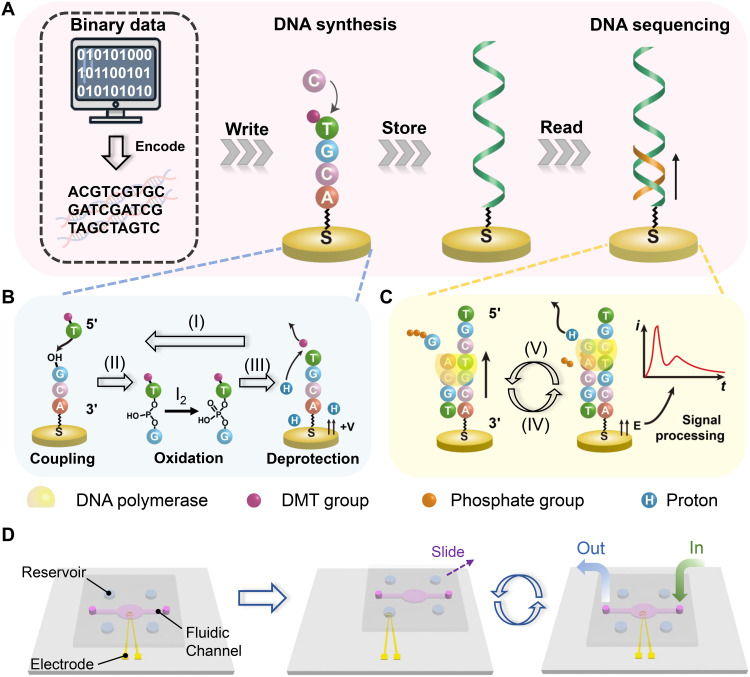
DNA synthesis and sequencing on a single electrode enables integrated data storage using a sliding microarray chip.
Abstract
DNA has been considered as a compelling candidate for digital data storage due to advantages such as high coding density, long retention time, and low energy consumption. Despite many works reported, the development of a DNA-based database of full integration, high efficiency, and practical applicability is still challenging. In this work, we report the synthesis and sequencing of DNA on a single electrode with scalability for an integrated DNA-based data storage system. The synthesis of DNA is based on phosphoramidite chemistry and electrochemical deprotection. The sequencing relies on charge redistribution originated from polymerase-catalyzed primer extension, leading to a measurable current spike. By regeneration of the electrode after sequencing, repeated sequencing can be achieved to improve the accuracy. A SlipChip device is developed to simplify the liquid introduction involved in DNA synthesis and sequencing. As the proof-of-concept experiment, text information is stored in the system and then accurately retrieved.
INTRODUCTION
With the development of information technology that leads to overwhelming demands for data storage capacity, people are increasingly interested in new data storage media. The new storage medium is expected to be of higher efficiency, coding density, and stability than the conventional semiconductive and magnetic media. As a promising candidate, DNA has attracted considerable attention during the past several years (1–4). DNA is a naturally selected molecule for storing our gene information, which serves as the blueprint to construct and maintain the most intricate biological system. DNA-related biotechnologies such as synthesis (5, 6), polymerase chain reaction (PCR) (7, 8), and sequencing (9, 10) can facilitate efficient writing, reading, editing, and random access of data encoded in DNA sequences (11, 12). The theoretical data density in DNA is as high as ~455 EB (exabyte) g−1 (13–15), and DNA is also very stable when preserved under proper conditions, so the information encoded in it can be still readable after a long time (e.g., over 2 million years in Global Seed Vault) (16, 17). It has been previously reported that 200-megabyte digital files have been successfully encoded into DNA sequences (18), which equals to the storage capacity of the most popular hard disk drives probably 20 years ago.
The operation of almost all the previously reported DNA-based databases generally involves several steps, including data writing, locating, and reading (2, 19). Briefly, in the data-writing step, the binary information stream is converted to quaternary (A, T, C, and G) base sequences, which are then synthesized using a chemical or enzymatic method (20–23). For the data locating, specific DNA strands in the data pool are selected and/or amplified (e.g., by PCR) for subsequent data reading (24, 25). In the reading step, a sequencing technology is used for the identification of the quaternary bases, which are then transformed back to the original digital data (23, 26).
Most of these methods reported so far are based on commercially available instrumentations (e.g., synthesis, PCR, and sequencing apparatuses) that are sophisticated and require highly trained personnel to operate. Although they are very powerful for biological research, they are not devised or optimized for data storage and, therefore, may lead to problems. For example, current methods usually involve complicated liquid manipulations in each step and manual operations in between. Adding one phosphoramidite nucleotide monomer in the synthesis step generally requires the introduction of at least four kinds of liquid solutions, not to mention the sequencing step. These limit the scale-up capability of this technique and increase the error probability. Correcting data errors requires a considerable amount of data redundancy, which restricts the efficiency of data storage. In addition, random access of data may involve the addition of address tags into the DNA sequences, which can become markedly cumbersome with scaling up (27). Random access of data using PCR can also cause data loss owing to biased amplification (28, 29). Therefore, it is still challenging to develop a DNA-based database of full integration, high efficiency, and practical applicability, even using an automatic fluidic system (30).
Here, we report a fully integrated DNA data storage system based on electrochemical DNA synthesis and sequencing on a single electrode, as shown in Fig. 1A. Binary data are first transformed into DNA sequences using Reed-Solomon (RS) codes (31, 32). For the synthesis, the addition of nucleotide monomers is realized on a Au electrode based on phosphoramidite chemistry with electrochemical deprotection (Fig. 1B). The sequencing is implemented on the same electrode based on a sequencing-by-synthesis method, in which the charge redistribution caused by the polymerase-catalyzed primer extension leads to a measurable current spike (Fig. 1C). All of the liquid manipulations for the synthesis and sequencing are accomplished on the basis of the principle of SlipChip, which can be fully integrated for the scale-up of the database (Fig. 1D). By writing and reading data on a single electrode, an address sequence is not required so that the data volume is only limited by the number of electrodes. The strategy can also eliminate some random errors by multiple sequencing and base identification using the principle of plurality voting.
Fig. 1. Schematic illustration of a data storage system based on DNA synthesis and sequencing on the same Au electrode.
(A) Schematic showing the generation procedure for the DNA-based data storage. (B) Schematic of the electrochemically triggered phosphoramidite chemistry for the synthesis of DNA on the electrode. (I) A phosphoramidite nucleotide monomer with a dimethyltrityl (DMT) protecting group reacts with the free-hydroxyl group on the electrode/DNA molecule, forming a phosphite linkage. (II) The phosphite bond is oxidized by iodine to a more stable phosphate bond. (III) A positive potential is then applied to the electrode to generate protons. The acid-labile DMT protecting group on the nucleotide is removed to expose another free hydroxyl group for the addition of the next cycle. (C) Schematic of the DNA sequencing method on the same electrode based on charge redistribution in the sequencing-by-synthesis process. (IV) A known deoxyribonucleoside triphosphate (dNTP) and the DNA polymerase are added. Then, the polymerase binds to the template DNA, which is complementary with a primer DNA. (V) A proton is removed in the coupling reaction, and diffusion of the proton induces a charge redistribution and thus a transient current signal for base identification. (D) Schematic illustration of the principle of the SlipChip device for integrated DNA synthesis and sequencing. Four phosphoramidite nucleotide monomers for synthesis (or four dNTPs for sequencing) are preloaded in the reservoirs. A washing solution and other reagents are introduced using the fluidic channel. Liquid manipulation is accomplished by sliding the top plate.
