RECS1, a member of the TMBIM family, triggers lysosomal membrane permeabilization in cells undergoing lysosomal stress.
Abstract
Programmed cell death is regulated by the balance between activating and inhibitory signals. Here, we have identified RECS1 (responsive to centrifugal force and shear stress 1) [also known as TMBIM1 (transmembrane BAX inhibitor motif containing 1)] as a proapoptotic member of the TMBIM family. In contrast to other proteins of the TMBIM family, RECS1 expression induces cell death through the canonical mitochondrial apoptosis pathway. Unbiased screening indicated that RECS1 sensitizes cells to lysosomal perturbations. RECS1 localizes to lysosomes, where it regulates their acidification and calcium content, triggering lysosomal membrane permeabilization. Structural modeling and electrophysiological studies indicated that RECS1 is a pH-regulated calcium channel, an activity that is essential to trigger cell death. RECS1 also sensitizes whole animals to stress in vivo in Drosophila melanogaster and zebrafish models. Our results unveil an unanticipated function for RECS1 as a proapoptotic component of the TMBIM family that ignites cell death programs at lysosomes.
INTRODUCTION
Apoptosis is an evolutionarily conserved mechanism of regulated cell death. In mammals, apoptosis execution is tightly regulated at the level of mitochondria by the proteins of the B cell lymphoma 2 (BCL-2) family (1, 2). The BCL-2 family is functionally classified into antiapoptotic [i.e., BCL-2, BCL-XL, and myeloid cell leukemia 1 (MCL-1)] and proapoptotic proteins, which are further subdivided into “multidomain” [i.e., BCL-2–associated X protein (BAX) and BCL-2 antagonist or killer (BAK)] and BH3-only proteins [i.e., BCL-2 homology 3 (BH3)–interacting domain death agonist (BID), BCL-2–interacting mediator of cell death (BIM), and p53 upregulated modulator of apoptosis (PUMA)]. Activator BH3-only proteins trigger cell death through the conformational activation of the multidomain proteins BAX and BAK, resulting in their homo-oligomerization at the mitochondrial surface. This event triggers mitochondrial outer membrane permeabilization (MOMP), followed by the release of cytochrome c and other mitochondrial intermembrane factors to engage the caspase cascade and execute apoptotic cell death (1).
Although the attention of the field has been historically centered on the regulation of MOMP by the BCL-2 family, other organelles, such as lysosomes and the endoplasmic reticulum (ER), play important roles in the initiation and control of cell death triggered by several intra- and extracellular stimuli (3). For example, a pool of anti- and proapoptotic proteins of the BCL-2 family localizes to the ER membrane, where they regulate apoptosis in response to acute or sustained ER stress (4). Moreover, cell death triggered by lysosomal membrane permeabilization (LMP) contributes to a wide array of pathophysiological conditions, including cancer, neurodegenerative diseases, lysosomal storage disorders, and the involution of mammary glands, one of the main cell death events in the adult mammalian organism (5, 6).
The transmembrane BAX inhibitor motif containing (TMBIM) superfamily is composed of at least six widely conserved proteins, with homologs in mammals, plants, yeast, and viruses (7, 8). The founder member of the family, BAX inhibitor 1 (BI-1), was found in a yeast screen as a potent inhibitor of cell death induced by the human proapoptotic protein BAX (9). BI-1 localizes to the ER membrane where it inhibits cell death triggered by a wide variety of cytotoxic stimuli (9, 10) [reviewed in (8)]. In addition to BI-1, the TMBIM superfamily includes the following members: responsive to centrifugal force and shear stress 1 (RECS1/TMBIM1), lifeguard (LFG/TMBIM2), glutamate receptor ionotropic N-methyl-d-aspartate receptor 1 (GRINA/TMBIM3), Golgi anti-apoptotic protein (GAAP/TMBIM4), and growth hormone–inducible protein (GHITM/TMBIM5) (8). These proteins are characterized by seven transmembrane domains and localize to intracellular membranes, where they regulate calcium homeostasis and stress-induced cell death (8, 11), possibly through an ion channel activity (11, 12). Antiapoptotic functions have been reported for all six members of the TMBIM family; however, the molecular mechanisms for the antiapoptotic activity of each individual member are not completely understood and may involve the control of Golgi and ER calcium homeostasis, in addition to the regulation of the unfolded protein response (UPR) and autophagy, among others (10, 13–17). For example, RECS1 and LFG are regarded as inhibitors of the extrinsic pathway of apoptosis as they block cell death induced by Fas ligand (18, 19), while BI-1, GRINA, and GAAP repress apoptosis induced by intracellular cytotoxic stimuli (10, 17). Despite recent advances in our understanding of the structure and function of the TMBIM family, the molecular mechanisms linking the function of its members with the control of apoptosis are still poorly defined.
The evolutionary origin of the cell death machinery involves a rheostat model where the balance between activating and repressing signals determines the induction of apoptosis (1, 20). In yeast, the sole putative ortholog of BI-1 (termed YNL305C, Ybh3p, or Bxi1p) can repress or induce cell death depending on the context (21–23). However, no proapoptotic molecules have been identified in the mammalian TMBIM family. Sequence analyses of RECS1 revealed the presence of a conserved C-terminal domain that contains a putative BH3-like motif that is also present in the yeast Ybh3p/Bxi1p protein (22). RECS1 (also known as TMBIM1, LFG3, and PP1201) is a 35-kDa protein first identified as a gene up-regulated by shear stress in embryonic endothelial cells (24). At the cellular level, RECS1 inhibits cell death triggered by the Fas ligand through an interaction with the Fas receptor at the Golgi apparatus (19). Additional sequence analyses revealed the presence of a conserved di-aspartyl sensor and an arginine latch, shown to be critical for the regulation of the calcium channel function of the bacterial (Bacillus subtilis) homolog BsYetJ and human GAAP (11, 12, 25). To date, the function of RECS1 in the regulation of different modalities of cell death has not been investigated.
Here, we determined the significance of RECS1 to the regulation of cell death initiated at different intracellular compartments, including lysosomes and the ER. Using gain- and loss-of-function approaches, we found that, in contrast to other members of the TMBIM family, RECS1 expression results in cell death via the mitochondrial pathway of apoptosis. Unbiased screening with a large panel of cytotoxic agents revealed that RECS1 expression highly sensitizes cells to lysosomotropic agents and microtubule-destabilizing drugs. RECS1 localizes to the lysosomal membrane, where it rapidly induces cell death through LMP, involving the recruitment of active BAX to lysosomes. RECS1 also sensitizes cells to ER stress possibly via its transcriptional up-regulation. Under basal conditions, RECS1 promotes lysosomal acidification and increases lysosomal calcium concentration. At the molecular level, we show that RECS1 operates as a pH-dependent calcium and sodium channel, where a single mutation in the critical di-aspartyl sensor (D295Q) is sufficient to disrupt its calcium and sodium permeability, reducing cell death upon lysosomal stress. Last, we provide evidence indicating that RECS1 expression induces cell death and sensitizes whole animals to stress in fly and zebrafish models. Overall, our data indicate that RECS1 is a proapoptotic factor controlling the initiation of cell death programs at the lysosomal membrane.
RESULTS
RECS1 induces cell death through the mitochondrial apoptosis pathway
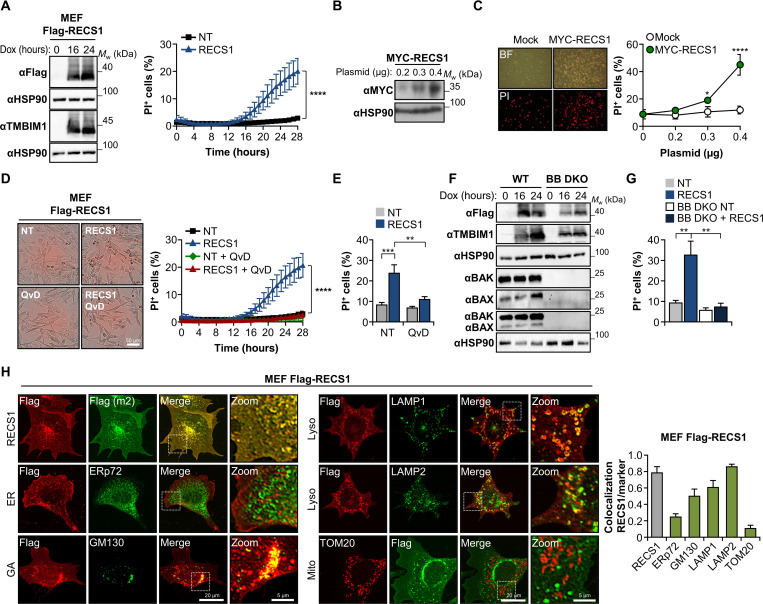
To define the significance of RECS1 in the regulation of cell death, we generated mouse embryonic fibroblast (MEF) cells expressing a doxycycline-inducible full-length human RECS1 tagged with FLAG (Flag-RECS1) (Fig. 1A, left). Unexpectedly, the sole expression of RECS1 in the absence of additional stressors resulted in cell death as determined by propidium iodide (PI) and SYTO16 staining, followed by automated live microscopy analysis (Fig. 1A, right). Under these conditions, cell death reached a plateau of around 20% after 28 hours of doxycycline stimulation (Fig. 1A). We were able to validate these results by transient transfection of MEFs with increasing concentrations of a RECS1 expression vector (Fig. 1, B and C). These observations suggest that RECS1 overexpression is sufficient to induce cell death.
Fig. 1. RECS1 induces cell death through the mitochondrial pathway of apoptosis.
(A) Left: MEFs expressing doxycycline-inducible Flag-RECS1 were stimulated with doxycycline (DOX), and the levels of RECS1 were determined by Western blot. Right: Cell death was determined by PI and SYTO16 costaining followed by automated microscopy (n = 7). Mw, weight-average molecular weight; NT, not treated. (B) MEFs were transiently transfected with the indicated concentrations of MYC-RECS1 followed by Western blot analysis. (C) Left: Bright-field (BF) and PI fluorescent images of MEFs transfected with 0.4 μg of MYC-RECS1 for 24 hours. Right: Cell death was determined by PI staining followed by FACS (n = 3). (D) Left: Images of MEF Flag-RECS1 cells cultured with doxycycline in the presence or absence of the caspase inhibitor QvD-OPh (QvD; 40 μM) for 24 hours (red, PI staining). Right: Corresponding cell death kinetics (n = 3). (E) PI staining and FACS of cells treated as in (D) (n = 4). (F) MEF Flag-RECS1 wild-type (WT) or BAX and BAK (BB) double-knockout (DKO) cells were treated with doxycycline, and indicated proteins were assessed by Western blot. (G) Indicated cells were cultured with doxycycline for 38 hours, and cell death was determined by FACS (n = 3). (H) Images (left) and quantification (right) of RECS1 (Flag) colocalization with indicated organelle markers in Flag-RECS1 MEFs treated with doxycycline. GA, Golgi apparatus. Data represent means ± SD (A, C, and D) or means ± SEM. Statistically significant differences were determined comparing the best individual fit for each curve using the extra-sum-of-squares F test (A and D) or two-way analysis of variance (ANOVA) followed by Holm-Sidak’s multiple comparisons test (C, E, and G). *P < 0.05; **P < 0.01; ****P < 0.0001).
