Abstract
Listeria monocytogenes and Salmonella spp. are considered important foodborne pathogens that are commonly associated with foods of animal origin. The aim of this study was to perform molecular characterization of L. monocytogenes and Salmonella spp. isolated from biofilms of cattle and poultry slaughterhouses located in the Federal District and State of Goiás, Brazil. Fourteen L. monocytogenes isolates and one Salmonella sp. were detected in poultry slaughterhouses. No isolates were detected in cattle slaughterhouses. All L. monocytogenes isolates belonged to lineage II, and 11 different pulsotypes were detected. Pulsed-field gel electrophoresis analysis revealed the dissemination of two strains within one plant, in addition to the regional dissemination of one of them. The Salmonella isolate was identified via whole genome sequencing as Salmonella enterica serovar Minnesota ST548. In the sequence analysis, no premature stop codons were detected in the inlA gene of Listeria. All isolates demonstrated the ability to adhere to Caco-2 cells, while 50% were capable of invading them. Antimicrobial resistance was detected in 57.1% of the L. monocytogenes isolates, and resistance to sulfonamide was the most common feature. The tetC, ermB, and tetM genes were detected, and four isolates were classified as multidrug-resistant. Salmonella sp. was resistant to nine antimicrobials and was classified as multidrug-resistant. Resistance genes qnrB19, blaCMY-2, aac(6’)-Iaa, sul2, and tetA, and a mutation in the parC gene were detected. The majority (78.5%) of the L. monocytogenes isolates were capable of forming biofilms after incubation at 37°C for 24 h, and 64.3% were capable of forming biofilms after incubation at 12°C for 168 h. There was no statistical difference in the biofilm-forming capacity under the different evaluated conditions. Salmonella sp. was capable of forming biofilms at both tested temperatures. Biofilm characterization was confirmed by collecting the samples consistently, at the same sampling points, and by assessing biofilm formation in vitro. These results highlight the potential risk of cross-contamination in poultry slaughterhouses and the importance of surveillance and pathogen control maintenance programs within the meat production industry.
Introduction
The hygienic and sanitary conditions of the industrial environment are essential for ensuring food safety and quality [1–3]. One of the current challenges encountered in slaughtering and meat processing is the prevention of cross-contamination by microorganisms [4, 5]. Cross-contamination can occur via contact with bacterial biofilms due to the performance of improper hygiene and sanitization practices at the slaughter system. This leads to a decrease in food quality, resulting in pathogen transmission and the occurrence of recurring contamination of food [5, 6]. Listeria monocytogenes and Salmonella spp. are deemed important pathogens whose dissemination can be controlled to ensure food quality and safety. Their presence is a criterion for evaluation within the various aspects of quality control of the meat production process [7].
L. monocytogenes is recognized as the main etiological agent of listeriosis in humans [8]. It is a psychrophilic bacterium, that can develop on surfaces of meat production industries [9]. The species is subdivided into 13 serotypes, of which 1/2a, 1/2c, and 4b are the most commonly reported serotypes involved in cases of human listeriosis [10]. Owing to the difficulties and limitations encountered with methods of traditional serology, different subtyping methods have been developed, such as the differentiation of the main lineages assessed via polymerase chain reaction (PCR) [11]. Additionally, molecular typing methods with high discriminatory power, such as pulsed-field gel electrophoresis (PFGE), are widely used to assess clonal dissemination within industries [10, 12, 13]. There is considerable pathogenic potential among the different L. monocytogenes clones. [14]. The role of internalin A in Listeria, encoded by the inlA gene, is exemplary as its functionality is necessary for pathogen internalization [15–17]. The presence of premature stop codons (PMSCs) in this gene has been associated with clones with an attenuated invasion phenotype [18].
The genus Salmonella, which is responsible for salmonellosis, includes the Salmonella enterica species. It is a microorganism that is often identified as an etiological agent of food outbreaks, presenting a complex epidemiology regarding its transmission and distribution, and is therefore important for investigation in the assessment of risks to public health. [19]. The species demonstrates adherence to surfaces such as metals, glass and rubber, and exhibits the ability of biofilm formation [20–22].
Antimicrobial resistance (AMR) is considered a grave concern and risk to human health [23, 24]. To mitigate its risks, the World Health Organization established a global action plan [25]. One of the main strategies outlined in the plan is to monitor the spread of AMR. Considering the importance of monitoring L. monocytogenes and Salmonella spp. dissemination and the necessity of ensuring public health and safety, and owing to the limited investigations related to the presence of biofilms in Brazil, the objectives of this study were to detect biofilm-induced contamination points and the presence of these microorganisms in processing plants of cattle and poultry slaughterhouses, to characterize these strains by performing serotyping, AMR analysis, and to evaluate the in vitro biofilm formation capability of all isolates. Additionally, whole genome sequencing (WGS) of Salmonella spp. was performed in addition to the conduction of PFGE, sequencing of the inlA gene, and cell adhesion and invasion assays in Caco-2 cells for all Listeria strains.
Material and methods
Sample collection
Samples were collected from three poultry slaughterhouses (A, B and C) and two cattle slaughterhouses (D and E) located in the Federal District area (A, B and D) and in the State of Goiás (C and E). All slaughterhouses availed an official inspection service. All slaughterhouses agreed to participate voluntarily in the study. Sterile swabs (Absorve®, Brazil) and 25 cm2 molds were used to examine the surfaces of the installations, equipment, and utensils in cattle and poultry slaughterhouses [26]. The sampling locations were separated into the following two groups: installations (floors, walls, and drains of dirty and clean areas), and equipment (mats, evisceration tables, chutes, and different machinery), and this method was based on protocols described by Barros et al. 2007 [27] and Nicolau and Bolocan 2014 [26]. Sixteen visits (eight in cattle slaughterhouses and eight in poultry slaughterhouses) were conducted between March 2017 and September 2018, for the collection of 287 swabs. Of the swabs collected, 118 were obtained from cattle slaughterhouses and 169 were obtained from poultry slaughterhouses. The swabs were individually added to tubes containing a sterile transport solution (peptone water 0.1%; Himedia®, India) and transported in isothermal boxes to the Food Microbiology Laboratory at the University of Brasília for culturing and microbiological isolation. Sample processing was performed within 24 h of sample collection. Biofilms were characterized within a processing plant by repetitively collecting samples at the same locations within each plant but on different visits, to observe the recurrence of the investigated microorganisms [28], and by assessing their in vitro biofilm-forming capability. Studies have correlated the permanence of microorganisms on surfaces after subjection to cleaning and sanitization practices to the existence of biofilms on such surfaces [29, 30]. Thus, sample collections were conducted at different times, with the surfaces treated hygienically and sanitized between visits.
Isolation of L. monocytogenes and Salmonella spp
For the isolation of L. monocytogenes, swab samples were analyzed according to the methodology described by Ryser and Donnely 2015 [31]. Transport tubes containing swabs were homogenized in a tube shaker, with the subsequent addition of 1 mL of transport solution to 9 mL of UVM broth (Acumedia®, USA). After 24 h of incubation at 35°C, the culture (0.1 mL) was transferred to 9 mL of Fraser broth (Acumedia®, USA). Esculin hydrolysis was performed after incubation at 35°C for 24–48 h. Each sample was plated on Modified Oxoid (MOX) agar (Difco®, France). Presumptive colonies were incubated in Brain Heart Infusion (BHI) broth (Acumedia®, -USA) at 35°C for 24 h and were biochemically analyzed. Isolates with characteristics compatible with the selected genus were examined by PCR to confirm the species using the primers LIP1 [32] and LIP2A [33] for amplification of the prfA gene. PCR was performed as per methods described by Kérouanton et al. 2010 [33]. Positive controls were provided by Dr. Ernesto Hofer of the Oswaldo Cruz Foundation located in Rio de Janeiro.
To identify species of Salmonella, swab samples were analyzed as per protocols described by Ryser and Donnely 2015 [31] and ISO 6579/2002 [34]. After homogenization of transport tubes containing the collected swab samples, each homogenate was individually transferred to tubes containing 1% buffered peptone water (Acumedia®, USA). Samples were incubated at 37°C for 24 h. Aliquots were transferred to tetrathionate broth (Merck®, Germany), selenite cystine broth (Merck®, Germany), and Rappaport-Vassiliadis broth (Merck®, Germany). After incubation in selective media, samples were plated on Brilliant Green Phenol Red Agar (Acumedia®, USA), Xylose Lysine Deoxycholate (Merck®, Germany), and Bismuth Sulfite agars (Acumedia®, USA). Presumptive colonies were cultured on Nutrient agar (Acumedia®, USA) and were biochemically analyzed using appropriate tests [34]. This was followed by serology analysis using polyvalent anti-Salmonella somatic and flagellar sera (Probac®, Brazil). PCR amplification of the ompC gene was performed using the OMPCF and OMPCR primers [35] for confirmation of the genus [36]. Positive controls were provided by the Oswaldo Cruz Foundation.
Molecular serotyping of L. monocytogenes and Salmonella
L. monocytogenes subtyping was performed using multiplex PCR and primers lmo0737, lmo1118, ORF2819 and ORF2110, as per methods described by Doumith et al. [11] for differentiation into the four main lineages.
Salmonella spp. serovars were identified via WGS at the Laboratory of Bacterial Resistance and Therapeutic Alternatives at the Institute of Biomedical Sciences of the University of São Paulo (USP–São Paulo/SP). Genomic DNA extraction was performed using the PureLinkTM Genomic DNA Mini Kit (Life Technologies, CA, USA), and used for library preparation using the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA). Subsequently, the DNA quantified by Qubit 2.0 fluorometer (Life Technologies) was sequenced using the Illumina NextSeq PE instrument (Illumina Inc., San Diego, CA) using a paired-end (75bp) library. The short reads were trimmed with TrimGalore v0.6.5 (https://github.com/FelixKrueger/TrimGalore) and de novo assembly was performed using Unicycler v.0.4.8 [37]. Genome annotations were performed using NCBI PGAP v.3.2 (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/), and serovar was predicted via SeqSero2 analysis [38].
Pulsed-field gel electrophoresis of L. monocytogenes
PFGE was performed as per guidelines recommended by the Centers for Disease Control and Prevention [39]. The restriction enzyme AscI (Invitrogen®, Lithuania) was used at a concentration of 10 U/μL. Salmonella ser. Braenderup H9812 subjected to digestion with XbaI (Roche®, Germany) at a concentration of 10 U/μL was used as the standard, and was provided by the Enterobacteria Laboratory of the Oswaldo Cruz Foundation. The BioNumerics 7.7. software (Applied Maths) was used to analyze the fragments, with a Dice similarity coefficient of 1.5%. The dendrogram was constructed by analyzing the unweighted pair group method with arithmetic mean (UPGMA) clusters.
Analysis of the inlA gene sequence of L. monocytogenes
The sequencing of the inlA gene (2400bp) was performed following the protocol described by Poyart et al. 1996 [17]. Two gene fragments of 1157 bp and 760 bp were amplified, using primers O1, O2, O3, and O4 [17]. PCR products were purified using the PureLink kit (Invitrogen®, USA), following which they were quantified by using high mass marker (Invitrogen®, Lithuania) and were sequenced using the ABI3500 sequencer (Applied Biosystems).
