction (MI; triangles, down), and corresponding sham mice (open circles) with 95% confidence ellipses of clusters marked in gray (n=4 per group, except n=3 for 8-wk-old WT mice, all male). Information on genes contributing to variance in PC1 (x axis) and PC2 (y axis) is shown in Figure VII in the Data Supplement. C and D, Venn diagrams of significantly upregulated (C) and downregulated (D) genes in 3-, 5-, and 8-wk-old PLN-R14Δ/Δ mice and mice 3 d post-I/R or 8 wk post-MI as compared with corresponding controls. E, Top 10 most significantly upregulated and downregulated gene ontology biological processes in 3-, 5-, and 8-wk-old PLN-R14Δ/Δ mice and mice 3 d post-I/R or 8 wk post-MI as compared with corresponding controls. For response to wounding, positive regulation of immune response, and fatty acid β-oxidation, heat maps of mRNA levels of the lead-edge genes are depicted in Figure VIII in the Data Supplement. F, Gene ontology biological processes that are significantly upregulated or downregulated in 3-, 5-, and 8-wk-old PLN-R14Δ/Δ mice but not in hearts 3 d post-I/R or 8 wk post-MI. For proteasomal protein catabolic process, a heat map of mRNA levels of the lead-edge genes is depicted in Figure VIII in the Data Supplement. An overview of the enrichment data can be found in Tables I through V in the Data Supplement. HF indicates heart failure; LAD, left anterior descending; n.s., not significant; and NES, normalized enrichment score.
We performed PC analysis and found that transcriptomes of PLN-R14Δ/Δ hearts were comparable to WT at 3 weeks of age, while diverging from age-matched controls over time, resulting in distinct clusters at the age of 5 and 8 weeks (Figure 3B). Similarly, I/R and MI hearts had a distinct gene expression pattern compared with sham hearts. Genes that contributed to the variance in PC 1, which accounts for 50% of the total variance, included HF-associated genes Nppa, Nppb (BNP [B-type natriuretic peptide]), and Myh7 (β-MHC [myosin heavy chain 7/beta]) and cardiac remodeling genes such as Col8a1, Timp1, Gdf15, Mmp3, Lgals3 (galectin-3), and Postn (Figure VII in the Data Supplement). Interestingly, 5- and 8-week-old PLN-R14Δ/Δ mice and mice post-I/R and post-MI formed separate clusters, indicating that, although both PLN-R14del and ischemic events lead to HF, there are clear transcriptional differences between the different etiologies.
The number of genes that was significantly differentially expressed in PLN-R14Δ/Δ mice as compared with age-matched controls increased over time (Figure 3C and 3D). While there was substantial overlap with genes that were differentially expressed post-I/R or post-MI, a considerable number of genes was uniquely altered in PLN-R14Δ/Δ mice. We performed functional enrichment analysis to identify gene programs that were differentially regulated between disease states and their matched controls. Comparison of the most significantly enriched gene ontology (GO) terms for biological processes demonstrated shared features that are universally affected in HF, including activation of inflammatory (myeloid leukocyte mediated immunity, positive regulation of immune response) and fibrotic (response to wounding) pathways (Figure 3E). Processes that were decreased in all disease states were involved in mitochondrial energy production: branched chain amino acid metabolic process, fatty acid β-oxidation, tricarboxylic acid cycle, and ATP synthesis coupled proton transport. Indeed, looking at the leading-edge genes of these GO terms, a similar differential expression pattern was observed in both PLN-R14del–related cardiomyopathy and ischemic HF (Figure VIII in the Data Supplement).
Interestingly, comparing all significantly enriched GO terms between PLN-R14Δ/Δ hearts and post-I/R or post-MI hearts, we observed a striking enrichment for proteostasis (proteasomal protein catabolic process, protein folding, response to topologically incorrect protein) in PLN-R14Δ/Δ mice at every time point, while not (significantly) altered upon I/R or MI (Figure 3F). Comparing the leading-edge genes of these GO terms, we found that all upregulated GO terms, except for lung morphogenesis, consisted of genes involved in proteostasis, including endoplasmic reticulum stress genes (Atf4, Ddit3, Derl1, Derl3, Dnajb4, Dnajb9, and Edem3), heat shock proteins (Hspa1a, Hspa1b, and Hspa5), proteasomal genes (Psma2, Psmb1, Psmd5, and Psmd13), and a ubiquitin-conjugating E2 enzyme gene (Ube2g2) that are only identified in PLN-R14Δ/Δ mice but not post-I/R or post-MI (Figure VIII in the Data Supplement; Tables I through V in the Data Supplement). Notably, these processes were already significantly enriched in PLN-R14Δ/Δ mice at the age of 3 weeks, before functional deficits have developed.
