HHi reverses the CA-MSC driven tumor immune cell exclusion restoring the response to ICI.
Abstract
We investigated the impact of cancer-associated mesenchymal stem cells (CA-MSCs) on ovarian tumor immunity. In patient samples, CA-MSC presence inversely correlates with the presence of intratumoral CD8+ T cells. Using an immune “hot” mouse ovarian cancer model, we found that CA-MSCs drive CD8+ T cell tumor immune exclusion and reduce response to anti–PD-L1 immune checkpoint inhibitor (ICI) via secretion of numerous chemokines (Ccl2, Cx3cl1, and Tgf-β1), which recruit immune-suppressive CD14+Ly6C+Cx3cr1+ monocytic cells and polarize macrophages to an immune suppressive Ccr2hiF4/80+Cx3cr1+CD206+ phenotype. Both monocytes and macrophages express high levels of transforming growth factor β–induced (Tgfbi) protein, which suppresses NK cell activity. Hedgehog inhibitor (HHi) therapy reversed CA-MSC effects, reducing myeloid cell presence and expression of Tgfbi, increasing intratumoral NK cell numbers, and restoring response to ICI therapy. Thus, CA-MSCs regulate antitumor immunity, and CA-MSC hedgehog signaling is an important target for cancer immunotherapy.
INTRODUCTION
The introduction of immune checkpoint inhibitor (ICI) therapy to the clinic has revolutionized cancer care. However, even in the most responsive of tumor types, such as melanoma and lung cancer, only a minority (20 to 40%) of patients respond to single-agent ICIs (1). In epithelial ovarian cancer, there is strong evidence of naturally occurring, antitumor immune responses. Tumor-reactive T cells can be found in the circulation of patients with ovarian cancer (2). Many tumors having tumor-infiltrating lymphocytes (TILs), with patients whose TILs can penetrate into tumor islets (immune “hot”), have a better prognosis than patients whose tumors either lack TILs or have TILs that are restricted to the tumor stroma surrounding tumor islets (tumor immune excluded) (3). Despite this, ICI response rates in ovarian cancer remain disappointingly low, with only 10 to 15% of patients treated with single-agent ICI showing a favorable clinical outcome (4, 5).
The tumor microenvironment (TME) likely dictates response to ICI. In ovarian cancer, patients with an immune-infiltrated molecular signature have a better prognosis compared to patients with a desmoplastic/mesenchymal signature (6). Desmoplasia and intratumoral fibrosis drive resistance to chemotherapy in ovarian, breast, lung, pancreas, and head and neck cancers (7–12). Transcriptional profiling of ICI-responsive and ICI-nonresponsive tumors has linked genes suggestive of a mesenchymal signature, monocyte and macrophage chemotaxis, and activation of the hedgehog (HH) pathway with ICI resistance (5).
Mesenchymal stem cells (MSCs) are multipotent stromal stem cells known to be involved in the desmoplastic response and immune modulation. MSCs are critical for wound healing and immunomodulation (13, 14). In cancer, their role remains controversial, with evidence for both protumorigenic and tumor-suppressive roles previously reported (15–17). Our studies suggest that these contrasting findings may relate to the source of the MSCs being used [cell line, bone marrow (BM), adipose, or tumor derived] and the degree of “cancer education,” as we found that local tissue MSCs can be epigenetically reprogrammed by the TME into cancer-associated MSCs (CA-MSCs) (18).
Our group and others have demonstrated that CA-MSCs can differentiate into cancer-associated fibroblasts (CAFs) and myofibroblasts to increase tumor desmoplasia (8, 17, 19, 20). Ovarian CA-MSCs are strongly protumorigenic and induce chemotherapy resistance when co-engrafted with ovarian cancer cell lines and primary ovarian tumor cells (8, 17).
The role of the CA-MSCs in the modulation of antitumor immunity in epithelial tumors is less defined. Studies in pancreatic cancer have suggested that CA-MSCs and CAFs can recruit monocytes and macrophages and induce their differentiation to a protumorigenic phenotype through secretion of interleukin-1 receptor (IL-1R) antagonist (21), transforming growth factor–β1 (TGF-β1) (22), and IL-6 (23). In addition, CA-MSCs block the cytotoxic activity of natural killer (NK) cells in lung cancer and melanoma (24, 25). These events result in a CA-MSC–dependent immunosuppression and inhibition of the functional activities of antitumor cytotoxic cells in many types of cancer (26, 27).
In this study, using an immune hot, anti–PD-L1 (Programmed death-ligand 1)–responsive model of ovarian cancer, we explored the mechanisms by which CA-MSCs and HH signaling modulate tumor immune surveillance and response to ICI. Our results demonstrate that CA-MSCs induce CD8+ T cell tumor immune exclusion and inhibit response to ICI. The effect was mediated by recruitment of myeloid cells that expressed high levels of TGF-β induced (Tgfbi). We find that, by inhibiting HH signaling, which we previously demonstrated plays a role in desmoplasia-associated chemotherapy resistance (8), we were able to reduce the number of tumor-associated monocytes and macrophages, reverse the CD8+ T cell tumor immune exclusion, and increase the influx of NK cells into the TME, ultimately restoring the anti–PD-L1 response. Together, our data demonstrate that CA-MSCs are a critical driver of the immune-suppressive TME and an important therapeutic target to overcome resistance to ICI therapy in ovarian cancer.
RESULTS
CA-MSCs induce an immune-suppressive TME
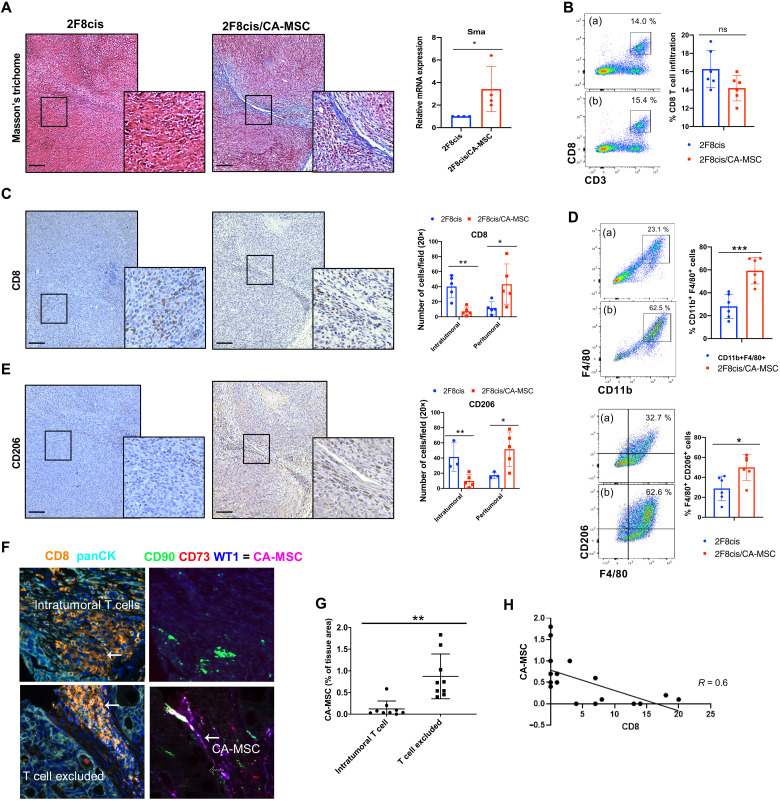
In ovarian cancer, CA-MSCs induce tumor desmoplasia and drive chemotherapy resistance (8, 28). Here, we assessed the impact of CA-MSCs on the tumor immune microenvironment, using the murine cisplatin-resistant 2F8 (2F8cis) ovarian cancer model. In immune-competent syngeneic mice, 2F8cis cells generate tumors abundantly infiltrated by CD3+ T cells and CD8+ T cells (29). To generate murine CA-MSCs, murine adipose-derived MSCs (a-MSCs) were cocultured with 2F8cis mouse ovarian cancer cells at 1:1 ratio for 7 days to allow MSC reprogramming (18). Flow cytometry analysis at day 7 indicated that MSCs represented 10 to 15% of the mixed population. A total of 1 × 106 2F8cis/CA-MSC mixed cells or 1 × 106 of 2F8cis cells alone were then subcutaneously injected into syngeneic mice. Masson’s trichrome staining indicated that, as we previously reported (8), the presence of CA-MSCs induces significant increases in tumor desmoplasia (Fig. 1A). Real-time quantitative polymerase chain reaction (RT-qPCR) showed that the addition of CA-MSCs significantly increases the gene expression of the mesenchymal stromal cell marker α–smooth muscle actin (α-Sma) (Fig. 1A). Flow cytometric evaluation of the abundance of tumor-isolated T lymphocytes showed that the absolute number of CD45+CD3+CD8+ T cells was not significantly different between 2F8cis and 2F8cis/CA-MSC tumors (Fig. 1B). However, immunohistochemical evaluation of the spatial distribution of CD8+ T cells within the tumor tissue demonstrated that, while the control 2F8cis tumors were highly infiltrated by CD8+ T cells, the 2F8cis/CA-MSC tumors showed immune exclusion, with T cells restricted to areas of peritumoral desmoplasia (Fig. 1C).
Fig. 1. The presence of CA-MSCs induces tumor immune cell exclusion.
(A) Representative images of Masson’s trichrome staining in 2F8cis and 2F8cis/CA-MSC tumor tissue sections and RT-qPCR analysis of Sma-1 (a marker associated with desmoplasia) mRNA expression in the indicated tumor groups. Gapdh was used as housekeeping gene. (B) Flow cytometric evaluation and summary of CD3+CD8+ T cells in 2F8cis (a) and 2F8cis/CA-MSC (b) tumors (n = 6 tumors per group). ns, not significant. (C) Representative IHC images and quantification of CD8+ T cells in 2F8cis and 2F8cis/CA-MSC tumor tissue sections. (D) Flow cytometric evaluation of CD11b+F4/80+ (top) and F4/80+CD206+ macrophages (bottom) in 2F8cis (a) and 2F8cis/CA-MSC (b). (E) Representative IHC images and quantification of anti-CD206 immunostaining in 2F8cis and 2F8cis/CA-MSC tumor tissue sections. (F) Representative multispectral imaging of the indicated cell markers in primary tumor samples from patients with high-grade serous ovarian cancer. (G) Percent of CA-MSCs present in patient tissue samples and scoring for intratumoral CD8+ T cells versus CD8+ T cell–excluded tissues. (H) The proportion of CA-MSCs plotted against the proportion of intratumoral CD8+ T cells (n = 18 patients evaluated). A close correlation was noted (R2 = 0.66). Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001. All results are representative of three independent experiments. Scale bars, 100 μm.
Tumor-resident MSCs have been reported to promote an M2-like tumor-associated macrophage (TAM) phenotype in lymphoma (30), and in lung cancer, M2-like TAMs can reduce CD8+ T cell motility and limit their trafficking into the tumor islets (31). We therefore evaluated the abundance and localization of TAMs in control and CA-MSC–containing tumors. The total number of infiltrating macrophages, defined as CD45+CD11c−CD11b+F4/80+ cells, was increased in the CA-MSC+ tumors (Fig. 1D, top). Of these, the percentage of CD11b+F4/80+CD206+ (M2-like) macrophages was significantly higher in 2F8cis/CA-MSC tumors versus control tumors (Fig. 1D, bottom). Similarly to T cell localization, immunohistochemistry (IHC) revealed that macrophages were mainly distributed in the desmoplastic tumor stroma (Fig. 1E). These data indicate that CA-MSCs increase the presence of stromal TAMs and drive CD8+ T cell tumor immune exclusion.
To determine whether CA-MSCs could be driving tumor immune exclusion in human tumors, we performed multiplex immunofluorescence for CA-MSCs [CD73+, CD90+, and WT1+ (Wilms tumor 1)] and T cells (CD8) in serial sections (Fig. 1F). Using a tissue microarray (TMA) of 18 therapy-naïve high-grade serous ovarian cancer, we evaluated the correlation between the presence of CA-MSC and CD8+ T cells. Using a modified H-score based on the expression of WT1 (which histologically distinguishes CA-MSC from a-MSC), T cell–excluded tumors had a CA-MSC H-score of 250 (SEM, 7.3) versus 50.6 (SEM, 13) in tumors with intratumoral T cells (P < 0.001). The presence of CA-MSCs was strongly inversely correlated with the presence of intratumoral T cells, with T cell–excluded tumors demonstrating a significantly higher abundance of CA-MSCs (Fig. 1, G and H).