Cell adhesion and invasion assays for L. monocytogenes
The Caco-2 human colon adenocarcinoma cell line was used. The cells were provided by Prof. Elaine Cristina Pereira De Martinis, from the Faculty of Pharmaceutical Sciences of Ribeirão Preto, USP. Cells were cultured as per methods described by Gaillard et al. 1987 [40]. Wells in polypropylene plates (Kasvi®) were seeded with cells at a final density of 1.0 × 105 cells per well. Adhesion assays were conducted as per protocols described by Moroni et al. 2006 [41]. The wells seeded with the cells were inoculated with a bacterial suspension using a volume adjusted to obtain a multiplicity of infection (MOI) of 100. After adding the Dulbecco’s modified Eagle medium (DMEM) (Gibco®, USA) supplemented with 10% fetal bovine serum (Gibco®, USA), the plates were incubated at 37°C for 2 h in a 5% CO2 atmosphere. The wells were rinsed three times with 1× phosphate-buffered saline (PBS; Laborclin®, Brazil). The cells were then subjected to treatment with a lysis buffer solution containing 0.1% Triton-X100 (Sigma-Aldrich®, USA), and incubated for 10 min under the aforementioned conditions. Viable bacterial cells were counted by seeding serial dilutions on BHI agar (Difco®, France) incubated at 37°C for 24 h. The counts were expressed as colony forming units (CFU)/mL. All tests were performed in triplicate. Adhesion of bacteria to Caco-2 cells was calculated (%) as (number of adherent cells × 100) / number of cells adhering to the well [41]. Invasion tests were performed as per methods described by Gaillard et al. 1987 [40] and Moroni et al. 2006 [41]. Cells were added to each well at an MOI of 100. Following incubation, the cells were subjected to washing steps with PBS as described above for the adhesion assay. After subjection to washing steps, 1 mg/mL gentamicin (Sigma-Aldrich®, Israel) was added to the wells and incubation was performed at 37°C for 1 h in a 5% CO2 atmosphere. The lysis buffer solution was added to each well, and viable bacterial cells were counted as described above. All tests were performed in triplicate. The percentage of bacterial invasion in Caco-2 cells was determined by using the following formula: % adhesion = (number of internalized recovered cells × 100) / number of cells adhering to the well.
Antibiogram and assessment of antimicrobial resistance genes
The AMR of L. monocytogenes isolates was evaluated by performing the disk diffusion assay using Mueller Hinton agar (Acumedia®, USA), as per methods described by the Clinical and Laboratory Standards Institute [42]. The antibiotics tested were ampicillin (10 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), doxycycline (30 μg), erythromycin (15 μg), gentamicin (10 μg), sulfonamides (300 μg), and tetracycline (30 μg). The interpretation of the zone of inhibition diameters was based on the standards prescribed for Staphylococcus spp. [43] which were defined by CLSI M100 [42], with the exception of standards for erythromycin and ampicillin, for which the standards defined by the European Committee on Antimicrobial Susceptibility Testing [44] were used for L. monocytogenes. The identification of resistance genes in L. monocytogenes isolates was performed using PCR. Genes related to the resistance to tetracycline (tetA, tetB, tetC, tetM), macrolides (ermA, ermB, ermC, ereA), amphenicols (cat1, cmlA), sulfonamides (sull), beta-lactams (ampC, blaSHV), and aminoglycosides (aac (3)-I) were investigated. Reactions were performed in a final volume of 25 μL, according to the conditions described in the studies reported in Table 1 for each pair of primers.
Table 1. Primers used to investigate antimicrobial resistance genes in Listeria monocytogenes isolates.
| Gene | Primer | Nucleotide sequence (5’-3’) | Size (pb) | Reference |
|---|---|---|---|---|
| aac(3)-I | aac(3)-I-F | ACCTACTCCCAACATCAGCC | 157 | Van et al., 2008 [45] |
| aac(3)-I-R | ATATAGATCTCACTACGCGC | |||
| ampC | AmpC-For | TTCTATCAAMACTGGCARCC | 550 | Schwartz et al., 2003 [46] |
| AmpC-Rev | CCYTTTTATGTACCCAYGA | |||
| bla SHV | blaSHV-F | TCGCCTGTGTATTATCTCCC | 768 | Van et al., 2008 [45] |
| blaSHV-R | CGCAGATAAATCACCACAATG | |||
| ermA | ermA-F | TCTAAAAAGCATGTAAAAGAA | 645 | Sutcliffe et al., 1996 [47] |
| ermA-R | CTTCGATAGTTTATTAATATTAGT | |||
| ermB | ermB-F | GAAAAGGTACTCAACCAAATA | 639 | Sutcliffe et al., 1996 [47] |
| ermB-R | AGTAACGGTACTTAAATTGTTTAC | |||
| ermC | ermC-F | TCAAAACATAATATAGATAAA | 642 | Sutcliffe et al., 1996 [47] |
| ermC-R | GCTAATATTGTTTAAATCGTCAAT | |||
| ereA | ere(A)-F | GCCGGTGCTCATGAACTTGAG | 419 | Van et al., 2008 [45] |
| ere(A)-R | CGACTCTATTCGATCAGAGGC | |||
| cat1 | CATIF | AGTTGCTCAATGTACCTATAACC | 547 | Van et al., 2008 [45] |
| CATIR | TTGTAATTCATTAAGCATTCTGCC | |||
| cmlA | cmlA-F | CCGCCACGGTGTTGTTGTTATC | 698 | Van et al., 2008[45] |
| cmlA-R | CACCTTGCCTGCCCATCATTAG | |||
| sul | sull-F | TTCGGCATTCTGAATCTCAC | 822 | Van et al., 2008 [45] |
| sulI-R | ATGATCTAACCCTCGGTCTC | |||
| tetA | tet(A)-F | GTGAAACCCAACATACCCC | 887 | Van et al., 2008 [45] |
| tet(A)-R | GAAGGCAAGCAGGATGTAG | |||
| tetB | tet(B)-F | CCTTATCATGCCAGTCTTGC | 773 | Van et al., 2008 [45] |
| tet(B)-R | ACTGCCGTTTTTTCGCC | |||
| tetC | tet(C)-F | ACTTGGAGCCACTATCGAC | 880 | Van et al., 2008 [45] |
| tet(C)-R | CTACAATCCATGCCAACCC | |||
| tetM | tet(M)-1 | GTTAAATAGTGTTCTTGGAG | 700 | Aarestrup et al., 2000 [48] |
| tet(M)-2 | CTAAGATATGGCTCTAACAA |
The AMR of Salmonella spp. was evaluated by performing the disk diffusion assay using Mueller Hinton agar (Acumedia®, USA), according to the CLSI protocol [42]. The tested antibiotics were nalidixic acid (30 μg), amoxicillin (10 μg), ampicillin (10 μg), cephalothin (30 μg), cefazolin (30 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), colistin (10 μg), doxycycline (30 μg), gentamicin (10 μg), sulfonamides (300 μg), and tetracycline (30 μg). To interpret the results, the standards prescribed for Enterobacteriaceae defined by the CLSI M100 [42] were used. The presence of resistance genes was evaluated via WGS using the ResFinder v.4.0. The presence of genes related to the resistance to the following antimicrobials was investigated: quinolones, tetracycline, nitroimidazole, sulfonamides, macrolides, rifampicin, glycopeptides, colistin, trimethroprim, fusidic acid, aminoglycosides, beta-lactams, oxazolidinone, and fosfomycin. Chromosomal mutations related to AMR in the gyrA, gyrB, pmrA, pmrB, parC, parE, and 16s_rrsD genes were investigated.
In vitro biofilm formation capacity
In vitro biofilm formation capacity of L. monocytogenes and Salmonella spp. isolates was evaluated by performing incubation at 37°C for 24 h and incubation at 12°C for 168 h (7 days). Tests were performed using polystyrene titration microplates as per methods described by Djordjevic et al. 2002 [49] and modified by Borges et al. 2018 [50]. TSB broth (Kasvi®, Italy) was used to prepare bacterial suspensions at a final density of 3 x 108 CFU/mL. All tests were performed in triplicate. After the performance of preparation, incubation, washing steps, staining procedures, and resuspension of each sample, the optical density (OD) of each well was measured using an ELx800 enzyme-linked immunosorbent assay reader (Biotek Instruments) at 490 nm. The OD value for each isolate was determined as the arithmetic mean of the absorbance readings obtained from the three wells. This value was compared with that of the negative control (ODn). The classification proposed by Stepanović et al. 2000 [51] was used to determine the capacity and intensity of biofilm formation. Statistical analysis of data for comparison of the capacity of biofilm formation in the two conditions tested was performed using the SAS software (SAS Inc.).Normality was analyzed using the Shapiro-Wilk test followed by the paired t-test for means comparison between the two groups.
Results and discussion
Detection of microorganisms in poultry and cattle slaughterhouses
Detection of L. monocytogenes
A total of 14 L. monocytogenes isolates were detected from 287 swabs collected from poultry and cattle slaughterhouses (4.87%). All isolates were detected in poultry slaughterhouses (A, B, and C), which represented 169 swabs (8.28%). L. monocytogenes was not detected in any of the 118 samples collected from the cattle slaughterhouses (D and E). The detection points for the isolates are listed in Table 2. In the Federal District and State of Goiás region, reports have described the presence of L. monocytogenes isolated from different sources [52, 53]. These findings support the results of the present study. The absence of L. monocytogenes in cattle slaughterhouses differs from the findings reported by Palma et al. 2016 [53], who described the presence of L. monocytogenes in samples obtained from cattle slaughterhouses. The difference between the studies may be related to the small number of industries available and the sampling visits that were conducted, which was due to the hesitation of the facilities to participate in the study. The difference may also reflect the adequate hygiene and sanitation conditions prevalent in these establishments.