Cell death initiated by intracellular signals usually proceeds through the intrinsic mitochondrial pathway of apoptosis, a process involving the conformational activation of BAX and BAK (1, 2). To determine the requirement of caspases, we treated RECS1-overexpressing cells with the pan-caspase inhibitor QvD-OPh (QvD), followed by cell death kinetics and fluorescence-activated cell sorting (FACS) analysis. Caspase inhibition completely abrogated cell death induced by RECS1 (Fig. 1, D and E). We then generated a doxycycline-inducible Flag-RECS1 cell line in a BAX and BAK double-knockout (DKO) background (Fig. 1F) and observed full resistance to cell death induced by RECS1 overexpression (Fig. 1G). These results support the idea that RECS1 overexpression induces cell death through the mitochondrial pathway of apoptosis.
RECS1 C-terminal domain is required for its pro-dead function
RECS1 has been traditionally classified within the TMBIM superfamily (8). However, detailed sequence and phylogenetic analyses suggest that RECS1, together with GRINA, LFG, GAAP, and Tmbim1b, forms a distinct and evolutionarily conserved protein family, termed the “LFG family,” characterized by a shared common ancestry and the lack of the signature BI-1 motif (fig. S1A) (7, 26). Previous sequence comparison using the BH3 motif consensus from Prosite (PS01259) identified one putative BH3 motif on the C-terminal domain of the human RECS1 and LFG proteins (22). The BH3-like motif in the C-terminal region of RECS1 contains the characteristic minimal Leu-X3-Gly-Asp/Glu sequence, but no additional BH3-like motifs of this kind were found in the other members of the LFG and BI-1 protein families (fig. S1B). However, an additional conserved BH3-like sequence consisting of Leu-X4-Asp is also present in an adjacent sequence of RECS1, in addition to GRINA and LFG (fig. S1B). These putative “BH3 motifs” are conserved in humans and in several species of non-human primates (fig. S1, C and D). However, only one such motif is present in rodents, suggesting that they represent a late evolutionary event.
To determine the role of each putative BH3-like motif on cell death induced by RECS1, we generated two deletion mutants: RECS1Δ910, lacking the C-terminal BH3 motif, and RECS1Δ868, lacking both (fig. S1, E and F). Ablation of one BH3-like motif was sufficient to abrogate RECS1-induced cell death after transient transfection in MEFs (fig. S1G). Notably, both RECS1 deletion mutants also lack conserved C-terminal residues that have been shown to be necessary for the channel function of other TMBIM proteins, including BI-1 and GAAP (11, 25, 27). Specific structural and functional criteria are required to validate a BH3 motif, since definitive bioinformatic procedures that rely purely on sequence analyses are currently lacking (28). To this end, we synthesized two peptides: RECS1280–299 and RECS1296–311, based on the N- and C-terminal BH3-like motifs, respectively. These peptides were predicted to fold into a single amphipathic α helix, similar to those of other BH3 motifs (fig. S1H). We then determined whether the peptides were able to depolarize mitochondria in a BAX- and BAK-dependent manner. We loaded wild-type (WT) and BAX/BAK DKO cells with the mitochondrial potential (ΔΨm)–sensitive dye JC-1, followed by treatment with increasing concentrations of each peptide. However, neither of RECS1-derived BH3 peptides triggered mitochondrial membrane depolarization, suggesting that they do not act as direct activator BH3 motifs (fig. S1I). As positive and negative controls, peptides derived from BIM or a PUMA nonfunctional BH3 mutant (PUMA2A) were used. Second, we assessed whether RECS1 peptides could displace the interaction between BIM and antiapoptotic proteins of the BCL-2 family by fluorescence polarization immunoassay. As shown in fig. S1J, peptides derived from RECS1 failed to compete with the interaction between BIM and BCL-2, BCL-XL, or MCL-1. Together, these results suggest that the C-terminal region of RECS1 is essential to induce cell death, but this sequence does not bear functional bona fide BH3 motifs.
RECS1 localizes to lysosomes and triggers cell death upon lysosomal stress
To assess the possible role of RECS1 in the regulation of cell death triggered by stress conditions, we performed an unbiased screening using a library of 77 cytotoxic and chemotherapeutic compounds (fig. S2A). Unexpectedly, RECS1 overexpression sensitized cells to the death triggered by ~14 compounds, whereas it repressed the activity of 12 compounds (fig. S2A). The top sensitizing drugs were chloroquine (CQ) and hydroxychloroquine (HCQ), which are known to induce lysosomal stress by inhibiting their acidification (Fig. 2, A and B). In addition, RECS1 expression also protected against the toxicity of a subgroup of pharmacological agents, where the top inhibited compounds were the antifolate and anti-inflammatory drug methotrexate (MTX), in addition to several DNA damage–inducing agents, including the purine analogs floxuridine and cladribine (fig. S2, C and D). We decided to focus our validation experiments in the sensitizing effects of RECS1 to the exposure to lysosomotropic agents. Dose-response experiments showed that CQ and HCQ triggered cell death exclusively in RECS1 overexpression cells, with a median effective concentration of 33.6 and 36.8 μM, respectively (fig. S2B). RECS1 overexpression also sensitized cells to drugs that target the cytoskeleton, including taxanes such as docetaxel and paclitaxel (Fig. 2, A and B, and fig. S2C).
Fig. 2. RECS1 triggers cell death upon lysosomal stress.
(A) Flag-RECS1 MEFs were stimulated with doxycycline for 16 hours and treated with a library of 77 cytotoxic compounds for 24 hours. Cell death was determined by PI staining using an ImageXpress Micro XL automated microscope. ETO, etoposide; NVB, vinorelbine tartrate; DTX, docetaxel. (B) Heatmap of the top 10 sensitizing compounds in (A). For each compound, heatmap colors depict doxycycline-treated PI values after the subtraction of cell death induced by doxycycline alone and by the compound in the absence of doxycycline. *CQ and HCQ were used at 50 μM. **Cisplatin (CDDP) was used at 150 μM. (C) Left: Representative images of MEF Flag-RECS1 cells pretreated with doxycycline and treated with 50 μM CQ for 24 hours (red, PI staining). Right: Cell death kinetic analyses of cells treated as in (B) (n = 3). (D) Cells were cultured as in (C) and then treated with indicated concentrations of bafilomycin A1 for 24 hours. Cell death was determined by FACS (n = 4). (E) Left: Representative images of HeLa Flag-RECS1 cells treated as in (C). Right: Corresponding cell death kinetic analyses (n = 3). (F) HeLa Flag-RECS1 cells were treated as in (E), and cell death was assessed by FACS (n = 3). Data are shown as means ± SD (C and E) or means ± SEM (A, D, and F). Statistically significant differences were determined using the extra-sum-of-squares F test (C and E) or two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. *P < 0.05; **P < 0.01; ****P < 0.0001.
To gain further mechanistic insights into the function of RECS1 under lysosomal stress, we measured cell death in MEF expressing Flag-RECS1 after lysosomal pH alkalization. We found that CQ treatment strongly induces cell death in cells overexpressing RECS1, whereas control cells were fully resistant (fig. S3A). Kinetic analyses confirmed the rapid sensitization of cells to CQ treatment upon RECS1 expression, detecting 80% of cell death after 24 hours of treatment (Fig. 2C, fig. S3B, and movie S1). Similar results were obtained with the V-ATPase (vacuolar H+-ATPase) inhibitor bafilomycin A1 (Fig. 2D). Last, we also confirmed these observations in HeLa cells expressing Flag-RECS1 (Fig. 2, E and F). These results indicate that RECS1 potentiates cell death triggered by lysosomal stress.
Previous studies have mapped RECS1 localization to different intracellular compartments including late endosomes and lysosomes, multivesicular bodies, and, to a lesser extent, the Golgi apparatus (19, 29–32). We confirmed RECS1 localization to lysosomes and the Golgi apparatus using immunofluorescence and confocal microscopy, followed by quantitative colocalization analysis using MEF and HeLa cells expressing Flag-RECS1 (Fig. 1H and fig. S1L). We also performed immunogold staining followed by transmission electron microscopy (TEM) and observed that endogenous RECS1 localizes to the membrane and lumen of intracellular vesicles (fig. S1K). Together, these results support a role of RECS1 at the lysosome as a modulator of cell death in response to several cytotoxic agents (fig. S2F).
We then explored the effects of lysosomal stress on RECS1 localization. Analysis of the distribution of endogenous mouse RECS1 indicated that, under basal conditions, RECS1 colocalizes mainly in lysosomes and late endosomes (around ~20%) (fig. S3, C to E). However, in cells undergoing starvation or starvation and treated with CQ, RECS1 was further recruited to lysosomes but not to late endosomes, suggesting that RECS1 distribution is highly dynamic (fig. S3, C to E). Notably, the levels of Recs1 mRNA were up-regulated in cells undergoing nutrient starvation (fig. S3F). In addition, RECS1 overexpression induced autophagosome formation, an effect that was increased by CQ treatment (fig. S3G). Overall, our results indicate that RECS1 is located at lysosomes and regulates the susceptibility of cells to lysosomal stress.
RECS1 expression enhances LMP
LMP has emerged as a relevant cell death routine involved in several pathophysiological processes (5, 33). During LMP, lysosomal intraluminal cathepsins and hydrolases leak into the cytosol, where they act as cell death proteases (5, 33). The relocation of the cytosolic β-galactoside–binding proteins LGALS1/galectin-1 and LGALS3/galectin-3 to the lysosomal glycocalyx is a hallmark of LMP and a direct measure of lysosomal membrane damage (34). Thus, to investigate the effects of RECS1 expression on LMP, we monitored their distribution by immunofluorescence. Although RECS1 overexpression had no effects on LGALS1/galectin-1 localization at basal levels, treatment with CQ resulted in a dose-dependent increase in the number of LGALS1/galectin-1 puncta (Fig. 3, A and B). We observed an increase in LMP events in cells overexpressing RECS1 following treatment with a higher concentration of CQ, even when at this concentration LMP was also induced under basal conditions (fig. S4, A to C). We validated these results using LGALS3/galectin-3, although this protein exhibited translocation to the lysosomes under basal conditions (fig. S4, D to F). Notably, LGALS3/galectin-3–positive lysosomes were smaller under basal conditions when compared with permeabilized lysosomes in cells overexpressing RECS1 (fig. S4D, arrow versus arrowhead). LGALS1/galectin-1 translocation to the lysosomal lumen occurred before caspase activation (Fig. 3, A and B, and fig. S4, A and B), indicating that RECS1-induced LMP is not a secondary process downstream of MOMP. In addition, we measured the enzymatic activities of three lysosomal enzymes—cathepsin D, acid phosphatase, and hexosaminidase—in cytosolic lysosomal-free fractions. In line with previous results, we found that the activity of these three enzymes is increased in the cytosol of MEFs expressing Flag-RECS1 after CQ treatment but not in control cells (Fig. 3C). Last, we defined the requirement of BAX and BAK in cell death induced by lysosomal stress. As expected, BAX and BAK DKO cells were almost completely resistant to cell death induced by treatment with 25 μM CQ (Fig. 3D). However, we observed 30% of cell death in BAX and BAK DKO Flag-RECS1 cells treated with the higher CQ dose (50 μM), an effect that could not be reversed by inhibition of mitochondrial transition permeability pore (mPTP) (cyclosporin A) or necroptosis (necrostatin-1s) (Fig. 3D and fig. S4G).