RESULTS
DNA synthesis and characterization on a Au electrode
The synthesis of DNA on the Au electrode was based on electrochemically initiated acid deprotection, so the electrode was first modified with bis-aniline–cross-linked Au nanoparticles (NPs) for the generation of sufficient protons at a relatively low overpotential (fig. S1) (33). For the NP modification, the Au electrode was immersed in a solution of thioaniline-functionalized Au NPs, and then the electrode potential was scanned for electropolymerization. The surface area of the electrode increased after the modification, which was confirmed by the increased characteristic Au peak (~4.2 times of a naked electrode) at ~0.97 V (versus AgCl/Ag) in the cyclic voltammogram in 0.50 M H2SO4 (fig. S2B). The larger surface area can provide more active sites for DNA synthesis so as to enhance the current signals for the subsequent DNA sequencing. The modified electrode was then immersed in a solution containing 6-mercapto-1-hexanol (MCH), which led to the formation of a self-assembled monolayer having terminal hydroxyl groups on the electrode to link with reactive phosphoramidite nucleotide monomers for DNA synthesis.
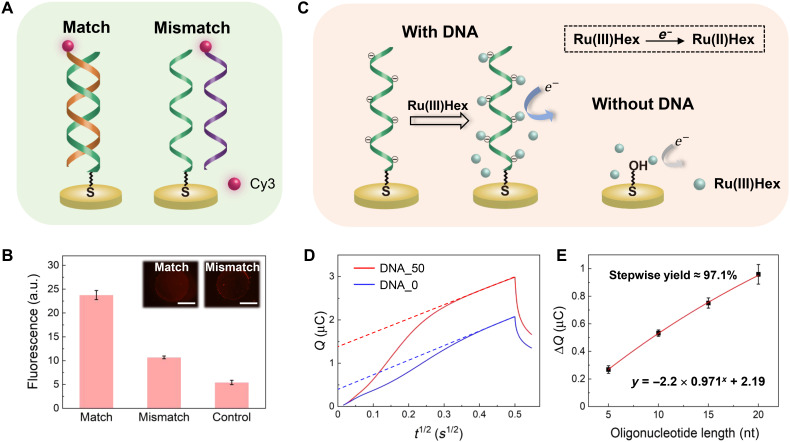
Conventional DNA synthesis based on the phosphoramidite chemistry relies on an acidic reagent (e.g., trichloroacetic acid) to remove the protecting group [e.g., dimethyltrityl (DMT)] at the 5′ end to activate the nucleotide monomer (34). It has been reported that proton-coupled redox reactions, such as the electrochemical oxidation of hydroquinone to benzoquinone, can create the required acidic condition to remove the protecting groups (35, 36). In this work, the bis-aniline units used to cross-link the Au NPs on the electrode exhibited similar redox properties. As the electrochemical reaction was reversible, the electrode could quickly switch between the ON and OFF states to increase the speed of the DNA synthesis. Furthermore, as the redox groups were tethered on the electrode surface, the deprotection was more localized, which could help to avoid cross-talk in a microelectrode array. These features can be beneficial for the large-scale application of the technique. To demonstrate the applicability of the electrode for DNA synthesis, an oligonucleotide with a sequence of 3′-TTTTTTGAACCTT-5′ (Oligo-1) was synthesized using our method. The synthesized oligonucleotide on the electrode was characterized by fluorescence measurements, in which a Cy3-labeled complementary probe (Oligo-c1, 5′-ACTTGGAA-Cy3-3′) and a mismatched probe (Oligo-mis1, 5′-ACTTAGAA-Cy3-3′) were used to hybridize with the surface Oligo-1, respectively (Fig. 2A). After washing to remove the nonspecifically adsorbed oligonucleotide, the fluorescence measured for Oligo-c1 was higher than that for Oligo-mis1, as shown in Fig. 2B, which indicated that the oligonucleotide with the predetermined sequence was available on the electrode.
Fig. 2. Synthesis and characterization of DNA on the electrode.
(A) Schematic of the fluorescent determination of the synthesized oligonucleotide on the electrode using Cy3-labeled probes. (B) Fluorescence intensity measured for determination of the synthesized oligonucleotide on the electrode (insets, fluorescence micrographs of the electrodes). For the control experiment, the fluorescence from a blank Au electrode was measured without the hybridization reaction. The error bars represent the SD of three replicated measurements. Scale bars, 1 mm. a.u., arbitrary units. (C) Schematic of quantitative determination of the oligonucleotide based on chronocoulometric measurements. For the quantification, positively charged RuHex was stoichiometrically adsorbed on the electrode owing to electrostatic interaction with the DNA backbone and was then reduced so that the electric charges were correlated to the number of surface oligonucleotides. (D) Electric charges passed as a function of the square root of time for electrodes having the synthesized DNA in 10 mM tris-HCl (pH 8.3) containing 50 μM RuHex (red) or not (blue). The linear parts of the curves were fitted and marked using dash lines. (E) Electric charges attributed to the reduction of the stoichiometrically bound RuHex on the electrode as a function of the oligonucleotide length. The charges were calculated on the basis of the difference in the intercepts of the blue and red dash curves in (D) (37, 43). The error bars represent the SD of three replicated measurements, and the plots were fitted using an exponential attenuation model. Photo credit: Chengtao Xu, Southeast University.
A previously reported method based on chronocoulometry was used to quantitatively analyze the oligonucleotide on the surface (37, 38). Briefly, the phosphate backbone of DNA was negatively charged, which can electrostatically attract redox cations [e.g., ruthenium hexaammine (RuHex)] in the solution. The RuHex on the electrode surface can be quantitatively determined on the basis of chronocoulometry and stoichiometrically correlated to the surface density of charge on the electrode (Fig. 2C). As illustrated in Fig. 2D and fig. S3, the difference in intercepts of the two curves was used to calculate the charge that was attributed to the reduction of the stoichiometrically bound RuHex on the electrode. The charge exhibited a correlation with the length of the oligonucleotides synthesized [in nucleotides (nt)] on the electrode (Syn-5nt, 3′-TGACT-5′; Syn-10nt, 3′-TGACTGACTG-5′; Syn-15nt, 3′-TGACTGACTGACTGA-5′; Syn-20nt, 3’-TGACTGACTGACTGACTGAC-5′), as shown in Fig. 2E. However, the charge and the oligonucleotide length were not linearly correlated, and this is reasonable because the stepwise yield for the chemical DNA synthesis was less than 100%. The nonideal yield was probably owing to incomplete deprotection and the insufficient reaction between the phosphoramidite and the hydroxyl group, leading to deletion errors in the DNA synthesis (39). The errors could accumulate, so a larger deviation from the linear relationship with increasing oligonucleotide length was observed in Fig. 2E. An exponential attenuation model was used to fit the data points in Fig. 2E so that an averaged stepwise yield of ~97.1% was obtained from the equation, which was acceptable compared with previously reported yields for chemical synthesis (90 to 98%) (40, 41) and enzymatic synthesis (~97.7%) (42).