Protein Quality Control Is Also Differentially Regulated on the Protein Level
Finally, we performed mass spectrometry on LVs of PLN-R14Δ/Δ, PLN-R14Δ/+, and WT mice at the same time points to determine how the differentially regulated genes resulted in changes in the proteome. Importantly, although PLN protein expression decreased over time for all genotypes, total PLN levels were not significantly different between WT, PLN-R14Δ/+, and PLN-R14Δ/Δ mice of the same age, except for a downregulation in 8-week-old PLN-R14Δ/Δ mice, which is known to occur in HF (Figure IX in the Data Supplement).30 An abundance of the PLN peptide containing the Arg14 amino acid (PLN-WT peptide) was approximately half in PLN-R14Δ/+ mice compared with WT mice, whereas the PLN-WT peptide was absent in PLN-R14Δ/Δ mice, confirming the anticipated expression levels of PLN-WT and PLN-R14del proteins in this mouse model.
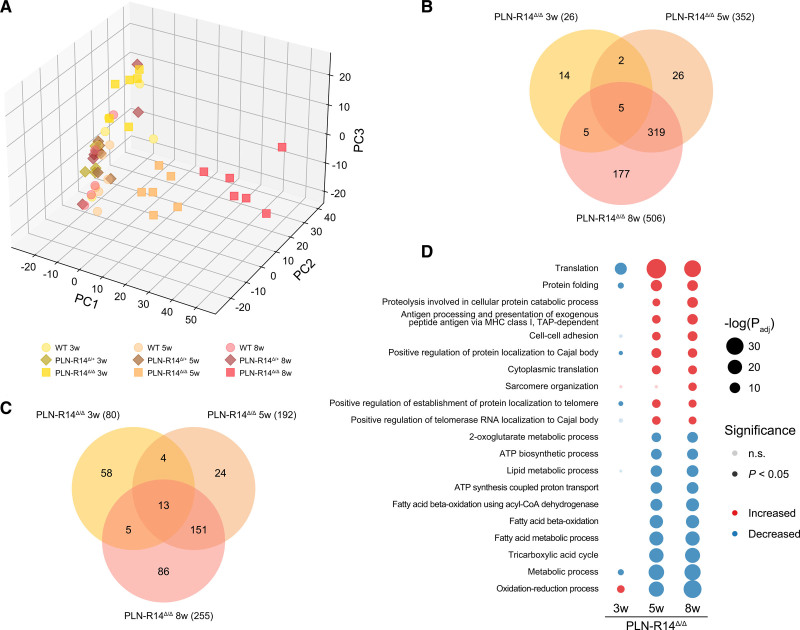
Proteomics analysis identified 3255 proteins with 1075 consistently quantified in >70% of samples and used for statistical and PC analysis analysis (Table VI in the Data Supplement). Similar to RNA-Seq, PC analysis of LV protein levels demonstrated that PLN-R14Δ/Δ mice clustered with WT mice at 3 weeks of age but formed separate clusters at 5 and 8 weeks of age, diverging from control over time (Figure 4A). Consistently, as PLN-R14Δ/Δ mice aged and developed HF, an increasing number of proteins was significantly (P<0.05) upregulated (Figure 4B) or downregulated (Figure 4C) relative to age-matched WT mice. At the age of 5 and 8 weeks, proteins that were elevated were most significantly enriched for GO terms that indicate myocardial remodeling (cell-cell adhesion, sarcomere organization), while significantly reduced proteins showed enrichment mainly for metabolism and mitochondrial functions (oxidation-reduction process, metabolic process, tricarboxylic acid cycle, ATP biosynthetic process), corroborating the observed LV dysfunction. Importantly, the most significantly enriched biological processes included terms of proteostasis, for example, protein folding and proteolysis involved in cellular protein catabolic process (Figure 4D). In fact, in the majority of enriched biological processes, many of the proteins that induced the observed enrichment play a role in proteostasis (Tables VII through IX in the Data Supplement). For example, enrichment of the process called “antigen processing and presentation of exogenous peptide antigen via MHC (myosin heavy chain) class I, transporter associated with antigen processing (TAP)-dependent” was solely driven by proteins of the proteasome complex (Psma3, Psmb4, Psmb7, and Psma4). Additionally, proteins of TRiC (T-complex protein ring complex; corresponding genes: Cct4, Cct2, Cct7, and Cct8)—a chaperonin complex that aids protein folding31—led to enrichment of the processes termed positive regulation of protein localization to Cajal body, positive regulation of establishment of protein localization to telomere, and positive regulation of telomerase RNA localization to Cajal body. Thus, changes of mRNA levels were generally well translated to the protein level, confirming the HF phenotype of PLN-R14Δ/Δ mice at 5 and 8 weeks of age and differential regulation of proteostatic systems, even before disease onset.