CA-MSCs inhibit response to anti–PD-L1 checkpoint immunotherapy
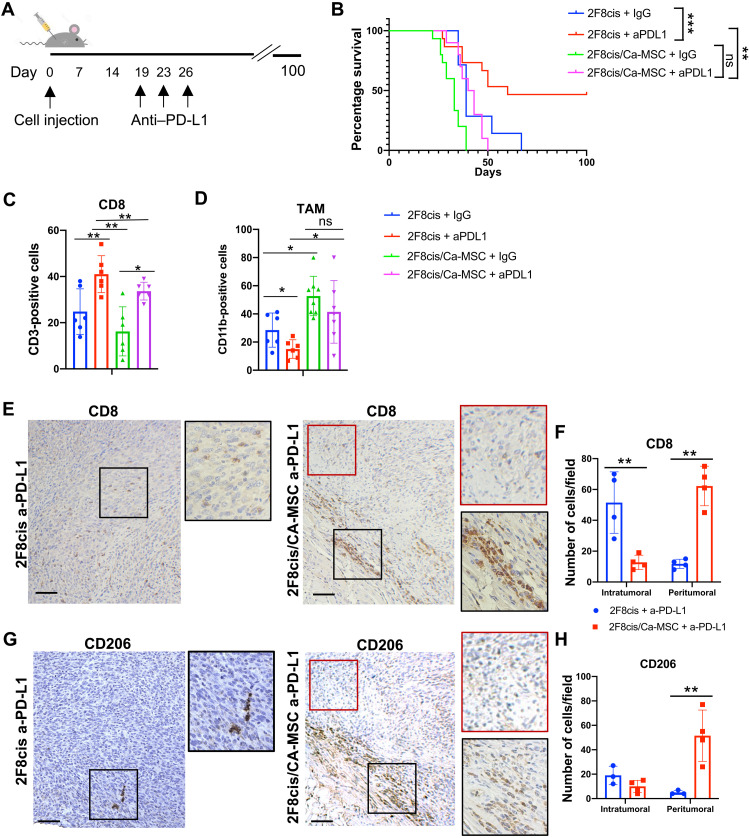
The 2F8cis tumor model is greatly infiltrated by CD8+ T cells, and ~40% of tumors can be eradicated with anti–PD-L1 treatment (29). We used this model to evaluate the impact of CA-MSCs on the therapeutic efficacy of anti–PD-L1 immune therapy. The 2F8cis cells and 2F8cis/CA-MSCs were engrafted and allowed to establish for 19 days before tumor-bearing mice were treated with three intraperitoneal anti–PD-L1 injections as illustrated (Fig. 2A). Control mice received isotype control immunoglobulin (rat IgG2b). Consistently with our previous study (29), while control animals all succumbed to cancer, ~50% of 2F8cis tumor-bearing mice had their tumors eradicated by anti–PD-L1 treatment (Fig. 2B). Notably, for tumors co-inject with CA-MSCs, anti–PD-L1 treatment had no impact on increasing overall survival (Fig. 2B).
Fig. 2. CA-MSC–induced resistance to anti–PD-L1 therapy.
(A) Tumor treatment schema. (B) Kaplan-Meier overall survival analysis for the indicated tumor and treatment groups showing the time of injection and effect of anti–PD-L1 (a-PD-L1) treatment in 2F8cis or 2F8cis/CA-MSC tumors. ***P < 0.0001, 2F8cis versus 2F8cis + a-PD-L1; **P = 0.0042, 2F8cis + a-PD-L1 versus 2F8cis/CA-MSC + a-PD-L1; P > 0.9, 2F8cis/CA-MSC versus 2F8cis/CA-MSC + a-PD-L1. Survival statistics were calculated using log-rank analysis from Kaplan-Meier survival plots. (C and D) Summary of flow cytometry analysis (n = 6 tumors per group) for CD45+CD3+CD8+ T cells and CD45+CD11b+F4/80+CD206+ macrophages, respectively, in the indicated tumors/treatment groups. (E and F) Representative IHC images and quantification of CD8+ T cell localization in 2F8cis and 2F8cis/CA-MSC tumor tissue sections. (G and H) Representative IHC images and quantification of CD206+ cells in a-PD-L1–treated 2F8cis and 2F8cis/CA-MSC tumor tissue sections.
As expected, analysis of the immune cell composition in anti–PD-L1–treated 2F8cis tumors versus IgG-treated 2F8cis or 2F8cis/CA-MSC tumor controls showed an increased number of CD8+ T cells. 2F8cis/CA-MSC tumors treated with anti–PD-L1 also had an increased total number of CD8+ T cells compared to the 2F8cis/CA-MSC control tumors (Fig. 2C). We similarly evaluated the effect of anti–PD-L1 treatment on the abundance of protumorigenic F4/80+CD206+ TAMs. While the number of TAMs was reduced in anti–PD-L1–treated versus control-treated 2F8cis tumors, the TAM numbers did not change in anti–PD-L1–treated 2F8cis/CA-MSC tumors compared to their untreated control group (Fig. 2D). Comparing anti–PD-L1–treated 2F8cis and anti–PD-L1–treated 2F8cis/CA-MSC tumors, the number of TAMs was higher in the 2F8cis/CA-MSC tumors (Fig. 2D).
IHC analysis of CD8 immune cells demonstrated that 2F8cis hot tumors treated with anti–PD-L1 became even more densely infiltrated with T cells throughout the tumor. In contrast, CD8+ T cells in the anti–PD-L1–treated 2F8cis/CA-MSC tumors predominantly remained in desmoplastic peritumoral areas and were rarely found in tumor islets (Fig. 2, E and F). A similar trend was observed for CD206+ TAMs (Fig. 2, G and H), which were rare in anti–PD-L1–treated control tumors yet highly abundant in the peritumoral stroma of anti–PD-L1 2F8cis/CA-MSC tumors. These results suggest that, while anti–PD-L1 increases the frequency of CD8 T cells in both tumor models, the presence of CA-MSCs restricts T cells to the peritumoral stroma and inhibits response to anti–PD-L1 treatment.
CA-MSCs induce a protumorigenic macrophage phenotype via secretion of inflammatory cytokines
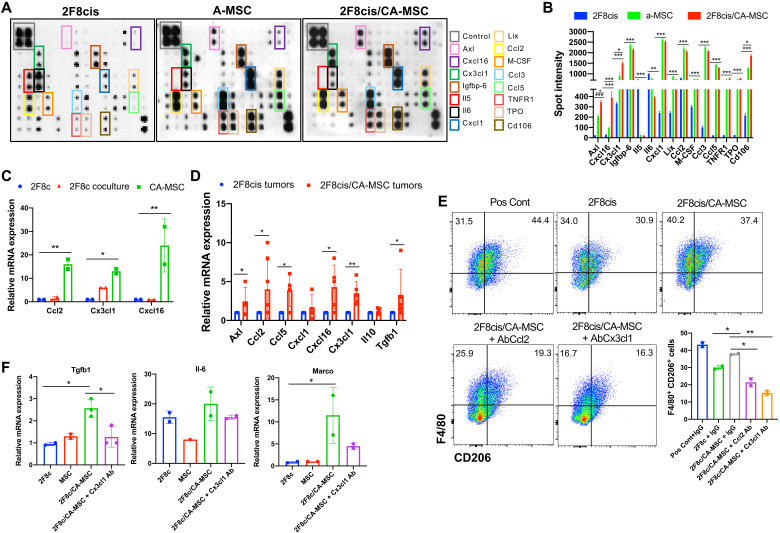
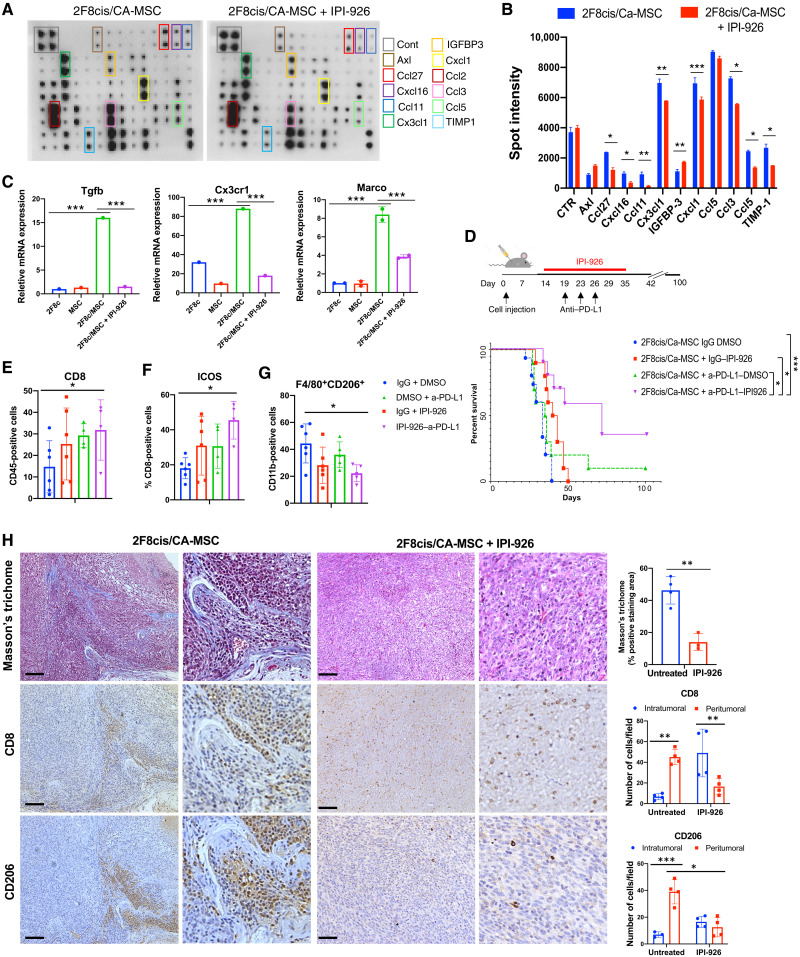
To address the potential mechanism by which CA-MSCs promote tumor immune exclusion and increase protumorigenic/immunosuppressive macrophages, we evaluated a 62-panel array that measured cytokine and chemokine proteins with important functions across several immune pathways. The array was conducted using the conditioned medium (CM) of 2F8cis/a-MSC cocultures to generate CA-MSCs and is subsequently referred to as 2F8cis/CA-MSC. As references, we used the CM from a-MSC and 2F8cis cells cultured alone (Fig. 3A). Cxcl16 and Cx3cl1 emerged as the most up-regulated proteins in the CM of 2F8cis/CA-MSC cocultures versus a-2F8cis and a-MSC cultured alone (Fig. 3, A and B). Both Cxcl16 and Cx3cl1 have been shown to induce macrophage polarization toward an M2-like phenotype (32, 33). In addition, Axl and Cd106 (vascular cell adhesion molecule–1), Ccl2 (C-C Motif Chemokine Ligand 2), and Ccl3 factors, known to recruit monocytes and macrophages; Ccl5, a chemotactic cytokine; and macrophage colony-stimulating factor (M-CSF), which can promote monocyte and macrophage differentiation and activation, were highly secreted in both a-MSC and 2F8cis/CA-MSC cultures (Fig. 3, A and B). These findings are consistent with the increase in macrophage accumulation seen with addition of CA-MSCs in vivo (Fig 1D). To determine which cell type secreted these cytokines, we repeated the assay using CellTrace Violet (CTV)–labeled CA-MSCs. RT-qPCR of fluorescence-activated cell sorting (FACS)–isolated CA-MSCs indicated that Ccl2, Cx3cl1, and Cxcl16 were mainly expressed by CA-MSCs (Fig. 3C).
Fig. 3. 2F8cis/CA-MSC–secreted inflammatory cytokines induce a tumor-promoting macrophage phenotype.