To estimate the surface density of oligonucleotide on the electrode, we used a previously reported equation with some modification (43)
where ΓDNA is the surface density of the synthesized oligonucleotide (in molecules per square centimeter), Γ0 is the surface density of adsorbed RuHex (in moles per square centimeter), m is the length of the oligonucleotide, z is the charge of RuHex (z = 3), NA is the Avogadro’s number (6.022 × 1023 molecules mol−1), n is the number of electrons transferred for reduction of one RuHex (n = 1), ΔQ′is the corrected charge attributed to the reduction of the surface RuHex cations, F is the Faraday constant (96,485 C mol−1), A is the surface area (in square centimeters), ΔQ is the experimentally measured charge in chronocoulometry, and Y is the stepwise yield for the oligonucleotide synthesis (details are available in the Supplementary Materials). By calculation, the average surface density of the Syn-10nt on a bare Au electrode was 1.644 × 1013 molecules cm−2. After cross-linking Au NPs on the electrode to increase the surface area, the surface density increased to 4.062 × 1013 molecules cm−2, as shown in fig. S4A, which was higher than that reported for conventional surface-immobilized DNA (~6 × 1012 molecules cm−2) (38, 43). This is probably because the larger surface area provided more active sites for the DNA synthesis on the electrode surface. Besides, with the bis-aniline–cross-linked Au NPs on the surface, a larger anodic current for the proton generation was observed than that for a bare Au electrode, as shown in fig. S4B.
The potential applied for the proton-generation reaction was optimized to not only ensure complete acidic deprotection for DNA synthesis but also avoid unwanted side reactions leading to synthetic errors. As shown in the Supplementary Materials, the anodic current monotonically increased with increasing potentials from 1.2 to 1.7 V (fig. S5A), but the resulting oligonucleotide density of Syn-10nt on the electrode first increased and then decreased with increasing potentials (fig. S5B). The result indicated that a potential of 1.4 V was the optimal potential for the electrochemical deprotection step for DNA synthesis, which was used for future experiments.
DNA sequencing based on charge redistribution
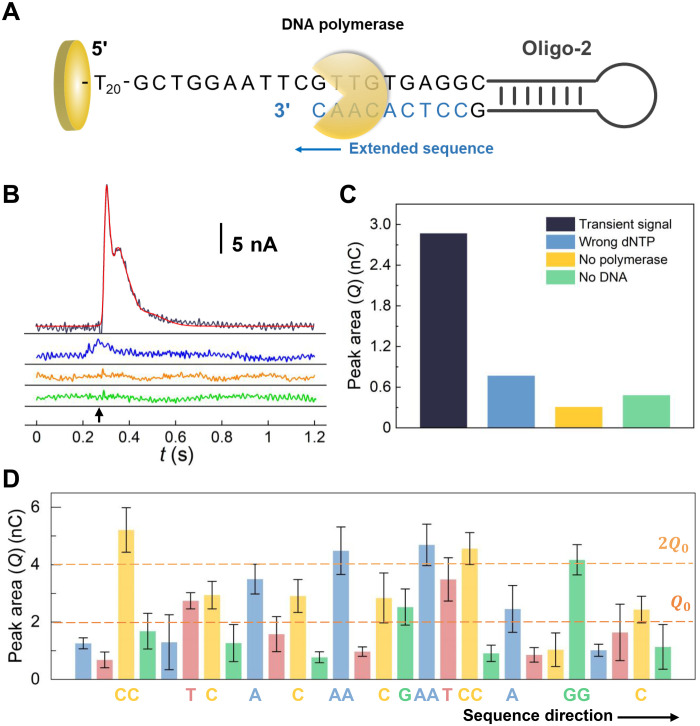
To demonstrate the DNA sequencing on the electrode based on charge redistribution in the sequencing-by-synthesis process (44, 45), we immobilized a self-priming single-stranded oligonucleotide (Oligo-2, 5′-thiol-TTTTTTTTTTTTTTTTTTTTGCTGGAATTCGTTGTGAGGCCGTC GTTTTACAACGGAACCGTTGTAAAACGACGG-3′, commercially synthesized and purified) on the electrode as the target for sequencing. The self-priming Oligo-2 spontaneously formed a hairpin secondary structure after incubation so that a DNA polymerase could directly bind to its 3′ end without an additional primer (Fig. 3A and fig. S6). When a complementary deoxyribonucleoside triphosphate (dNTP) coupled with the oligonucleotide strand, a proton was released and diffused to the bulk solution. This induced a charge redistribution on the electrode so that a transient current could be recorded. As shown in Fig. 3B, when the complementary dNTP was introduced to the electrode with Oligo-2 and the polymerase, a transient current of ~20 nA was observed within 300 ms. In contrast, the addition of a noncomplementary dNTP only brought about a negligible transient current over the background. For the experiments without either the polymerase or Oligo-2, only background current was observed, which indicated that the perturbation of the interface owing to the fluid introduction was negligible. Therefore, the transient current in the sequencing-by-synthesis process could be attributed to the selective addition of a complementary dNTP to the self-priming Oligo-2. For quantitative analysis, the electric charges (the integral of current over time) were used instead of current signals (fig. S7), because the charges directly reflected the number of protons released during the polymerase-catalyzed reaction, and measurement of charges was more reproducible than current for the transient process. It was found that for a valid signal of a dNTP addition, the averaged charge was approximately 2.87 nC, which corresponded to a surface density of 5.7 × 1011 DNA molecules cm−2. We compared the signal with those in the control experiments, as shown in Fig. 3C, the number of charges generated in a valid dNTP addition was distinguishable.
Fig. 3. DNA sequencing on the electrode.
(A) Schematic illustration of the DNA sequencing-by-synthesis process for the self-priming Oligo-2. (B) Current as a function of time measured in the sequencing-by-synthesis process. Different reagents were introduced to electrodes having the self-priming Oligo-2 at the time marked using the black arrow, including polymerase and a complementary dNTP (black), polymerase and a noncomplementary dNTP (blue), and a complementary dNTP without polymerase (yellow), respectively. For the control experiment (green), there was no oligonucleotide on the Au electrode. A Gaussian fitting of the black curve was also shown (red). (C) Peak area/charge (Q) of the transient signals in response to the dNTP addition. The colors correspond to those of curves in (B). (D) Peak area/charge (Q) measured for sequencing of the Oligo-2. The sequencing was carried out by sequential introduction of different dNTPs in the order of A, T, C, and G to the electrode, while measuring the current. The number of dNTPs coupled for each time was determined on the basis of the magnitude of the peak area relative to Q0 and 2Q0, as indicated by the brown dash lines in the figure. Error bars represent the SD of five replicated measurements.
The Oligo-2 strand on the electrode was sequenced by sequentially adding dNTPs into the solution (in the order of A, T, C, and G) and simultaneously measuring the electric signals. By analysis of the data, a statistic value Q0, which equals 2 nC, was defined. An electric signal with charge less than Q0 was regarded as negative, which indicated that no dNTP was added to the DNA strand (or the added dNTP was not complementary to the target strand). A signal whose charge was between Q0 and 2Q0 was considered as a positive signal for the selective addition of one complementary dNTP. Note that repeated nucleotides in the sequence would generate charges in proportion to their lengths, and a signal between 2Q0 and 3Q0 was considered as the signal for the addition of two identical dNTPs at the same time. The self-priming Oligo-2 included a sequence of 5′-GCTGGAATTCGTTGTGAGG-3′, which was sequenced multiple times using our method, and the resulting data were shown in Fig. 3D and fig. S8. To evaluate the sequencing accuracy, we used the Needleman-Wunsch alignment in DNAMAN software to compare the sequencing result with the actual sequence (46, 47). Sixteen of 19 sites were matched in the final alignment, which indicated that an accuracy of 84.21% was achieved. The accuracy was acceptable compared with the previously reported results (48–50). Now both the synthesis and sequencing of an oligonucleotide could be accomplished on the same electrode for data writing and reading.