Table 2. Detection of Listeria monocytogenes in swabs collected from poultry slaughterhouse facilities, equipment and utensils located in the Federal District and State of Goiás, Brazil.
| Region and establishment | DF | DF | DF | DF | DF | GO | GO | DF | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identification | A | A | A | A | A | C | C | B | ||||
| Swab collection points | Collection 1 | Collection 2 | Collection 3 | Collection 4 | Collection 5 | Collection 6 | Collection 7 | Collection 8 | Total Listeria monocytogenes strains | Total no. of swabs obtained from each collection point | Percentage of positive samples at each collection point (%) | Identification of isolates detected at each collection point |
| No. of swabs = 16 | No. of swabs = 16 | No. of swabs = 16 | No. of swabs = 15 | No. of swabs = 25 | No. of swabs = 25 | No. of swabs = 25 | No. of swabs = 31 | |||||
| Facilities | 4 | 56 | 7.14 | |||||||||
| Floor in clean area | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0 | |
| Drains in clean area | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 3 | 38 | 7.89 | 63A-1;69A-2; 117A-3 |
| Drains in dirty area | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.0 | |
| Walls in clean area | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 13 | 0.0 | 76A-2 |
| Walls in dirty area | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.0 | |
| Equipment and utensils | 10 | 113 | 8.84 | |||||||||
| Evisceration table | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 35 | 5.71 | 74A-2; 77A-2 |
| Mats in clean area | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 33 | 9,09 | 45A-2; 52A-2; 59A-2 |
| Chutes of meat | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | 5 | 13 | 38.46 | 42A-2; 54A-2; 72A-2; 78A-2; 88A-2 |
| Chutes of carcass | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0.0 | |
| Chutes of bones and viscera | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0.0 | |
| Hooks | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0.0 | |
| Machinery | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0.0 | |
| Total no. of L. monocytogenes isolates obtained in each collection | 0 | 0 | 1 | 0 | 0 | 5 | 7 | 1 | 14 | |||
| Total no. of swabs obtained in each collection | 16 | 16 | 16 | 15 | 25 | 25 | 25 | 31 | 169 |
The detection of L. monocytogenes in poultry slaughterhouses is similar to the findings reported in previous studies described in Brazil [4, 54–56]. The detection of L. monocytogenes at the sampling points described in the present study agrees with previously reported detection on stainless steel tables [4, 55], mats [4], chutes [56], and drains [12, 28]. The usual definition of persistence in the context of foodborne pathogens is the repeated isolation of strains that are identical subtypes identified on different days [2]. In this study, strains were considered persistent when detected at the same sampling location after at least one sanitation process. All collections were performed after the environment was subjected to sanitation procedures at least once, suggesting the persistence of the microorganism, possibly due to the ability to adhere to surfaces and to form biofilms [57–59]. At one collection point (a chute in slaughterhouse C), L. monocytogenes was detected in two samples collected on different days, suggesting the presence of biofilms at the site. In two different studies conducted in the United States, Berrang et al. [28, 60] performed serial collection from the same points within the slaughter plant at different times to characterize the presence of persistent L. monocytogenes isolates in the environments of poultry slaughterhouses. The authors concluded that the persistence of this microorganism in drains was related to the formation of biofilms [60]. Similar to the methods described in the present study, Sereno et al. 2019 [61] performed serial swab collections of walls, drains, tables, mats, and floors in a swine slaughterhouse in the southern region of Brazil. The persistence of L. monocytogenes strains was detected at the same sampling point over time and was related to the presence of biofilms at said location. Areas of bacterial persistence include drains [28, 60] and a mat [61]. The present study also detected the presence of L. monocytogenes at these sites, although there was no detection at the same site more than once, except for the chute.
Detection of Salmonella spp.
One Salmonella sp. was detected in 287 (0.34%) swabs collected from the cattle and poultry slaughterhouses. The isolate originated from a drain sample that was collected from a dirty area of poultry slaughterhouse B. This is the first investigation of Salmonella spp. in a processing plant of a poultry slaughterhouse located in the Federal District. However, other reports have described the occurrence of Salmonella spp. in poultry carcasses/viscera produced and/or sold in this region [36]. The low detection in poultry slaughterhouses and absence in cattle slaughterhouses may reflect the efficiency of self-control, disinfection, and sanitation programs in these industries, the reduced number of visits resulting from the hesitation of industries to allow internal access for sample collection, and the implementation of control programs by the government with the aim of reducing the incidence of Salmonella spp. to protect consumers from considerable risks [62–64].
Serotyping analysis by PCR and evaluation of genetic variability of Listeria monocytogenes by PFGE
All 14 isolates were classified via PCR as 1/2a or 3a serotypes, belonging to lineage II [65–67], with amplification of the lmo0737 gene (691pb). These results corroborate reports available in the literature that highlight serotype 1/2a, along with serotypes 1/2b and 1/2c, as one of the most commonly detected serotypes in food samples and food production environments of animal origin [68]. The polymorphic results obtained via PFGE identified 11 distinct pulsotypes that were grouped into four clusters (I, II, III and IV) with a similarity threshold of 80% [69] (Figs 1 and S1). The sample collection points of origin from each pulsotype are shown in Fig 2. To analyze strain dissemination within industries and in the studied regions, clonal variations were considered only for those isolates that presented 100% identity with each other [70]. Pulsotype 9 was the most frequently detected (3/14 isolates), followed by pulsotype 5 (2/14 isolates). The same pulsotype was detected at different sampling points within the same industry (pulsotype 5, detected in a meat chute (72A-2) and an evisceration table (77A-2); and pulsotype 9, detected on a mat in the clean area (59A-2) and a meat chute (78A-2), both present on slaughterhouse C located in Goiás State; Table 2), suggesting the dissemination of these two clones within this establishment. Pulsotype 9 was detected at different sites and at different visits within this slaughterhouse. Though sanitization was performed between sample collections, the fact that the same strain was detected in different collections within the same establishment indicates the presence of biofilms. This finding corroborates those reported by Berrang et al. 2005 [28], Berrang et al. 2010 [60], Camargo et al. 2015 [13], and Sereno et al. 2019 [61]. Pulsotype 9 was also detected in a drain sample obtained from slaughterhouse B, located in the Federal District, suggesting regional dissemination of the strain. These results highlight the importance of using molecular typing methods with high discriminatory power, such as PFGE, to investigate the dissemination and persistence of strains within food processing plants [12, 13, 71, 72]. These typing methods are valuable when WGS analysis cannot be performed. Similar to the results obtained in the present study in relation to the use of PFGE, Camargo et al. 2015 [13] described the serial collection of samples during different visits from environmental locations, cuts of meat, and the hands of employees working in a cattle slaughterhouse in the state of Minas Gerais. This aided the identification of cross-contamination between these points and helped determine the persistence of specific pulsotypes. The results of the present study confirm the persistence of two pulsotypes (5 and 9) in slaughterhouse C and possible regional strain dissemination (pulsotype 9). However, due to the low number of isolates identified in slaughterhouses A and B, it was not possible to confirm the persistence of isolates within these establishments. However, the detection of isolates before the commencement of operations in the slaughterhouses (pre-sanitization) and after slaughter (post-sanitization) suggests the continuous presence of this microorganism in existing biofilms in these facilities.
Fig 1. Dendrogram and PFGE patterns of 14 isolates of Listeria monocytogenes restricted with AscI.
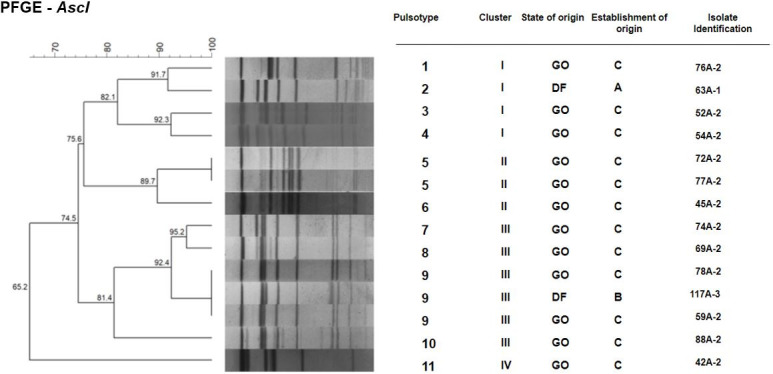
The data were analyzed using the BioNumerics software. Discrimination of pulsotypes, clusters, origins of isolates (Federal District, DF; or Goiás, GO), establishment of origin (A, B or C), and isolate identification are presented.
Fig 2.
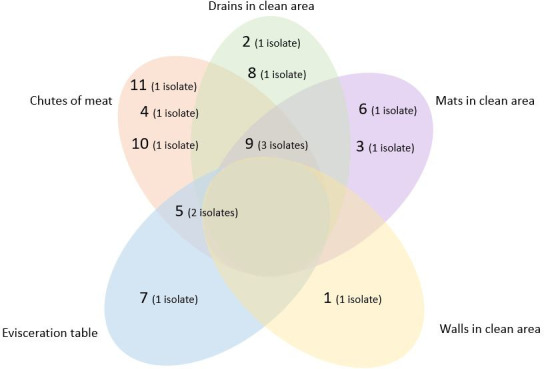
Sample collection points of the 11 L. monocytogenes pulsotypes detected in poultry slaughterhouses A, B and C located in Federal District and Goiás. The numbers in parenthesis represent the number of isolates belonging to each pulsotype, totaling 14 isolates.
Salmonella sp. serotyping by WGS analysis
WGS analysis of Salmonella sp. isolate identified it as Salmonella enterica serovar Minnesota belonging to ST548 [73]. This finding differs from those reported in other studies in the investigated regions, which indicated a higher occurrence of serovar Enteritidis [36, 74, 75]. S. Minnesota ST548 derived from chicken carcasses, chicken feet, and mechanically recovered meat have been reported in Brazil (São Paulo and Minas Gerais, São Paulo, and Federal District, respectively) [76]. On the other hand, this is the first report of its presence in a poultry slaughterhouse located in the Federal District.
Sequencing analysis of the inlA gene in L. monocytogenes isolates
PMSCs were not detected in any of the 14 L. monocytogenes isolates investigated. These results are similar to the results reported by Medeiros et al. 2020 [77]. The authors reported the absence of PMSCs in L. monocytogenes isolates in samples collected from poultry slaughterhouse drains in the Federal District. However, results of the present study differ from those reported previously by Manuel et al. 2015 [78] in the United States and by Camargo et al. 2019 [79] in Brazil. Both studies reported a higher occurrence of PMSCs in strains isolated from food samples and food production environments. The occurrence of PMSCs in L. monocytogenes isolates is associated with the occurrence of attenuated invasion phenotypes [18, 80]. Isolates from human listeriosis have less frequent PMSCs in the inlA gene than isolates obtained from food [78, 79, 81]. The present report indicating the absence of PMSCs is an important finding for public health, since the integrity of the inlA gene is necessary to promote the internalization of this pathogen in host cells [18, 82].
Cell invasion and adhesion assays
All 14 L. monocytogenes isolates demonstrated adherence to the Caco-2 cell surface, with an adhesion capacity that varied from 17.38% to 57.14%. Seven of the fourteen isolates (50%) showed ability to invade Caco-2 cells, with an invasion capacity varying from 5.16% to 34.27% (Fig 3). The isolates that exhibited invasion ability belonged to clusters I and III, and pulsotypes 1 (76A-2), 4 (54A-2), 7 (74A-2), 9 (59A-2, 78A-2 and 117A-3), and 10 (88A-2) (Fig 3). The three isolates belonging to pulsotype 9 could invade Caco-2 cells, although presenting with different capacities. The invasion capacity observed in seven of the fourteen isolates, in which the presence of PMSCs was not detected, corroborates with the findings reported by Nightingale et al. 2005 [18]. The latter authors reported that isolates without PMSCs could invade Caco-2 cells with an ability that was significantly more than isolates with PMSCs in the inlA gene. However, the results differed from those reported for the other seven isolates. Despite the absence of PMSCs, these isolates could not invade Caco-2 cells. Several studies have demonstrated the central role of inlA in cell invasion [18, 81, 83]. However, other elements such as internalin B [84] and listeriolysin O [85] may also be important for the internalization of L. monocytogenes in different cell types. Furthermore, differences in invasion capacity among isolates with the complete gene inlA have been described, indicating that other factors may be related to the efficiency of invasion of this pathogen [86].
Fig 3. Results of invasion and cell adhesion tests using Caco-2 cells for 14 Listeria monocytogenes isolates.
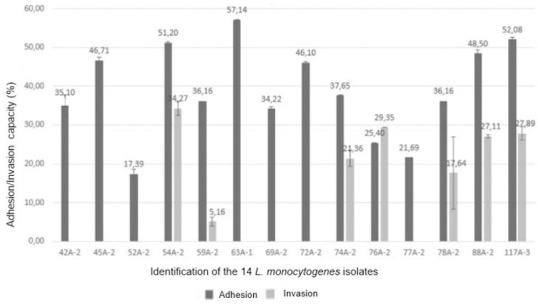
The values shown represent the average of the results, and the bars represent the standard deviation.