(A and B) Cytokine array and densitometry of the CM of 2F8cis cells, a-MSCs, and 2F8cis/CA-MSCs. Spot intensities were calculated using ImageJ software and compared using two-way analysis of variance (ANOVA). (C) Summary of RT-qPCR analysis of mRNA expression of the indicated genes in CTV-labeled a-MSCs directly cocultured with 2F8cis cancer cells for 7 days. The CTV-labeled CA-MSCs were sorted via FACS from 2F8cis coculture. 2F8cis cells cultured alone were used as control (2F8c). (D) Summary of RT-qPCR analysis of mRNA expression of the indicated genes in the indicated tumors. Results are averages of two independent experiments, each performed in triplicate. (E and F) Flow cytometric and RT-qPCR analyses of F4/80+CD206+ cells generated from BM-derived and differentiated macrophages cultured with the CM of 2F8cis cells or 2F8cis/CA-MSCs with or without neutralizing antibodies to Ccl2 or Cx3cl1. Macrophages stimulated with GM-CSF and IL-4 were used as positive controls (Pos Cont). In all RT-PCR experiments, Gapdh was used as housekeeping gene. P values were calculated using one-way or two-way ANOVA with Tukey posttest for multiple comparisons. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001. All results are representative of three independent experiments.
To confirm the in vitro findings and to evaluate the expression of immune modulatory cytokines and chemokines in the TME, we performed RT-qPCR, which confirmed the increased expression for Axl, Ccl5, Cxcl1, and Cx3cl1 in 2F8cis/CA-MSC and 2F8cis tumor tissues. In addition, we evaluated the expression of Il10 and Tgfβ1, both of which can promote the recruitment of monocytes and macrophages into the tumor and/or enhance the immunosuppressive phenotype of macrophages (34–37). CA-MSC–containing tumors displayed higher levels of Ccl2, Cxcl16, and Tgfβ1 (Fig. 3D).
To further understand whether and how the presence of CA-MSCs affects macrophage biology, we used freshly isolated BM cells, which were differentiated into mature myeloid cells with M-CSF, granulocyte G-CSF, and granulocyte-macrophage CSF (GM-CSF) for 4 days. The cells were then washed and cultured with the CM of 2F8cis or 2F8cis/CA-MSCs for 48 hours and evaluated by flow cytometry for F4/80 and CD206. As a positive control, macrophages were stimulated with IL-4 and GM-CSF. Compared to CM from the 2F8cis monoculture, the CM from the 2F8cis/CA-MSC coculture significantly increased the number of F4/80+CD206+ cells (Fig. 3E). Suggesting a critical role for both Ccl2 and Cx3cl1, neutralizing antibodies (versus IgG control) blocked 2F8cis/CA-MSC CM-mediated increase in CD11b+F4/80+CD206+ cells (Fig. 3E). To characterize the phenotype of macrophages cultured with 2F8cis/CA-MSC CM, we evaluated the expression of genes associated with macrophage immunosuppressive activities, including Il6, macrophage receptor with collagenous structure (Marco), and Tgfβ1 (38). CA-MSCs enhanced the expression of Marco and Tgfβ1 in macrophages, and this activity was inhibited in the presence of Cx3cl1-neutralizing antibody (Fig. 3F).
CA-MSC–enriched tumors displayed an increased number of Ccr2+, Tgfbi-expressing monocytes and macrophages
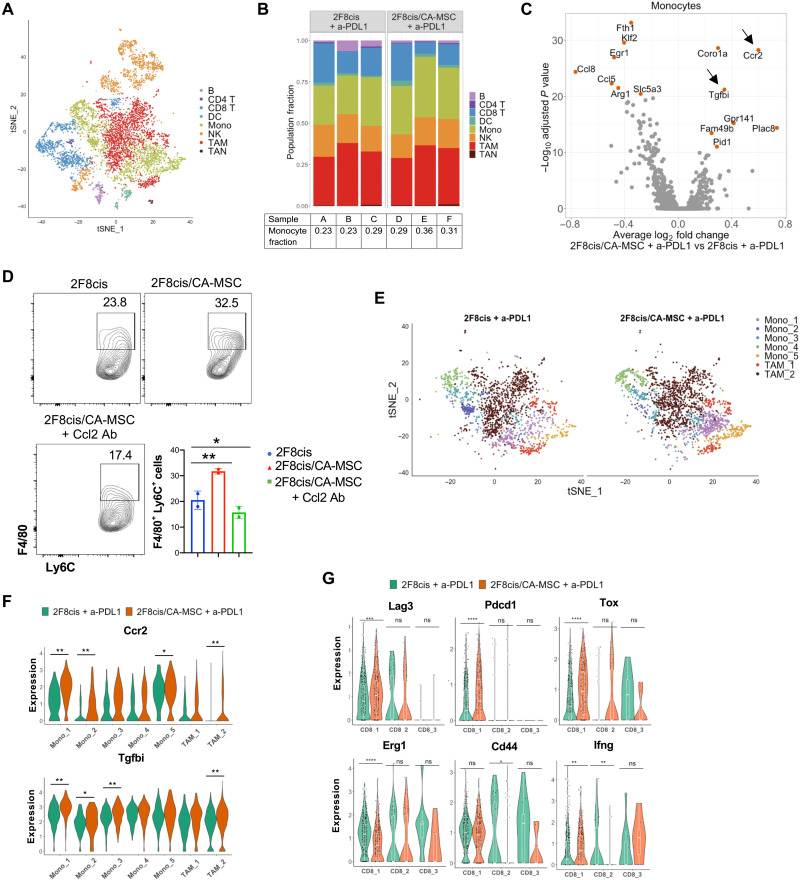
For a comprehensive assessment of the immune cell composition of the TME in anti–PD-L1–treated 2F8cis and 2F8cis/CA-MSC tumors, we performed single-cell RNA sequencing (scRNA-seq) on CD45+ cells isolated from each tumor type (n = 3, each). Clustering (39, 40) revealed numerous distinct cell populations (Fig. 4A). Comparison with the Immunological Genome Project database and assessment of known cell type markers resulted in elaboration of eight distinct immune cell clusters: B lymphocytes [Cd19, Ms4a1 (Membrane Spanning 4-Domains A1), and Cd79b], CD4 T cells, (Cd3d and Cd4), CD8 T cells (Cd3d and Cd8a), dendritic cells [DCs; Itgax (Integrin Subunit Alpha X), Cd209a, Cd40,and Cd83], NK cells [Fcgr4 (Fc receptor, IgG, low affinity IV)], monocytes [Mono; Fcgr1, Cd14, and Ccr2 (C-C Motif Chemokine Receptor 2)], TAMs [Cx3cr1, Mrc1, Cd68, and Adgre (Adhesion G Protein-Coupled Receptor)], and tumor-associated neutrophils (TANs; Itgam, S100a8, S100a9, and matrix metalloproteinase 9). The relative expression of the 10 most highly expressed genes across each population cluster is presented in fig. S1. scRNA-seq analysis revealed that, compared to anti–PD-L1–treated 2F8cis tumors, anti–PD-L1–treated 2F8cis/CA-MSC tumors demonstrated a trend toward an increase in the number of infiltrating monocytes (Fig. 4B). Specifically, the average monocyte cell fraction was 0.25 and 0.32 in anti–PD-L1–treated 2F8cis and 2F8cis/CA-MSC tumors, respectively. Differential gene expression analysis indicated that the monocytes in CA-MSC–enriched tumors overexpress multiple factors, including Tgfbi and Ccr2 genes (Fig. 4C).
Fig. 4. Anti–PD-L1–treated CA-MSC–enriched tumors displayed a high number of monocytes and immature macrophages.
(A) tSNE (t-distributed stochastic neighbor embedding) plot of tumor-derived CD45+ cells from merged data of a-PD-L1–treated 2F8cis and 2F8cis/CA-MSC tumors. (B) Stacked bar graphs showing the proportion of cell types in each tumor sample. (C) Volcano plot showing differentially expressed genes (DEGs) between monocytes of a-PD-L1–treated 2F8cis/CA-MSC versus 2F8cis tumors. The average log2 fold change of gene expression between a-PD-L1–treated 2F8cis/CA-MSC and 2F8cis tumors is plotted on the x axis, and −log10 FDR–adjusted P values using t test are plotted on the y axis. DEGs are colored in red. (D) Flow cytometry showing the abundance of F4/80+Ly6C+ cells in BM-derived monocytes cultured with the indicated CMs. (E) tSNE scRNA-seq plot of monocyte- and macrophage-specific data from a-PD-L1–treated 2F8cis and 2F8cis/CA-MSC tumors. (F) Violin plots showing the expression of Ccr2 and Tgfbi genes in monocytes and TAM clusters. (G) Violin plots showing the expression of Lag3, Pdcd1, Tox, Erg1, Cd44, and Ifng in each CD8 T cell cluster of the indicated a-PD-L1–treated tumors. Results were analyzed using two-way ANOVA or Wilcoxon signed-rank test. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
Ccr2 is a receptor for Ccl2, which is highly expressed by CA-MSCs (Fig 3, A and B). Ccl2 recruits Ccr2+ monocytes to tumor sites and promotes their differentiation into immunosuppressive TAMs (41). To confirm a direct effect of CA-MSCs on monocytes, we cultured freshly isolated BM cells in the presence of the CM of 2F8cis or 2F8cis/CA-MSC cultures for 3 days. The CA-MSC CM enhanced the proliferation of monocytes, defined as F4/80+Ly6C+ cells (Fig. 4D). Directly implicating the Ccl2/Ccr2 axis, the number of monocytes was markedly reduced when Ccl2-neutralizing antibody was added in the CM of 2F8cis/CA-MSCs, pointing to the significant role of Ccl2 in meditating monocyte/macrophage accumulation in our model (Fig. 4D).
Little is known about the role of Tgfbi in ovarian cancer immunity. Tgfbi is normally up-regulated by Tgf-β1 (42). Consistent with Tgfbi induction, Tgfβ1 is up-regulated both in 2F8cis cells and CA-MSCs from coculture (fig. S2A) and in 2F8cis/CA-MSC tumors (Fig. 3D). Higher-resolution analysis of monocyte and macrophage subpopulations, using the scRNA-seq data of anti–PD-L1–treated 2F8cis and 2F8cis/CA-MSC tumors, identified five distinct monocyte subclusters (Fig. 4E and fig. S2B). Monocyte subclusters 1 and 5 expressed Ly6c2 (Lymphocyte antigen 6C2), whereas Mono 2, Mono 3, and Mono 4 were Ly6c2 negative. Compared to anti–PD-L1–responsive 2F8cis tumors, the monocyte populations 1, 2, and 4 in anti–PD-L1–nonresponsive 2F8cis/CA-MSC tumors showed significantly increased expression of Ccr2, while populations 1, 2, and 3 overexpressed Tgfbi (Fig. 4F). Since Ccr2 and Tgfbi have been reported to be expressed not only by monocytes but also by macrophages (43, 44), we evaluated their expression in macrophage clusters. Two distinct TAM subpopulations were identified: TAM1 (H2-Aa+ and Nos2+) and TAM2 (Cx3cr1Hi and Mrc+), both of which expressed Ccr2 and Tgfbi (Fig. 4F). The expression levels were higher in TAM2 from anti–PD-L1–treated 2F8cis/CA-MSC tumors (Fig. 4F).
We similarly evaluated the characteristics of CD8+ T cells in the anti–PD-L1–treated tumors. High-resolution analyses of gene expression levels revealed three subclusters of CD8+ T cells (Fig. 4G). Consistent with the poor treatment response rates, immunosuppressive genes Pdcd, Lag3, and Tox were overexpressed in 2F8cis/CA-MSC tumors, while genes associated with CD8+ T cell activation, including Erg1, Cd44, and Infg, were down-regulated in 2F8cis/CA-MSC tumors (Fig. 4G). Together with the above scRNA-seq data, these results point to a tumor environment characterized by monocyte/macrophage chemoattraction, enriched in immune-suppressive Tgfbi-secreting M2-dominant TAMs, with depletion in T cell activation.
HH inhibition reverses CA-MSC–driven tumor immune exclusion and restores response to anti–PD-L1 therapy
We previously demonstrated that HH inhibitors (HHis) could reverse CA-MSC–driven desmoplasia and resistance to chemotherapy (8). To determine whether HHi could similarly affect CA-MSC immune function, we evaluated the impact of IPI-926, a HHi, on CA-MSC–driven cytokine secretion. We repeated the cytokine array assay using the CM of 2F8cis + A-MSC cocultured for 7 days (2F8cis/CA-MSC) with and without HHi. HHi treatment reduced the secretion of numerous chemokines induced in CA-MSC by tumor cells (Fig. 3, B to D), including Ccl27, Cxcl16, Ccl11, and Cx3cl1 (Fig. 5, A and B). We then tested whether HHi treatment could modulate the activity of 2F8cis/CA-MSC–dependent BM-derived myeloid cell differentiation toward an immunosuppressive phenotype. HHi treatment of 2F8cis/CA-MSCs markedly reduced the 2F8cis/CA-MSC–CM induction of Tgfb1, Cx3cr1, and Marco in myeloid cells (Fig. 5C), suggesting that HHi may reduce some of the observed immune-suppressive phenotype of CA-MSCs.