DNA synthesis and sequencing for data storage
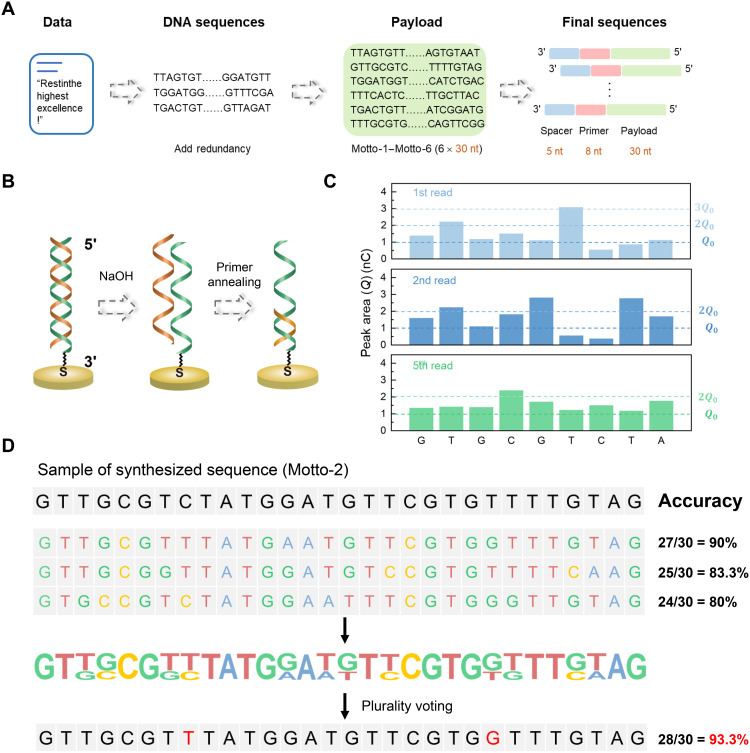
A text of the motto of Southeast University, “Restinthehighestexcellence!”, was translated into binary data (27 bytes) and then to quaternary DNA sequences by converting 00, 01, 10, and 11 to A, T, G, and C, respectively (Fig. 4A). The resulting sequence was first divided into three pieces and then processed in MATLAB to add redundant information based on the RS encoding algorithm for error correction (overall redundancy ratio, 6/15 = 40%). The sequences were further separated into six data sequences of 30 nt (Motto-1 to Motto-6; sequences are available in table S2). As illustrated in Fig. 4A, an additional primer sequence (3′-TGAACCTT-5′, 8 nt) and a polyT spacing sequence (5 nt) were added to the 3′ end of each data sequence. Oligonucleotide strands of these sequences (43 nt) were synthesized on different Au electrodes, respectively, which were subsequently sequenced using the same electrode and decoded to obtain the encoded data. The length of the oligonucleotides (43 nt) was much greater than that we have previously synthesized (10 nt), so the surface density of the oligonucleotides we measured (3.2 × 1011 DNA molecules cm−2) was much lower, which was reasonable considering the nonideal yield of the DNA synthesis.
Fig. 4. Data storage based on the DNA synthesis and sequencing.
(A) Schematic illustration of the procedure to transform the text data into DNA sequences with data redundancy, a polyT spacer, and a primer. (B) Schematic illustration of the regeneration process for repeated sequencing to improve the accuracy. For the regeneration, the DNA strand was removed from the target DNA by denaturalizing the double-strand DNA in 0.20 M NaOH for 5 min. Then, the next round of sequencing started by primer reannealing. (C) Peak area/charge (Q) measured in the replicated sequencing (first, second, and fifth attempts) of the oligonucleotide on the electrode (negative signals not shown). The number of dNTPs coupled each time was determined on the basis of the magnitude of the peak area relative to Q0 indicated by dash lines in the figure. (D) The process to obtain the final sequence based on plurality voting using the multiple sequencing results, which can improve the accuracy.
The accuracy of the sequencing step was found to be 84.21% (Fig. 3), which can be representative of the sequencing accuracy of our method. The accuracy accumulated for the integrated DNA synthesis and sequencing was 75.56%, as obtained from experiments shown in Fig. 4 and calculation in table S3. So the accuracy of the synthesis could be estimated by 75.56/84.21% = 89.73%. Note that this is a rough estimation. The accuracy also depends on specific sequence and length.
Because the synthesized oligonucleotide with encoded information was covalently linked to the electrode, replicated sequencing can be carried out after regeneration of the electrode using 0.20 M NaOH (Fig. 4B). The alkaline solution denaturized the double-stranded DNA and removed the extended primer so that the original DNA was able to bind with another primer for sequencing. With the multiple-sequencing results, some random errors can be eliminated on the basis of the principle of plurality voting to improve the sequencing accuracy (Fig. 4, C and D) (51, 52). However, excessive sequencing would increase the cost and time notably and would not necessarily further improve the accuracy. As shown in fig. S9, with increasing cycles of DNA sequencing, the accuracy decreased gradually, which was probably because of incomplete electrode regeneration and DNA degradation. It was estimated that the accuracy would decrease to 50% after ~21 cycles.
It was found that the accuracy for data recovery was improved to ~87.22% on average after three repeated reading and plurality voting, and errors were usually correlated to the synthesis and sequencing of homopolymer regions, as shown in table S2. The homopolymer can lead to low coupling efficiency in DNA synthesis (especially for G and C), and quantification of the number of identical nucleotides in DNA sequencing can also be problematic (39, 53). As for the remaining mistakes, error correction based on data redundancy added in the encoding step served to correct them to achieve 100% data recovery. The time required for the data writing and reading process was calculated and shown in table S4. It took about 9 hours to write and read 4.5 bytes of data on the single electrode, which can be further improved by scaling up using an electrode array as shown below.
Liquid manipulation based on a SlipChip for integrated data storage system
The synthesis and sequencing of DNA on the electrode involve repeated manipulation of liquids, which can become prohibitively complicated for a really large database. As a microfluidic device for handling a large array of microdroplets, SlipChip is a platform that is especially appropriate for the development of an integrated DNA-based data storage system. All the microdroplets can be moved simultaneously on a SlipChip by simply sliding the device, so no valves, pumps, or tubes are required (54–56). This is beneficial for developing a fully integrated DNA-based database with low reagent consumption and cost. In this work, we developed a SlipChip-based microfluidic device for integrated DNA-based data storage. Briefly, a 2 × 2 Au electrode array was fabricated on a glass slide by standard photolithography and physical vapor deposition. A block of polydimethylsiloxane (PDMS) with a fluidic channel and reservoirs was fabricated and then assembled with the glass slide (Fig. 5A and fig. S10). The reservoirs were preloaded with reagents for DNA synthesis and sequencing on the electrode. To seal the reagent droplets in reservoirs and avoid cross-contamination, we used amphiphobic fluorinated fluid FC-40. As the proof-of-concept experiment, a piece of text information “DNAStoresFutureWorld” (Oligo-slip-1 to Oligo-slip-4, 30 nt; sequences are available in table S2) was stored in this integrated device using our method. Similarly, an auxiliary primer sequence (8 nt) and a polyT spacing sequence (5 nt) were added to the 3′ end of each data sequence.