Figure 4.
Mass spectrometry revealed differential regulation of proteins involved in protein quality control in hearts of PLN-R14Δ/Δ mice. A, Principal component (PC) analysis plot of mass spectrometry results of 3-, 5-, and 8-wk-old WT (wild-type; circles), PLN-R14Δ/+ (diamonds), and PLN-R14Δ/Δ (squares) mice (n=6 per group, except n=5 for 8-wk-old WT mice, all male). B and C, Venn diagrams of significantly upregulated (B) and downregulated (C) proteins in 3-, 5-, and 8-wk-old PLN-R14Δ/Δ mice as compared with age-matched WT mice. D, Top 10 most significantly upregulated and downregulated gene ontology biological processes in 3-, 5-, and 8-wk-old PLN-R14Δ/Δ mice as compared with age-matched WT mice. An overview of the enrichment data can be found in Tables VII through IX in the Data Supplement. MHC indicates myosin heavy chain; n.s., not significant; Padj, adjusted P; and TAP, transporter associated with antigen processing.
Discussion
In this study, we describe in detail the progression of cardiomyopathy in PLN-R14Δ/Δ mice (summarized in Figure 5). These results provide important information on the sequence of pathophysiological manifestations of PLN-R14del–related cardiomyopathy. Importantly, we demonstrate that PLN aggregates are formed early during the disease, before functional impairment occurs. RNA-Seq and mass spectrometry of cardiac tissues show that genes and proteins that are part of protein quality control pathways are indeed differentially regulated. Interestingly, these pathways were not, or to a far lesser extent, activated in models of ischemic HF, and, therefore, we conclude that these pathways are specific to PLN-R14del–related cardiomyopathy.
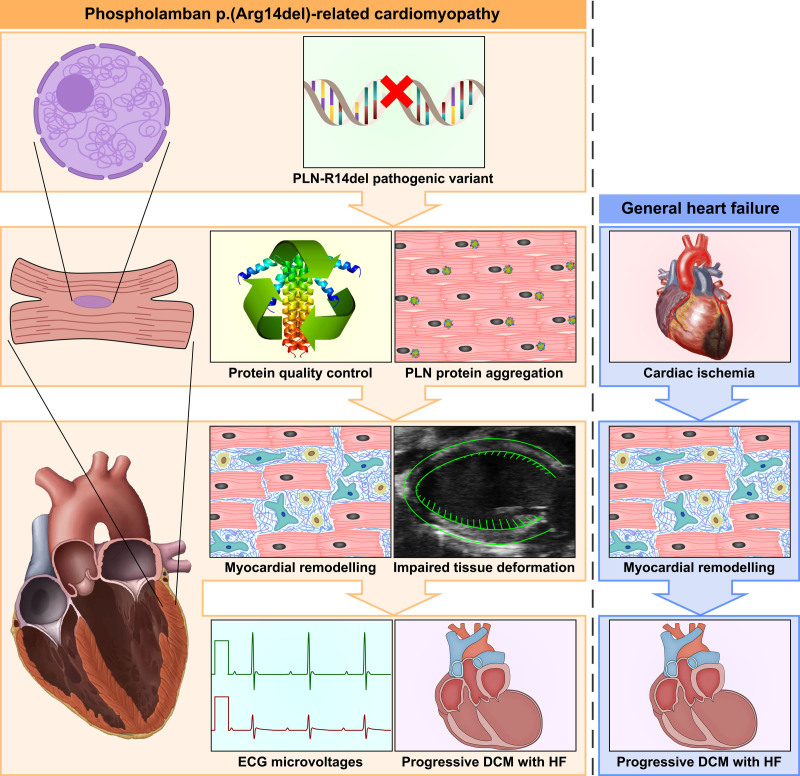
Figure 5.
Graphic abstract summarizing the main findings of the current study. In PLN-R14Δ/Δ mice, the PLN-R14del pathogenic variant resulted in intracardiomyocyte PLN (phospholamban) protein aggregation together with alterations in protein homeostasis systems at 3 wk of age, before onset of other abnormalities or functional deficits. At 4 wk of age, slightly increased amounts of myocardial fibrosis were observed, and left ventricular tissue deformation was impaired. At the age of 5 wk, ventricular dilatation, impaired contractility, and reduced ECG voltages became apparent, which progressively worsened in the weeks thereafter. Ultimately, volume overload resulted in atrial hypertrophy after 7 wk and premature death around the age of 2 mo. While myocardial remodeling and contractile dysfunction were also observed in models of ischemic heart failure (HF), aberrations in protein quality control were exclusively found in PLN-R14Δ/Δ mice. DCM indicates dilated cardiomyopathy.