Antibiograms and detection of resistance genes
L. monocytogenes
Resistance or intermediate sensitivity was detected for seven of the eight antimicrobials tested (Table 3). Six of the fourteen isolates (42.9%) were sensitive to all the tested antibiotics. Eight isolates (57.1%) displayed resistance or intermediate sensitivity. The absence of resistance to ampicillin and chloramphenicol corroborates the results observed by other authors in isolates obtained from food sources and food production environments [53, 87–89]. Surveillance of the occurrence of ampicillin resistance is important for public health, since this drug is a treatment of choice for human listeriosis [90]. The sensitivity to chloramphenicol can be explained by the ban on the use of this antibiotic for the production of food of animal origin in Brazil since 2003 [91]. The results of the present study on the resistance and intermediate sensitivity towards ciprofloxacin, erythromycin, and gentamicin (42.8%, 35.7%, and 35.7% of L. monocytogenes isolates, respectively) contrasted with those observed by other authors, such as Teixeira et al. 2020 [89], who reported the absence of resistance to the three antibiotics in isolates obtained from beef cuts in the state of Mato Grosso, Brazil. The findings of the present study on resistance to gentamicin and erythromycin indicate a public health concern, since these are the drugs of choice for the treatment of listeriosis in specific cases, such as the use of gentamicin associated with penicillin as an alternative to ampicillin, and the use of erythromycin in the treatment of infections in pregnant women [90, 92]. The low occurrence of resistance to doxycycline detected in this study corroborates the findings reported by Vitas et al. 2007 [93], who reported a low occurrence of resistance in isolates of L. monocytogenes obtained from food and human clinical cases in Spain. The low detection of resistance to tetracycline (7.1% of the isolates) is in agreement with the findings reported by Haubert et al. 2016 [88] for isolates obtained from food and food production environments in the southern region of Brazil. However, the findings of the present study differ from those described by Palma et al. 2016 [53] and Camargo et al. 2015 [87], in which resistance to tetracycline was not detected in isolates obtained from samples of beef cuts, cattle slaughterhouse environments, and clinical cases in Brazil. The highest occurrence of AMR detected in this study was in relation to sulfonamides, a characteristic which was found in eight isolates (57.1%). These findings corroborate the results reported by other authors, who also reported a pronounced prevalence of resistance to this class of antimicrobials in isolates obtained from food and food production environments [53, 89, 94, 95]. Sulfonamides associated with trimethoprim are considered as the second-choice of treatment for human listeriosis [90]. Therefore, the detection of resistance to this class of antimicrobials may indicate a risk to human health.
Table 3. Antibiogram constructed for 14 Listeria monocytogenes isolates obtained from poultry slaughterhouses located in the region of the Federal District and State of Goiás, based on the results obtained via disk diffusion assay for antimicrobial resistance (CLSI, 2020).
| Antimicrobial and class | No. of resistant isolates (%) | No. of isolates with an intermediate sensitivity (%) | No. of sensitive isolates (%) | Total no. of intermediate resistance-displaying isolates (%) |
|---|---|---|---|---|
| Sulfonamides (sulfonamide) | 8 (57.1) | 0 (0.0) | 6 (42.9) | 8 (57.1) |
| Erythromycin (macrolide) | 4 (28.6) | 1 (7.1) | 9 (64.3) | 5 (35.7) |
| Ciprofloxacin (quinolone) | 3 (21.4) | 3 (21.4) | 8 (57.2) | 6 (42.9) |
| Gentamicin (aminoglycoside) | 4 (28.6) | 1 (7.1) | 9 (64.3) | 5 (35.7) |
| Chloramphenicol (chloramphenicol) | 0 (0.0) | 4 (28.6) | 10 (71.4) | 4 (28.6) |
| Tetracycline (tetracycline) | 1 (7.1) | 1 (7.1) | 12 (85.8) | 2 (14.3) |
| Doxycycline (tetracycline) | 1 (7.1) | 0 (0.0) | 13 (92.9) | 1 (7.1) |
| Ampicillin (beta-lactamases) | 0 (0.0) | 0 (0.0) | 14 (100) | 0 (0.0) |
The results of the detection of AMR genes are shown in Table 4. The ermA, ermC, cat1, sulI, aaC (3)-1, cmlA, ereA, SHV, and ampC genes were not detected.
Table 4. Results of antibiogram susceptibility test and antimicrobial resistance genes for the 14 L. monocytogenes isolates.
| L. monocytogenes isolate identification | Pulsotype | Region and establishment identification | Swab collection point in the industry | Antibiogram results of resistance or intermediate sensibility | Antimicrobial resistance genes |
|---|---|---|---|---|---|
| 42A-2 | 11 | GO/C | Chutes of meat | Sensible to all tested bases | No gene targeted in the study detected |
| 45A-2 | 6 | GO/C | Mats in clean area | CIP* ERI GEN* SUL VAN* | tet(M) tet(C) |
| 52A-2 | 3 | GO/C | Mats in clean area | CIP* CLO* ERI GEN SUL TET* VAN | ermB tet(C) |
| 54A-2 | 4 | GO/C | Chutes of meat | CIP CLO* DOX ERI GEN SUL TET VAN* | ermB tet(M) tet(C) |
| 59A-2 | 9 | GO/C | Mats in clean area | Sensible to all tested bases | tet(B) tet(C) |
| 63A-1 | 2 | DF/A | Drains in clean area | Sensible to all tested bases | tet(C) |
| 69A-2 | 8 | GO/C | Drains in clean area | Sensible to all tested bases | tet(B) |
| 72A-2 | 5 | GO/C | Chutes of meat | CIP CLO* ERI* GEN SUL VAN | tet(C) |
| 74A-2 | 7 | GO/C | Evisceration table | SUL | tet(B) |
| 76A-2 | 1 | GO/C | Walls in clean area | Sensible to all tested bases | tet(C) |
| 77A-2 | 5 | GO/C | Evisceration table | SUL | tet(C) |
| 78A-2 | 9 | GO/C | Chutes of meat | Sensible to all tested bases | tet(C) |
| 88A-2 | 10 | GO/C | Chutes of meat | CIP* CLO* ERI GEN SUL VAN* | tet(B) tet(C) |
| 117A-3 | 9 | DF/B | Drains in clean area | CIP SUL TEC* | tet(B) tet(C) |
* Antimicrobial agents with intermediate sensitivity.
The detection of the ermB gene was in accordance with the finding reported by Haubert et al. [88], who reported the presence of this gene in an isolate of L. monocytogenes obtained from a poultry slaughterhouse environment in the southern region of Brazil. Only two of the four isolates of L. monocytogenes that presented with an erythromycin resistance phenotype in the disk diffusion assay (Table 3) harbored one of the erythromycin-related resistance genes investigated. It could be possible that the isolates that did not harbor ermA, ermB or ermC genes might have harbored genes related to the development of other mechanisms of resistance to this antimicrobial, such as those pertaining to the presence of msr(A) or mef(A) genes [8]. Alternatively, the resistance of these isolates may be attributable to chromosomal mutations [95].
The presence of the tetM gene in two isolates (14.28%) was similar to the findings reported by Bertrand et al. [96], who reported the presence of this gene in isolates exhibiting phenotypic resistance to tetracycline in strains obtained from human clinical samples and swine and poultry slaughterhouse environments from Belgium and France. Several studies have highlighted the tetM gene as the genotype that is most commonly associated with the development of tetracycline resistance in Listeria isolates [96, 97]. These findings differ from findings of the present study that suggested that tetC was the most prevalent gene related to tetracycline resistance. Despite the detection of tetC in eleven of the fourteen isolates (78.6%), only two isolates displayed resistance or intermediate sensitivity to tetracycline in the disk diffusion assay (Table 3). One of these two isolates harbored two of the four genes related to the development of resistance to this drug (tetC and tetM), and was classified as a resistant isolate based on the antibiogram. The other isolate harbored only the tetC gene and showed intermediate sensitivity to this antimicrobial. The other nine isolates with the tetC gene did not demonstrate resistance to tetracycline in the disk diffusion assay. Therefore, the unique presence of tetC in the isolates did not confer a resistance phenotype to tetracycline, suggesting that the genes did not necessarily trigger the resistance phenotype, which might be related to the presence of mutations that could lead to gene dysfunction [98].
Despite the occurrence of AMR to gentamicin in five of the fourteen isolates in the disk diffusion assay (Table 3), the presence of the aac(3)-1 gene was not detected in any isolate. It could be possible that resistance to gentamicin in these isolates was related to the presence of other genes not investigated in this study, since more than 170 genes related to resistance to aminoglycosides have been described in bacteria [8]. In the present study, resistance to sulfonamides was observed in eight of fourteen isolates in the disk diffusion assay, but no isolates harbored the sulI gene. It could be possible that this resistance was associated with other mechanisms, such as the presence of other genes related to sulfonamide resistance, namely sul2, folP, or thyA genes, with the description of the last two genes reported in L. monocytogenes [99].
Only intermediate sensitivity to chloramphenicol was detected in four of fourteen isolates in the disk diffusion assay. The cmlA and cat1 genes were not detected. The intermediate sensitivity observed in this study could be related to cross-resistance due to the presence of other genes that were not investigated, such as floR [100]. Notably, in Brazil, this antimicrobial has been banned from use in animal production since 2003 [91]. Selection pressure was not a factor. The absence of the ampC and blaSHV genes is consistent with the absence of the ampicillin resistance phenotype in the disk diffusion assay.
Four of the fourteen isolates (28.5%) showed resistance to three or more classes of antimicrobials and were classified as multidrug-resistant isolates [101]. All were isolated from establishment C. Two isolates (54A-2 and 88A-2) showed adherence and invasion in Caco-2 cells. Reports of the presence of L. monocytogenes multidrug-resistant isolates have increased in the literature [88, 97, 102]. All multidrug-resistant isolates detected in the present study showed resistance to antimicrobials used in the treatment of human listeriosis (gentamicin and erythromycin) [90]. There is a possibility of the transfer of genes from multidrug-resistant isolates to others. Further studies are warranted to elucidate the resistance mechanisms observed in these isolates and their potential as a source of transfer of resistance to other microorganisms.
Salmonella enterica Minnesota
Resistance or intermediate sensitivity of Salmonella Minnesota was detected in nine of thirteen antimicrobials tested (Table 5). The resistance to tetracycline and ampicillin, and the sensitivity to ciprofloxacin and gentamicin were similar to the results obtained by Dantas et al. [103]. The authors reported the same pattern of resistance in isolates of Salmonella spp. obtained from a poultry slaughterhouse environment in the state of São Paulo. The detection of resistance to ampicillin, cephalothin, and sulfonamide bases, in addition to sensitivity to ciprofloxacin, is comparable to the findings reported by Cunha-Neto et al. 2018 [104] who investigated isolates of Salmonella spp. derived from chicken carcasses in a slaughterhouse in the state of Mato Grosso. However, these authors reported the detection of resistance to gentamicin and chloramphenicol, and sensitivity to nalidixic acid, which differed from the results of the present study. The detection of resistance to nalidixic acid in Salmonella spp. isolates has been widely reported [105–107]. Meta-analyses of studies published over a period of 20 years in Brazil have revealed the increased occurrence of resistance to quinolone in isolates derived from human samples and chicken meat [108]. Resistance to nalidixic acid may be attributable to the prolonged and widespread use of this antimicrobial in human and veterinary medicine, since this was the first drug in the quinolone class to have clinical use [109]. Similar to nalidixic acid resistance, resistance to tetracycline, sulfonamides, and beta-lactams may be related to the overuse of these antimicrobials in the different stages of the chicken meat production chain in Brazil [110].