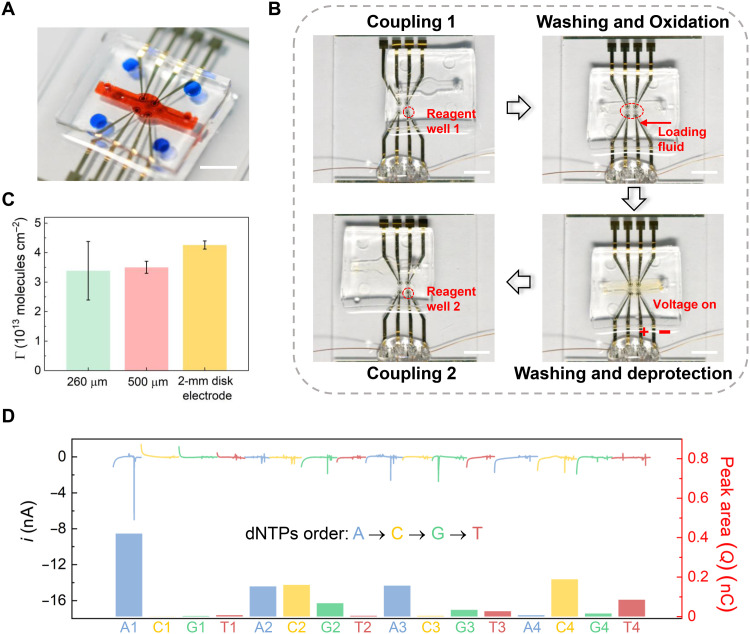
Fig. 5. Integrated data storage based on an electrode array with a SlipChip device.
(A) A photograph of the SlipChip device with a fluidic channel (orange) and reagent reservoirs (blue) on the top PDMS plate as well as a 2 × 2 Au electrode array on the bottom glass slide. Scale bar, 5 mm. (B) Photographs showing the DNA synthesis process using the SlipChip device. Phosphoramidite coupling, washing, oxidation, and deprotection steps were performed by aligning the reagent reservoirs or fluidic channel with the electrodes, respectively. For electrochemical deprotection, a potential was applied to the electrodes using a CHI900D workstation. Scale bars, 5 mm. (C) Surface densities of the synthesized DNA on microfabricated electrodes of two sizes (d = 260 and 500 μm) and a commercial 2-mm disk electrode. (D) Current signals and corresponding peak area/charge (Q) for sequencing. Photo credit: Chengtao Xu, Southeast University.
To initiate the DNA synthesis, we slipped the top PDMS relative to the bottom glass slide to expose the electrodes to phosphoramidite nucleotide monomers in the reagent reservoirs (Fig. 5B). Then, washing, oxidation, and electrochemical deprotection steps were sequentially conducted by aligning the electrodes with corresponding reagent reservoirs and the fluidic channel, respectively. As shown in fig. S11, similar current signals were observed in the electrochemical deprotection step. Compared with the chronocoulometric signal of a blank Au electrode, it indicated that the oligonucleotide was available on the electrode after the synthesis (fig. S12). Then, the surface densities of the synthesized oligonucleotide were obtained for the microfabricated electrodes of two sizes (d = 260 and 500 μm), which were 3.386 × 1013 and 3.501 × 1013 DNA molecules cm−2, respectively (Fig. 5C). The slightly decreased surface densities could arise from slipping operations but were sufficient for the subsequent DNA sequencing.
Sequencing on the same electrode was accomplished by incubating the electrodes in the polymerase solution in the fluidic channel and then sequentially exposing the electrode to four reservoirs containing different dNTPs. During the process, the current from the electrode was measured for sequencing (Fig. 5D). It was found that the overall accuracy for Oligo-slip-1 to Oligo-slip-4 was ~89.17% after three replicated sequencing and plurality voting (table S2). After error correction with the RS algorithm, the original data, DNAStoresFutureWorld, was properly recovered, which demonstrated the scalability of the proposed SlipChip-based DNA data storage device. Similarly, the time required for the data writing and reading process on the electrode array was calculated and shown in table S5. It took about 14 hours to write and read 20 bytes of data on the four-electrode array.
DISCUSSION
Here, we have reported the DNA synthesis and sequencing on the same electrode with scalability for integrated DNA data storage system. The electrochemical reactions for the DNA synthesis and sequencing were first investigated and optimized, and then a four-electrode array was used with a SlipChip microfluidic device to demonstrate the scalability of the system. Parallel DNA synthesis and sequencing can be accomplished almost simultaneously using the electrode array, which can markedly increase the volume of data stored in a given time in the future. By writing and reading data on the same electrode, there are potentially several advantages. First, the increased electrode surface by Au NP modification can lead to the synthesis of high-density DNA molecules on the electrode, which can enhance the charge redistribution signal in the sequencing process. Second, the address information, which can be very cumbersome for a really large database, is no longer required in the DNA sequences. The location of data is correlated to the identity of electrodes so that the volume of the database is now limited by the number of electrodes. Fabrication of a microelectrode array of high-density is technically feasible, which we are working on now for practically applicable large-scale data storage. The microelectrode array of high density is also fully compatible with the paralleled liquid manipulation technique based on SlipChip demonstrated in this work. Last, by regeneration of the electrode after the synthesis-by-sequencing process, the electrode can be sequenced multiple times so that the reading accuracy can be improved.
By the SlipChip device, parallel liquid manipulation involved in the complicated DNA synthesis and sequencing processes can be substantially simplified (57, 58). By simply slipping the device, a large array of microdroplets can be manipulated simultaneously without requiring any pump or valve, which is beneficial for developing a highly integrated miniaturized data storage device. Automation of the whole process can also be feasible using mechanical devices that are similar to those of conventional hard disks and CDs. Besides, because of the small size of the microfluidic channels, reservoirs, and the microelectrodes, mass transfer can be accelerated to improve the reaction kinetics, which can potentially reduce the time required for the data storage. Last, the small dead volume of the SlipChip device can save the expensive reagents to make it more cost-effective to write and read a large volume of data compared with conventional systems (30).
We envision the future developments of the proposed DNA data storage systems. By scaling up the numbers of electrodes and reaction reservoirs on the microchip, the proposed method can be potentially used for development of a really large DNA-based database with high efficiency, integration, and automation in the future. Therefore, we believe the proposed system is a promising platform for DNA-based data storage.