Fig. 5. IPI-926 reduces CA-MSC–associated desmoplasia and restores anti–PD-L1 therapy response.
(A and B) Cytokine array and densitometry of the CM of a-MSCs stimulated with IPI-926 or left untreated and then cultured for 7 days with tumor 2F8cis cells. Spot intensities were calculated using ImageJ software. (C) RT-qPCR analysis of the mRNA expression of the indicated genes in BM-derived macrophages cultured with the CM of 2F8cis, a-MSC monoculture, and 2F8cis/CA-MSC coculture, with and without IPI-926. (D) Schematic representation of the treatment schedule in 2F8cis/CA-MSC tumors and Kaplan-Meier survival analysis for the indicated treatment groups. ***P = 0.0003, a-PD-L1 + IPI-926 versus IgG + DMSO (dimethyl sulfoxide); *P = 0.02, a-PD-L1 + IPI-926 versus IgG + IPI-926; *P = 0.013, a-PD-L1 + IPI-926 versus a-PD-L1 + DMSO. Survival statistics were calculated using log-rank analysis from Kaplan-Meier survival plots. (E to G) Graph showing the abundance of CD45+CD3+CD8+ (E) CD3+CD8+ ICOS+ T cells (F) and CD11b+F4/80+CD206+ macrophages (G), determined by flow cytometry, in the indicated treatments of 2F8cis/CA-MSC tumors (n = 6 tumors per group). (H) Representative images of Masson’s trichrome and IHC staining, showing CD8+ T cell and CD206+ macrophage localization in control and IPI-926–treated 2F8cis/CA-MSC tumor tissue sections. Graphs represent the abundance of intratumor and peritumoral CD8+ and CD206+ immune cells in control or IPI-926–treated 2F8cis/CA-MSC tumors. Scale bars, 100 μm. Results were analyzed using two-way ANOVA. Error bars, SEMs. *P < 0.05, **P < 0.01, ***P < 0.001.
We therefore explored the impact of HHi on CA-MSC–driven resistance to ICI. We repeated the tumor treatment study above with and without HHi as outlined (Fig. 5D). While treatment with HHi or anti–PD-L1 alone had no significant effect, the HHi/anti–PD-L1 combination significantly improved overall survival, with 40% of the animals demonstrating tumor eradication (Fig. 5D). Confirming that mice treated with HHi and anti–PD-L1 had developed antitumor immune memory, the animals that appeared tumor-free at 100 days were rechallenged with 1 × 106 2F8cis tumor cells, and no tumors developed within 6 months of tumor cell rechallenge.
To further validate the effect of HHi on the growth of CA-MSC+ tumors, we tested the effect of the U.S. Food and Drug Administration–approved agent GDC-0449 (vismodegib) in 2F8cis tumors with and without CA-MSCs. Similar to IPI-926, GDC-0449 overcame the resistance to anti–PD-L1 therapy in 2F8cis/CA-MSC tumors (fig. S3A). Indicating that the HHi is specifically affecting CA-MSCs, (i) HHi alone had no impact on tumor growth, and (ii) HHi did not enhance the response to anti–PD-L1 treatment in tumors lacking CA-MSCs (fig. S3B).
Next, we assessed whether HHis modulate the phenotype and distribution of tumor immune cells in CA-MSC–enriched tumors. Flow cytometry analyses indicated that the dual HHi–anti–PD-L1 therapy increased the number of CD8+ T cells and their expression of ICOS (Inducible T Cell Costimulator), a marker of T cell activation (Fig. 5, E and F), and decreased the number of CD206+ TAMs (Fig. 5G). Histological analysis of tumor tissue demonstrated that HHis significantly reduce CA-MSC–associated desmoplasia (Fig. 5H). IHC confirmed that HHi restored CD8+ T cell infiltration in tumors and suppressed CA-MSC–driven increases in CD206+ TAMs (Fig. 5H). Together, these results demonstrate that HHi treatment inhibits the secretion of cytokines involved in the induction of a TAM phenotype, reduces tumor desmoplasia, and prevents T cell tumor immune exclusion.
HH inhibition combined with anti–PD-L1 reduces Ccr2+Tgfbi+ myeloid cells and increases NK cell presence in tumors
To identify the tumor-infiltrating immune cells potentially responsible for the improved antitumor response observed in mice receiving the anti–PD-L1/HHi combination, we once again performed scRNA-seq. We compared the immune profile of CD45+ cells infiltrating 2F8cis/CA-MSC tumors treated with anti–PD-L1 alone or anti–PD-L1/HHi. Analysis of CD8+ T cell phenotype suggested that, while T cell exhaustion markers Lag3 and Pdcd1 were only minimally decreased (fig. S4), T cells were not significantly different in the two treatment groups. We therefore evaluated other cell types.
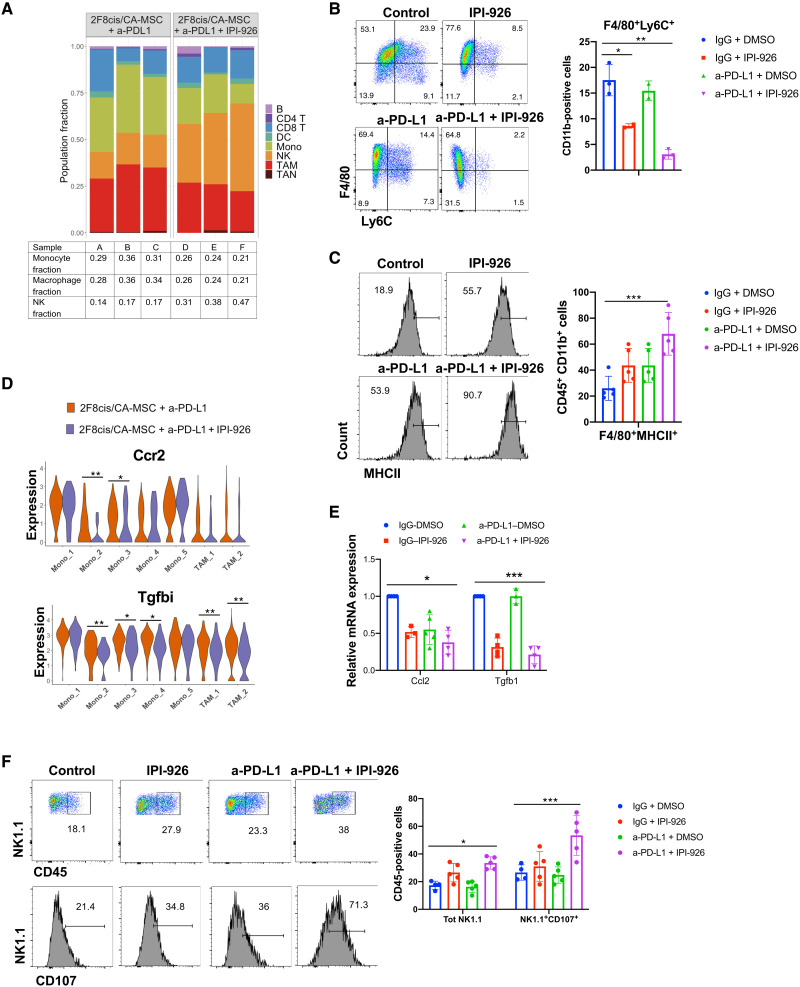
In contrast to T cells, myeloid cell populations differed significantly in the treatment groups. While 2F8cis-CA-MSC tumors treated with anti-PD-L1 alone had a high percentage of monocytes (31.6%) and macrophages (32.6%), the addition of HHi resulted in a significant reduction in the percentage of monocytes (16.3%) and macrophages (23.6%) (Fig. 6A). Flow cytometry confirmed a significant reduction in F4/80+Ly6C+ monocytes when HHi was administered alone and further when in combination with anti–PD-L1 (Fig. 6B). Similarly, dual therapy was associated with a significant decrease in the abundance of CD206+ TAMs (Figs. 5G and 6A) and an increase of F4/80+major histocompatibility complex II–positive (MHCII+) macrophages (Fig. 6C).
Fig. 6. Dual HHi/a-PD-L1 therapy reduces Ccr2+Tgfbi+ monocytes and macrophages and increases the number of NK cells.
(A) Stacked bar graphs of scRNAseq results showing the proportion of cell types in a-PD-L1–treated or a-PD-L1 + IPI-926–treated 2F8cis/CA-MSC tumor samples. (B and C) Flow cytometry showing the abundance of CD11b+F4/80+Ly6C+ monocytes (B) and the expression of MHCII in CD11b+F4/80+ cells.in the indicated 2F8cis/CA-MSC tumor treatment groups. (D) Violin plots showing the expression of Ccr2 and Tgfbi genes in monocytes and TAM clusters in the indicated tumor treatment groups. (E) RT-qPCR analysis of mRNA expression of Ccl2 and Tgfb1 gene expression in control, a-PD-L1–treated, IPI-926–treated, or a-PD-L1 + IPI-926–treated 2F8cis/CA-MSC tumor samples. (F) Flow cytometric analysis of NK cells, defined as CD45+NK1.1+ cells, and their expression of CD107 in the indicated 2F8cis/CA-MSC tumor treatment groups. Results were analyzed using two-way ANOVA or Wilcoxon signed-rank test. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
As CA-MSC–driven desmoplasia was associated with an increase in monocyte and macrophage expression of Ccr2 and Tgfbi, we also analyzed changes in these two markers following HHi/anti–PD-L1 combination. The addition of HHi therapy down-regulated Ccr2 expression in Mono 2 and Mono 3 subpopulations and Tgfbi in Mono 2, Mono 3, and Mono 4 subsets and both macrophage subpopulations (Fig. 6D). As monocyte-chemoattracting Ccl2 and immune-modulatory Tgf-β1 are both produced by CA-MSCs (Fig. 3C and fig. 2A) and Ccl2 binds Ccr2 while Tgf-β1 induces Tgfbi, we evaluated Ccl2 and Tgf-β1 levels in tumors in the treatment groups. RT-qPCR of tumors confirmed that the dual HHi/anti–PD-L1 treatment was associated with down-regulation of Ccl2 and Tgfβ1, with Tgfβ1 being most affected by HHi (Fig. 6E).
To better understand the induction of Tgfbi in TAMs and monocytes and the effect of HHi on this expression, BM-derived macrophages were cultured with the CM of 2F8cis/CA-MSCs or IPI-926–treated 2F8cis/CA-MSCs. Indicating that a secreted factor induces Tgfbi expression, the expression of Tgfbi was up-regulated in macrophages cultured with the CM of 2F8cis/CA-MSCs. Demonstrating that secretion of this factor is regulated by HH signaling, the effect was abrogated when 2F8cis/CA-MSCs were treated with the HHi (fig. S5A). Indicating that Tgf-β1 is at least partly responsible for the induction of Tgfbi, the addition of Tgf-β1–neutralizing antibody to the CM abrogated Tgfbi induction (fig. S5B). Furthermore, using a MotifMaps system, we have found that Tgfbi promoter contains the consensus site for Smad3, one of the downstream mediators of the Tgf-β1 signaling pathway (fig. S5C).
The most notably distinct antitumor immune effector cell population between treatment groups was the NK cell population. Dual therapy was associated with a significant increase in the overall abundance of NK cells (33.1% versus 18.73%) (Fig. 6A). Using flow cytometry, we confirmed that the combined HHi/anti–PD-L1 treatment increased the number of CD45+NK1.1+ cells and their expression of the NK cell activation marker CD107 (Fig. 6F).