Table 5. Antibiogram and detection of antimicrobial resistance genes from the Salmonella enterica serovar Minnesota isolate from a poultry slaughterhouse located in the Federal District, carried out using the disk diffusion assay (CLSI, 2020) and WGS, respectively.
| Antibiogram (tested drugs) | Antibiogram result | Antimicrobial resistance genes |
|---|---|---|
| Nalidixic acid | R | qnrB19 |
| Amoxicillin | R | bla CMY-2 |
| Ampicillin | R | bla CMY-2 |
| Cephalothin | R | bla CMY-2 |
| Cefazoline | R | bla CMY-2 |
| Ceftazidime | R | bla CMY-2 |
| Ciprofloxacin | S | - |
| Chloramphenicol | S | - |
| Colistin | S | - |
| Doxycycline | I | - |
| Gentamicin | S | aac(6’)-1aa |
| Sulfonamides | R | sul2 |
| Tetracycline | R | tetA |
S, sensitive; I, intermediate; R, resistant.
The present study detected resistance to three generations of cephalosporins. Cephalosporins, especially those of the third and fourth generations, are important for human and animal health [111]. Resistance to cephalosporins (including third- and fourth-generation drugs) has been reported in recent years [112, 113]. The detection of resistance, especially to ceftazidime, is a public health concern and highlights the need for vigilance and the reduction of the use of antimicrobials in veterinary medicine. In recent years, the Brazilian government has restricted the use of antimicrobials in the production of animal foods, such as via implementation of a ban on the use of chloramphenicol and nitrofurans [91], colistin sulfate [114], and more recently, via the enforcement of a ban on the use of tylosin, lincomycin, and tiamulin [115].
WGS analysis of Salmonella isolate (GenBank accession number JABBEB000000000.1) led to the detection of the tetA, sul2, aac (6’)-Iaa, blaCMY-2, and qnrB19 genes, as well as aided identification of a mutation in the parC gene (T57S, ACC →AGC, causing the mutation T→S). All detected resistance genes have been described in Salmonella spp. isolates in Brazil [76, 116]. The concomitant presence of plasmid-mediated quinolone resistance (PMQR) via the qnrB19 gene, and mutations in the region determining quinolone resistance (QRDR) in the parC gene of topoisomerase IV was also observed. These genetic characteristics were confirmed by ascertaining resistance to nalidixic acid, but resistance to ciprofloxacin was not detected in the antibiograms. Despite this, the presence of the qnrB19 gene and other genes related to PMQR may be responsible for the reduced susceptibility to quinolones, which may facilitate the selection of less susceptible isolates and may lead to consequent failure in the treatment with this class of drugs [117]. The detection of PMQR in this study highlights the need for surveillance of the persistence of quinolone resistance genes in the poultry production chain. The blaCMY-2 gene is a plasmid gene related to the production of beta-lactamases (pAmpC), and is a gene that is most commonly detected globally [118]. The presence of this gene corroborates the results of resistance to beta-lactams observed in the disk diffusion assay (ampicillin, cephalothin, cefazolin, and ceftazidime). Its presence is another public health concern considering the importance of beta-lactams in human and animal health, especially third-generation cephalosporins [111]. The presence of the aac-(6’)-Iaa gene related to the development of resistance to aminoglycosides did not confer resistance to gentamicin, the only drug from this class of antimicrobials evaluated in the disk diffusion assay. This discrepancy between phenotype and genotype was also observed in Salmonella spp. isolates obtained from different samples in the poultry production chain in the states of São Paulo, Bahia, and Minas Gerais [76]. In contrast, the occurrence of resistance to tetracycline and sulfonamides is related to the presence of the tetA and sul2 genes, respectively. The absence of colistin resistance genes corroborates findings reported by Monte et al. [76], who reported the absence of these genes in Salmonella spp. isolates obtained from samples of swine and poultry production chains from different regions of Brazil, including the Federal District.
The resistance profile observed in this Salmonella sp. isolate enabled its classification as a multidrug-resistant isolate [101]. This characteristic, associated with the presence of mobile genetic elements related to resistance development, such as the presence of PMQR and pAmpC, suggests a potential public health risk. Additionally, the occurrence of resistance to quinolones and cephalosporins, associated with the presence of genetic elements related to this phenotype, is of special concern, considering the importance of these drugs for human health [111].
In vitro biofilm formation capacity
L. monocytogenes
During incubation at 37°C for 24 h, 11 of 14 isolates (78.57%) were capable of biofilm formation and were classified as weak biofilm formers. During incubation at 12°C for 168 h, nine of 14 isolates (64.3%) were capable of biofilm formation. All nine were deemed weak biofilm formers (Fig 4). The biofilm formation capacity of L. monocytogenes isolates corroborated findings reported in other studies indicating their ability to adhere to abiotic surfaces and to form biofilms on such surfaces [119–121]. The poor biofilm formation capacity of most isolates (78.57%) at 37°C corroborated the findings reported by Harvey et al. 2007 [119]. The authors reported that 90% of the tested L. monocytogenes isolates were weak biofilm-forming bacteria. There was no significant difference (p = 0.2450) in the biofilm formation capacity of the isolates when incubated at 37°C for 24 h and at 12°C for 168 h. The finding that nine of the 14 isolates can form biofilms during incubation at 12°C highlights the risks that these isolates can pose to cattle and poultry slaughter facilities in the event of failures of the hygiene and sanitation practices adopted for the environments, since this is a common temperature documented inside the industry.
Fig 4. Results of the in vitro biofilm formation capacity test performed using polystyrene microplates (Djordjevic et al., 2002) and 12 Listeria monocytogenes isolates exhibiting biofilm formation capacity at 37°C and/or 12°C.
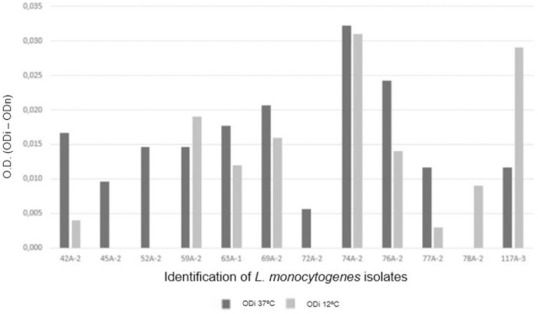
The bars represent the average value of the optical density of each test for each isolate (ODi), all performed in triplicate, subtracted from the average of the optical density of the negative control for each repetition (ODn).
There was no association between cluster/pulsotype and in vitro biofilm formation capacity. Pulsotypes 9 and 5, which were the most commonly detected, did not present better in vitro biofilm formation capacity than the other pulsotypes (Table 6). These results are similar to the findings of other studies that reported no difference in the in vitro ability to form biofilms between persistent and non-persistent strains [49, 119], although other studies report persistent strains as better in vitro biofilm formers [57, 58]. The association between adherence capacity and strain persistence in the industry remains unclear.
Table 6. Results of antimicrobial resistance, antimicrobial resistance genes and in vitro biofilm formation capacity of the 14 L. monocytogenes isolates.
| L. monocytogenes isolate identification | Pulsotype | Antimicrobial resistance | Antimicrobial resistance genes | In vitro biofilm formation capacity | Classification at 37°C | Classification at 12°C |
|---|---|---|---|---|---|---|
| 42A-2 | 11 | Sensible to all tested bases | No gene targeted in the study detected | Yes | Weak | Weak |
| 45A-2 | 6 | CIP* ERI GEN* SUL VAN* | tet(M) tet(C) | Yes | Weak | Non-forming |
| 52A-2 | 3 | CIP* CLO* ERI GEN SUL TET* VAN | ermB tet(C) | Yes | Weak | Non-forming |
| 54A-2 | 4 | CIP CLO* DOX ERI GEN SUL TET VAN* | ermB tet(M) tet(C) | No | Non-forming | Non-forming |
| 59A-2 | 9 | Sensible to all tested bases | tet(B) tet(C) | Yes | Weak | Weak |
| 63A-1 | 2 | Sensible to all tested bases | tet(C) | Yes | Weak | Weak |
| 69A-2 | 8 | Sensible to all tested bases | tet(B) | Yes | Weak | Weak |
| 72A-2 | 5 | CIP CLO* ERI* GEN SUL VAN | tet(C) | Yes | Weak | Non-forming |
| 74A-2 | 7 | SUL | tet(B) | Yes | Weak | Weak |
| 76A-2 | 1 | Sensible to all tested bases | tet(C) | Yes | Weak | Weak |
| 77A-2 | 5 | SUL | tet(C) | Yes | Weak | Weak |
| 78A-2 | 9 | Sensible to all tested bases | tet(C) | Yes | Non-forming | Weak |
| 88A-2 | 10 | CIP* CLO* ERI GEN SUL VAN* | tet(B) tet(C) | Yes | Weak | Non-forming |
| 117A-3 | 9 | CIP SUL TEC* | tet(B) tet(C) | Yes | Weak | Weak |
* Antimicrobial agents with intermediate sensitivity.
Three of the four multidrug-resistant isolates (52A-2, 72A-2 and 88A-2) were capable of in vitro biofilm formation at 37°C, and none of them formed biofilm at 12°C. In contrast, 13 out of the 14 isolates that harbored at least one of the resistance genes targeted in this study were capable of biofilm formation in the in vitro assay, at least, at one of the tested temperatures (Table 6). These results highlight the possibility of adherence of these isolates to the surfaces of these industries, posing the risk of acting as resistance gene reservoirs for other strains and species, ultimately posing a threat to public health and causing dissemination of AMR in various strains.
Most isolates investigated in the present study demonstrated the ability to form biofilms under at least one of the conditions tested. These findings, along with the findings of repeated detection of clonal variations within the same industry at different points and different visits, in addition to the identification of the same pulsotype in another slaughterhouse in another region, support the possible existence of L. monocytogenes biofilms in poultry slaughterhouses located in the Federal District and State of Goiás. The detection of the biofilm formation capacity of the isolates, especially the ability documented at 12°C and observed inside slaughterhouses, indicates the possibility that these isolates can adhere to surfaces, including those of equipment and utensils, which suggests the possibility of biofilm formation in the event of failures in appropriate implementation of hygiene procedures. This creates a potential public health risk because of possible food-associated cross-contamination.
Salmonella spp.
The Salmonella sp. isolate was capable of biofilm formation, albeit weakly, under both conditions tested. These results corroborate those reported by Yin et al. 2018 [122]. The authors reported the ability of biofilm formation of Salmonella spp. isolates obtained from beef production environments in China at different temperatures, including 12°C and 37°C. The present results suggest the potential risk to public health by the Salmonella sp. isolate in the slaughterhouse environment, since, in addition to its multidrug-resistance characteristic, this isolate may adhere to abiotic surfaces, and may be considered a potential contaminant of processed foods.