MATERIALS AND METHODS
Chemicals and materials
Tris-HCl, DMT-dC(bz) phosphoramidite, DMT-dG(ib) phosphoramidite, potassium chloride (KCl), polyvinylpyrrolidone (PVP; molecular weight ≈ 40,000), and hexaammine ruthenium (III) chloride were purchased from Sigma-Aldrich. Monosodium dihydric phosphate, disodium monohydric phosphate, chloroauric acid, magnesium chloride (MgCl2), concentrated sulfuric acid, p-aminothiophenol, ethanol (EtOH), methanol (MeOH), glacial acetic acid, acetonitrile, sodium borohydride, sodium hydroxide (NaOH), tetrahydrofuran (THF), ethylenediamine (EDA), EDTA, sulfuric acid, hydrogen peroxide (H2O2), and dichloromethane (DCM) were obtained from Sinopharm Chemical Reagent Co. Ltd. Dithiothreitol (DTT), 5-ethylthio-1H-tetrazole (ETT), MCH, iodine, benzoquinone, methyl glutaronitrile, and 1H,1H,2H,2H-perfluorodecyltriethoxysilane (PFDTES) were purchased from Aladdin Reagent Co. Ltd. Sodium 2-mercaptoethane sulfonate and hydroquinone were purchased from Macklin Reagent. Co. Ltd. DMT-dA(bz) phosphoramidite, DMT-dT phosphoramidite, and 3-methoxypropionitrile were obtained from Bidepharm Technology Co. Ltd. Hepes was purchased from Alfa Aesar Co. Inc. DNA polymerase [Klenow (exo-) Fragment] was obtained from Takara Biomedical Technology Co. Ltd. Tetrabutylammonium hexafluorophosphate was purchased from Innochem Technology Co. Ltd. dNTP solution was obtained from Beyotime Institute of Biotechnology. Bovine serum albumin (BSA) was purchased from Biofroxx Biotechnology Co. Ltd. Pyridine was obtained from Maya Reagent Co. Ltd. All reagents were used as received. Ultrapure water was produced by a Milli-Q system and used in all experiments. Au plate electrodes, platinum wire electrodes, and Ag/AgCl reference electrodes were purchased from Gaoss Union Technology Co. Ltd. PDMS (Sylgard 184) was obtained from Dow Corning Co. Ltd. Fluorinated fluid FC-40 was purchased from 3M company. All experiments were carried out at a room temperature of 25°C unless otherwise specified. In the Supplementary Materials, DNA sequences without data information were summarized in table S1, and DNA sequences with data information were summarized in table S2.
Instrumentation
The electrochemical measurements were implemented using a CHI900D electrochemical workstation from Shanghai Chenhua Instrument Co. Ltd. A three-electrode system was used in the electrochemical measurements. Optical and fluorescent micrographs were taken using an Olympus SZX16 microscope system. A field-emission scanning electron microscopy and elemental mapping via energy-dispersive spectrometry were performed using an UltraPlus Zeiss microscope system.
Synthesis of the modified Au NPs
Au NPs functionalized with p-aminothiophenol and 2-mercaptoethane sulfonate were synthesized as previously reported (33). Briefly, 15 ml of an EtOH solution containing 296 mg of HAuCl4 was mixed with 7.5 ml of a MeOH solution containing 63 mg of sodium 2-mercaptoethane sulfonate and 12 mg p-aminothiophenol. Glacial acetic acid (3.1 ml) was added dropwise into the mixture, which was stirred in an ice bath for at least 1 hour. Then, 7.5 ml of 1.0 M sodium borohydride aqueous solution was added dropwise into the solution. An immediate color change from yellow to black was observed in this process. The solution was stirred in an ice bath for 1 hour and then for 12 hours. The Au NPs were subsequently centrifuged and washed with EtOH twice. The as-prepared Au NPs were then dissolved in 10 mM Hepes buffer (pH 7.2) so that a solution containing Au NPs (2.0 mg ml−1) was lastly prepared. The solution was diluted sixfold with 100 mM phosphate solution (pH 7.4) and used for electropolymerization.
Modification of the Au electrode with Au NPs and thiol
A Au electrode was first polished, followed by sonication in EtOH for 1 min and then in water for another 1 min. Subsequently, the Au electrode was cleaned by repeated scanning of the applied potential from −0.3 to 1.5 V (versus Ag/AgCl; scan rate, 0.1 V s−1) in an aqueous solution containing 0.50 M H2SO4 until the characteristic peak at ~0.97 V was sharp and reproducible. The electropolymerization was accomplished by repeated scanning of the applied potential from −0.2 to 1.2 V for 200 cycles (versus Ag/AgCl; scan rate, 0.1 V s−1). After the electropolymerization, the Au NP–modified electrode was rinsed with water and then incubated in an aqueous solution containing 20 mM MCH overnight so that a self-assembled monolayer formed on the Au surface.
Converting data to DNA sequences
Original binary data were first transformed into DNA sequences. In this step, RS code was used, and redundancy information was added to the sequences (31, 32). The redundancy information can be used for the correction of errors resulted from DNA synthesis and sequencing to avoid data recovery failure.
DNA synthesis on the modified electrode
To initiate the synthesis of DNA, an acetonitrile solution containing 0.10 M phosphoramidite nucleotide and 0.50 M ETT as an activator was dropcast on the modified Au electrode and allowed for reaction of 120 s (the coupling step in Fig. 1B). After rinsed with anhydrous acetonitrile, an oxidation solution of 10% water, 20% pyridine, and 70% THF containing 0.10 M iodine was dropcast on the electrode and allowed to react for 40 s (the oxidation step in Fig. 1B). After rinsing with anhydrous acetonitrile, the Au electrode was immersed in an anhydrous acetonitrile solution containing 25 mM hydroquinone, 25 mM benzoquinone, and 25 mM tetrabutylammonium hexafluorophosphate, and a positive potential of 1.4 V was applied to the Au electrode versus the other Au electrode for 10 s to generate acid for deprotection (the deprotection step in Fig. 1B). DNA was synthesized on the electrode by repeating those three steps and alternating selected phosphoramidite in each coupling step. After the synthesis, the protecting groups on the bases were removed by incubating the electrode in EDA/EtOH [anhydrous, 1:1 (v/v)] to react for 2 hours. Last, the electrode was rinsed with water at least three times.
Fluorescent characterization of the synthesized DNA
A probe DNA with a Cy3 label at its 3′ end for the fluorescent detection of the synthesized DNA on the electrode was obtained from Sangon Biotech Co. Ltd.. The hybridization buffer of the probe DNA was an aqueous solution containing 20 mM tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 1.0 mM EDTA, 0.10% (w/v) PVP, and 0.10% (w/v) BSA. For the fluorescent measurement, 5.0 μl of 25 μM fluorescent probe was dropcast on the electrode and then incubated for at least 2 hours. The electrode was subsequently rinsed with the hybridization buffer at least five times to remove nonspecifically adsorbed fluorescent probe. Last, fluorescence images of the electrode were obtained using the Olympus SZX16 microscope. ImageJ software was applied to calculate the relative fluorescence intensity.
Quantitative characterization of the synthesized DNA by chronocoulometry
For the quantitative characterization, the three electrodes were immersed into 20 ml of 10 mM tris-HCl (pH 8.3), which had been previously deoxygenated by bubbling nitrogen for 10 min, and then the chronocoulometric measurement was accomplished (initial potential, 0.2 V; final potential, −0.5 V; the number of steps, 2; pulse width, 0.25 s; sensitivity, 1 × 10−4 A V−1). Next, 50 μl of an aqueous solution containing 20 mM hexaammine ruthenium (III) chloride was added into the buffer, which had a final concentration of 50 μM, and the chronocoulometric measurement was conducted again.
Immobilization of DNA
A total of 0.10 M phosphate buffer (pH 7.4) containing 10 μM DTT and 10 μM 5′-thiolated Oligo-2 was prepared and allowed to react for 1 hour. Five microliters of the as-prepared DNA solution was dropcast on a cleaned Au electrode and incubated for 12 hours. Last, the modified Au electrode was incubated in an aqueous solution containing 20 mM MCH and reacted for 4 hours.