Tgfbi suppresses NK cell functions and correlates with poor prognosis in patients with ovarian cancer
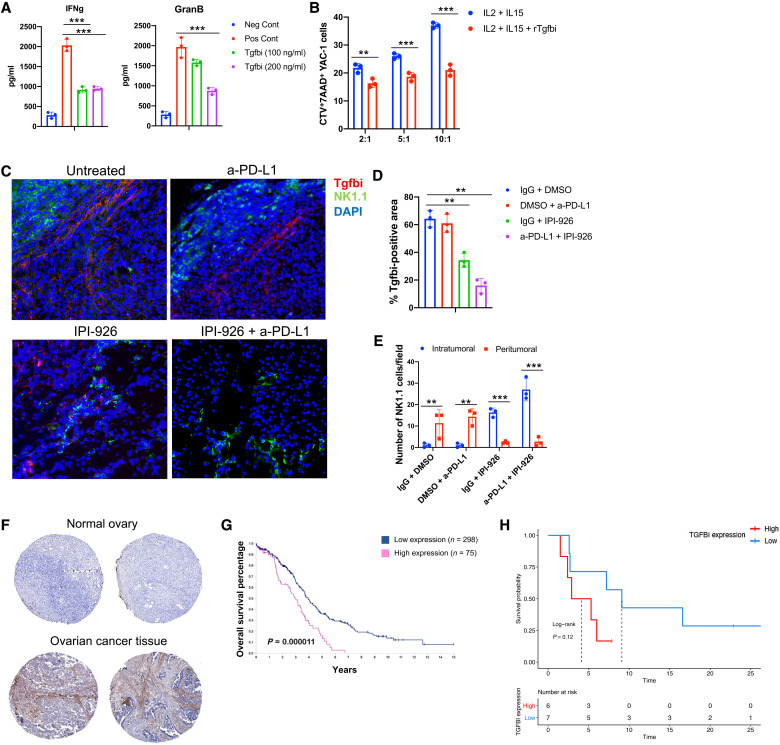
scRNA-seq data indicated that Tgfbi is up-regulated in anti–PD-L1–resistant CA-MSC–containing tumors and reduced by HHi, which is associated with a concomitant increase in NK cells. We therefore evaluated the ability of Tgfbi to directly modulate NK functions. Splenic NK cells were stimulated for 72 hours with IL-2 (20 ng/ml) and IL-15 (50 ng/ml) with or without recombinant Tgfbi (rTgfbi; 100 and 200 ng/ml) protein. Using an enzyme-linked immunosorbent assay (ELISA), we observed that treatments with rTgfbi reduced NK cell production of both interferon-γ (IFN-γ) and granzyme B, two known effectors of NK cell cytotoxic function (Fig. 7A). To test the role of Tgfbi in inhibiting NK cytotoxicity, we evaluated the impact of Tgfbi on NK target cell killing. Activated splenic NK cells were cultured with CTV-stained YAC-1 lymphoma cells at effector-to-target ratios of 2:1, 5:1, and 10:1 for 4 hours. Tgfbi reduced NK cell killing at all effector/target ratios, with a 60% reduction in YAC-1 cell killing at the highest effector:target cell ratio (Fig. 7B).
Fig. 7. Tgfbi reduces NK tumor killing activity, and its expression negatively correlates with ovarian cancer patient prognosis and response to ICI.
(A) Summary of ELISA analysis of activated (Il-2/Il-15–treated) splenic NK cell IFN-γ or granzyme B secretion in control and rTgfbi-treated cells. Unstimulated NK cells were used as negative control (Neg Cont). (B) Activated NK tumor cell killing activity of YAC-1 target cells, with the indicated effector:tumor cell ratios in the presence or absence of Tgfbi. Results were analyzed by two-way ANOVA. (C) Representative IF (immunofluorescence) images showing Tgfbi expression and NK cell localization in control or indicated treated 2F8cis/CA-MSC tumor tissue sections. DAPI, 4′,6-diamidino-2-phenylindole. (D) Graphs represent the percentage of positive stained areas for Tgfbi in control or indicated treated 2F8cis/CA-MSC tumors. (E) Graph shows the abundance of NK1 cells in the indicated treatments of 2F8cis/CA-MSC tumors (n = 3 tumors per group). (F) Representative images from The Human Protein Atlas of TGFBI IHC staining in normal and ovarian carcinoma tissues. (G) Kaplan-Meier overall survival analysis based on high versus low TGFBI expression. (H) Progression-free survival analysis of patients with ovarian cancer (n = 14) based on expression of TGFBI, comparing those patients expressing above the median expression to those expressing at or below the median. Results were analyzed using two-way ANOVA. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
Combined, these data indicate a direct inhibitory role of Tgfbi on NK cytolytic function. To analyze the expression and spatial distribution of Tgfbi and NK cells in vivo, we performed tissue immunofluorescence staining of control and treated 2F8cis/CA-MSC tumors. We observed that, in control and anti–PD-L1–treated tumors, Tgfbi was highly expressed and NK cells were limited in the peritumoral area (Fig. 7, C to E). In HHi-treated tumors, the expression of Tgfbi was reduced compared to control tumors, whereas the number of tumor-infiltrating NK cells was increased. However, the combined HHi/anti–PD-L1 treatment abrogated the expression of Tgfbi and further increased NK cell tumor trafficking (Fig. 7, C to E).
Consistent with our murine studies, evaluation of TGFBI in The Human Protein Atlas shows that the protein is commonly restricted to the human desmoplastic tumor stroma (Fig. 7F). Furthermore, increased TGFBI expression is linked with a poor prognosis in ovarian cancer (Fig. 7G) (www.proteinatlas.org/ENSG00000120708-TGFBI/pathology/ovarian+cancer). Last, we analyzed TGFBI mRNA expression in a small dataset of patients with ovarian cancer treated with ICI therapy. We observed a trend toward TGFBI mRNA expression level being inversely correlated with progression-free survival in patients with ovarian cancer following ICI treatment (Fig. 7H). Together, our results show that Tgfbi secreted by monocytes and macrophages localizes in stromal tumor tissues and critically impairs NK cell cytotoxicity.
DISCUSSION
In this study, we demonstrate the critical role of CA-MSCs in driving tumor immune exclusion and resultant resistance to anti–PD-L1 ICI. We find that (i) CA-MSCs produce numerous factors, such as Ccl2, Cx3cl1, and Tgf-β1, that can recruit Ccr2+ monocytes to the TME and promote their differentiation toward a protumorigenic M2-like phenotype; (ii) CA-MSC–exposed monocytes and TAMs overexpress Tgfbi, which, in turn, inhibits NK cell cytolytic function; and (iii) CA-MSC–mediated immunosuppressive effects can be overcome via inhibition of the HH pathway.
Our findings linking CA-MSC–driven desmoplasia to tumor immune exclusion and resistance to ICI therapy are consistent with human studies demonstrating that highly fibrotic cancers, in which CD8+ T cells are primarily localized to the peritumoral stroma, respond poorly to anti–PD1/PD-L1 immunotherapy (45–47). We found that, in human ovarian tumors, the presence of CA-MSCs was inversely correlated with the presence of intratumoral CD8+ T cells, with T cells primarily relegated to peritumoral stroma, where they colocalized with immunosuppressive myeloid cells. scRNA-seq data revealed that, in CA-MSC–enriched tumors, both monocytes and macrophages overexpress Ccr2 and Tgfbi. This is in line with work in lymphoma showing that MSCs recruit Ccr2+ myeloid cells (30) and that the Ccl2/Ccr2 signaling axis promotes monocyte recruitment to the tumor and enhances M2-like macrophage differentiation (48, 49). Similarly, studies from pancreatic cancer suggest that CA-MSCs increase recruitment of protumorigenic myeloid cells (50, 51). While our studies indicate an important role for CA-MSC–produced chemokines in creating an immunosuppressive TME, we cannot rule out a role for CA-MSC–produced exosomes. Recent work suggests that exosomes produced from tumor-associated MSCs drive macrophage reprogramming, enhancing their immunosuppressive activity via expression of Tgf-β1 (52).
One of the factors that Tgf-β1 regulates is Tgfbi. We show here that Tgfbi was up-regulated in monocytes and TAMs in the presence of CA-MSCs and down-regulated by combination HHi/anti–PD-L1 treatment. Tgfbi is an extracellular matrix–interacting protein that contains an arginine-glycine-aspartic acid motif. Tgfbi stimulation of tumor cells promotes the activation of AKT and focal adhesion kinase signaling pathways, consequently enhancing tumor cell migration, invasion, and metastatic spread (42, 44, 53). The role of Tgfbi in tumor immune surveillance is essentially unstudied. However, Tgfbi has been inversely correlated with response to ICI therapy in lung cancer (54). We similarly find Tgfbi to be inversely correlated with both overall survival in all patients and in patients treated with ICI therapy. This can be partly explained by our results showing that Tgfbi can directly lower the cytolytic activity of NK cells and inhibit their intratumoral trafficking. Whether NK cells are the main effectors responsible for the tumor rejection or whether they serve to promote CD8+ T cell activity (55) remains to be determined.
The main inducer of Tgfbi is Tgf-β1, an immunosuppressive factor that inhibits the cytolytic function and impairs expression of activating receptors of CD8+ T and NK cells (56–59). We found that both ovarian tumor cells and CA-MSCs express Tgf-β1, which can drive Tgfbi expression in monocytes and macrophages, thus contributing to resistance to anti–PD-L1 therapy. Further supporting a potential role for this signaling pathway, data from a murine breast cancer model showed that coadministration of neutralizing antibodies against TGF-β1 and PD-L1 reduced TGF-β1 signaling in stromal cells, facilitated intratumoral T cell penetration, and induced antitumor immunity (47). In light of our data, further studies, focused on the interplay between TGF-β1 and TGFBI in solid tumors with various levels of immune infiltration, will further our understanding of tumor immune surveillance and the development of future immune therapies.
HH inhibition effectively reversed tumor desmoplasia and tumor immune exclusion in our model. HH inhibition also reprogrammed macrophage phenotype, favoring a proinflammatory CD206LowMHCIIHi M1-like phenotype. High expression of MHCII on the surface of macrophages is associated with T cell proliferation and activation (60). Thus, HHi overcome CA-MSC–driven resistance to ICI therapy. This is supported by a recent report that HHi could increase ICI response in models of lung cancer and liver cancer (61). While HH signaling and desmoplasia have a controversial role in other cancers (62, 63), in ovarian cancer, numerous studies have linked desmoplasia with poor prognosis and chemotherapy resistance, with HHi reversing chemotherapy resistance in a stroma-dependent manner (8, 64, 65).
In animal models, Smo (Smoothened, Frizzled Class Receptor) deletion to block HH signaling has been linked with a loss of desmoplasia but increased rates of metastasis (62). This points to a potentially biphasic role for HH signaling during cancer initiation as opposed to established cancers (15). In addition, these studies have not assessed the impact of Smo deletion on the quality and quantity of the immune response in these tumors. We show here that reducing desmoplasia increases intratumoral immune effector cell infiltration, an independent prognostic marker in ovarian cancer. Several HHi have received approval for treatment of conditions such as metastatic basal cell carcinoma and acute myeloid leukemia, although clinical activity remains relatively modest. Evidence from ovarian cancer shows that vismodegib, administered to patients in second or third complete remission, showed no effect on progression-free survival (66). Similarly, our studies demonstrate that single-agent HHi fail to control tumor growth. However, when combined with ICI, the HHi compound rendered the tumors responsive to immunotherapy, supporting their use in combination treatment regimens (67).
In summary, we find that CA-MSCs promote tumor desmoplasia and tumor immune exclusion. This is associated with (i) an increase in Ccr2+ monocytes and TAMs that express high levels of Tgfbi and (ii) resistance to ICI therapy. All of these CA-MSC–driven immune-suppressive effects could be reversed by HHi, thereby increasing response to ICI therapy. This work points to previously unknown mechanisms behind desmoplasia-induced immune exclusion and supports the use of a combination of HH signaling inhibitor and anti–PD-L1 immunotherapy to improve the clinical outcome of ovarian cancer.
MATERIALS AND METHODS
Cell culture
The 2F8 mouse ovarian cancer cell line was derived in-house, from Cre-encoding adenovirus-induced orthotopic ovarian tumors (29, 68). The 2F8cis cells were initially obtained by exposing 2F8 cells in vitro to increasing concentrations (up to 10 μM) of cisplatin (Sigma-Aldrich). The 2F8cis cells were then maintained in cultured Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/ml), streptomycin (100 mg/ml), 2 mM l-glutamine (Corning Life Sciences), and 1 μM cisplatin. Cells were regularly tested for Mycoplasma contamination. Mouse adipose-derived MSCs (a-MSC) were purchased from Cyagen and frozen upon initial expansion (typically up to three passages). Cells were cultured for a maximum of six passages in OriCell Mouse Adipose-Derived Mesenchymal Stem Cell Growth Medium (Cyagen). In the coculture experiments, 2F8cis cells were plated with a-MSCs at a 1:1 ratio (1 × 106 cells) and maintained in culture for 7 days. Cells were cultured in DMEM and mouse adipocyte-derived stem cell medium at a 1:1 ratio. In some experiments, cells were treated with IPI-926 (10 nM) or DMSO (dimethyl sulfoxide) as control for 72 hours and then processed for qPCR or immunoblotting.