Conclusions
The presence of L. monocytogenes and Salmonella spp. was identified for the first time in poultry slaughterhouses located in the Federal District and in the State of Goiás, Brazil. The bacterial species were detected on equipment and utensils, especially chutes, which were the main routes of contamination by L. monocytogenes within the evaluated plants. Species could not be detected in cattle slaughterhouses. The consistent and repeated collection at the sampling points enabled the characterization of biofilms in the environments of the industries, which was confirmed in 12 of the 14 L. monocytogenes isolates and in Salmonella sp. via the in vitro biofilm formation capacity test that was performed at 12°C and/or at 37°C. The PFGE patterns aided the evaluation of the dissemination of specific pulsotypes both within an industry and in the studied regions. Sequencing of the inlA gene derived from L. monocytogenes isolates demonstrated the absence of PMSCs in all isolates. These results, along with the adhesion and cell invasion assay results, suggest their virulence potential. The detection of resistance to antimicrobials of public health importance in L. monocytogenes and Salmonella sp. isolates, in addition to the detection of multidrug-resistant isolates, also underlines the potential risk for the consumer due to the possibility of food-associated cross-contamination via contact with biofilms formed on abiotic surfaces. The results of this study indicate the importance of conducting research that supports surveillance in slaughterhouse environments for possible adjustments in environmental hygiene and sanitation procedures. Additionally, the detection of isolates with pathogenic potential related to the capability of cell invasion, the ability to form biofilms, and AMR development indicates the necessity of assessing the risk for the population as an instrument of public health protection.
Nucleotide sequence accession number
Information on this Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JABBEB000000000 (SRA: SRR12606957). The version described in this paper is version JABBEB000000000.1.
Supporting information
(PDF)
Acknowledgments
We thank the staff at the Laboratory of Veterinary Microbiology and Laboratory of Genic Therapy for providing assistance when necessary.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
APS was supported by FAPDF (spontaneous demand n.3/2016).EFAD was funded as well by CAPES, via PROAP and DPG of University of Brasília. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Møretrø T, Langsrud S. Residential Bacteria on Surfaces in the Food Industry and Their Implications for Food Safety and Quality. Compr Rev Food Sci Food Saf. 2017. Sep 1;16(5):1022–41. doi: 10.1111/1541-4337.12283 [DOI] [PubMed] [Google Scholar]
- 2.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. Vol. 77, Journal of Food Protection. J Food Prot; 2014. p. 150–70. doi: 10.4315/0362-028X.JFP-13-150 [DOI] [PubMed] [Google Scholar]
- 3.Carrascosa C, Raheem D, Ramos F, Saraiva A, Raposo A. Microbial biofilms in the food industry—a comprehensive review. Int J Environ Res Public Health. 2021. Feb 2;18(4):1–31. doi: 10.3390/ijerph18042014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues L, Dos Santos L, Tagliari V, Rizzo N, Trenhago G, de Oliveira A, et al. Quantification of biofilm production on polystyrene by Listeria, Escherichia coli and Staphylococcus aureus isolated from a poultry slaughterhouse. Braz J Microbiol. 2010;41(4). doi: 10.1590/S1517-838220100004000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen MH, Dalmasso M, Ingmer H, Langsrud S, Malakauskas M, Mader A, et al. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control. 2014. Oct 1;44:92–109. [Google Scholar]
- 6.Kasnowski MC, Mantilla SPS, Oliveira LAT, Franco RM. Formação de biofilme na indústria de alimentos e métodos de validação de superfícies. Rev Científica Eletrônica da Med Veterinária. 2010;VIII(15). [Google Scholar]
- 7.FAO F and AO of the UN, WHO WHO. Principles and Guidelines for the Conduct of Microbiological Risk Assessment. FAO Agric Consum Prot. 2007;63:68.
- 8.Luque-Sastre L, Arroyo C, Fox EM, McMahon BJ, Bai L, Li F, et al. Antimicrobial Resistance in Listeria Species. In: Antimicrobial Resistance in Bacteria from Livestock and Companion Animals. Washington, DC, USA: ASM Press; 2018. p. 237–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpentier B, Cerf O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Vol. 145, International Journal of Food Microbiology. Elsevier; 2011. p. 1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 10.Datta AR, Burall LS. Serotype to genotype: The changing landscape of listeriosis outbreak investigations. Vol. 75, Food Microbiology. Academic Press; 2018. p. 18–27. doi: 10.1016/j.fm.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 11.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004. Aug;42(8):3819–22. doi: 10.1128/JCM.42.8.3819-3822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berrang ME, Meinersmann RJ, Northcutt JK, Smith DP. Molecular characterization of Listeria monocytogenes isolated from a poultry further processing facility and from fully cooked product. J Food Prot. 2002. Oct 1;65(10):1574–9. doi: 10.4315/0362-028x-65.10.1574 [DOI] [PubMed] [Google Scholar]
- 13.Camargo AC, Dias MR, Cossi MVC, Lanna FGPA, Cavicchioli VQ, Vallim DC, et al. Serotypes and pulsotypes diversity of Listeria monocytogenes in a beef-processing environment. Foodborne Pathog Dis. 2015. Apr 1;12(4):323–6. doi: 10.1089/fpd.2014.1875 [DOI] [PubMed] [Google Scholar]
- 14.Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016. Mar 1;48(3):308–13. doi: 10.1038/ng.3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, et al. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science (80-) [Internet]. 2001. Jun 1 [cited 2020 Dec 6];292(5522):1722–5. Available from: https://science.sciencemag.org/content/292/5522/1722 doi: 10.1126/science.1059852 [DOI] [PubMed] [Google Scholar]
- 16.Orsi RH, Ripoll DR, Yeung M, Nightingale KK, Wiedmann M. Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology. 2007. Aug;153(8):2666–78. doi: 10.1099/mic.0.2007/007310-0 [DOI] [PubMed] [Google Scholar]
- 17.Poyart C, Trieu-Cuot P, Berche P. The inlA gene required for cell invasion is conserved and specific to Listeria monocytogenes. Microbiology. 1996;142(1):173–80. doi: 10.1099/13500872-142-1-173 [DOI] [PubMed] [Google Scholar]
- 18.Nightingale KK, Windham K, Martin KE, Yeung M, Wiedmann M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial ce. Appl Environ Microbiol. 2005. Dec;71(12):8764–72. doi: 10.1128/AEM.71.12.8764-8772.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrari RG, Rosario DKA, Cunha-Neto A, Mano SB, Figueiredo EES, Conte-Juniora CA. Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl Environ Microbiol. 2019. Jul 1;85(14). doi: 10.1128/AEM.00591-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph B, Otta SK, Karunasagar I, Karunasagar I. Biofilm formation by Salmonella spp. On food contact surfaces and their sensitivity to sanitizers. Int J Food Microbiol. 2001. Mar 20;64(3):367–72. doi: 10.1016/s0168-1605(00)00466-9 [DOI] [PubMed] [Google Scholar]
- 21.Leriche V, Carpentier B. Limitation of adhesion and growth of Listeria monocytogenes on stainless steel surfaces by Staphylococcus sciuri biofilms. J Appl Microbiol. 2000;88(4):594–605. doi: 10.1046/j.1365-2672.2000.01000.x [DOI] [PubMed] [Google Scholar]
- 22.Sinde E, Carballo J. Attachment of Salmonella spp. and Listeria monocytogenes to stainless steel, rubber and polytetrafluorehtylene: The influence of free energy and the effect of commercial sanitizers. Food Microbiol. 2000. Aug 1;17(4):439–47. [Google Scholar]
- 23.McEwen SA, Collignon PJ. Antimicrobial Resistance: a One Health Perspective. In: Antimicrobial Resistance in Bacteria from Livestock and Companion Animals. American Society of Microbiology; 2018. p. 521–47. [Google Scholar]
- 24.World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report Early implementation 2017–2018. 2018.
- 25.World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015. [DOI] [PubMed]
- 26.Nicolau AI, Bolocan AS. Sampling the processing environment for listeria. In: Methods in Molecular Biology [Internet]. Humana Press Inc.; 2014 [cited 2020 Dec 8]. p. 3–14. Available from: https://pubmed.ncbi.nlm.nih.gov/24792544/ [DOI] [PubMed]
- 27.Barros MAF, Nero LA, Silva LC, D’Ovidio L, Monteiro FA, Tamanini R, et al. Listeria monocytogenes: Occurrence in beef and identification of the main contamination points in processing plants. Meat Sci. 2007. Aug;76(4):591–6. doi: 10.1016/j.meatsci.2007.01.016 [DOI] [PubMed] [Google Scholar]
- 28.Berrang ME, Meinersmann RJ, Frank JF, Smith DP, Genzlinger LL. Distribution of Listeria monocytogenes subtypes within a poultry further processing plant. J Food Prot. 2005;68(5):980–5. doi: 10.4315/0362-028x-68.5.980 [DOI] [PubMed] [Google Scholar]
- 29.Pan Y, Breidt F, Kathariou S. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl Environ Microbiol. 2006. Dec;72(12):7711–7. doi: 10.1128/AEM.01065-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vestby LK, Møretrø T, Langsrud S, Heir E, Nesse LL. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet Res. 2009. May 27;5. doi: 10.1186/1746-6148-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryser ET, Donnelly CW. 35. Listeria. In: Salfinger Y, Tortorello M Lou, editors. Compendium of Methods for the Microbiological Examination of Foods. 5th ed. American Public Health Association; 2015. [Google Scholar]
- 32.D’Agostino M, Wagner M, Vazquez-Boland JA, Kuchta T, Karpiskova R, Hoorfar J, et al. A validated PCR-based method to detect Listeria monocytogenes using raw milk as a food model—Towards an international standard. J Food Prot. 2004;67(8):1646–55. doi: 10.4315/0362-028x-67.8.1646 [DOI] [PubMed] [Google Scholar]
- 33.Kérouanton A, Marault M, Petit L, Grout J, Dao TT, Brisabois A. Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. J Microbiol Methods. 2010. Feb;80(2):134–7. doi: 10.1016/j.mimet.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 34.ISO—ISO 6579:2002—Microbiology of food and animal feeding stuffs—Horizontal method for the detection of Salmonella spp. 2002. p. 27.
- 35.Alvarez J, Sota M, Vivanco AB, Perales I, Cisterna R, Rementeria A, et al. Development of a Multiplex PCR Technique for Detection and Epidemiological Typing of Salmonella in Human Clinical Samples. J Clin Microbiol. 2004; doi: 10.1128/JCM.42.4.1734-1738.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Freitas CG, Santana ÂP, da Silva PHC, Gonçalves VSP, Barros M de AF, Torres FAG, et al. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int J Food Microbiol. 2010. Apr;139(1–2):15–22. doi: 10.1016/j.ijfoodmicro.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 37.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017. Jun 1;13(6). doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, et al. SeqSero2: Rapid and improved salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol. 2019. Dec 1;85(23). doi: 10.1128/AEM.01746-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.CDC (Centers for Disease Control and Prevention). Standard Operating Procedure for PulseNet PFGE of Listeria monocytogenes [Internet]. 2017 [cited 2020 Dec 8]. Available from: https://www.cdc.gov/pulsenet/PDF/listeria-pfge-protocol-508c.pdf
- 40.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun [Internet]. 1987. [cited 2020 Dec 8];55(11):2822–9. Available from: /pmc/articles/PMC259983/?report=abstract doi: 10.1128/iai.55.11.2822-2829.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moroni O, Kheadr E, Boutin Y, Lacroix C, Fliss I. Inactivation of adhesion and invasion of food-borne Listeria monocytogenes by bacteriocin-producing Bifidobacterium strains of human origin. Appl Environ Microbiol. 2006. Nov;72(11):6894–901. doi: 10.1128/AEM.00928-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. CLSI M100 ED30:2020. 2020.