DNA sequencing based on the sequence-by-synthesis method
For the sequencing, 25 μl of 5.0 mM tris-HCl aqueous solution (pH 8.3) containing 25 mM KCl, 1.3 mM MgCl2, and 1 U of DNA polymerase [Klenow (exo-) Fragment from Takara Biomedical Technology Co. Ltd] was dropcast onto the Au electrode. A AgCl/Ag wire electrode was inserted into the liquid droplet as the quasi-reference electrode. After incubation for at least 2 min, the measurement was initiated by adding 1.5 μl of aqueous solution containing 25 mM dNTP solution. For the measurement, the potential of the Au electrode was held at 0 V, and the current signal was measured simultaneously [sample interval, 0.005 s; sensitivity, 1 × 10−7 A V−1). After each measurement, the electrode was rinsed with the 10 mM tris-HCl solution (pH 8.3) to remove unreacted dNTP for subsequent addition of the next dNTP.
Integration of DNA synthesis and sequencing for data storage
The integrated DNA synthesis and sequencing experiments started from synthesizing DNA on the modified Au electrode. The as-synthesized DNA was then hybridized with 10 μM primer DNA of 8 nt for at least 2 hours (Motto-1 to Motto-6; sequences are available in table. S2). After rinsing with the 10 mM tris-HCl aqueous solution (pH 8.3) at least five times, the sequencing measurement was then carried out as mentioned above.
Repeated sequencing based on electrode regeneration
After a sequencing measurement, regeneration of the electrode was performed by dropcasting 10 μl of an aqueous solution containing 0.20 M NaOH on the electrode for 5 min. The electrode was then rinsed with a 10 mM tris-HCl aqueous solution (pH 8.3) at least five times and then incubated with 5 μl of 10 μM primer DNA for at least 2 hours. The above-mentioned sequencing measurement was repeated to improve accuracy.
Fabrication of the SlipChip-based device
A piece of glass slide was cleaned by the following three steps. First, the glass slide was cleaned in a piranha solution (H2SO4:H2O2 = 3:1) for 30 min. Second, it was rinsed with water and EtOH, respectively. Third, it was sonicated in EtOH and water, respectively, and lastly dried in an oven at 80°C for 3 hours. Au electrodes were fabricated on the cleaned glass slide by standard lithography, physical vapor deposition, and lift-off technique. The Au electrode was modified as mentioned above. A piece of PDMS plate was fabricated by pouring a prepolymer solution into a positive mold, and then the polymerization was performed in an oven at 80°C for 2 hours. On the PDMS plate, there were four cylindrical reagent reservoirs of 2 mm in diameter and 100 μm in depth and a fluidic channel with a dimension of 12 mm by 1.5 mm by 100 μm. The surfaces of the glass slide and the PDMS plate were cleaned by treatment in oxygen plasma (150 W for 1 min). Then, 5.0 ml of 2% (v/v) PFDTES/DCM solution in an ampoule was placed alongside the glass slide and the PDMS plate in an oven at 80°C to react for 3 hours to obtain an amphiphobic surface by the silanization. A mixture of methyl glutaronitrile and 3-methoxypropionitrile (1:1, v/v) was used to replace the volatile acetonitrile solvent used in the previous experiment for the on-chip experiments. After loading reagents into the four reservoirs, the top PDMS plate was sealed using amphiphobic FC-40. Then, the bottom glass slide was immersed into 5-mm-deep FC-40 in a petri dish and assembled with the top PDMS plate. The DNA synthesis and sequencing were performed by aligning the electrode with the reagent reservoirs and the microfluidic channel. For the DNA synthesis, phosphoramidite nucleotide monomers were preloaded in the four reservoirs, and for the DNA sequencing experiments, dNTP solutions were preloaded in the reservoirs.
Acknowledgments
Funding: We acknowledge financial support from the National Natural Science Foundation of China (21635001), the Key Project, and Open Research Fund of State Key Laboratory of Bioelectronics, Southeast University.
Author contributions: C.X. and H.L. conceived the research idea. C.X. performed experiments. Z.G. designed the RS code–based software. C.X. and H.L. wrote the paper. C.X., B.M., Z.G., X.D., C.Z., and H.L. analyzed the data. All authors discussed the results and reviewed the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S12
Tables S1 to S5
REFERENCES AND NOTES
- 1.Song X., Reif J., Nucleic acid databases and molecular-scale computing. ACS Nano 13, 6256–6268 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Ceze L., Nivala J., Strauss K., Molecular digital data storage using DNA. Nat. Rev. Genet. 20, 456–466 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Goldman N., Bertone P., Chen S., Dessimoz C., LeProust E. M., Sipos B., Birney E., Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church G. M., Gao Y., Kosuri S., Next-generation digital information storage in DNA. Science 337, 1628 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Caruthers M. H., A brief review of DNA and RNA chemical synthesis. Biochem. Soc. Trans. 39, 575–580 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Hughes R. A., Ellington A. D., Synthetic DNA synthesis and assembly: Putting the synthetic in synthetic biology. Cold Spring Harb. Perspect. Biol. 9, a023812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R., Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Garibyan L., Avashia N., Polymerase chain reaction. J. Invest. Dermatol. 133, 1–4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin S., McPherson J. D., McCombie W. R., Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shendure J., Balasubramanian S., Church G. M., Gilbert W., Rogers J., Schloss J. A., Waterston R. H., DNA sequencing at 40: Past, present and future. Nature 550, 345–353 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Lin K. N., Volkel K., Tuck J. M., Keung A. J., Dynamic and scalable DNA-based information storage. Nat. Commun. 11, 2981 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Y., Kim S., Heller M. J., Huang X., DNA multi-bit non-volatile memory and bit-shifting operations using addressable electrode arrays and electric field-induced hybridization. Nat. Commun. 9, 281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anavy L., Vaknin I., Atar O., Amit R., Yakhini Z., Data storage in DNA with fewer synthesis cycles using composite DNA letters. Nat. Biotechnol. 37, 1229–1236 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Erlich Y., Zielinski D., DNA fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Organick L., Chen Y. J., Dumas Ang S., Lopez R., Liu X., Strauss K., Ceze L., Probing the physical limits of reliable DNA data retrieval. Nat. Commun. 11, 616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grass R. N., Heckel R., Puddu M., Paunescu D., Stark W. J., Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. Engl. 54, 2552–2555 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Koch J., Gantenbein S., Masania K., Stark W. J., Erlich Y., Grass R. N., A DNA-of-things storage architecture to create materials with embedded memory. Nat. Biotechnol. 38, 39–43 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Organick L., Ang S. D., Chen Y. J., Lopez R., Yekhanin S., Makarychev K., Racz M. Z., Kamath G., Gopalan P., Nguyen B., Takahashi C. N., Newman S., Parker H. Y., Rashtchian C., Stewart K., Gupta G., Carlson R., Mulligan J., Carmean D., Seelig G., Ceze L., Strauss K., Random access in large-scale DNA data storage. Nat. Biotechnol. 36, 242–248 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Meiser L. C., Antkowiak P. L., Koch J., Chen W. D., Kohll A. X., Stark W. J., Heckel R., Grass R. N., Reading and writing digital data in DNA. Nat. Protoc. 15, 86–101 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Caruthers M. H., The chemical synthesis of DNA/RNA: Our gift to science. J. Biol. Chem. 288, 1420–1427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matteucci M. D., Caruthers M. H., Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 103, 3185–3191 (1981). [PubMed] [Google Scholar]