Fig. 3. RECS1 causes LMP in cells undergoing stress.
(A) HeLa Flag-RECS1 cells were cultured with doxycycline and then treated with indicated concentrations of CQ, QvD, or both for 16 hours. Immunofluorescence against galectin-1 was performed. (B) Quantification of galectin-1 puncta per field (n = 2; 6 to 27 fields in total). (C) MEF Flag-RECS1 cells were cultured with doxycycline and treated with 50 μM CQ for indicated time points. The enzymatic activities of cathepsin D, acid phosphatase, and hexosaminidase were assessed from cytosolic, lysosomal-free fractions (n = 3). (D) MEF Flag-RECS1 WT and BAX and BAK DKO cells were cultured with doxycycline and then treated with indicated concentrations of CQ for 24 hours. Cell death was determined by PI staining followed by FACS (n = 3). (E) MEF Flag-RECS1 cells were cultured with doxycycline and treated with CQ for 16 hours. Immunofluorescence against LAMP-2 (green), BAX N-terminal domain (red, BAX 6A7 antibody), and Flag-RECS1 (blue, anti-Flag antibody) was performed. The triple colocalization of Flag-RECS1, LAMP-2, and active BAX is visualized as white color. (F) Representative immunofluorescence images of Flag-RECS1 MEF cells treated as in (E). Bottom panels correspond to zoomed white boxes (marked as z) from the top panels. (G) Intensity profiles for LAMP-2, BAX, and Flag-RECS1 signals along the line shown with an L1 (left) or an L2 (right) in (F). Data are shown as means ± SEM. Statistically significant differences were determined by two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
BAX has been shown to translocate to lysosomes where it is suggested to trigger LMP and cell death under different pathophysiological conditions, including Parkinson’s disease, oxidative stress, and autophagic cell death (35–37). To assess BAX translocation to the lysosomal membrane and its colocalization with RECS1, we detected active BAX using the 6A7 conformational antibody. Analysis of RECS1 (using the Flag antibody) and the distribution of the lysosomal protein LAMP-2 in MEFs indicated that under basal conditions (no RECS1), BAX remained mainly cytosolic (fig. S4H). However, BAX colocalized with RECS1 in LAMP-2–positive vesicles following RECS1 induction (Fig. 3, E to G). After CQ treatment, BAX was redistributed into large LAMP-2–positive vesicles, an effect that was dependent on RECS1 expression (Fig. 3E). Active BAX was present preferentially in Flag-RECS1–positive lysosomes in cells treated with CQ (Fig. 3, F and G). Together, these results suggest that RECS1 induces cell death through LMP in response to lysosomal stress, correlating with the translocation of BAX to lysosomes.
RECS1 overexpression sensitizes cell death induced by ER stress
Several proteins of the TMBIM family, such as BI-1 and GRINA, inhibit apoptosis triggered by ER stress, probably through the modulation of ER calcium (10, 13, 15, 17, 38). To gain further insights into the role of RECS1 in the control of cell death under cellular stress, we measured its expression in cells undergoing ER stress induced by the treatment with the N-glycosylation inhibitor tunicamycin (Tm). We found that Recs1 mRNA levels were up-regulated under ER stress, whereas Bi-1 mRNA levels remained unaltered (fig. S5A). This expression profile parallels that of certain BCL-2 proteins known to be induced by ER stress (i.e., Puma), while antiapoptotic components were unchanged (i.e., Bcl-2) (fig. S5B). Recs1 mRNA was also up-regulated in cells undergoing ER stress induced by the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) calcium pump inhibitor thapsigargin (Tg) (fig. S5C). The kinetics of the up-regulation of Recs1 mRNA under ER stress paralleled the up-regulation of the ER chaperone Bip/Grp78 and the proapoptotic factor Chop/Gadd153 (fig. S5D).
Given that RECS1 is a highly hydrophobic protein, we evaluated whether its overexpression was sufficient to perturb ER proteostasis. To this end, we induced Flag-RECS1 expression in MEFs and determined the levels of XBP1s mRNA splicing, an early indicator of the activation of the UPR (39). We found that, although Flag-RECS1 rapidly accumulated after doxycycline treatment (fig. S5E), the levels of Xbp1 mRNA splicing were not induced (fig. S5, F and G). As positive controls, cells were stimulated with Tm. Furthermore, RECS1 overexpression did not result in the up-regulation of Bip/Grp78 and Chop/Gadd153 (fig. S5H), two sensitive markers of ER stress (13, 15). We also evaluated the effect of RECS1 overexpression in the activating transcription factor 6 (ATF6) and PERK [protein kinase R (PKR)–like endoplasmic reticulum kinase] branches of the UPR. We determined the protein levels of CHOP [CCAAT/enhancer-binding protein (C/EBP) homologous protein] and ATF4 (downstream PERK pathway effectors) and BiP (binding immunoglobulin protein, regulated by ATF6/ATF4) in MEFs treated with Tm. We also assessed the activation of PERK by detecting its phosphorylation measured as a mobility shift in Western blot analysis. In contrast to ER stress induced by Tm treatment, we found that RECS1 overexpression did not trigger the activation of PERK or the up-regulation of CHOP, ATF4, or BiP, suggesting that the PERK pathway is not engaged (fig. S5J). We also measured the levels of two downstream genes, Sel1L and Pdia4, which are regulated by ATF6, and found that RECS1 overexpression failed to up-regulate any of these genes (fig. S5I). These results indicate that RECS1 overexpression alone does not cause ER stress. We also determined whether CQ treatment could induce ER stress. Treatment of Flag-RECS1–overexpressing MEFs with CQ failed to induce Xbp1 mRNA splicing (fig. S5, K and L), Bip/Grp78, or Chop/Gadd153 (fig. S5L). These results suggest that cell death induced by RECS1 under lysosomal stress conditions is not associated with the activation of the UPR.
We then determined the rate of cell death in MEF expressing Flag-RECS1 under ER stress. We found that RECS1 expression sensitized cells to Tm, evidenced by an increased number of annexin V and PI double-positive cells (Fig. 4A). Since ER stress affects protein trafficking through the secretory pathway and the expression of RECS1 alone does not trigger ER stress, we allowed RECS1 to accumulate for 16 hours before challenging cells with Tm or other ER stressors (Fig. 4B). Using this protocol, we found that RECS1 overexpression hastened cell death to ER stress, as evidenced in cell death kinetic experiments (Fig. 4C). Moreover, we confirmed these observations in HeLa cells in dose-response experiments (fig. S5, M and N). We further validated these results in cells stimulated with Tg, followed by cell death measurements using both FACS and live microscopy (Fig. 4, D and E). Long-term survival assessment by clonogenic assays indicated that RECS1 expression reduced the growth of cells under ER stress (Fig. 4F). Last, we treated MEFs expressing Flag-RECS1 with QvD together with Tm and determined cell death by live microscopy. As expected, QvD treatment reduced cell death under ER stress (Fig. 4G). Similarly, BAX and BAK double deficiency fully inhibited cell death triggered by RECS1 overexpression under ER stress (Fig. 4H).
Fig. 4. RECS1 sensitizes cells to ER stress.
(A) Annexin V versus PI FACS density plots from MEF Flag-RECS1 cells cultured with doxycycline for 16 hours and then treated with Tm (100 ng/ml) for 16 hours followed by FACS (n = 9). (B) Experimental design for cell death kinetic experiments. (C) Cell death kinetic analyses of MEFs treated as in (A) (n = 6). (D) Cell death of MEF Flag-RECS1 cells incubated with doxycycline and then treated with 100 nM Tg for 24 hours (n = 3). (E) Corresponding cell death kinetic analysis (n = 3). (F) Clonal assay of MEF Flag-RECS1 cells treated with doxycycline alone or in the presence of Tm (50 ng/ml) or 25 nM Tg. (G) Cell death kinetics of MEF Flag-RECS1 cells cultured with doxycycline and then treated with Tm (100 ng/ml) in the presence or absence of 40 μM QvD (n = 3). (H) MEF Flag-RECS1 WT and BAX and BAK DKO cells were cultured with doxycycline and treated with Tm (100 ng/ml) or 50 nM Tg for 24 hours. Cell death was determined by FACS (n = 2 to 3). Data are shown as means ± SD (C, E, and G) or means ± SEM. Statistical differences were determined by the extra-sum-of-squares F test (C, E, and G) or two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To assess the function of endogenous RECS1 in the control of cell death under ER stress, we transiently transfected MEF with two different pools of small interfering RNAs (siRNAs) directed against Recs1 mRNA (siRecs1#1 and siRecs1#2) or control (siSCR) (Fig. 5A). Silencing endogenous RECS1 in MEFs had no effects in basal cell viability (fig. S5, O and P), but it conferred protection against ER stress induced by Tm and Tg (Fig. 5, B and C). Virtually identical results were observed when HeLa cells were transfected with siRNA against the endogenous human RECS1 (fig. S5, Q and R). We confirmed these results by silencing endogenous mouse Recs1 in MEFs through the stably delivery of a short hairpin RNA (shRNA), although the levels of RECS1 down-regulation were not optimal (fig. S5S). These results suggest that the basal expression of RECS1 regulates cell death under ER stress. Although more studies are needed to fully assess the significance of RECS1 expression in the global control of intrinsic cell death, these studies suggest a major role in injuries to the lysosome and the ER.
Fig. 5. Inhibition of the endogenous RECS1 protects cells against ER stress.
(A) Knockdown of the endogenous mouse RECS1 using two different siRNA pools was confirmed by quantitative polymerase chain reaction (qPCR). (B) Left: Cells transfected with RECS1 siRNAs were treated with Tm (50 ng/ml), and cell death was determined by SYTOX green staining. A representative experiment from four independent experiments is shown. Right: Quantification of cell death at 24 hours (n = 4). (C) Cell death kinetics of cells transfected as in (B) and treated with 25 nM Tg. Left: Representative kinetics. Right: Quantification at 24 hours (n = 3). Data are shown as means ± SD. Statistical differences were determined by one-way ANOVA followed by Holm-Sidak’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001.