Isolation and differentiation of murine BM-derived macrophages
BM cell suspensions were isolated by flushing femurs and tibias of 8- to 10-week-old C57BL/6 mice (the Jackson Laboratory) with complete DMEM. Cell debris was removed by passaging the suspension through a 70-μm nylon sterile strainer (Greiner Bio-One). After two washes with medium, 2 × 106 cells were seeded on six-well plates (Corning Costar). Cells were supplemented with recombinant mouse GM-CSF (50 ng/ml) and mouse M-CSF (R&D Systems) and cultured for 3 days in a humidified incubator at 37°C and 5% CO2. Following this initial differentiation, cells were washed with phosphate-buffered saline (PBS) and incubated with the CM of 2F8cis, a-MSC cultured alone, or 2F8cis cocultured with a-MSC (2F8cis/CA-MSC) for two additional days. Positive control cells were cultured with GM-CSF and IL-4 (R&D Systems) cytokines. In certain experiments, Cx3cl1-, Tgf-β1–, and Ccl2-neutralizing antibodies (R&D Systems) were added at 2 μg/ml.
NK cell culture and cytotoxicity assays
Murine NK cells were isolated from red blood cell–depleted spleens, using NK cell microbeads (Miltenyi Biotec). In the proliferation assay, NK cells were labeled with CTV and stimulated with IL-2 (20 ng/ml), IL-15 (20 ng/ml), and rTgfbi (200 ng/ml) for 3 to 5 days. NK cells stimulated only with IL-2/IL-15 were used as control. Following stimulation, NK cell supernatants were collected and analyzed for cytokine production by ELISA as described below. For the cytotoxicity assays, 3 × 105 CTV-labeled YAC-1 (American Type Culture Collection) target cells were plated in 24-well microplates. IL-2/IL-15–stimulated splenic effector NK cells, treated with and without rTgfbi, were added to YAC-1 cells at 2:1, 5:1, and 10:1 effector/target ratios and incubated at 37°C for 4 hours. Cells were stained with 7-AAD (7-aminoactinomycin D) (BD Biosciences) and analyzed by flow cytometry.
Enzyme-linked immunosorbent assay
IFN-γ and granzyme B concentrations in the supernatants of NK cells were measured using a mouse ELISA kit (BioLegend) following the manufacturer’s protocol. IFN-γ and granzyme B concentrations were within the range of the standard curve. All points were done in triplicate, and the experiments were repeated three times. Samples were read in a microplate reader (Infinite 200 PRO, Tecan).
Ovarian cancer mouse model
C57BL/6 female mice were purchased from the Jackson Laboratory. Mice (8 to 10 weeks old) were subcutaneously injected with 2 to 5 × 106 2F8cis cells or 2F8cis/CA-MSCs. Mice were sacrificed when tumor burden exceeded 2000 mm3. Animals were monitored for weight gain and body conditioning every 2 to 3 days and were maintained in accordance with institutional policies. All studies were performed with approval of the University Committee on Use and Care of Animals of the University of Pittsburgh.
In vivo treatment protocols
Tumor-bearing mice were randomly divided into groups (10 mice per group) and treated intraperitoneally with 150 μg of anti–PD-L1 monoclonal antibody (10F.9G2, Bio X Cell, Japan) or IPI-926 (20 mg/kg; Active Biochem), used alone or in combination. Control mice received isotype rat IgG2b (Bio X Cell) or vehicle control (DMSO for IPI-926). Anti–PD-L1 and isotype control antibodies were administered three times every 2 days, starting at day 19 after tumor cell injection. IPI-926 and vehicle control were administered for 21 days, starting at day 14 after tumor cell injection, as previously described by Lee et al. (69). GDC-0449 (20 mg/kg; Selleckchem), alone or in combination with anti–PD-L1, was administered for 7 days, starting when the tumor size was approximately 70 to 100 mm3.
Patients and tissue samples
Multispectral IHC staining
Eighteen biopsies of patients with high-grade serous ovarian cancer were selected and collected at the Department of Obstetrics, Gynecology, and Reproductive Science, University of Pittsburgh. The study was approved by the institutional review board of the University. Fresh tissues were fixed in formalin and embedded in paraffin. TMA was created using formalin-fixed paraffin-embedded (FFPE) tissue sections from selected patient samples. Slides were deparaffinized using xylene and ethanol. Tissues were then fixed in neutral buffered formalin for 15 min before antigen retrieval. Briefly, the TMA was subjected to heat-induced epitope retrieval cycles in AR6 or AR9 citrate buffers (Akoya Biosciences), followed by blocking for 10 min before incubation with primary antibody for 30 min. Secondary horseradish peroxidase (HRP) antibody was added after washing off excess primary and incubated for 10 min. Opal 570 was used to detect CD8 T cells. A final round of antigen retrieval was carried out for counterstaining nuclei with spectral 4′,6-diamidino-2-phenylindole (DAPI). Tissues were then sealed using Diamond Antifade mountant (Thermo Fisher Scientific, catalog no. P36970). Imaging was performed using the Vectra platform and unmixed using Akoya Biosciences Inform software. To detect CA-MSCs, FFPE TMA slides were deparaffinized, rehydrated, and processed for heat-induced antigen retrieval. Samples were then washed with PBS and blocked against nonspecific binding using universal blocking buffer for 1 hour at room temperature. Conjugated antibodies CD73 (BD Pharmingen), CD90 (BD Pharmingen), and WT1 (R&D Systems) were diluted in 10% universal blocking buffer (5 μg/ml) and applied for 1 hour at room temperature. Samples were then washed with PBS, mounted with antifade media, and left to dry overnight at 4°C. All images were acquired on a Nikon A1 confocal microscope and analyzed using Nikon elements NIS.
Cancer-associated MSCs
ImageJ was used to quantify the proportion of CA-MSCs (coexpression of CD73/CD90/WT1+) versus total cellularity per image. A modified H-score was also calculated on the basis of the expression of WT1 in MSCs within each image. MSCs were identified by coexpression of CD73/CD90, and WT1 expression within the MSCs was divided into “negative expression,” “weak expression,” “moderate expression,” and “strong expression.” The modified H-score was calculated as [0 × percentage of WT1 negative expression MSCs] + [1 × percentage of WT1 weak expression MSCs] + [2 × percentage of WT1 moderate expression MSCs] + [3 × percentage of WT1 strong expression MSCs], ranging from 0 to 300. T cells were counted in 10 to 15 high-power fields. T cell exclusion was defined as ≤3 intra-islet CD8 T cells (3).
Next-generation sequencing
Fourteen patients with epithelial ovarian cancer who received ICI therapy in recurrent setting were included. Baseline patients’ characteristics are summarized in table S1. FFPE tumor samples were collected in this retrospective study with Cleveland Clinic institutional review board approval. FFPE specimens were processed and sequenced by MedGenome only when there was sufficient tumor for sequencing. Samples were included only if they passed RNA extraction QC and RNA library prep QC and proceeded for mRNA sequencing. Data quality check was performed using FastQC (v0.11.8). Alignment was performed using STAR (v2.7.3a). Read count data were normalized, and gene expression analysis was performed using R/Bioconductor package DESeq2 (v1.28.1). Overall survival was calculated from initiation of immunotherapy to last follow-up or death. Progression-free survival was calculated from initiation of immunotherapy to disease progression or last follow-up or death. Survival analyses were estimated using Kaplan-Meier survival curves with log-rank tests. Kaplan-Meier estimator was used for time-to-event analyses comparing between two groups based on the TGFBI gene expression with median as cut-off.
Mouse cytokine array
The supernatants of 2F8cis cells and a-MSCs and the CM of 2F8cis cocultured with a-MSC for 7 days (2F8cis/CA-MSC) were tested for cytokines and chemokines using a cytokine array (RayBiotech, AAH-CYT-C3). The procedure was performed according to the manufacturer’s instructions. Then, the membranes were developed, and the dots were quantified using ImageJ.
Isolation of single cells from murine tumors and flow cytometry analysis
Fresh tumors were isolated, minced in a petri dish on ice, and then enzymatically dissociated into a single-cell suspension according to the protocol for the mouse Tumor Dissociation Kit (Miltenyi Biotec), followed by mechanical dissociation using gentleMACS Dissociator. Cell suspensions were filtered through a 70-μm cell strainer. The ACK (Ammonium-Chloride-Potassium) lysing buffer (Gibco) was used for the lysis of red blood cells, and the resulting suspension was refiltered through a cell strainer to produce a single-cell suspension. Cells were washed with PBS and then stained for flow cytometry analysis. Single cells were stained with fluorescently labeled antibodies and incubated at 4°C for 40 min. The primary antibodies were as follows: Pe-Cy7–F4/80 (clone BM8, eBioscience), Pacific Blue–CD45 (clone 30-F11, BioLegend), APC/Fire750-CD11b (clone M1/70, BioLegend), fluorescein isothiocyanate–CD206 (clone MMR, BioLegend), APC/Fire750-CD3 (clone 17-A2, BioLegend), PE-MHCII (clone M5/114.15.2, BioLegend), BV-510-NK1.1 (clone PK136, BioLegend), APC/Fire 750-CD107 (clone 1D4B, BioLegend), BV510-CD8 (clone 53-6.7, BioLegend), and BV510-Ly6C (clone HK 1.4, BioLegend). Multicolor FACS analysis was performed on a BD LSRII analyzer. All data analysis was performed using the flow cytometry analysis program FlowJo (Tree Star).
Immunohistochemistry
Slides were deparaffinized by baking overnight at 59°C. Endogenous peroxidase activity was eliminated by treatment with 30% H2O2 for 15 min at room temperature. Antigen retrieval was performed by microwave heating in 0.1% citrate buffer (pH 6), for 10 min at 850 V. Nonspecific binding sites were blocked with 2% bovine serum albumin (BSA). Reaction with anti-CD206 (Abcam) anti-CD8 (eBioscience) was for 16 hours at 4°C. Staining was performed by the avidin-biotin-peroxidase complex method with a commercial kit (VECTASTAIN ABC HRP; Vector Laboratories), according to the manufacturer’s protocol. Positive signals were visualized by a DAB Substrate Kit (BD Pharmingen) according to the manufacturer’s protocol. Masson’s trichrome staining was conducted using a ready-to-use kit from Abcam, following the manufacturer’s protocol. Histology sections were observed using a Leica DM4 microscope. Images were acquired using a Leica DFC7000T camera and Leica Application Suite X.
Immunofluorescence
Murine frozen tissue sections were fixed in 4% paraformaldehyde for 20 min and permeabilized in 0.5% Triton X-100 for 20 min. The fixed tissues were incubated with NK1.1 (Tonbo Biosciences), Ly6C (Abcam), or Tgfbi (Novus Biologicals) antibodies overnight at 4°C, followed by secondary anti-mouse Alexa Fluor 598, anti-rat Alexa Fluor 488, or anti-rabbit Alexa Fluor 488 antibodies (Invitrogen Life Technologies) for 1 hour at room temperature. Antibodies were diluted in 1% BSA. Nuclei were stained with mounting medium containing DAPI (Vector). Confocal images were captured on a Leica microscope.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from BM-derived macrophages and murine frozen tumor tissues, using a QIAshredder and RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions. A total of 500 to 1000 μg of RNA were reverse-transcribed (70) using the SuperScript III First-Strand Synthesis System from Invitrogen. A total of 2 μl of RT products were used to amplify Axl, Cxcl16, Ccl2, Ccl5, Il6, Tgfb1, and Tgfbi. Gapdh was used as an internal control. RT-qPCR was performed using a SYBR Green PCR kit (Bio-Rad) and a CFX384 Real PCR system (Bio-Rad).
scRNA-seq library generation
Anti–PD-L1–treated 2F8cis (n = 3), a-PD-L1–treated 2F8cis/CA-MSC (n = 3), and a-PD-L1 + IPI-926–treated 2F8cis/CA-MSC (n = 3) tumor-bearing C57BL/6 mice were sacrificed 2 days after the last treatment. CD45+ cells were isolated from fresh tumors as indicated above. Dead cells were removed using the Dead Cell Removal Kit (Miltenyi). Samples were labeled with different TotalSeq oligo-conjugated antibodies (BioLegend) and then resuspended in 1× PBS containing 0.04% BSA. Two thousand cells per sample were loaded into the Chromium instrument (10x Genomics), and the resulting barcoded cDNAs were used to construct libraries according to the manufacturer’s instructions. Single-cell cDNA libraries were then processed for RNA sequencing, which was performed at the University of Pittsburgh Genomics Core, using an Illumina NovaSeq 6000 platform.
scRNA-seq data analysis
Raw sequence data were processed via Cell Ranger 3.1.0 (10x Genomics) and aligned to mm10 to generate a unique molecular identifier matrix for the downstream analysis. Normalization and explanatory analysis of scRNA-seq data were performed using the Seurat R package (71). During quality control, we excluded cells with <400 expressed genes and genes expressed in <3 cells. Then, cells were demultiplexed to their original samples of origin, and only singlet cells were kept. A basic filtering was performed on the singlets, which removed those with fewer than 500 expressed genes or a mitochondrial percentage higher than 35%. The filtered data contained 7896 singlets, normalized with a scale factor of 1 × 104, scaled, and centered on the basis of 2000 variable features.