At 3 weeks of age, PLN-R14Δ/Δ mice had normal cardiac function and morphology, but altered proteostasis and PLN protein distribution were already observed. One week later, marginally increased myocardial fibrosis, elevated ANP levels, and compromised GLS became apparent. Thereafter, PLN-R14Δ/Δ mice progressively developed ventricular dilatation, impaired contractility, and reduced ECG voltages. Finally, overt HF resulted in volume overload and atrial enlargement at 7 weeks of age. The unbiased approach of combined transcriptomics and proteomics revealed a progressive increase of myocardial stress response, remodeling, and inflammation together with dysfunctional energy production in PLN-R14Δ/Δ hearts, which is in accordance with the cardiomyopathy progression in vivo. PLN-R14Δ/+ littermates demonstrated no expression alterations, in line with the absence of a functional phenotype at this age.8 However, since these are generic processes that are consequential to HF in general, we compared our findings to models of I/R and MI. As anticipated, 3 days after myocardial I/R, we detected activated remodeling and inflammatory pathways, whereas a sustained response to established damage was observed 8 weeks post-MI. Although there was substantial overlap between PLN-R14del–related cardiomyopathy and ischemic HF, we found that proteostasis was exclusively affected in PLN-R14Δ/Δ mice. Of note, ischemic models may not provide an ideal comparison as it involves more localized cardiac injury, but clearly protein homeostasis is an important hallmark of PLN-R14del–related cardiomyopathy. These findings are consistent with a study of te Rijdt et al,16 which showed that PLN protein aggregates were only found in ventricular tissue specimens of PLN-R14del carriers and not in other genetic cardiomyopathies, and, therefore, are specific to this disease. Furthermore, 2 recent studies in human induced pluripotent stem cell–derived cardiomyocytes, with validation in human end-stage HF specimens, have linked PLN-R14del–related cardiomyopathy to the unfolded protein response and disturbed endoplasmic reticulum.32,33
An important strength of this study is the investigation of cardiac tissues at different disease stages. Generally, using patient material is hindered by the limited availability of tissue samples.34 Furthermore, most human tissue specimens are collected from surgical or postmortem biopsies and are mostly derived from end-stage HF.34 Analyses of advanced disease states are greatly influenced by myocardial injury and extensive fibrosis. Moreover, appropriate control tissues are even scarcer. Availability of PLN-R14del mice—a clinically translatable model—allows for temporal examinations, which provide insights into the molecular changes that underlie the disease. This way, we demonstrated that aberrations in proteostasis and PLN protein aggregation were already present before functional impairment or other tissue abnormalities were observed, implicating that these cellular perturbations may lead to cardiomyocyte death and subsequent tissue injury, organ dysfunction, and mortality. Together with the finding that protein aggregation was unique to PLN-R14del–related cardiomyopathy, these data suggest that targeting PLN protein aggregation may provide a promising target for therapy. We have previously demonstrated that PLN-R14Δ/Δ mice do not benefit from standard HF therapy,8 further underlining the importance of problem-targeting therapeutic approaches.
It has become increasingly recognized that strain analysis can accurately quantify myocardial mechanics and identify subclinical LV dysfunction, thereby providing a better prognostic value than LVEF.35 Also in this study, we found that GLS impairment preceded reduced LVEF. Notably, we observed an increased amount of collagen deposition at the same time point. Several studies have shown that fibrosis is an early hallmark of PLN-R14del–related cardiomyopathy.28,36 As fibrotic tissue is stiffer than myocardium,37 these observations are likely related. Taken together, quantification of GLS and fibrosis can have great prognostic value for PLN-R14del carriers.