- 22.Lee H., Wiegand D. J., Griswold K., Punthambaker S., Chun H., Kohman R. E., Church G. M., Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage. Nat. Commun. 11, 5246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H. H., Kalhor R., Goela N., Bolot J., Church G. M., Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat. Commun. 10, 2383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yazdi S., Gabrys R., Milenkovic O., Portable and error-free DNA-based data storage. Sci. Rep. 7, 5011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazdi S. M., Yuan Y., Ma J., Zhao H., Milenkovic O., A rewritable, random-access DNA-based storage system. Sci. Rep. 5, 14138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez R., Chen Y. J., Dumas Ang S., Yekhanin S., Makarychev K., Racz M. Z., Seelig G., Strauss K., Ceze L., DNA assembly for nanopore data storage readout. Nat. Commun. 10, 2933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomek K. J., Volkel K., Simpson A., Hass A. G., Indermaur E. W., Tuck J. M., Keung A. J., Driving the scalability of DNA-based information storage systems. ACS Synth. Biol. 8, 1241–1248 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Chen Y. J., Takahashi C. N., Organick L., Bee C., Ang S. D., Weiss P., Peck B., Seelig G., Ceze L., Strauss K., Quantifying molecular bias in DNA data storage. Nat. Commun. 11, 3264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kebschull J. M., Zador A. M., Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 43, e143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi C. N., Nguyen B. H., Strauss K., Ceze L., Demonstration of end-to-end automation of DNA data storage. Sci. Rep. 9, 4998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guruswami V., Sudan M., Improved decoding of Reed-Solomon and algebraic-geometry codes. IEEE Trans. Inf. Theory 45, 1757–1767 (1999). [Google Scholar]
- 32.Reed I. S., Solomon G., Polynomial codes over certain finite fields. J. Soc. Ind. Appl. Math. 8, 300–304 (1960). [Google Scholar]
- 33.Frasconi M., Tel-Vered R., Elbaz J., Willner I., Electrochemically stimulated pH changes: A route to control chemical reactivity. J. Am. Chem. Soc. 132, 2029–2036 (2010). [DOI] [PubMed] [Google Scholar]
- 34.LeProust E. M., Peck B. J., Spirin K., McCuen H. B., Moore B., Namsaraev E., Caruthers M. H., Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 38, 2522–2540 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egeland R. D., Southern E. M., Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication. Nucleic Acids Res. 33, e125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer K., Cooper J., Caraballo M., Crye J., Suciu D., Ghindilis A., Leonetti J. A., Wang W., Rossi F. M., Stover A. G., Larson C., Gao H., Dill K., McShea A., Electrochemically generated acid and its containment to 100 micron reaction areas for the production of DNA microarrays. PLOS ONE 1, e34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steel A. B., Herne T. M., Tarlov M. J., Electrochemical quantitation of DNA immobilized on gold. Anal. Chem. 70, 4670–4677 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Song S., Zhang L., Wang L., Wu H., Pan D., Fan C., Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): Effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J. Am. Chem. Soc. 128, 8575–8580 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Heckel R., Mikutis G., Grass R. N., A characterization of the DNA data storage channel. Sci. Rep. 9, 9663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGall G. H., Barone A. D., Diggelmann M., Fodor S. P., Gentalen E., Ngo N., The efficiency of light-directed synthesis of DNA arrays on glass substrates. J. Am. Chem. Soc. 119, 5081–5090 (1997). [Google Scholar]
- 41.Chow B. Y., Emig C. J., Jacobson J. M., Photoelectrochemical synthesis of DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 106, 15219–15224 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palluk S., Arlow D. H., de Rond T., Barthel S., Kang J. S., Bector R., Baghdassarian H. M., Truong A. N., Kim P. W., Singh A. K., Hillson N. J., Keasling J. D., De novo DNA synthesis using polymerase-nucleotide conjugates. Nat. Biotechnol. 36, 645–650 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Zhang J., Song S., Wang L., Pan D., Fan C., A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat. Protoc. 2, 2888–2895 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Rothberg J. M., Hinz W., Rearick T. M., Schultz J., Mileski W., Davey M., Leamon J. H., Johnson K., Milgrew M. J., Edwards M., Hoon J., Simons J. F., Marran D., Myers J. W., Davidson J. F., Branting A., Nobile J. R., Puc B. P., Light D., Clark T. A., Huber M., Branciforte J. T., Stoner I. B., Cawley S. E., Lyons M., Fu Y., Homer N., Sedova M., Miao X., Reed B., Sabina J., Feierstein E., Schorn M., Alanjary M., Dimalanta E., Dressman D., Kasinskas R., Sokolsky T., Fidanza J. A., Namsaraev E., McKernan K. J., Williams A., Roth G. T., Bustillo J., An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Anderson E. P., Daniels J. S., Yu H., Karhanek M., Lee T. H., Davis R. W., Pourmand N., A system for multiplexed direct electrical detection of DNA synthesis. Sens. Actuators B 129, 79–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgenstern B., DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15, 211–218 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Wilbur W. J., Lipman D. J., Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. U.S.A. 80, 726–730 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harismendy O., Ng P. C., Strausberg R. L., Wang X., Stockwell T. B., Beeson K. Y., Schork N. J., Murray S. S., Topol E. J., Levy S., Frazer K. A., Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 10, R32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wick R. R., Judd L. M., Holt K. E., Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox E. J., Reid-Bayliss K. S., Emond M. J., Loeb L. A., Accuracy of next generation sequencing platforms. Next Gener. Seq. Appl. 1, 1000106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi Y., Bae H. J., Lee A. C., Choi H., Lee D., Ryu T., Hyun J., Kim S., Kim H., Song S. H., Kim K., Park W., Kwon S., DNA micro-disks for the management of DNA-based data storage with index and write-once–read-many (WORM) memory features. Adv. Mater. 32, 2001249 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Bimboim H., Doly J., A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7, 1513–1523 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross M. G., Russ C., Costello M., Hollinger A., Lennon N. J., Hegarty R., Nusbaum C., Jaffe D. B., Characterizing and measuring bias in sequence data. Genome Biol. 14, R51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen F., Davydova E. K., Du W., Kreutz J. E., Piepenburg O., Ismagilov R. F., Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal. Chem. 83, 3533–3540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du W., Li L., Nichols K. P., Ismagilov R. F., SlipChip. Lab Chip 9, 2286–2292 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen F., Du W., Kreutz J. E., Fok A., Ismagilov R. F., Digital PCR on a SlipChip. Lab Chip 10, 2666–2672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreutz J. E., Munson T., Huynh T., Shen F., Du W., Ismagilov R. F., Theoretical design and analysis of multivolume digital assays with wide dynamic range validated experimentally with microfluidic digital PCR. Anal. Chem. 83, 8158–8168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen F., Sun B., Kreutz J. E., Davydova E. K., Du W., Reddy P. L., Joseph L. J., Ismagilov R. F., Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load. J. Am. Chem. Soc. 133, 17705–17712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S12
Tables S1 to S5