RECS1 promotes lysosomal acidification and calcium accumulation
The resolution of the crystal structure of the bacterial TMBIM ortholog BsYetJ from B. subtilis revealed a seven-transmembrane pH-sensitive calcium channel, providing a possible functional model for some members of the BI-1 and LFG protein families (11, 12). Since calcium-binding affinities of many fluorescent probes vary with pH, it is critical to have a precise knowledge of lysosomal luminal pH and its effect on the calcium-binding affinity (Kd) of the probe to accurately measure the concentration of lysosomal intraluminal calcium (40). Accordingly, we simultaneously measured these two parameters in RECS1-overexpressing cells using ratiometric calcium (Fura-2Dx) and pH (OGDx) probes, as previously described (40). We first confirmed the specific endosomal localization of these probes in our cellular models (fig. S6A). One pH and calcium calibration curves were performed per experimental day using calibration solutions from pH 3.0 to 7.0 and calcium concentrations of 0 and 10 mM, respectively (fig. S6B). To obtain precise lysosomal calcium measurements, we used our pH values to convert lysosomal Fura-2Dx signals to calcium concentration, taking into account the effects of pH on the Fura-2Dx Kd. RECS1 overexpression resulted in lysosomal acidification in HeLa cells, with lysosomal pH (pHL) decreasing from 4.77 to 4.38 (Fig. 6A). The acidification of lysosomes correlated with an increase in lysosomal intraluminal calcium concentration ([Ca2+]lys) from an average of 149 to 286 μM (1.9-fold increase) (Fig. 6B), while ER calcium content was not altered (Fig. 6C). In agreement with these results, expression of RECS1 in MEFs lowered lysosomal pH from 4.4 to 4.1, an effect that was dependent on BAX and BAK expression (Fig. 6D). In cells overexpressing RECS1, lysosomes exhibited a near 3.7-fold increase in calcium content, from 463 μM to 1.4 mM (Fig. 6E). Again, the consequences of RECS1 expression on lysosomal calcium levels were dependent on BAX and BAK expression (Fig. 6E). In agreement with these results, knocking down Recs1 with siRNAs (Fig. 6F) led to an increase in lysosomal pH (Fig. 6G). However, inhibition of Recs1 expression only slightly altered lysosomal luminal calcium content (Fig. 6H).
Fig. 6. RECS1 increases lysosomal acidification and calcium content.
(A) Lysosomal pH of HeLa Flag-RECS1 loaded with FuraDx and OGDx and cultured in the absence or presence of doxycycline for 24 hours (n = 5, 15 to 16 total coverslips). (B) Corresponding lysosomal calcium concentration. (C) Left: Cytosolic calcium levels were determined in HeLa Flag-RECS1 cells treated with doxycycline. Cells were treated with 5 μM Tg to stimulate ER calcium release. Right: Maximum ER calcium peak (n = 4, 10 to 11 coverslips). ns, not significant. (D) MEF WT and BAX and BAK DKO Flag-RECS1 cells were loaded as in (A), and lysosomal pH was determined after 24 hours of doxycycline treatment (n = 6, 12 to 17 coverslips per condition). (E) Lysosomal calcium measurements (n = 6). (F) Knockdown of the endogenous mouse RECS1 by siRNA was confirmed by qPCR after 48 hours of transient transfection. (G) Cells transfected as described in (F) were loaded as described in (A), and lysosomal pH was determined after 48 hours of transfection (n = 5, 14 to 15 coverslips). (H) Lysosomal calcium measurements (n = 5, 14 to 15 coverslips). (I) MEF Flag-RECS1 WT and BAX and BAK DKO cells were treated with doxycycline for 16 hours and analyzed by TEM. Image magnification, ×4000 (n = 13 to 17 cells per condition). (J) Diameter of primary and secondary lysosomes (n = 8 to 13 cells per condition). Data represent means ± SEM. Statistically significant differences were detected using ratio-paired two-tailed t test (A, B, D, E, G, and H) or two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To investigate the possible consequences of RECS1 expression on lysosomal morphology and content, we quantified the number of primary and secondary lysosomes in WT and BAX and BAK DKO MEFs using TEM. We found that RECS1 overexpression increased the amount of secondary lysosomes, an effect that was dependent on BAX and BAK expression, but it had no effect on lysosomal size (Fig. 6, I and J). Since secondary lysosomes are characterized by increased catabolic activity, these results suggest that RECS1 may boost lysosomal function. To define the possible consequences of CQ treatment in lysosomal morphology, we performed TEM in MEF cells expressing Flag-RECS1 treated with CQ. Although RECS1 had no apparent effects on lysosomal size or morphology, CQ treatment resulted in the accumulation of enlarged lysosomes (fig. S6C). As a complementary approach, we stained MEF overexpressing RECS1 cells with lysotracker and evaluated lysosomal size and number (fig. S6D). Overexpression of RECS1 did not have any significant effect in lysosomal size and total lysosomal number per cell (fig. S6, E to G). In addition, treatment with CQ triggered lysosome enlargement, while it also reduced lysosomal number (fig. S6, E to H). We also assessed the effects of RECS1 overexpression on the number of large lysosomes (area, >1.5 μm2), which usually correspond to endolysosomes characterized by a more acidic intraluminal pH and higher catabolic activity (41), and found no significant differences (fig. S6H). Together, these results suggest that RECS1 localizes to the lysosomes where it regulates acidification and luminal calcium concentration.
RECS1 is a pH-dependent Ca2+ and Na+ channel
To define the possible biochemical activity of human RECS1, we constructed homology models of the three-dimensional (3D) structure in closed and open conformations based on the bacterial BsYetJ protein using structure-based sequence alignment and the MODELLER platform (11). BsYetJ has 27% identity with GAAP and 36% with RECS1 (fig. S7). In the bacterial BsYetJ, the closed channel conformation is maintained through the interaction of the Arg60 with a conserved di-aspartyl group formed by Asp171-Asp195, which are hydrogen-bonded to one another through their carboxylate groups (11, 12). The di-aspartyl sensor is conserved in all six TMBIM proteins, while the arginine latch is only present in the LFG subfamily (LFG1 to LFG4) but not in BI-1, where it has been replaced by His78 (Fig. 7A) (11). Our human RECS1 model predicts an interaction between Arg158 with the Asp268-Asp295 di-aspartyl sensor in the closed conformation (Fig. 7B, green). In the open state, this latch becomes dislodged, and the second transmembrane is displaced out, leading to the opening of the channel (Fig. 7B, magenta). Our model suggests that RECS1, as demonstrated for other proteins of the LFG and BI-1 families (11, 12, 25), could operate as an ion channel. Our bioinformatic analysis also suggests that Asp295 (D295) is a strong candidate to regulate the channel activity of RECS1 and was therefore selected for mutagenesis and functional studies.
Fig. 7. RECS1 is a pH-dependent calcium and sodium channel.
(A) Alignment showing the conservation of the critical Arg60 residue from BsYetJ. Right: Comparison of the conserved di-aspartyl sensor. (B) Tridimensional structural model of the human RECS1 in closed and open conformations. Zoom: Key conserved C-terminal residues involved in pH sensing and channel opening. (C) X. laevis oocytes were injected with mRNA for Flag-RECS1 WT or D295Q, and ionic single-channel currents were recorded in the cell-attached mode using patch clamp at pH 7 or 6.5. Inset: Amplified plots depicting three opening states for RECS1 (O1, O2, and O3). (D) Quantification of the probability to find the channel in the open state (NPo) for RECS1 WT and D295Q (left). The NPo for open states O1 (middle) and O2 (right) is also shown [n = 4 (RECS1 WT) or n = 3 (RECS1 D295Q)]. (E) Images of X. laevis oocytes injected with different concentrations of Flag-RECS1 (top) and oocyte survival (bottom). Arrowhead indicates morphological alterations (n = 3). NI, not injected. (F) Recordings of macroscopic currents using the cut-open technique. (G) Permeability ratios with respect to K+ for indicated cations (n = 3 to 7). (H) Top: Indicated MEF Flag-RECS1 cells were treated with doxycycline, and protein levels were monitored by Western blot. Dashed lines indicate Western blot splicing. Bottom: Cells were treated with doxycycline and 25 μM CQ for 24 hours. Cell death was determined by FACS and normalized to WT (100%) (n = 3). Data represent means ± SEM. Statistical differences were determined by two-way (D, E, and G) or one-way ANOVA followed by Holm-Sidak’s multiple comparisons test. *P < 0.05; ***P < 0.001; ****P < 0.0001.
To directly test the electrophysiological properties of RECS1, we expressed WT and D295Q mutant human RECS1 in Xenopus laevis oocytes and recorded ionic single-channel currents in the cell-attached mode. Recordings at constant voltage (−100 mV) and pH 7 showed spontaneous, single-channel current spikes with three different opened states in both RECS1 WT and D295Q mutant. Lowering the pH to 6.5 caused a marked decrease in the number of current spikes in both channels (Fig. 7C), consistent with the hypothesis of RECS1 as a pH-regulated channel. Quantification of the total open probability (NPo) in addition to the probability for each independent opening state (NPo Ox) showed that the channel activity of RECS1 is heavily dependent on pH (Fig. 7D). Mutation of the key Asp295 from the putative di-aspartyl sensor significantly reduced RECS1 opening probability at pH 7, while it only slightly affected the ability to respond to pH changes (Fig. 7D). Notably, overexpression of RECS1 triggered cell death and structural abnormalities in the oocytes in a dose-dependent manner, consistent with its role as a cell death regulator (Fig. 7E). We were not able to obtain reliable recordings at pH lower than 6.5 because of increased basal permeability of the oocyte’s membrane.
We then assessed the permeability of RECS1 for different cations using the cut-open voltage clamp technique with potassium as the reference ion. Analyses of permeability ratios indicated that RECS1 allows the flux of Ca2+ and, to a lesser extent, Na+ (Fig. 7F). Mutation of the di-aspartyl sensor results in a marked decrease in the channel’s permeability to Ca2+ and Na+ but not to Ba2+ or Cs+ (Fig. 7G). Together, these results suggest that RECS1 is a pH-dependent cationic channel, with a main preference for calcium.
Last, to define the relationship between the channel activity of RECS1 and the regulation of cell death induced under lysosomal stress, we generated a doxycycline-inducible MEF cell line expressing a single-point mutant version of RECS1 (Fig. 7H). When compared to WT RECS1, the overexpression of the D295Q mutant diminished cell death, behaving similarly to the mock conditions (Fig. 7H). These results suggest that RECS1 channel function is required for the regulation of cell death under conditions of lysosomal stress.
RECS1 regulates cell death and organismal resistance to stress in Drosophila melanogaster
Members of the TMBIM superfamily, including BI-1 and GRINA, have a conserved activity in the control of stress-induced cell death in fly models. To validate the role of RECS1 in the regulation of apoptosis in vivo, we searched for putative ortholog genes within the TMBIM family in D. melanogaster and found CG9722 as a strong candidate, sharing a 34% amino acid sequence identity with the human RECS1 (fig. S8A). CG9722 belongs to the LFG subfamily of proteins and, based on a structural modeling, exhibits a conserved 3D structure together with the presence of critical channel residues (fig. S8, B and C). To assess the possible function of CG9722 (here termed dRECS1 as a candidate RECS1 ortholog) in cell death during fly development, we created a transgenic fly overexpressing the full-length dRECS1 protein using the GAL4/UAS system and monitored larvae viability by counting the number of eclosed pupae. Unexpectedly, we found that dRECS1 overexpression (dRECS1OE) completely abrogated animal development, with none of them reaching the adult stage (Fig. 8A). In sharp contrast, a mutant line where a transposon is inserted in the coding region (dRECS1KO) exhibited a slightly slower eclosion rate but was completely viable (Fig. 8A). This mutation was predicted to generate a truncated and nonfunctional protein.