- 43.Chen BY, Pyla R, Kim TJ, Silva JL, Jung YS. Antibiotic resistance in Listeria species isolated from catfish fillets and processing environment. Lett Appl Microbiol. 2010. Jun;50(6):626–32. doi: 10.1111/j.1472-765X.2010.02843.x [DOI] [PubMed] [Google Scholar]
- 44.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. 2019.
- 45.Van T, Chin J, Chapman T, Tran L, Coloe P. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol. 2008;124(3):217–23. doi: 10.1016/j.ijfoodmicro.2008.03.029 [DOI] [PubMed] [Google Scholar]
- 46.Schwartz T, Kohnen W, Jansen B, Obst U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol. 2003;43(3):325–35. doi: 10.1111/j.1574-6941.2003.tb01073.x [DOI] [PubMed] [Google Scholar]
- 47.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40(11):2562–6. doi: 10.1128/AAC.40.11.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aarestrup FM, Agersø Y, Ahrens P, Jørgensen JCØ, Madsen M, Jensen LB. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet Microbiol. 2000; doi: 10.1016/s0378-1135(00)00197-8 [DOI] [PubMed] [Google Scholar]
- 49.Djordjevic D, Wiedmann M, McLandsborough LA. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol. 2002;68(6):2950–8. doi: 10.1128/AEM.68.6.2950-2958.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borges KA, Furian TQ, Souza SN, Menezes R, Tondo EC, Salle CTP, et al. Biofilm formation capacity of Salmonella serotypes at different temperature conditions. Pesqui Vet Bras. 2018. Jan 1;38(1):71–6. [Google Scholar]
- 51.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–9. doi: 10.1016/s0167-7012(00)00122-6 [DOI] [PubMed] [Google Scholar]
- 52.Cesar APR, Mesquita AJ de, Prado CS, Nunes IA, Almeida Filho ES de. Listeria spp. e Listeria monocytogenes na produção de salsichas tipo hot dog. Ciência Anim Bras. 2011. Jun 27;12(2):339–52. [Google Scholar]
- 53.Palma JM, Lisboa RC, Rodrigues DP, Santos AFM, Hofer E, Santana AP. Caracterização molecular de Listeria monocytogenes oriundas de cortes cárneos bovinos e de abatedouros frigoríficos de bovinos localizados no Distrito Federal, Brasil. Pesqui Vet Bras [Internet]. 2016. [cited 2020 Dec 5];36(10):957–64. Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0100-736X2016001000957&lng=en&nrm=iso&tlng=pt [Google Scholar]
- 54.Chiarini E, Tyler K, Farber JM, Pagotto F, Destro MT. Listeria monocytogenes in two different poultry facilities: Manual and automatic evisceration. Poult Sci. 2009. Apr 1;88(4):791–7. doi: 10.3382/ps.2008-00396 [DOI] [PubMed] [Google Scholar]
- 55.Loura CAC, Almeida RCC, Almeida PF. The incidente and level of Listeria spp. and Listeria monocytogenes contamination in processed poultry at a poultry processing plant. J Food Saf. 2005. Feb 1;25(1):19–29. [Google Scholar]
- 56.Schäfer DF, Steffens J, Barbosa J, Zeni J, Paroul N, Valduga E, et al. Monitoring of contamination sources of Listeria monocytogenes in a poultry slaughterhouse. LWT. 2017. Dec 1;86:393–8. [Google Scholar]
- 57.Borucki MK, Peppin JD, White D, Loge F, Call DR. Variation in Biofilm Formation among Strains of Listeria monocytogenes. Appl Environ Microbiol. 2003. Dec 1;69(12):7336–42. doi: 10.1128/AEM.69.12.7336-7342.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lunden JM, Miettinen MK, Autio TJ, Korkeala HJ. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J Food Prot. 2000;63(9):1204–7. doi: 10.4315/0362-028x-63.9.1204 [DOI] [PubMed] [Google Scholar]
- 59.Møretrø T, Langsrud S. Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms. 2004. Apr;1(2):107–21. [Google Scholar]
- 60.Berrang ME, Meinersmann RJ, Frank JF, Ladely SR. Colonization of a newly constructed commercial chicken further processing plant with Listeria monocytogenes. J Food Prot. 2010;73(2):286–91. doi: 10.4315/0362-028x-73.2.286 [DOI] [PubMed] [Google Scholar]
- 61.Sereno MJ, Viana C, Pegoraro K, da Silva DAL, Yamatogi RS, Nero LA, et al. Distribution, adhesion, virulence and antibiotic resistance of persistent Listeria monocytogenes in a pig slaughterhouse in Brazil. Food Microbiol. 2019. Dec 1;84:103234. doi: 10.1016/j.fm.2019.05.018 [DOI] [PubMed] [Google Scholar]
- 62.Ministério da Agricultura Pecuária e Abastecimento, Secretaria de Defesa Agropecuária. Instrução Normativa No 70, de de 2003—Dispõe sobre o Programa de redução de Patógenos, monitoramento microbiológico e controle de Salmonella sp. em carcaças de frangos e perus. 2003.
- 63.Agência Nacional de Vigilância Sanitária. Programa Nacional de Monitoramento da Prevalência e da Resistência Bacteriana em Frango-PREBAF. 2004.
- 64.Ministério da Agricultura Pecuária e Abastecimento, Secretaria de Defesa Agropecuária. INSTRUÇÃO NORMATIVA No 20, DE 21 DE OUTUBRO DE 2016. 2016.
- 65.Orsi RH, Bakker HC de, Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. International Journal of Medical Microbiology. 2011. Feb;301(2):79–96. doi: 10.1016/j.ijmm.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 66.Piffaretti JC, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser JM, et al. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A. 1989;86(10):3818–22. doi: 10.1073/pnas.86.10.3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol. 2008. Dec;74(24):7629–42. doi: 10.1128/AEM.01127-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Vol. 9, Microbes and Infection. Microbes Infect; 2007. p. 1236–43. doi: 10.1016/j.micinf.2007.05.011 [DOI] [PubMed] [Google Scholar]
- 69.Tenover FC, Arbeit RD, Goering R V., Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13(SUPPL. 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x [DOI] [PubMed] [Google Scholar]
- 71.Wiedmann M. Molecular Subtyping Methods for Listeria monocytogenes. J AOAC Int. 2002. Mar 1;85(2):524–32. [PubMed] [Google Scholar]
- 72.Magalhães VD, Ferreira JC, Barelli C, Darini ALC. Eletroforese em campo pulsante em bacteriologia—uma revisão técnica. Rev Inst Adolfo Lutz. 2005;64(2):155–61. [Google Scholar]
- 73.Larsen M V., Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012. Apr;50(4):1355–61. doi: 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moraes DMC, Andrade MA, Minafra-Rezende CS, Barnabé AC de S, Jayme V de S, Nunes IA, et al. Fontes de infecção e perfil de suscetibilidade aos antimicrobianos de Salmonella sp. isoladas no fluxo de produção de frangos de corte. Arq Inst Biol (Sao Paulo) [Internet]. 2014. Sep [cited 2021 Mar 3];81(3):195–201. Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1808-16572014000300195&lng=en&nrm=iso&tlng=pt [Google Scholar]
- 75.Moreira G do N, e Rezende CSM, Carvalho RN, de Mesquita SQP, de Oliveira AN, Arruda MLT. Occurrence of Salmonella sp. in carcasses of broilers slaughtered and marketed in cities of state of Goiás, Brazil. Rev Inst Adolfo Lutz. 2008;126–30. [Google Scholar]
- 76.Monte DF, Lincopan N, Berman H, Cerdeira L, Keelara S, Thakur S, et al. Genomic Features of High-Priority Salmonella enterica Serovars Circulating in the Food Production Chain, Brazil, 2000–2016. Sci Rep. 2019. Dec 1;9(1):1–12. doi: 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Medeiros M, Castro VHL de, Mota ALA de A, Pereira MG, De Martinis ECP, Perecmanis S, et al. Assessment of Internalin A Gene Sequences and Cell Adhesion and Invasion Capacity of Listeria monocytogenes Strains Isolated from Foods of Animal and Related Origins. Foodborne Pathog Dis. 2020. Dec 18;18(4):243–52. doi: 10.1089/fpd.2020.2855 [DOI] [PubMed] [Google Scholar]
- 78.Manuel CS, Stelten A Van, Wiedmann M, Nightingale KK, Orsi RH. Prevalence and distribution of Listeria monocytogenes inlA alleles prone to phase variation and inlA alleles with premature stop codon mutations among human, food, animal, and environmental isolates. Appl Environ Microbiol. 2015;81(24):8339–45. doi: 10.1128/AEM.02752-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Camargo AC, Moura A, Avillan J, Herman N, McFarland AP, Sreevatsan S, et al. Whole-genome sequencing reveals Listeria monocytogenes diversity and allows identification of long-term persistent strains in Brazil. Environ Microbiol. 2019. Dec 1;21(12):4478–87. doi: 10.1111/1462-2920.14726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nightingale KK, Ivy RA, Ho AJ, Fortes ED, Njaa BL, Peters RM, et al. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl Environ Microbiol. 2008. Nov;74(21):6570–83. doi: 10.1128/AEM.00997-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferreira da Silva M, Ferreira V, Magalhães R, Almeida G, Alves A, Teixeira P. Detection of premature stop codons leading to truncated internalin A among food and clinical strains of Listeria monocytogenes. Food Microbiol. 2017. May 1;63:6–11. doi: 10.1016/j.fm.2016.10.033 [DOI] [PubMed] [Google Scholar]
- 82.Lecuit M, Ohayon H, Braun L, Mengaud J, Cossart P. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect Immun. 1997;65(12):5309–19. doi: 10.1128/iai.65.12.5309-5319.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olier M, Garmyn D, Rousseaux S, Lemaître JP, Piveteau P, Guzzo J. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: In vivo investigation by allelic exchange. Infect Immun [Internet]. 2005. Jan [cited 2020 Dec 6];73(1):644–8. Available from: /pmc/articles/PMC538994/?report=abstract doi: 10.1128/IAI.73.1.644-648.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quereda JJ, Rodríguez-Gómez IM, Meza-Torres J, Gómez-Laguna J, Nahori MA, Dussurget O, et al. Reassessing the role of internalin B in Listeria monocytogenes virulence using the epidemic strain F2365. Clin Microbiol Infect. 2019. Feb 1;25(2):252.e1-252.e4. doi: 10.1016/j.cmi.2018.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phelps CC, Vadia S, Arnett E, Tan Y, Zhang X, Pathak-Sharma S, et al. Relative roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect Immun. 2018. Oct 1;86(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ward TJ, Evans P, Wiedmann M, Usgaard T, Roof SE, Stroika SG, et al. Molecular and phenotypic characterization of listeria monocytogenes from U.S. department of agriculture food safety and inspection service surveillance of ready-to-eat foods and processing facilities. J Food Prot. 2010;73(5):861–9. doi: 10.4315/0362-028x-73.5.861 [DOI] [PubMed] [Google Scholar]
- 87.Camargo AC, De Castilho NPA, Da Silva DAL, Vallim DC, Hofer E, Nero LA. Antibiotic resistance of listeria monocytogenes isolated from meat-processing environments, beef products, and clinical cases in Brazil. Microb Drug Resist [Internet]. 2015. Aug 1 [cited 2020 Dec 5];21(4):458–62. Available from: https://pubmed.ncbi.nlm.nih.gov/25756759/ doi: 10.1089/mdr.2014.0270 [DOI] [PubMed] [Google Scholar]
- 88.Haubert L, Mendonça M, Lopes G V., de Itapema Cardoso MR, da Silva WP. Listeria monocytogenes isolates from food and food environment harbouring tetM and ermB resistance genes. Lett Appl Microbiol. 2016. Jan 1;62(1):23–9. doi: 10.1111/lam.12516 [DOI] [PubMed] [Google Scholar]
- 89.Teixeira LAC, Carvalho FT, Vallim DC, Pereira RCL, Neto AC, Vieira BS, et al. Listeria monocytogenes in export-approved beef from mato grosso, brazil: Prevalence, molecular characterization and resistance to antibiotics and disinfectants. Microorganisms. 2020. Jan 1;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Charpentier E, Courvalin P. Antibiotic resistance in Listeria spp. Vol. 43, Antimicrobial Agents and Chemotherapy. American Society for Microbiology; 1999. p. 2103–8. [DOI] [PMC free article] [PubMed]
- 91.Ministério da Agricultura Pecuária e Abastecimento. Instrução Normativa no 9 de 27 de junho de 2003. 2003.