An important limitation of this study is that, although the employed -omics technologies provide great insights on mRNA and protein levels, these techniques do not capture the full complexity of cellular dynamics. Indeed, PLN regulates cardiomyocyte calcium handling via SERCA inhibition, and it has been reported that the PLN-R14del pathogenic variant results in superinhibition of the SERCA activity.6,38 Although we did not observe aberrant calcium handling in the data presented here, it may be difficult to detect adaptations in calcium homeostasis solely based on mRNA or protein levels. To study the effect of the PLN-R14del pathogenic variant on calcium cycling, calcium imaging would be required. Therefore, it is important to note that, based on our current findings, dysregulation of calcium homeostasis cannot be excluded from the pathophysiology of PLN-R14del–related cardiomyopathy. Second, whole tissue sequencing is inherently limited by the presence of nonmyocyte cells.34 Single-cell sequencing could possibly provide specific insight into the molecular perturbations in individual cardiomyocytes. Finally, it has to be noted that this study was performed with homozygous PLN-R14del mice, whereas until now all carriers were found heterozygotes.3,4 Since this model exerts the same characteristics of human patients, we believe it is an accurate representation of the disease, and that in the absence of PLN-WT proteins, the PLN-R14del pathogenic variant leads to an accelerated phenotype in PLN-R14Δ/Δ mice.8 It is often observed that mouse models of genetic dilated cardiomyopathy exert a fast-forward phenotype in homozygotes compared with heterozygotes.39,40
In conclusion, alterations in proteostasis and aggregation of PLN proteins are among the first hallmarks of PLN-R14del–related cardiomyopathy and can be observed before the onset of HF. Additionally, these manifestations are unique to the PLN-R14del pathogenic variant. Therefore, targeting PLN protein aggregates or boosting proteostatic systems could provide novel therapeutic targets for this disease.
Article Information
Acknowledgments
We acknowledge E. Marloes Schouten, Canxia Shi, and Pablo I. Sánchez Aguilera for their excellent technical assistance and the Utrecht Sequencing Facility for providing sequencing service and data.
Sources of Funding
This work was supported by the Netherlands Cardiovascular Research (CVON) and Dutch CardioVascular Alliance (DCVA) initiatives of the Dutch Heart Foundation (2020B005 DCVA-DOUBLE-DOSE; 2018-30 CVON-PREDICT2; 2017-21 CVON-SHE-PREDICTS-HF; 2017-11 CVON-RED-CVD; 2014-40 CVON-DOSIS), the Cure PhosphoLambaN-induced cardiomyopathy (Cure-PLaN) initiative of the Leducq Foundation, and the European Research Council Consolidator grant (ERC CoG) 818715 (SECRETE-HF). Generation of PLN-R14del mice was supported by the de Boer Foundation, Ubbo Emmius Fund, Netherlands Heart Institute, and PLN Foundation. Dr Boogerd was supported by the European Union Horizon 2020 research and innovation program (Marie Skłodowska-Curie grant 751988). The Utrecht Sequencing Facility is subsidized by the University Medical Center Utrecht, Hubrecht Institute, University of Utrecht, and Netherlands X-Omics Initiative (NWO project 184.034.019).
Disclosures
The University Medical Center Groningen, which employs several of the authors, has received research grants and fees from Abbott, AstraZeneca, Boehringer Ingelheim, Cardior Pharmaceuticals, Ionis Pharmaceuticals, Novo Nordisk, and Roche. Dr de Boer has received speaker fees from Abbott, AstraZeneca, Bayer, Novartis, and Roche. Dr van der Meer received consultancy fees from Novartis, Corvidia, Servier, Vifor Pharma, AstraZeneca, Pfizer, Pharmacosmos, and Ionis Pharmaceuticals. The other authors report no conflicts.
Supplemental Materials
Expanded Methods
Figures I–IX
Tables I–IX
References 41-50
Supplementary Material
Nonstandard Abbreviations and Acronyms
- β-MHC
- myosin heavy chain 7/beta
- ANP
- atrial natriuretic peptide
- BNP
- B-type natriuretic peptide
- GLS
- global longitudinal strain
- GO
- gene ontology
- HF
- heart failure
- I/R
- ischemia-reperfusion
- LV
- left ventricle
- LVEF
- left ventricular ejection fraction
- MHC
- myosin heavy chain
- MI
- myocardial infarction
- PC
- principal component
- PLN
- phospholamban
- RNA-Seq
- RNA sequencing
- SERCA
- sarco/endoplasmic reticulum Ca2+-ATPase
- TRiC
- T-complex protein ring complex
- WGA
- wheat germ agglutinin
- WT
- wild-type
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCHEARTFAILURE.121.008532.
For Sources of Funding and Disclosures, see page 1258.
Contributor Information
Tim R. Eijgenraam, Email: t.r.eijgenraam@umcg.nl.
Cornelis J. Boogerd, Email: k.boogerd@hubrecht.eu.
Nienke M. Stege, Email: n.m.stege@umcg.nl.
Vivian Oliveira Nunes Teixeira, Email: v.oliveira.nunes.teixeira@umcg.nl.
Martin M. Dokter, Email: m.m.dokter@umcg.nl.