Fig. 8. The overexpression of the RECS1-like/CG9722 protein causes lethality and cell death during Drosophila development.
(A) D. melanogaster mutant RECS1 (dRECS1KO), ubiquitously expressed transgenic (dRECS1OE), and isogeneic control flies (CTRL) were grown in normal food, and the percentage of eclosed pupae was measured daily (n = 4, >50 animals per experiment). (B) Left: Diagram of the wing imaginal disc (V, ventral; D, dorsal; A, anterior; P, posterior). Right: Immunofluorescence image of the adult fly wing imaginal disc where the Flag-dRECS1 construct was expressed (Flag, blue). Cleaved caspase-3 stain (CC3, green) is shown. (C) WT Flag-dRECS1 or a C-terminal domain deletion mutant (dRECS1ΔC) was overexpressed in the wing imaginal disc, and adult fly wings were imaged. A fly overexpressing two copies of dRECS1 (Nub>2xdRECS1) was also generated. (D) Images of wing phenotypes associated with the overexpression of Flag-dRECS1. The number of wing notches was quantified. (E) Quantification of wing notches for male (left) and female (right) flies overexpressing Flag-dRECS1 in combination with different constructs, including an unrelated fusion protein CD8-GFP as a control, an additional copy of dRECS1, the human version of RECS1 (hRECS1), or a dRECS1 RNA interference (dRECS1IR) (n = 3 to 5, >80 wings per conditions). Data represent means ± SEM. Statistically significant differences were determined by two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. ****P < 0.0001.
To bypass the developmental lethality, we restricted dRECS1 overexpression to the wing imaginary discs with the nub>Gal4 driver. dRECS1 overexpression resulted in the activation of caspase-3 in the center of the imaginal wing discs (Fig. 8B). We then evaluated the wing phenotype in adult flies expressing either one or two copies of the dRECS1 transgene. In line with our previous results, dRECS1 overexpression induced notches and reduced wing size in a dose-dependent manner (Fig. 8C), a morphological phenotype previously associated with increased cell death. Expression of a 22-residue C-terminal deletion mutant (dRECS1ΔC) failed to induce notches, suggesting that, as with human RECS1 in mammalian cells, this domain is required for the proapoptotic function of dRECS1 (Fig. 8C).
To quantify the pro-death effects of dRECS1 dosage and its potentially conserved function with human RECS1 on cell death induction, we counted the number of notches per wing and the number of blistered wings in adult flies expressing one copy of dRECS1 together with a control transgene [CD8–green fluorescent protein (GFP)], an additional copy of dRECS1, or the human RECS1 (hRECS1) counterpart (Fig. 8, D and E). We found that overexpression of two dRECS1 transgenes resulted in a complete blistered wing phenotype in male and female wings (Fig. 8E). Moreover, human RECS1 overexpression increased the severity of overexpressing one copy of the dRECS1 transgene (Fig. 8E), an effect that was more evident in male flies since the nub>Gal4 driver is inserted in the X chromosome. As control, we expressed a siRNA against dRECS1 mRNA (dRECS1IR), which completely reversed the adverse effects of dRECS1 overexpression (Fig. 8E). Together, these data suggest that dRECS1 induces cell death in vivo on a fly model.
Last, we evaluated the consequences of dRECS1 expression on cell survival and the adaptation of whole animals to adverse stress conditions. To this end, we assessed the wing phenotype of flies overexpressing one copy of dRECS1 after exposure to CQ or Tm in the food. Overexpression of dRECS1 strongly sensitizes to lysosomal perturbations and ER stress, resulting in exacerbated wing abnormalities (Fig. 9A). Moreover, dRECS1 deficiency conferred resistance to Tm in male and female flies, increasing life span under ER stress (Fig. 9, B and C). In addition, dRECS1KO female animals were more resistant to death induced by nutrient starvation, an effect that was still observed but not as prominent in male flies or larvae (Fig. 9, D and E). Last, given our observations showing that dRECS1KO flies are more resistant to starvation, we evaluated the effects of dRECS1 expression in autophagy. To this end, we used the fly fat body as a model, an organ that is well known for its prominent autophagic activity in response to starvation (42). We found that dRECS1KO cells exhibited increased formation of autophagosomes and autolysosomes together with higher overall autophagy flux (fig. S8, D to F). Together, these results suggest that CG9722 has a proapoptotic activity in vivo and sensitizes flies to lysosomal- and ER stress–induced cell death.
Fig. 9. RECS1-like/CG9722 modulates viability in response to intracellular stress in D. melanogaster.
(A) One copy of dRECS1 was overexpressed in the wing imaginal discs of animals fed with control food or food supplemented with CQ (100 μg/ml) or Tm (10 μg/ml). The phenotype of adult male (right) and female (left) wings was quantified (n = 3 to 7, >80 wings per condition). (B) Control (yw) and mutant (dRECS1KO) male and female flies were treated with Tm (10 μg/ml), and the animal death rate was quantified daily (n = 2 to 3). (C) WT, dRECS1KO, or dRECS1IR pupae were treated with Tm, and the percentage of eclosion was quantified (n = 2 to 4). (D) WT and dRECS1KO female flies were fed a normal diet or starved, and the death rate was daily quantified (n = 3). (E) WT and dRECS1KO knockout male flies were fed a normal diet or starved, and the death rate was daily quantified (n = 2 to 3). Data represent means ± SEM. Statistically significant differences were determined by two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. ****P < 0.0001.
RECS1 expression induces cell death and developmental abnormalities in zebrafish
There is a high degree of functional conservation in the mechanisms regulating cell death between vertebrates, with almost all the members of the BCL-2 and the TMBIM families conserved in zebrafish (17, 43, 44). We were able to detect the expression of the endogenous zebrafish RECS1 in different tissues in the zebrafish embryo using an antibody directed against the mouse and human RECS1 protein (fig. S9A). To investigate the role of RECS1 on the control of cell death in vivo in a vertebrate model, we overexpressed the full-length human RECS1 in zebrafish by injecting in vitro synthetized mRNA to one-cell stage embryos, followed by the analysis of animal viability and morphology 24 hours post-fertilization (hpf). RECS1 overexpression significantly decreased animal survival, a phenotype that was independent of its C-terminal domain (Fig. 10A). As a control, we overexpressed the TMBIM protein GRINA, which, under the same conditions, did not result in any developmental defects (Fig. 10A). A fraction of the viable embryos injected with RECS1 (around 52%) or two C-terminal deletion mutants exhibited morphological defects, such as the formation of cyclops (embryos with one eye), ocular fusion, and anterior-posterior shortening, among other abnormalities (Fig. 10B).
Fig. 10. RECS1 induces developmental alterations and cell death during zebrafish development.
(A) One-cell stage zebrafish embryos were microinjected with WT or C-terminal deletion mutant versions of RECS1, GRINA, or left uninjected (NT). Viability was assessed by visual inspection after 24 hours [n = 3 (Δ910) or n = 4 (NT, WT, Δ868, and GRINA)]. (B) Morphological alterations were quantified 24 hours after fertilization [n = 3 (Δ910 and Δ868) or n = 4 (NT, WT, and GRINA)]. (C) Viability was monitored after 24 hours by acridine orange (AO) staining in full-mounted embryos. Fluorescence (top) and bright-field (bottom) images of the embryos’ lateral views are shown. Middle panels show higher magnification of the inset depicted in the top panels (white box), highlighting the front section of the head. (D) Quantification of AO-positive spots in the head of the embryos from the middle panel of (C) (n = 3). (E) One-cell zebrafish embryos were microinjected with WT human RECS1 or the D295Q mutant RNA or left uninjected. Viability was assessed 24 hpf (n = 3). (F) Quantification of morphological alterations (n = 3). (G) Percentage of viable, dead, and altered embryos. (H) Zebrafish embryos were injected as in (A), treated with indicated concentrations of Tg for 4 hours, and stained with AO as described in (C). (I) Quantification of AO-positive dots from the head of embryos (n = 3). (J) Working model. Data represent means ± SEM. Statistically significant differences were determined by one-way ANOVA followed by Dunnett’s multiple comparisons test (A, B, and D to F) or by two-way ANOVA followed by Holm-Sidak’s multiple comparisons test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine the possible role of RECS1 in the regulation of cell death in zebrafish, we stained injected embryos with acridine orange (AO) and quantified the number of AO-positive spots corresponding to dead cells in the embryo. As expected, RECS1 overexpression induced a high number of AO spots, distributed along the head and, to a lesser extent, the body of the embryo (Fig. 10, C and D). Deletion of the C-terminal domain of RECS1 completely abrogated its cytotoxic effects (Fig. 10, C and D). Together, these results suggest that the overexpression of RECS1 induces cell death in zebrafish embryos, involving its C-terminal region; however, RECS1’s detrimental effects in embryonic development may involve other regions and/or functions of the protein.
Next, we determined the importance of RECS1 channel activity to cell death control during zebrafish development. We injected single-cell embryos with RECS1 WT or the D295Q mutant and assessed animal viability 24 hpf. In line with our in vitro results, overexpression of the RECS1 D295Q mutant failed to induce cell death in these embryos (Fig. 10E). Moreover, while the injection of RECS1 WT resulted in a high proportion of morphological-altered embryos, expression of the mutant protein did not have any adverse effect (Fig. 10, F and G). We determined the mRNA levels of human RECS1 in a pool of viable and altered embryos by real-time polymerase chain reaction (PCR). Dysmorphic embryos exhibited around 3.8-fold more RECS1 mRNA levels than embryos with normal appearance (i.e., without apparent phenotype) (fig. S9B), suggesting that the differences observed in viability are dose-dependent. Last, we determined the consequences of RECS1 expression to the susceptibility of animals to ER stress. RECS1 overexpression resulted in reduced survival when zebrafish were exposed to Tg in the water, while overexpression of the mouse GRINA conferred protection (Fig. 10, H and I) as reported (17). We were unable to set up the conditions to use CQ in this animal model. Together, these results indicate that RECS1 induces cell death in zebrafish and sensitizes this vertebrate to ER stress.
DISCUSSION
Most mechanisms controlling cell death involve the presence of activating and inhibitory signals (rheostat model) to determine cell fate (1). RECS1 is a member of the evolutionarily conserved TMBIM superfamily and, more specifically, the LFG family of cell death regulators. Although the role of these proteins as inhibitors of cell death is well documented, the possible existence of proapoptotic members remained elusive (8). Here, we provide evidence suggesting that, in contrast to other proteins of the TMBIM family, RECS1 promotes apoptosis in response to different intracellular cytotoxic stimuli probably through a mechanism involving the regulation of lysosomal calcium content, its permeabilization, and BAX translocation to the lysosomal membrane (Fig. 10J). Using gain- and loss-of-function approaches, we demonstrate that (i) RECS1 overexpression alone is sufficient to induce cell death and that (ii) RECS1 expression sensitizes to apoptosis induced by lysosomal pH neutralization and ER stress.