For dimensionality reduction and visualization, principal components analysis was run for the normalized counts of variable features, excluding genes related to hemoglobin, mitochondria, and ribosomes. The first 40 principal components were selected. The data were visualized with t-distributed stochastic neighbor embedding (40). Seurat “FindClusters” was applied to the first 40 principal components, with the resolution parameter set to 6.5. Cell labels were assigned using marker gene expression levels.
Statistical analysis
Differences between two conditions were analyzed by Student’s t test and one-way or two-way analysis of variance (ANOVA), with Tukey posttests for multiple pairwise comparisons. All survival statistics were calculated using log-rank analysis from Kaplan-Meier survival plots. In all cases, P < 0.05 was considered statistically significant. Statistics were calculated using Prism software (GraphPad). For the modified CA-MSC H-score, with a sample size of 18 (nine per group), we calculate a >90% power to detect an H-score difference of at least 100 with an SD of 20 (PS Power and Sample Size Calculations, version 3.0).
Acknowledgments
We would like to acknowledge the Lafyatis laboratory single-cell sequencing core for assistance in immediately processing the samples. This project used the following core facilities: (i) the UPMC Hillman Cancer Center and Tissue and Research Pathology/Pitt Biospecimen Core shared resource, which is supported, in part, by award P30CA047904; (ii) UPMC Genome Center, with funding from UPMC’s Immunotherapy and Transplant Center; (iii) the University of Pittsburgh Single-Cell Core, Single-Cell 3′ v3, with cell-hashing service; and (iv) the University of Colorado Human Immune Monitoring Shared Research Facility.
Funding: This work was supported by an Ovarian Cancer Research Alliance Foundation award to L.G.C., A.M.V., and R.J.B. and a grant from National Institutes of Health (R00 CA207871) to H.U.O.
Author contributions: S.C. performed the mouse and human IHC and analysis, ELISAs, flow cytometry, cytokine array, neutralization assays, and data analysis. S.C. and C.C. performed mouse cohort management and tumor sample collection. S.S. performed the isolation of BM cells. H.U.O. and L.Z. performed the scRNA-seq analysis and all computational studies. S.O. performed the multiplex immunofluorescence staining. H.M. provided the TGFBI data from ICI-treated patients. S.C., A.M.V., L.G.C., and R.J.B. assembled the data and wrote the manuscript, with advice from C.C., H.U.O., D.A.A.V., and T.C.B. All authors reviewed and approved the final version of the manuscript.
Competing interests: D.A.A.V., cofounder and stock holder: Novasenta, Tizona, and Potenza; stock holder: Tizona, Oncorus, and Werewolf; patents licensed and royalties: Astellas and BMS; scientific advisory board member: Tizona, Werewolf, F-Star, and Bicara; consultant: Astellas, BMS, Almirall, Incyte, and G1 Therapeutics; research funding: BMS, Astellas, and Novasenta. T.C.B. receives research support from Alkermes and Pfizer. The authors declare that they have no other competing interests.
Data and materials availability: All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The scRNA-seq data is available in GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185013).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.Hamanishi J., Mandai M., Konishi I., Immune checkpoint inhibition in ovarian cancer. Int. Immunol. 28, 339–348 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Martin S. D., Wick D. A., Nielsen J. S., Little N., Holt R. A., Nelson B. H., A library-based screening method identifies neoantigen-reactive T cells in peripheral blood prior to relapse of ovarian cancer. Onco. Targets. Ther. 7, e1371895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Conejo-Garcia J. R., Katsaros D., Gimotty P. A., Massobrio M., Regnani G., Makrigiannakis A., Gray H., Schlienger K., Liebman M. N., Rubin S. C., Coukos G., Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 348, 203–213 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Galluzzi L., Vacchelli E., Pedro J. M. B. S., Buqué A., Senovilla L., Baracco E. E., Bloy N., Castoldi F., Abastado J. P., Agostinis P., Apte R. N., Aranda F., Ayyoub M., Beckhove P., Blay J. Y., Bracci L., Caignard A., Castelli C., Cavallo F., Celis E., Cerundolo V., Clayton A., Colombo M. P., Coussens L., Dhodapkar M. V., Eggermont A. M., Fearon D. T., Fridman W. H., Fučíková J., Gabrilovich D. I., Galon J., Garg A., Ghiringhelli F., Giaccone G., Gilboa E., Gnjatic S., Hoos A., Hosmalin A., Jäger D., Kalinski P., Kärre K., Kepp O., Kiessling R., Kirkwood J. M., Klein E., Knuth A., Lewis C. E., Liblau R., Lotze M. T., Lugli E., Mach J. P., Mattei F., Mavilio D., Melero I., Melief C. J., Mittendorf E. A., Moretta L., Odunsi A., Okada H., Palucka A. K., Peter M. E., Pienta K. J., Porgador A., Prendergast G. C., Rabinovich G. A., Restifo N. P., Rizvi N., Sautès-Fridman C., Schreiber H., Seliger B., Shiku H., Silva-Santos B., Smyth M. J., Speiser D. E., Spisek R., Srivastava P. K., Talmadge J. E., Tartour E., van der Burg S. H., van den Eynde B. J., Vile R., Wagner H., Weber J. S., Whiteside T. L., Wolchok J. D., Zitvogel L., Zou W., Kroemer G., Classification of current anticancer immunotherapies. Oncotarget 5, 12472–12508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hugo W., Zaretsky J. M., Sun L., Song C., Moreno B. H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., Seja E., Lomeli S., Kong X., Kelley M. C., Sosman J. A., Johnson D. B., Ribas A., Lo R. S., Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tothill R. W., Tinker A. V., George J., Brown R., Fox S. B., Lade S., Johnson D. S., Trivett M. K., Etemadmoghadam D., Locandro B., Traficante N., Fereday S., Hung J. A., Chiew Y. E., Haviv I.; Australian Ovarian Cancer Study Group, Gertig D., deFazio A., Bowtell D. D. L., Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 14, 5198–5208 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Amornsupak K., Insawang T., Thuwajit P., O-Charoenrat P., Eccles S. A., Thuwajit C., Cancer-associated fibroblasts induce high mobility group box 1 and contribute to resistance to doxorubicin in breast cancer cells. BMC Cancer 14, 955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffman L. G., Choi Y. J., McLean K., Allen B. L., di Magliano M. P., Buckanovich R. J., Human carcinoma-associated mesenchymal stem cells promote ovarian cancer chemotherapy resistance via a BMP4/HH signaling loop. Oncotarget 7, 6916–6932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farmer P., Bonnefoi H., Anderle P., Cameron D., Wirapati P., Becette V., André S., Piccart M., Campone M., Brain E., MacGrogan G., Petit T., Jassem J., Bibeau F., Blot E., Bogaerts J., Aguet M., Bergh J., Iggo R., Delorenzi M., A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 15, 68–74 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Su S., Chen J., Yao H., Liu J., Yu S., Lao L., Wang M., Luo M., Xing Y., Chen F., Huang D., Zhao J., Yang L., Liao D., Su F., Li M., Liu Q., Song E., CD10+GPR77+ cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 172, 841–856.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Tao L., Huang G., Wang R., Pan Y., He Z., Chu X., Song H., Chen L., Cancer-associated fibroblasts treated with cisplatin facilitates chemoresistance of lung adenocarcinoma through IL-11/IL-11R/STAT3 signaling pathway. Sci. Rep. 6, 38408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vennin C., Mélénec P., Rouet R., Nobis M., Cazet A. S., Murphy K. J., Herrmann D., Reed D. A., Lucas M. C., Warren S. C., Elgundi Z., Pinese M., Kalna G., Roden D., Samuel M., Zaratzian A., Grey S. T., Silva A. D., Leung W.; Australian Pancreatic Genome Initiative (APGI), Mathivanan S., Wang Y., Braithwaite A. W., Christ D., Benda A., Parkin A., Phillips P. A., Whitelock J. M., Gill A. J., Sansom O. J., Croucher D. R., Parker B. L., Pajic M., Morton J. P., Cox T. R., Timpson P., CAF hierarchy driven by pancreatic cancer cell p53-status creates a pro-metastatic and chemoresistant environment via perlecan. Nat. Commun. 10, 3637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Silva Meirelles L., Chagastelles P. C., Nardi N. B., Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 119, 2204–2213 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kolf C. M., Cho E., Tuan R. S., Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: Regulation of niche, self-renewal and differentiation. Arthritis Res. Ther. 9, 204 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandler C., Liu T., Buckanovich R., Coffman L. G., The double edge sword of fibrosis in cancer. Transl. Res. 209, 55–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes C. M., The dual role of mesenchymal stem cells in tumor progression. Stem Cell Res. Ther. 4, 42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean K., Gong Y., Choi Y., Deng N., Yang K., Bai S., Cabrera L., Keller E., McCauley L., Cho K. R., Buckanovich R. J., Human ovarian carcinoma-associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J. Clin. Invest. 121, 3206–3219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffman L. G., Pearson A. T., Frisbie L. G., Freeman Z., Christie E., Bowtell D. D., Buckanovich R. J., Ovarian carcinoma-associated mesenchymal stem cells arise from tissue-specific normal stroma. Stem Cells 37, 257–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon E. S., Moon H. J., Lee M. J., Song H. Y., Kim Y. M., Cho M., Suh D. S., Yoon M. S., Chang C. L., Jung J. S., Kim J. H., Cancer-derived lysophosphatidic acid stimulates differentiation of human mesenchymal stem cells to myofibroblast-like cells. Stem Cells 26, 789–797 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R., Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Luz-Crawford P., Djouad F., Toupet K., Bony C., Franquesa M., Hoogduijn M. J., Jorgensen C., Noël D., Mesenchymal stem cell-derived interleukin 1 receptor antagonist promotes macrophage polarization and inhibits B cell differentiation. Stem Cells 34, 483–492 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Melief S. M., Schrama E., Brugman M. H., Tiemessen M. M., Hoogduijn M. J., Fibbe W. E., Roelofs H., Multipotent stromal cells induce human regulatory T cells through a novel pathway involving skewing of monocytes toward anti-inflammatory macrophages. Stem Cells 31, 1980–1991 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Cho H., Seo Y., Loke K. M., Kim S. W., Oh S. M., Kim J. H., Soh J., Kim H. S., Lee H., Kim J., Min J. J., Jung D. W., Williams D. R., Cancer-stimulated CAFs enhance monocyte differentiation and protumoral TAM activation via IL6 and GM-CSF secretion. Clin. Cancer Res. 24, 5407–5421 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Balsamo M., Scordamaglia F., Pietra G., Manzini C., Cantoni C., Boitano M., Queirolo P., Vermi W., Facchetti F., Moretta A., Moretta L., Mingari M. C., Vitale M., Melanoma-associated fibroblasts modulate NK cell phenotype and antitumor cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 20847–20852 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galland S., Vuille J., Martin P., Letovanec I., Caignard A., Fregni G., Stamenkovic I., Tumor-derived mesenchymal stem cells use distinct mechanisms to block the activity of natural killer cell subsets. Cell Rep. 20, 2891–2905 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Davidson S., Efremova M., Riedel A., Mahata B., Pramanik J., Huuhtanen J., Kar G., Vento-Tormo R., Hagai T., Chen X., Haniffa M. A., Shields J. D., Teichmann S. A., Single-cell RNA sequencing reveals a dynamic stromal niche that supports tumor growth. Cell Rep. 31, 107628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H., Hegde S., DeNardo D. G., Tumor-associated fibrosis as a regulator of tumor immunity and response to immunotherapy. Cancer Immunol. Immunother. 66, 1037–1048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva I. A., Bai S., McLean K., Yang K., Griffith K., Thomas D., Ginestier C., Johnston C., Kueck A., Reynolds R. K., Wicha M. S., Buckanovich R. J., Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 71, 3991–4001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grabosch S., Bulatovic M., Zeng F., Ma T., Zhang L., Ross M., Brozick J., Fang Y. S., Tseng G., Kim E., Gambotto A., Elishaev E., P. Edwards R., Vlad A. M., Cisplatin-induced immune modulation in ovarian cancer mouse models with distinct inflammation profiles. Oncogene 38, 2380–2393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren G., Zhao X., Wang Y., Zhang X., Chen X., Xu C., Yuan Z. R., Roberts A. I., Zhang L., Zheng B., Wen T., Han Y., Rabson A. B., Tischfield J. A., Shao C., Shi Y., CCR2-dependent recruitment of macrophages by tumor-educated mesenchymal stromal cells promotes tumor development and is mimicked by TNFα. Cell Stem Cell 11, 812–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peranzoni E., Lemoine J., Vimeux L., Feuillet V., Barrin S., Kantari-Mimoun C., Bercovici N., Guérin M., Biton J., Ouakrim H., Régnier F., Lupo A., Alifano M., Damotte D., Donnadieu E., Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc. Natl. Acad. Sci. U.S.A. 115, E4041–E4050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allaoui R., Bergenfelz C., Mohlin S., Hagerling C., Salari K., Werb Z., Anderson R. L., Ethier S. P., Jirström K., Påhlman S., Bexell D., Tahin B., Johansson M. E., Larsson C., Leandersson K., Cancer-associated fibroblast-secreted CXCL16 attracts monocytes to promote stroma activation in triple-negative breast cancers. Nat. Commun. 7, 13050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Fu Y., Xue S., Ai A., Chen H., Lyu Q., Kuang Y., The M2 polarization of macrophage induced by fractalkine in the endometriotic milieu enhances invasiveness of endometrial stromal cells. Int. J. Clin. Exp. Pathol. 7, 194–203 (2014). [PMC free article] [PubMed] [Google Scholar]
- 34.Chuang Y., Hung M. E., Cangelose B. K., Leonard J. N., Regulation of the IL-10-driven macrophage phenotype under incoherent stimuli. Innate Immun. 22, 647–657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elgert K. D., Alleva D. G., Mullins D. W., Tumor-induced immune dysfunction: The macrophage connection. J. Leukoc. Biol. 64, 275–290 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Kitamura T., Qian B. Z., Soong D., Cassetta L., Noy R., Sugano G., Kato Y., Li J., Pollard J. W., CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med. 212, 1043–1059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruytinx P., Proost P., Van Damme J., Struyf S., Chemokine-induced macrophage polarization in inflammatory conditions. Front. Immunol. 9, 1930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgoudaki A. M., Prokopec K. E., Boura V. F., Hellqvist E., Sohn S., Östling J., Dahan R., Harris R. A., Rantalainen M., Klevebring D., Sund M., Brage S. E., Fuxe J., Rolny C., Li F., Ravetch J. V., Karlsson M. C. I., Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 15, 2000–2011 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Waltman L., van Eck N. J., A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 471 (2013). [Google Scholar]
- 40.L. van der Maaten, Barnes-Hut-SNE, arXiv:1301.3342 (2013).