- 92.Craig AM, Dotters-Katz S, Kuller JA, Thompson JL. Listeriosis in Pregnancy: A Review. Obstet Gynecol Surv [Internet]. 2019. Jun 1 [cited 2021 Mar 15];74(6):362–8. Available from: https://journals.lww.com/00006254-201906000-00019 doi: 10.1097/OGX.0000000000000683 [DOI] [PubMed] [Google Scholar]
- 93.Vitas AI, Sánchez RM, Aguado V, García-Jalón I. Antimicrobial susceptibility of Listeria monocytogenes isolated from food and clinical cases in Navarra, Spain. J Food Prot. 2007;70(10):2402–6. doi: 10.4315/0362-028x-70.10.2402 [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Yeh E, Hall G, Cripe J, Bhagwat AA, Meng J. Characterization of Listeria monocytogenes isolated from retail foods. Int J Food Microbiol. 2007. Jan 1;113(1):47–53. doi: 10.1016/j.ijfoodmicro.2006.07.010 [DOI] [PubMed] [Google Scholar]
- 95.Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, Lecuit M, et al. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob Agents Chemother. 2010. Jun;54(6):2728–31. doi: 10.1128/AAC.01557-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertrand S, Huys G, Yde M, D’Haene K, Tardy F, Vrints M, et al. Detection and characterization of tet(M) in tetracycline-resistant Listeria strains from human and food-processing origins in Belgium and France. J Med Microbiol. 2005. Dec;54(12):1151–6. doi: 10.1099/jmm.0.46142-0 [DOI] [PubMed] [Google Scholar]
- 97.Escolar C, Gómez D, Del Carmen Rota García M, Conchello P, Herrera A. Antimicrobial resistance profiles of listeria monocytogenes and listeria innocua isolated from ready-to-eat products of animal origin in Spain. Foodborne Pathog Dis. 2017. Jun 1;14(6):357–63. doi: 10.1089/fpd.2016.2248 [DOI] [PubMed] [Google Scholar]
- 98.Davis MA, Besser TE, Orfe LH, Baker KNK, Lanier AS, Broschat SL, et al. Genotypic-phenotypic discrepancies between antibiotic resistance characteristics of Escherichia coli isolates from calves in management settings with high and low antibiotic use. Appl Environ Microbiol. 2011. May;77(10):3293–9. doi: 10.1128/AEM.02588-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Y, Li X, Osmundson T, Shi L, Yan H. Comparative Genomic Analysis of a Multidrug-Resistant Listeria monocytogenes ST477 Isolate. Foodborne Pathog Dis. 2019. Sep 1;16(9):604–15. doi: 10.1089/fpd.2018.2611 [DOI] [PubMed] [Google Scholar]
- 100.Srinivasan V, Nam HM, Nguyen LT, Tamilselvam B, Murinda SE, Oliver SP. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog Dis. 2005. Sep;2(3):201–11. doi: 10.1089/fpd.2005.2.201 [DOI] [PubMed] [Google Scholar]
- 101.European Centre for Disease Prevention and Control. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. Vol. 11, EFSA Journal. Wiley-Blackwell Publishing Ltd; 2013 May.
- 102.Maung AT, Mohammadi TN, Nakashima S, Liu P, Masuda Y, Honjoh K ichi, et al. Antimicrobial resistance profiles of Listeria monocytogenes isolated from chicken meat in Fukuoka, Japan. Int J Food Microbiol. 2019. Sep 2;304:49–57. doi: 10.1016/j.ijfoodmicro.2019.05.016 [DOI] [PubMed] [Google Scholar]
- 103.Dantas STA, Camargo CH, Tiba-Casas MR, Vivian RC, Pinto JPAN, Pantoja JCF, et al. Environmental persistence and virulence of Salmonella spp. Isolated from a poultry slaughterhouse. Food Res Int. 2020. Mar 1;129:108835. doi: 10.1016/j.foodres.2019.108835 [DOI] [PubMed] [Google Scholar]
- 104.Da Cunha-Neto A, Carvalho LA, Carvalho RCT, Dos Prazeres Rodrigues D, Mano SB, De Souza Figueiredo EE, et al. Salmonella isolated from chicken carcasses from a slaughterhouse in the state of Mato Grosso, Brazil: Antibiotic resistance profile, serotyping, and characterization by repetitive sequence-based PCR system. Poult Sci [Internet]. 2018. Apr 1 [cited 2021 Mar 18];97(4):1373–81. Available from: https://pubmed.ncbi.nlm.nih.gov/29365208/ doi: 10.3382/ps/pex406 [DOI] [PubMed] [Google Scholar]
- 105.Yamatogi RS, Oliveira HC, Camargo CH, Fernandes SA, Hernandes RT, Pinto JP, et al. Clonal relatedness and resistance patterns of salmonella corvallis from poultry carcasses in a Brazilian slaughterhouse. J Infect Dev Ctries. 2015. Oct 1;9(10):1161–5. doi: 10.3855/jidc.5634 [DOI] [PubMed] [Google Scholar]
- 106.Vinueza-Burgos C, Baquero M, Medina J, De Zutter L. Occurrence, genotypes and antimicrobial susceptibility of Salmonella collected from the broiler production chain within an integrated poultry company. Int J Food Microbiol. 2019. Jun 16;299:1–7. doi: 10.1016/j.ijfoodmicro.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 107.Zhu Y, Lai H, Zou L, Yin S, Wang C, Han X, et al. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int J Food Microbiol. 2017. Oct 16;259:43–51. doi: 10.1016/j.ijfoodmicro.2017.07.023 [DOI] [PubMed] [Google Scholar]
- 108.Voss-Rech D, Potter L, Vaz CSL, Pereira DIB, Sangioni LA, Vargas ÁC, et al. Antimicrobial resistance in nontyphoidal salmonella isolated from human and poultry-related samples in Brazil: 20-year meta-analysis. Foodborne Pathog Dis. 2017. Feb 1;14(2):116–24. doi: 10.1089/fpd.2016.2228 [DOI] [PubMed] [Google Scholar]
- 109.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006. Oct;6(10):629–40. doi: 10.1016/S1473-3099(06)70599-0 [DOI] [PubMed] [Google Scholar]
- 110.PAMvet-PR. Levantamento do Uso e Comercialização de Medicamentos Veterinários em Frango de Corte. 2005. [Google Scholar]
- 111.World Health Organization. World Health Organization Model List of Essential Medicines, 21st List. 2019.
- 112.Jeon HY, Seo KW, Kim Y Bin, Kim DK, Kim SW, Lee YJ. Characteristics of third-generation cephalosporin-resistant Salmonella from retail chicken meat produced by integrated broiler operations. Poult Sci. 2019;98(4):1766–74. doi: 10.3382/ps/pey514 [DOI] [PubMed] [Google Scholar]
- 113.Ma Y, Li M, Xu X, Fu Y, Xiong Z, Zhang L, et al. High-levels of resistance to quinolone and cephalosporin antibiotics in MDR-ACSSuT Salmonella enterica serovar Enteritidis mainly isolated from patients and foods in Shanghai, China. Int J Food Microbiol. 2018. Dec 2;286:190–6. doi: 10.1016/j.ijfoodmicro.2018.09.022 [DOI] [PubMed] [Google Scholar]
- 114.Ministério da Agricultura Pecuária e Abastecimento. INSTRUÇÃO NORMATIVA No 45, DE 22 DE NOVEMBRO DE 2016. 2016.
- 115.Ministério da Agricutura Pecuária e Abastecimento. INSTRUÇÃO NORMATIVA No 1, DE 13 DE JANEIRO DE 2020. 2020.
- 116.Vilela FP, Gomes CN, Passaglia J, Rodrigues DP, Costa RG, Casas MRT, et al. Genotypic resistance to quinolone and tetracycline in Salmonella Dublin strains isolated from humans and animals in Brazil. Microb Drug Resist. 2019. Mar 1;25(2):143–51. doi: 10.1089/mdr.2017.0329 [DOI] [PubMed] [Google Scholar]
- 117.Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-Mediated Quinolone Resistance. Microbiol Spectr. 2014. Oct 3;2(5). doi: 10.1128/microbiolspec.PLAS-0006-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jacoby GA. AmpC Β-Lactamases. Clin Microbiol Rev. 2009. Jan 1;22(1):161–82. doi: 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harvey J, Keenan KP, Gilmour A. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 2007. Jun;24(4):380–92. doi: 10.1016/j.fm.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 120.Di Bonaventura G, Piccolomini R, Paludi D, D’Orio V, Vergara A, Conter M, et al. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J Appl Microbiol. 2008. Jun;104(6):1552–61. doi: 10.1111/j.1365-2672.2007.03688.x [DOI] [PubMed] [Google Scholar]
- 121.Magalhães R, Ferreira V, Biscottini G, Brandão TRS, Almeida G, Teixeira P. Biofilm formation by persistent and non-persistent Listeria monocytogenes strains on abiotic surfaces. Acta Aliment. 2017. Mar 1;46(1):43–50. [Google Scholar]
- 122.Yin B, Zhu L, Zhang Y, Dong P, Mao Y, Liang R, et al. The Characterization of Biofilm Formation and Detection of Biofilm-Related Genes in Salmonella Isolated from Beef Processing Plants. Foodborne Pathog Dis. 2018. Oct 1;15(10):660–7. doi: 10.1089/fpd.2018.2466 [DOI] [PubMed] [Google Scholar]