Lukas E. Schmidt, Email: lukas.schmidt@kcl.ac.uk.
Xiaoke Yin, Email: xiaoke.yin@kcl.ac.uk.
Konstantinos Theofilatos, Email: konstantinos.theofilatos@kcl.ac.uk.
Manuel Mayr, Email: manuel.mayr@kcl.ac.uk.
Peter van der Meer, Email: p.van.der.meer@umcg.nl.
Eva van Rooij, Email: E.vanrooij@hubrecht.eu.
Jolanda van der Velden, Email: j.vandervelden1@amsterdamumc.nl.
Herman H.W. Silljé, Email: h.h.w.sillje@umcg.nl.
References
- 1.Seferović PM, Polovina M, Bauersachs J, Arad M, Gal TB, Lund LH, Felix SB, Arbustini E, Caforio ALP, Farmakis D, et al. Heart failure in cardiomyopathies: a position paper from the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail. 2019; 21:553–576. doi: 10.1002/ejhf.1461 [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum AN, Agre KE, Pereira NL. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020; 17:286–297. doi: 10.1038/s41569-019-0284-0 [DOI] [PubMed] [Google Scholar]
- 3.van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, Cox MG, van Lochem LT, de Boer RA, Hofstra RM, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012; 14:1199–1207. doi: 10.1093/eurjhf/hfs119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Rijsingen IA, van der Zwaag PA, Groeneweg JA, Nannenberg EA, Jongbloed JD, Zwinderman AH, Pinto YM, Dit Deprez RH, Post JG, Tan HL, et al. Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ Cardiovasc Genet. 2014; 7:455–465. doi: 10.1161/CIRCGENETICS.113.000374 [DOI] [PubMed] [Google Scholar]
- 5.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003; 4:566–577. doi: 10.1038/nrm1151 [DOI] [PubMed] [Google Scholar]
- 6.Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA, Fan GC, Tsiapras D, Parekh RR, Dorn GW, 2nd, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci U S A. 2006; 103:1388–1393. doi: 10.1073/pnas.0510519103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Te Rijdt WP, van Tintelen JP, Vink A, van der Wal AC, de Boer RA, van den Berg MP, Suurmeijer AJ. Phospholamban p.Arg14del cardiomyopathy is characterized by phospholamban aggregates, aggresomes, and autophagic degradation. Histopathology. 2016; 69:542–550. doi: 10.1111/his.12963 [DOI] [PubMed] [Google Scholar]
- 8.Eijgenraam TR, Boukens BJ, Boogerd CJ, Schouten EM, van de Kolk CWA, Stege NM, Te Rijdt WP, Hoorntje ET, van der Zwaag PA, van Rooij E, et al. The phospholamban p.(Arg14del) pathogenic variant leads to cardiomyopathy with heart failure and is unreponsive to standard heart failure therapy. Sci Rep. 2020; 10:9819. doi: 10.1038/s41598-020-66656-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010; 8:e1000412. doi: 10.1371/journal.pbio.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du W, Piek A, Schouten EM, van de Kolk CWA, Mueller C, Mebazaa A, Voors AA, de Boer RA, Silljé HHW. Plasma levels of heart failure biomarkers are primarily a reflection of extracardiac production. Theranostics. 2018; 8:4155–4169. doi: 10.7150/thno.26055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zacchigna S, Paldino A, Falcão-Pires I, Daskalopoulos EP, dal Ferro M, Vodret S, Lesizza P, Cannatà A, Miranda-Silva D, Lourenço AP, et al. Toward standardization of echocardiography for the evaluation of left ventricular function in adult rodents: a position paper of the ESC working group on myocardial function. Cardiovasc Res. 2021; 117:43–59. doi: 10.1093/cvr/cvaa110 [DOI] [PubMed] [Google Scholar]
- 12.van der Pol A, Gil A, Tromp J, Silljé HHW, van Veldhuisen DJ, Voors AA, Hoendermis ES, Grote Beverborg N, Schouten EM, de Boer RA, et al. OPLAH ablation leads to accumulation of 5-oxoproline, oxidative stress, fibrosis, and elevated fillings pressures: a murine model for heart failure with a preserved ejection fraction. Cardiovasc Res. 2018; 114:1871–1882. doi: 10.1093/cvr/cvy187 [DOI] [PubMed] [Google Scholar]