- 41.Argyle D., Kitamura T., Targeting macrophage-recruiting chemokines as a novel therapeutic strategy to prevent the progression of solid tumors. Front. Immunol. 9, 2629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thapa N., Lee B. H., Kim I. S., TGFBIp/βig-h3 protein: A versatile matrix molecule induced by TGF-β. Int. J. Biochem. Cell Biol. 39, 2183–2194 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Sierra-Filardi E., Nieto C., Domínguez-Soto Á., Barroso R., Sánchez-Mateos P., Puig-Kroger A., López-Bravo M., Joven J., Ardavín C., Rodríguez-Fernández J. L., Sánchez-Torres C., Mellado M., Corbí Á. L., CCL2 shapes macrophage polarization by GM-CSF and M-CSF: Identification of CCL2/CCR2-dependent gene expression profile. J. Immunol. 192, 3858–3867 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Steitz A. M., Steffes A., Finkernagel F., Unger A., Sommerfeld L., Jansen J. M., Wagner U., Graumann J., Müller R., Reinartz S., Tumor-associated macrophages promote ovarian cancer cell migration by secreting transforming growth factor beta induced (TGFBI) and tenascin C. Cell Death Dis. 11, 249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ford K., Hanley C. J., Mellone M., Szyndralewiez C., Heitz F., Wiesel P., Wood O., Machado M., Lopez M. A., Ganesan A. P., Wang C., Chakravarthy A., Fenton T. R., King E. V., Vijayanand P., Ottensmeier C. H., al-Shamkhani A., Savelyeva N., Thomas G. J., NOX4 inhibition potentiates immunotherapy by overcoming cancer-associated fibroblast-mediated CD8 T-cell exclusion from tumors. Cancer Res. 80, 1846–1860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartmann N., Giese N. A., Giese T., Poschke I., Offringa R., Werner J., Ryschich E., Prevailing role of contact guidance in intrastromal T-cell trapping in human pancreatic cancer. Clin. Cancer Res. 20, 3422–3433 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Mariathasan S., Turley S. J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel III E. E., Koeppen H., Astarita J. L., Cubas R., Jhunjhunwala S., Banchereau R., Yang Y., Guan Y., Chalouni C., Ziai J., Şenbabaoğlu Y., Santoro S., Sheinson D., Hung J., Giltnane J. M., Pierce A. A., Mesh K., Lianoglou S., Riegler J., Carano R. A. D., Eriksson P., Höglund M., Somarriba L., Halligan D. L., van der Heijden M. S., Loriot Y., Rosenberg J. E., Fong L., Mellman I., Chen D. S., Green M., Derleth C., Fine G. D., Hegde P. S., Bourgon R., Powles T., TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giri J., Das R., Nylen E., Chinnadurai R., Galipeau J., CCL2 and CXCL12 derived from mesenchymal stromal cells cooperatively polarize IL-10+ tissue macrophages to mitigate gut injury. Cell Rep. 30, 1923–1934.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu R., Li Y., Yan H., Zhang E., Huang X., Chen Q., Chen J., Qu J., Liu Y., He J., Yi Q., Cai Z., CCL2 promotes macrophages-associated chemoresistance via MCPIP1 dual catalytic activities in multiple myeloma. Cell Death Dis. 10, 781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathew E., Brannon A. L., del Vecchio A. C., Garcia P. E., Penny M. K., Kane K. T., Vinta A., Buckanovich R. J., di Magliano M. P., Mesenchymal stem cells promote pancreatic tumor growth by inducing alternative polarization of macrophages. Neoplasia 18, 142–151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waghray M., Yalamanchili M., Dziubinski M., Zeinali M., Erkkinen M., Yang H., Schradle K. A., Urs S., Pasca di Magliano M., Welling T. H., Palmbos P. L., Abel E. V., Sahai V., Nagrath S., Wang L., Simeone D. M., GM-CSF mediates mesenchymal-epithelial cross-talk in pancreatic cancer. Cancer Discov. 6, 886–899 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biswas S., Mandal G., Roy Chowdhury S., Purohit S., Payne K. K., Anadon C., Gupta A., Swanson P., Yu X., Conejo-Garcia J. R., Bhattacharyya A., Exosomes produced by mesenchymal stem cells drive differentiation of myeloid cells into immunosuppressive M2-polarized macrophages in breast cancer. J. Immunol. 203, 3447–3460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costanza B., Rademaker G., Tiamiou A., de Tullio P., Leenders J., Blomme A., Bellier J., Bianchi E., Turtoi A., Delvenne P., Bellahcène A., Peulen O., Castronovo V., Transforming growth factor beta-induced, an extracellular matrix interacting protein, enhances glycolysis and promotes pancreatic cancer cell migration. Int. J. Cancer 145, 1570–1584 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Nakazawa N., Yokobori T., Kaira K., Turtoi A., Baatar S., Gombodorj N., Handa T., Tsukagoshi M., Ubukata Y., Kimura A., Kogure N., Ogata K., Maeno T., Sohda M., Yajima T., Shimizu K., Mogi A., Kuwano H., Saeki H., Shirabe K., High stromal TGFBI in lung cancer and intratumoral CD8-positive T cells were associated with poor prognosis and therapeutic resistance to immune checkpoint inhibitors. Ann. Surg. Oncol. 27, 933–942 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Wang F., Lau J. K. C., Yu J., The role of natural killer cell in gastrointestinal cancer: Killer or helper. Oncogene 40, 717–730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dasgupta S., Bhattacharya-Chatterjee M., O’Malley B. W. Jr., Chatterjee S. K., Inhibition of NK cell activity through TGF-β 1 by down-regulation of NKG2D in a murine model of head and neck cancer. J. Immunol. 175, 5541–5550 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Lazarova M., Steinle A., Impairment of NKG2D-mediated tumor immunity by TGF-β. Front. Immunol. 10, 2689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rook A. H., Kehrl J. H., Wakefield L. M., Roberts A. B., Sporn M. B., Burlington D. B., Lane H. C., Fauci A. S., Effects of transforming growth factor beta on the functions of natural killer cells: Depressed cytolytic activity and blunting of interferon responsiveness. J. Immunol. 136, 3916–3920 (1986). [PubMed] [Google Scholar]
- 59.Thomas D. A., Massague J., TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Wang B., Li Q., Qin L., Zhao S., Wang J., Chen X., Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 12, 43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petty A. J., Li A., Wang X., Dai R., Heyman B., Hsu D., Huang X., Yang Y., Hedgehog signaling promotes tumor-associated macrophage polarization to suppress intratumoral CD8+ T cell recruitment. J. Clin. Invest. 129, 5151–5162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin K., Lim A., Zhao C., Sahoo D., Pan Y., Spiekerkoetter E., Liao J. C., Beachy P. A., Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell 26, 521–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Theunissen J. W., de Sauvage F. J., Paracrine Hedgehog signaling in cancer. Cancer Res. 69, 6007–6010 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Steg A. D., Katre A. A., Bevis K. S., Ziebarth A., Dobbin Z. C., Shah M. M., Alvarez R. D., Landen C. N., Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol. Cancer Ther. 11, 1587–1597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCann C. K., Growdon W. B., Kulkarni-Datar K., Curley M. D., Friel A. M., Proctor J. L., Sheikh H., Deyneko I., Ferguson J. A., Vathipadiekal V., Birrer M. J., Borger D. R., Mohapatra G., Zukerberg L. R., Foster R., MacDougall J. R., Rueda B. R., Inhibition of Hedgehog signaling antagonizes serous ovarian cancer growth in a primary xenograft model. PLOS ONE 6, e28077 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaye S. B., Fehrenbacher L., Holloway R., Amit A., Karlan B., Slomovitz B., Sabbatini P., Fu L., Yauch R. L., Chang I., Reddy J. C., A phase II, randomized, placebo-controlled study of vismodegib as maintenance therapy in patients with ovarian cancer in second or third complete remission. Clin. Cancer Res. 18, 6509–6518 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Grund-Groschke S., Stockmaier G., Aberger F., Hedgehog/GLI signaling in tumor immunity—New therapeutic opportunities and clinical implications. Cell Commun. Signal 17, 172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mony J. T., Zhang L., Ma T., Grabosch S., Tirodkar T. S., Brozick J., Tseng G., Elishaev E., Edwards R. P., Huang X., Vlad A. M., Anti-PD-L1 prolongs survival and triggers T cell but not humoral anti-tumor immune responses in a human MUC1-expressing preclinical ovarian cancer model. Cancer Immunol. Immunother. 64, 1095–1108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee M. J., Hatton B. A., Villavicencio E. H., Khanna P. C., Friedman S. D., Ditzler S., Pullar B., Robison K., White K. F., Tunkey C., LeBlanc M., Randolph-Habecker J., Knoblaugh S. E., Hansen S., Richards A., Wainwright B. J., McGovern K., Olson J. M., Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc. Natl. Acad. Sci. U.S.A. 109, 7859–7864 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1