We showed that the RECS1 C-terminal domain is necessary for its pro-dead function, which contains two putative BH3-like domains, one of which was previously shown to trigger cell death by the yeast ortholog of BI-1 (22). However, in contrast to Ybh3, RECS1’s putative BH3-like motifs did not behave as bona fide BH3 motifs in vitro, since peptides containing such motifs were unable to induce mitochondrial permeabilization or to displace the interaction of a BH3-only protein with antiapoptotic proteins of the BCL-2 family. However, there are differences in the ability to induce apoptosis between 20- to 25-residue-long BH3 peptides and full-length BH3-only proteins. Short peptides derived from PUMA failed to induce cytochrome c release, while the full-length protein rapidly activates BAX and BAK, inducing cell death (45). Moreover, there may be posttranslational modifications that are required for the pro-dead function of BH3-peptides derived from RECS1, such as phosphorylation and ubiquitination (32). These caveats limit the conclusions from our assays using short BH3-derived peptides, and studies using the full-length protein and single–amino acid substitutions are necessary to fully address the role of this motif in cell death induced by RECS1. Notably, the yeast protein Ybh3p has been proposed to be an ancient member of the TMBIM family, as well as a putative BH3-only protein (21–23). This protein localizes to the ER and sensitizes cells to apoptosis induced by different cellular stresses including H2O2, acetic acid, and ER stress (22). However, other studies have found that Ybh3p is a prosurvival protein, questioning the functional relevance of this ancient BH3 motif (21, 23, 46). Such a BH3 sequence is present in 1834 different proteins in Saccharomyces cerevisiae and lacks conservation in orthologous proteins. Moreover, there are no BCL-2 homologs in yeast, making the function of this BH3-like motif in cell death very unlikely (28).
Indirect evidence has been shown for calcium leak activity for all members of the TMBIM family (10, 14, 17, 30, 47–50), although a direct proof of ion conductivity has only been demonstrated for bacterial BsYetJ, and human BI-1 and GAAP (11, 25); the permeability of BsYetJ or any other TMBIM family members to monovalent ions is currently unknown. Here, we provide evidence indicating that RECS1 is a pH-dependent calcium and sodium channel, an activity required to induce cell death. RECS1 contains a highly conserved di-aspartyl sensor localized within the TM7 domain, which is also disrupted in our BH3 deletion mutants. These residues are conserved across the TMBIM superfamily and play an important role in calcium signaling and their antiapoptotic functions (11, 12, 25). A recent study showed that mutations of either of the pH-sensing aspartates in the yeast BsYetJ protein resulted in defective calcium binding and reduced sensitivity to pH changes (12). In addition, mutation of Asp213 (corresponding to BsYetJ Asp195) completely inhibited the effects of BI-1 on ER calcium leak, while the analogous Asp219Asn mutation in GAAP increases its channel conductance and abrogates its antiapoptotic function (25, 27, 48). Thus, the activity of RECS1, as well as other proteins of the TMBIM superfamily, is linked to its calcium conductance activity and depends on key C-terminal residues.
RECS1 localizes in part at the lysosomal membrane, and RECS1 overexpression strongly sensitizes cells to CQ and HCQ, two lysosomal poisons. RECS1 expression resulted in lysosomal acidification together with increased lysosomal intraluminal calcium concentration. BI-1 has been recently shown to increase the pool of lysosomal releasable calcium through an indirect effect as an ER calcium channel leak (51). From the ER, BI-1 promotes the transfer of calcium to lysosomes, indirectly modulating lysosomal calcium content (51). Although we did not study the mechanisms of lysosomal acidification by RECS1, they may involve the regulation of the lysosomal H+-ATPase (V-ATPase), as it has been suggested for BI-1 (52). Low pH has differential effects on other lysosomal calcium-permeable channels: It activates transient receptor potential channel mucolipin-1 (TRPML1) and sensitizes two-pore channel-1 (TCP1) for activation by nicotinic acid adenine dinucleotide phosphate (NAADP), whereas it inhibits P2X purinoreceptor-4 and decreases TCP2 sensitivity for NAADP (53). Thus, it is difficult to predict the overall effect of hyperacidification on lysosomal calcium content. As an alternative hypothesis, under basal conditions, the acidic lysosomal intraluminal pH (pH <4.5) may keep RECS1 in a closed conformational state. Then, upon lysosomal pH neutralization or pathophysiological conditions, RECS1 switches to an open state, leading to LMP and the release of matured cathepsins into the cytosol, ultimately resulting in cell death (Fig. 10J). Loss of TRPML1 leads to lysosomal acidification and the translocation of cathepsin B to the cytosol to trigger apoptosis (54, 55). How does RECS1 trigger LMP? Evidence has emerged suggesting that the BCL-2 multidomain proteins BAX and BAK may induce LMP in a way analogous to their role at the mitochondria during apoptosis. BAX-mediated LMP has been suggested as the initial cell death signal in Parkinson’s disease, autophagic cell death, and some forms of oxidative stress, such as lysosomal permeabilization induced by free fatty acids or mitochondrial damage (35–37, 56). In line with this hypothesis, we provide evidence suggesting that RECS1 overexpression induces BAX activation at the lysosomal membrane. Moreover, BAX and BAK DKO cells are resistant to cell death triggered by CQ treatment, suggesting a direct role for these proteins in the initiation of cell death at the lysosomal membrane.
RECS1 was initially found as a gene up-regulated by shear stress in endothelial cells. Aged RECS1 knockout mice are prone to cystic medial degeneration of the aorta, a pathology characterized by basophilic depositions and smooth muscle loss (29, 57). Recent studies have also suggested that RECS1/TMBIM1 localizes to late endosomes and lysosomes, where it promotes the trafficking and lysosomal degradation of the Toll-like receptor 4, ameliorating the symptoms of nonalcoholic steatohepatitis in the liver of mice and monkeys (31, 32). RECS1 expression was shown to be protective in models of cardiac hypertrophy, where gene therapy to deliver the protein into the heart ameliorated disease features (32). Conversely, RECS1 deficiency accelerated high fat-induced metabolic disease in mice, exacerbating heart damage (58). These results suggest a dual role of RECS1 in the control of cell death and cell physiology depending on the context. Several TMBIM family members are deregulated in cancer. Consistent with this idea, the screening using a panel of cytotoxic agents suggested that overexpression of RECS1 has a wide impact in the regulation of cell death triggered by an array of anticancer drugs. RECS1 sensitizes cells to CQ, ER stress, and microtubule destabilizers, whereas it can block cell death induced by MTX and several pyrimidine analogs, among others. Recent studies identified genetic variants of RECS1 as a risk factor to develop colon cancer and the occurrence of inflammatory bowel disease (59, 60). Many cancer cells are resistant to lysosomotropic agents used for chemotherapy. Since RECS1 expression can reverse the resistance of immortalized HeLa or MEF cells to CQ, the analysis of its expression levels in cancer biopsies may serve as a predictor for the sensibility of tumors to the action of certain chemotherapies, a concept that needs to be further investigated.
In summary, here, we have uncovered a previously unanticipated function of the TMBIM member RECS1 as an inducer of cell death initiated by perturbations at the levels of the lysosome and the ER. Last, we provide evidence indicating that RECS1 is a previously unidentified lysosomal pH-dependent calcium channel that sensitizes cells to cell death induced by lysosomal stress resulting in LMP.
MATERIALS AND METHODS
Details are given in Supplementary Materials and Methods.
Cell culture, cell lines, and DNA constructs
All MEF, HeLa, and human embryonic kidney (HEK) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and nonessential amino acids and grown at 37°C and 5% CO2. The production of retroviruses using the HEK293 cell line and transduction of MEF and HeLa cells were performed according to standard protocols (61). Cells with stable expression of shRNA directed against the endogenous mouse Recs1 (shRECS1) or Luciferease genes (Broad Institute) as control (shLuc) were generated as previously described (62).
RNA isolation, reverse transcription PCR, and real-time PCR
Total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. The analysis of transcription targets was performed by real-time PCR using specific primers (see Supplementary Materials and Methods).
Western blot
Cells were collected and homogenized in radioimmunoprecipitation assay buffer. Total cell protein extracts were quantified and assessed by Western blot as described elsewhere (15). The following antibodies diluted in blocking solution [phosphate-buffered saline (PBS) and 0.1% Tween 20 containing 5% milk] were used: anti-HSP90 (1:5000; sc13119, Santa Cruz Biotechnology), anti-Flag (1:3000; F7425, Sigma-Aldrich), anti-TMBIM1 (1:1000; MBS1499661, MyBioSource), anti-BAX (1:1000; 06-499, EMD Millipore), anti-MYC (1:3000; ab9106, Abcam), and anti-actin (1:10,000; sc-8432, Santa Cruz Biotechnology). Bound antibodies were detected using peroxidase-coupled secondary antibodies and the enhanced chemiluminescence system.
Cell death assays
Cell death was monitored by PI and annexin V costaining, followed by flow cytometry using FACScan and FACSCalibur systems (BD Biosciences). Cell death kinetics were determined by either staining the cells with the cell-impermeable DNA binding dye SYTOX green (S7020, Thermo Fisher Scientific) or costaining them with PI and SYTO16 (S7578, Thermo Fisher Scientific), followed by imaging using a one-color FLR IncuCyte or a two-color ZOOM IncuCyte (Essen BioScience), respectively.
Indirect immunofluorescence analysis
Flag-RECS1, MYC-RECS1, BAX, ERp57, LAMP-1/2, LGALS1/galectin-1, and LGALS3/galectin-3 proteins were visualized by immunofluorescence. The following antibodies were used: anti-Flag (F7425, Sigma-Aldrich), anti-Flag M2 (F1804, Sigma-Aldrich), anti-TBMIM1 (MBS1499661, MyBioSource), anti-ERp57 (ab13506, Abcam), anti–LAMP-1 (an24170, Abcam), LAMP-2 (ab18528, Abcam), BAX (ab5714, Abcam), anti-LGAL1/galectin-1 (ab25138, Abcam), and LGAL3/galectin-3 (556904, BD Biosciences). All antibodies were diluted 1:1000. We used a sensitive method based on a confined displacement analysis algorithm to calculate colocalization coefficients between Flag-RECS1 and ERp57 (ER), GM130 (Golgi apparatus), and LAMP-1/2 (endosomes/lysosomes) (63). The colocalization of images was performed as previously described (63).
TEM and immunogold staining
For TEM analysis, cells were fixed in 2% glutaraldehyde (EM grade) in 0.1 M sodium cacodylate buffer (pH 7.4) for 30 min at room temperature. Samples were washed and postfixed with reduced 1% osmium tetroxide for 1 hour and infiltrated with Epon (catalog no. 812, TAAB). The images were randomly acquired using a Hitachi HT7800 microscope (Hitachi High-Technologies, Tokyo, Japan) and Rio9 complementary metal-oxide semiconductor camera (AMETEK Gatan Inc., Pleasanton, CA). For immunoelectron microscopy studies, MEF Flag-RECS1 cells (noninduced) were fixed with periodate-lysine-paraformaldehyde fixative for 2 hours at room temperature. Samples were washed, permeabilized with 0.01% saponin, and labeled with anti-TMBIM1 antibody (diluted 1:20; MBS149966, MyBioSource) for 1 hour and nano-gold–conjugated anti-rabbit FAB fragments for 1 hour before postfixation. Nano-gold particles were then intensified for 5 min using the HQ SILVER Enhancement Kit (catalog no. 2012, Nanoprobes) according to the manufacturer’s instructions, followed by gold toning in 2% sodium acetate, 0.05% HAuCl4, and 0.3% sodium thiosulfate. Samples were then osmicated, dehydrated, and flat-embedded into Epon as described above.