- 13.Piek A, Koonen DPY, Schouten EM, Lindtstedt EL, Michaëlsson E, de Boer RA, Silljé HHW. Pharmacological myeloperoxidase (MPO) inhibition in an obese/hypertensive mouse model attenuates obesity and liver damage, but not cardiac remodeling. Sci Rep. 2019; 9:18765. doi: 10.1038/s41598-019-55263-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagdorn QAJ, Bossers GPL, Koop AC, Piek A, Eijgenraam TR, van der Feen DE, Silljé HHW, de Boer RA, Berger RMF. A novel method optimizing the normalization of cardiac parameters in small animal models: the importance of dimensional indexing. Am J Physiol Heart Circ Physiol. 2019; 316:H1552–H1557. doi: 10.1152/ajpheart.00182.2019 [DOI] [PubMed] [Google Scholar]
- 15.Yu L, Ruifrok WP, Meissner M, Bos EM, van Goor H, Sanjabi B, van der Harst P, Pitt B, Goldstein IJ, Koerts JA, et al. Genetic and pharmacological inhibition of galectin-3 prevents cardiac remodeling by interfering with myocardial fibrogenesis. Circ Heart Fail. 2013; 6:107–117. doi: 10.1161/CIRCHEARTFAILURE.112.971168 [DOI] [PubMed] [Google Scholar]
- 16.Te Rijdt WP, van der Klooster ZJ, Hoorntje ET, Jongbloed JDH, van der Zwaag PA, Asselbergs FW, Dooijes D, de Boer RA, van Tintelen JP, van den Berg MP, et al. Phospholamban immunostaining is a highly sensitive and specific method for diagnosing phospholamban p.Arg14del cardiomyopathy. Cardiovasc Pathol. 2017; 30:23–26. doi: 10.1016/j.carpath.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 17.Yurista SR, Silljé HHW, Oberdorf-Maass SU, Schouten EM, Pavez Giani MG, Hillebrands JL, van Goor H, van Veldhuisen DJ, de Boer RA, Westenbrink BD. Sodium-glucose co-transporter 2 inhibition with empagliflozin improves cardiac function in non-diabetic rats with left ventricular dysfunction after myocardial infarction. Eur J Heart Fail. 2019; 21:862–873. doi: 10.1002/ejhf.1473 [DOI] [PubMed] [Google Scholar]
- 18.Meijers WC, Maglione M, Bakker SJL, Oberhuber R, Kieneker LM, de Jong S, Haubner BJ, Nagengast WB, Lyon AR, van der Vegt B, et al. Heart failure stimulates tumor growth by circulating factors. Circulation. 2018; 138:678–691. doi: 10.1161/CIRCULATIONAHA.117.030816 [DOI] [PubMed] [Google Scholar]
- 19.Yurista SR, Matsuura TR, Silljé HHW, Nijholt KT, McDaid KS, Shewale SV, Leone TC, Newman JC, Verdin E, van Veldhuisen DJ, et al. Ketone ester treatment improves cardiac function and reduces pathologic remodeling in preclinical models of heart failure. Circ Heart Fail. 2021; 14:e007684. doi: 10.1161/CIRCHEARTFAILURE.120.007684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Withaar C, Meems LMG, Markousis-Mavrogenis G, Boogerd CJ, Silljé HHW, Schouten EM, Dokter MM, Voors AA, Westenbrink BD, Lam CSP, et al. The effects of liraglutide and dapagliflozin on cardiac function and structure in a multi-hit mouse model of heart failure with preserved ejection fraction. Cardiovasc Res. 2021; 117:2108–2124. doi: 10.1093/cvr/cvaa256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov M, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019. doi: 10.1101/060012 [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. doi: 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 26.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. doi: 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grune J, Blumrich A, Brix S, Jeuthe S, Drescher C, Grune T, Foryst-Ludwig A, Messroghli D, Kuebler WM, Ott C, et al. Evaluation of a commercial multi-dimensional echocardiography technique for ventricular volumetry in small animals. Cardiovasc Ultrasound. 2018; 16:10. doi: 10.1186/s12947-018-0128-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Te Rijdt WP, Ten Sande JN, Gorter TM, van der Zwaag PA, van Rijsingen IA, Boekholdt SM, van Tintelen JP, van Haelst PL, Planken RN, de Boer RA, et al. Myocardial fibrosis as an early feature in phospholamban p.Arg14del mutation carriers: phenotypic insights from cardiovascular magnetic resonance imaging. Eur Heart J Cardiovasc Imaging. 2019; 20:92–100. doi: 10.1093/ehjci/jey047 [DOI] [PubMed] [Google Scholar]
- 29.Suthahar N, Meems LMG, Ho JE, de Boer RA. Sex-related differences in contemporary biomarkers for heart failure: a review. Eur J Heart Fail. 2020; 22:775–788. doi: 10.1002/ejhf.1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yano M, Ikeda Y, Matsuzaki M. Altered intracellular Ca2+ handling in heart failure. J Clin Invest. 2005; 115:556–564. doi: 10.1172/JCI24159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grantham J. The molecular chaperone CCT/TRiC: an essential component of proteostasis and a potential modulator of protein aggregation. Front Genet. 2020; 11:172. doi: 10.3389/fgene.2020.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feyen DAM, Perea-Gil I, Maas RGC, Harakalova M, Gavidia AA, Arthur Ataam J, Wu TH, Vink A, Pei J, Vadgama N, et al. Unfolded protein response as a compensatory mechanism and potential therapeutic target in PLN R14del cardiomyopathy. Circulation. 2021; 144:382–392. doi: 10.1161/CIRCULATIONAHA.120.049844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuello F, Knaust AE, Saleem U, Loos M, Raabe J, Mosqueira D, Laufer S, Schweizer M, van der Kraak P, Flenner F, et al. Impairment of the ER/mitochondria compartment in human cardiomyocytes with PLN p.Arg14del mutation. EMBO Mol Med. 2021; 13:e13074. doi: 10.15252/emmm.202013074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harakalova M, Asselbergs FW. Systems analysis of dilated cardiomyopathy in the next generation sequencing era. Wiley Interdiscip Rev Syst Biol Med. 2018; 10:e1419. doi: 10.1002/wsbm.1419 [DOI] [PubMed] [Google Scholar]
- 35.Kalam K, Otahal P, Marwick TH. Prognostic implications of global LV dysfunction: a systematic review and meta-analysis of global longitudinal strain and ejection fraction. Heart. 2014; 100:1673–1680. doi: 10.1136/heartjnl-2014-305538 [DOI] [PubMed] [Google Scholar]
- 36.Posch MG, Perrot A, Geier C, Boldt LH, Schmidt G, Lehmkuhl HB, Hetzer R, Dietz R, Gutberlet M, Haverkamp W, et al. Genetic deletion of arginine 14 in phospholamban causes dilated cardiomyopathy with attenuated electrocardiographic R amplitudes. Heart Rhythm. 2009; 6:480–486. doi: 10.1016/j.hrthm.2009.01.016 [DOI] [PubMed] [Google Scholar]
- 37.Eijgenraam TR, Silljé HHW, de Boer RA. Current understanding of fibrosis in genetic cardiomyopathies. Trends Cardiovasc Med. 2020; 30:353–361. doi: 10.1016/j.tcm.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 38.Haghighi K, Pritchard T, Bossuyt J, Waggoner JR, Yuan Q, Fan GC, Osinska H, Anjak A, Rubinstein J, Robbins J, et al. The human phospholamban Arg14-deletion mutant localizes to plasma membrane and interacts with the Na/K-ATPase. J Mol Cell Cardiol. 2012; 52:773–782. doi: 10.1016/j.yjmcc.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du CK, Morimoto S, Nishii K, Minakami R, Ohta M, Tadano N, Lu QW, Wang YY, Zhan DY, Mochizuki M, et al. Knock-in mouse model of dilated cardiomyopathy caused by troponin mutation. Circ Res. 2007; 101:185–194. doi: 10.1161/CIRCRESAHA.106.146670 [DOI] [PubMed] [Google Scholar]
- 40.Jiang H, Hooper C, Kelly M, Steeples V, Simon JN, Beglov J, Azad AJ, Leinhos L, Bennett P, Ehler E, et al. Functional analysis of a gene-edited mouse model to gain insights into the disease mechanisms of a titin missense variant. Basic Res Cardiol. 2021; 116:14. doi: 10.1007/s00395-021-00853-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012; 28:3211–3217. doi: 10.1093/bioinformatics/bts611 [DOI] [PubMed] [Google Scholar]
- 42.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012; 28:2184–2185. doi: 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 44.Daley T, Smith AD. Predicting the molecular complexity of sequencing libraries. Nat Methods. 2013; 10:325–327. doi: 10.1038/nmeth.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 46.Pedegrosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011; 12:2825–2830 [Google Scholar]
- 47.Cattell RB. The scree test for the number of factors. Multivariate Behav Res. 1966; 1:245–276. doi: 10.1207/s15327906mbr0102_10 [DOI] [PubMed] [Google Scholar]
- 48.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007; 9:90–95. doi: 10.1109/MCSE.2007.55 [Google Scholar]
- 49.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000; 25:25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The Gene Ontology Consortium.. The gene ontology resource: enriching a gold mine. Nucleic Acids Res. 2021; 49:D325–D334. doi: 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.