BH3 profiling and competition assays
The profiling of BH3 peptides derived from RECS1 C-terminal domain was performed as previously described (64). The ability of RECS1280–299 (SPEDYITGALQYRDIIYI) and RECS1296–311 (IYIFTFVLQLMGDRN) peptides to displace the interaction of the BH3-only protein BIM with BCL-2, BCL-XL, or MCL-1 was assessed by fluorescent polarization, as previously performed (65). BIM protein and a BH3-defective PUMA mutant (PUMA2A) were used as positive and negative controls, respectively.
Measurement of lysosomal pH and calcium
To measure lysosomal intraluminal calcium, lysosomal pH and calcium were measured simultaneously using two ratiometric dyes [see detailed protocol in (66)] according to a modified protocol based on the methods described by Christensen and colleagues (40).
Molecular modeling
Molecular models of human RECS1 in its open and closed conformations were generated by molecular similarity with MODELLER (67), using the crystal structure of BsYetJ as template (11). We used the chain A from 4PGS and 4PGW and from 4PGR and 4PGV Protein Data Bank structures as templates for the models of the open and closed conformations, respectively. The corresponding sequences were aligned with MAFFT using the linsi protocol (68, 69). One hundred models were generated, and the one with better discrete optimized protein energy was selected as the best model per conformation.
RECS1 expression in X. laevis oocytes
Full-length WT human RECS1 and RECS1 D295Q were subcloned into the pBSTA vector, which is optimized for expression in X. laevis oocytes. DNA was amplified and cleaned using a QIAprep kit (QIAGEN). Plasmid DNA (10 μg) was linearized with Not I, and 1 μg was used for in vitro transcription with a mMESSAGE mMACHINE kit (Ambion). Xenopus oocytes were injected with 50 nl of ∼5 ng of RECS1 WT or mutant complementary RNA per oocyte. Recordings were performed 1 to 3 days after injection.
Electrophysiology
Patch clamp
Using the patch-clamp technique, ionic single-channel currents were recorded in the cell-attached mode by a constant voltage or a ramp potential. Currents were recorded at −100 mV for constant voltage during 90 s, and the ramp was elicited between −250 and 150 mV during 40 s. Experiments were performed at room temperature (20°C).
Cut open
Macroscopic currents were recorded using the cut-open technique (Cut-open Oocyte Vaseline Gap voltage clamp), recording a wide fraction of the total oocyte membrane (20 to 25% of the total) (70). To obtain a low-resistance path to the intracellular side, the oocytes were permeabilized with 0.1% saponin during 15 to 30 s. Oocytes were dialyzed in internal solution during 30 min before individual recordings with different extracellular cations were performed. K+ was used as the internal reference ion. Tail current versus voltage relationships were measured from 150 to −350 mV, every −20 mV, and during 120 ms. Current recordings were sampled at 30 kHz and filtered at 5 kHz. The linear components were subtracted by P/-4 prepulse protocol (71).
Generation of D. melanogaster strains and cell death assays
Fly strains
All experiments were performed with D. melanogaster grown at 25°C unless otherwise indicated. We generated transgenic animals allowing the Gal4/UAS (72) guided expression of the full-length (dRECS1) or truncated version (dRECS1ΔC) of the CG9722/dRECS1 gene or the human RECS1 coding sequence (hRECS1), all of them tagged with a 3xFlag sequence at the N-terminal region. All additional Drosophila strains were obtained from Bloomington Drosophila Stock Center: w1118 (BDSC:3605), y1,w* (BDSC:1495), CG9722-KO [y1,w*; Mi{MIC}CG9722MI08483, BDSC:44979], UAS-CG9722-IR (TRiP.JF02887, BDSC:28052), UAS-CD8-GFP (BDSC:5130), UAS-GFP-mCherry-Atg8a (BDSC:37749), Cg-Gal4 (BDSC:7011), nub-Gal4 (BDSC:86108), and Tub-Gal4 (BDSC:5138).
Wing phenotypes
Flies (1 to 2 days) were immersed in 70% ethanol, and right-sided wings were dissected and transferred to a microscope slide for imaging. Animals with vestigial wings were mounted in glycerol for imaging. To determine the effect of lysosomal or ER stress, animals were grown in media supplemented with CQ (100 μg/ml) or Tm (10 μg/ml) throughout the development until hatching, where wing phenotypes were analyzed (N ≥ 3 independent experiments, n ≥ 80 wings per condition).
Survival assays
Depending on the case, animals were grown in vials containing normal food (Instant Drosophila Medium, #173200, Carolina) or starvation media (1% agarose in PBS), either alone or supplemented with CQ or Tm. To evaluate eclosion rate, embryos were collected in vials containing rich food alone or with Tm (10 μg/ml) for 6 hours, after which adults were discarded and eggs developed at 25°C until hatching.
Zebrafish studies
Zebrafish embryos were raised in E3 medium, kept at 28°C, and staged according to age (hpf). WT AB/Tübingen zebrafish were microinjected at one-cell stage with 39 pg of full-length human RECS1, C-terminal deletion mutants, RECS1 D295Q, or mouse GRINA or left uninjected as control. Embryo viability and morphology were quantified 24 hpf by visual inspection. In addition, at 24 hpf, apoptotic cells were visualized by AO (Sigma-Aldrich) staining, followed by epifluorescence microscopy in full-mounted embryos, as previously reported (73). The effect of RECS1 overexpression in ER stress–induced cell death was evaluated by incubating microinjected embryos with 1 to 5 mM Tg (Sigma-Aldrich) for 4 hours followed by AO staining. All of our animal experiments were previously approved by the appropriate ethics committee.
Statistics and reproducibility
Results were statistically compared using one-way analysis of variance (ANOVA) for unpaired groups, followed by Holm-Sidak’s multiple comparison test. When there were two independent variables, a two-way ANOVA followed by Holm-Sidak’s multiple comparison test was performed. For cell death kinetic experiments, the goodness of fit of the different curves was compared using the extra-sum-of-squares F test. When pertinent, two-tailed Student’s t test was performed for unpaired or paired groups. In all plots, P values are shown as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 and were considered significant. All results are presented as the means ± SD or means ± SEM as indicated in the figure legends. Analyses were performed using PRISM software.
Acknowledgments
We thank P. Fitzgerald from Saint Jude Children’s Research Hospital for helpful technical advice and assistance in the design and cloning of the different RECS1 plasmids used throughout this work. We thank M. J. Altamirano for veterinary assistance and C. Sepulveda for laboratory managing. We thank B. Dehay for collaborating in experiments to assess LMP and BAX activation. We thank D. Morales and D. Varela for technical assistance and collaboration in experiments assessing RECS1 channel function.
Funding: This work was supported by the following: FONDECYT 3150269 (to F.Li.); ANID/FONDAP program 15150012, Millennium Institute P09-015-F, FONDEF ID16I10223, FONDEF D11E1007, and FONDECYT 1180186 (to C.H.); and Ecos-Conicyt no. C17S02 (to C.H. and G.K.). In addition, we thank the support from the U.S. Air Force Office of Scientific Research FA9550-16-1-0384 and U.S. Office of Naval Research-Global (ONR-G) N62909-16-1-2003. We also thank the support from the following: FONDECYT 11180825 (to H.U.); FONDAP 15200002 and FONDECYT 1190119 (to A.G.), FONDECYT 3210294 (to P.P.), FONDECYT 1140522 (to A.G.), FONDECYT 3190738 (to A.Sag.), FODECYT 1190806, PIA ACT192015, P09-015-F, Nucleus Physics of Active Matter, FONDEQUIP EQM130051, and FONDAP 15150012 (to M.L.C.), and CONICYT fellowship (PCHA/Doctorado Nacional/21120667) (to P.P.). G.K. is supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR)–Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Chancelerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; the European Commission (ArtForce); European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); the European Research Council (ERC); Fondation Carrefour; Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumière; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI). This work was also supported by FONDECYT grant 1180999 (to K.C.), 1190203 (to R.L.), and CINV ICM-MINECOM P09-022-F and CONICYT doctoral fellowship (to D.D.G. and A.P.). The Centro Interdisciplinario de Neurociencia de Valparaíso (CINV) is a Millennium Institute supported by the Millennium Scientific Initiative of the Ministerio de Economía, Fomento y Turismo. P.N.-H. is funded by the Prof. Dr. Max Cloëtta Foundation Medical Research Award and the Swiss National Science Foundation (grant no. 310030_189094). N.D.’s research is supported by the Swiss National Science Foundation grant no. 310030_189042. This work was also supported by the Academy of Finland FIRI funding to E.J. Euro-BioImaging (www.eurobioimaging.eu) is acknowledged for providing access to services via Finland Node (FiALM, Helsinki, Finland) (to H.V., S.S., and E.J.). D.R.G. and D.A.R. were supported by grant CA231620 from the U.S. National Institutes of Health.
Author contributions: P.P. conceived and designed the project, carried out experimental work with cell culture models, interpreted the data, and wrote the manuscript. J.B. and M.L.C. carried out experimental in vivo work in zebrafish models, interpreted data, and prepared figures. P.N.-H., A.C.-S., and N.D. performed lysosomal calcium measurement experiments in cell models and interpreted the data. K.C., D.D.G., A.P., and R.L. performed bioinformatics and electrophysiological recordings of RECS1, interpreted data, and prepared figures. S.E.-J., F.L., and Á.G. carried out in vivo D. melanogaster experiments, designed experiments, interpreted the data, and contributed to the figures. O.K., S.F., A.S., and G.K. carried out in vitro cell death screening experiments. F.L., D.A.R., H.U., G.Q., and A.L. performed in vitro experiments and contributed to project design. S.S., H.V., and E.J. performed all electronic microscopy experiments. D.R.G. and C.H. contributed to project design, data interpretation, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S9
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Singh R., Letai A., Sarosiek K., Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tait S. W. G., Green D. R., Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Galluzzi L., Bravo-San Pedro J. M., Kroemer G., Organelle-specific initiation of cell death. Nat. Cell Biol. 16, 728–736 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Pihán P., Carreras-Sureda A., Hetz C., BCL-2 family: Integrating stress responses at the ER to control cell demise. Cell Death Differ. 24, 1478–1487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F., Gomez-Sintes R., Boya P., Lysosomal membrane permeabilization and cell death. Traffic 19, 918–931 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Sargeant T. J., Lloyd-Lewis B., Resemann H. K., Ramos-Montoya A., Skepper J., Watson C. J., Stat3 controls cell death during mammary gland involution by regulating uptake of milk fat globules and lysosomal membrane permeabilization. Nat. Cell Biol. 16, 1057–1068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu L., Smith T. F., Goldberger G., LFG: A candidate apoptosis regulatory gene family. Apoptosis 14, 1255–1265 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Rojas-Rivera D., Hetz C., TMBIM protein family: Ancestral regulators of cell death. Oncogene 34, 269–280 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Xu Q., Reed J. C., Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell 1, 337–346 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Chae H. J., Kim H. R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., Kitada S., Monosov E., Thomas M., Kress C. L., Babendure J. R., Tsien R. Y., Lipton S. A., Reed J. C., BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell 15, 355–366 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Chang Y., Bruni R., Kloss B., Assur Z., Kloppmann E., Rost B., Hendrickson W. A., Liu Q., Structural basis for a pH-sensitive calcium leak across membranes. Science 344, 1131–1135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo G., Xu M., Chang Y., Luyten T., Seitaj B., Liu W., Zhu P., Bultynck G., Shi L., Quick M., Liu Q., Ion and pH sensitivity of a TMBIM Ca2+ channel. Structure 27, 1013–1021.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly-Maitre B., Fondevila C., Kaldas F., Droin N., Luciano F., Ricci J. E., Croxton R., Krajewska M., Zapata J. M., Kupiec-Weglinski J. W., Farmer D., Reed J. C., Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 103, 2809–2814 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C., Xu W., Palmer A. E., Reed J. C., BI-1 regulates endoplasmic reticulum Ca2+ homeostasis downstream of Bcl-2 family proteins. J. Biol. Chem. 283, 11477–11484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J. H., Walter P., Reed J. C., Glimcher L. H., Hetz C., BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1α. Mol. Cell 33, 679–691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailly-Maitre B., Belgardt B. F., Jordan S. D., Coornaert B., von Freyend M. J., Kleinridders A., Mauer J., Cuddy M., Kress C. L., Willmes D., Essig M., Hampel B., Protzer U., Reed J. C., Brüning J. C., Hepatic Bax inhibitor-1 inhibits IRE1alpha and protects from obesity-associated insulin resistance and glucose intolerance. J. Biol. Chem. 285, 6198–6207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rojas-Rivera D., Armisen R., Colombo A., Martinez G., Eguiguren A. L., Diaz A., Kiviluoto S., Rodriguez D., Patron M., Rizzuto R., Bultynck G., Concha M. L., Sierralta J., Stutzin A., Hetz C., TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death Differ. 19, 1013–1026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtado de Mendoza T., Perez-Garcia C. G., Kroll T. T., Hoong N. H., O’Leary D. D. M., Verma I. M., Antiapoptotic protein Lifeguard is required for survival and maintenance of Purkinje and granular cells. Proc. Natl. Acad. Sci. U.S.A. 108, 17189–17194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla S., Fujita K., Xiao Q., Liao Z., Garfield S., Srinivasula S. M., A shear stress responsive gene product PP1201 protects against Fas-mediated apoptosis by reducing Fas expression on the cell surface. Apoptosis 16, 162–173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroemer G., Mitochondrial implication in apoptosis. Towards an endosymbiont hypothesis of apoptosis evolution. Cell Death Differ. 4, 443–456 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Chae H. J., Ke N., Kim H. R., Chen S., Godzik A., Dickman M., Reed J. C., Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene 323, 101–113 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Buttner S., Ruli D., Vogtle F. N., Galluzzi L., Moitzi B., Eisenberg T., Kepp O., Habernig L., Carmona-Gutierrez D., Rockenfeller P., Laun P., Breitenbach M., Khoury C., Frohlich K. U., Rechberger G., Meisinger C., Kroemer G., Madeo F., A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 30, 2779–2792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cebulski J., Malouin J., Pinches N., Cascio V., Austriaco N., Yeast Bax inhibitor, Bxi1p, is an ER-localized protein that links the unfolded protein response and programmed cell death in Saccharomyces cerevisiae. PLOS ONE 6, e20882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshisue H., Suzuki K., Kawabata A., Ohya T., Zhao H., Sakurada K., Taba Y., Sasaguri T., Sakai N., Yamashita S., Matsuzawa Y., Nojima H., Large scale isolation of non-uniform shear stress-responsive genes from cultured human endothelial cells through the preparation of a subtracted cDNA library. Atherosclerosis 162, 323–334 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Carrara G., Saraiva N., Parsons M., Byrne B., Prole D. L., Taylor C. W., Smith G. L., Golgi anti-apoptotic proteins are highly conserved ion channels that affect apoptosis and cell migration. J. Biol. Chem. 290, 11785–11801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariotti M., Smith T. F., Sudmant P. H., Goldberger G., Pseudogenization of testis-specific Lfg5 predates human/Neanderthal divergence. J. Hum. Genet. 59, 288–291 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Bultynck G., Kiviluoto S., Henke N., Ivanova H., Schneider L., Rybalchenko V., Luyten T., Nuyts K., De Borggraeve W., Bezprozvanny I., Parys J. B., De Smedt H., Missiaen L., Methner A., The C terminus of Bax inhibitor-1 forms a Ca2+-permeable channel pore. J. Biol. Chem. 287, 2544–2557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aouacheria A., Rech de Laval V., Combet C., Hardwick J. M., Evolution of Bcl-2 homology motifs: Homology versus homoplasy. Trends Cell Biol. 23, 103–111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H., Ito A., Kimura S. H., Yabuta N., Sakai N., Ikawa M., Okabe M., Matsuzawa Y., Yamashita S., Nojima H., RECS1 deficiency in mice induces susceptibility to cystic medial degeneration. Genes Genet. Syst. 81, 41–50 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Lisak D. A., Schacht T., Enders V., Habicht J., Kiviluoto S., Schneider J., Henke N., Bultynck G., Methner A., The transmembrane Bax inhibitor motif (TMBIM) containing protein family: Tissue expression, intracellular localization and effects on the ER CA2+-filling state. Biochim. Biophys. Acta 1853, 2104–2114 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Zhao G.-N., Zhang P., Gong J., Zhang X.-J., Wang P.-X., Yin M., Jiang Z., Shen L.-J., Ji Y. X., Tong J., Wang Y., Wei Q.-F., Zhu X.-Y., Zhang X., Fang J., Xie Q., She Z.-G., Wang Z., Huang Z., Li H., Tmbim1 is a multivesicular body regulator that protects against non-alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat. Med. 23, 742–752 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Deng K. Q., Zhao G. N., Wang Z., Fang J., Jiang Z., Gong J., Yan F. J., Zhu X. Y., Zhang P., She Z. G., Li H., Targeting transmembrane BAX inhibitor motif containing 1 alleviates pathological cardiac hypertrophy. Circulation 137, 1486–1504 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Repnik U., Hafner Cesen M., Turk B., Lysosomal membrane permeabilization in cell death: Concepts and challenges. Mitochondrion 19( Pt A), 49–57 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Aits S., Kricker J., Liu B., Ellegaard A. M., Hamalisto S., Tvingsholm S., Corcelle-Termeau E., Hogh S., Farkas T., Holm Jonassen A., Gromova I., Mortensen M., Jaattela M., Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bove J., Martinez-Vicente M., Dehay B., Perier C., Recasens A., Bombrun A., Antonsson B., Vila M., BAX channel activity mediates lysosomal disruption linked to Parkinson disease. Autophagy 10, 889–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan J. J., Zhang X. D., Sun W., Qi L., Wu J. C., Qin Z. H., DRAM1 regulates apoptosis through increasing protein levels and lysosomal localization of BAX. Cell Death Dis. 6, e1624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karch J., Schips T. G., Maliken B. D., Brody M. J., Sargent M. A., Kanisicak O., Molkentin J. D., Autophagic cell death is dependent on lysosomal membrane permeability through Bax and Bak. eLife 6, e30543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H.-R., Lee G.-H., Ha K.-C., Ahn T., Moon J.-Y., Lee B.-J., Cho S.-G., Kim S., Seo Y.-R., Shin Y.-J., Chae S.-W., Reed J. C., Chae H.-J., Bax inhibitor-1 is a pH-dependent regulator of Ca2+ channel activity in the endoplasmic reticulum. J. Biol. Chem. 283, 15946–15955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hetz C., Zhang K., Kaufman R. J., Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christensen K. A., Myers J. T., Swanson J. A., pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 115, 599–607 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Butor C., Griffiths G., Aronson N. N. Jr., Varki A., Co-localization of hydrolytic enzymes with widely disparate pH optima: Implications for the regulation of lysosomal pH. J. Cell Sci. 108( Pt. 6), 2213–2219 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Scott R. C., Schuldiner O., Neufeld T. P., Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Kratz E., Eimon P. M., Mukhyala K., Stern H., Zha J., Strasser A., Hart R., Ashkenazi A., Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ. 13, 1631–1640 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Jette C. A., Flanagan A. M., Ryan J., Pyati U. J., Carbonneau S., Stewart R. A., Langenau D. M., Look A. T., Letai A., BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ. 15, 1063–1072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H., Rafiuddin-Shah M., Tu H.-C., Jeffers J. R., Zambetti G. P., Hsieh J. J.-J., Cheng E. H.-Y., Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8, 1348–1358 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Teng X., Cheng W. C., Qi B., Yu T. X., Ramachandran K., Boersma M. D., Hattier T., Lehmann P. V., Pineda F. J., Hardwick J. M., Gene-dependent cell death in yeast. Cell Death Dis. 2, e188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Mattia F., Gubser C., van Dommelen M. M. T., Visch H.-J., Distelmaier F., Postigo A., Luyten T., Parys J. B., de Smedt H., Smith G. L., Willems P. H., van Kuppeveld F. J. M., Human Golgi antiapoptotic protein modulates intracellular calcium fluxes. Mol. Biol. Cell 20, 3638–3645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiviluoto S., Luyten T., Schneider L., Lisak D., Rojas-Rivera D., Welkenhuyzen K., Missaen L., De Smedt H., Parys J. B., Hetz C., Methner A., Bultynck G., Bax inhibitor-1-mediated Ca2+ leak is decreased by cytosolic acidosis. Cell Calcium 54, 186–192 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Saraiva N., Prole D. L., Carrara G., Maluquer de Motes C., Johnson B. F., Byrne B., Taylor C. W., Smith G. L., Human and viral Golgi anti-apoptotic proteins (GAAPs) oligomerize via different mechanisms and monomeric GAAP inhibits apoptosis and modulates calcium. J. Biol. Chem. 288, 13057–13067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westphalen B. C., Wessig J., Leypoldt F., Arnold S., Methner A., BI-1 protects cells from oxygen glucose deprivation by reducing the calcium content of the endoplasmic reticulum. Cell Death Differ. 12, 304–306 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Kim H.-K., Lee G.-H., Bhattarai K. R., Lee M.-S., Back S. H., Kim H.-R., Chae H.-J., TMBIM6 (transmembrane BAX inhibitor motif containing 6) enhances autophagy through regulation of lysosomal calcium. Autophagy , 761–778 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee G. H., Kim D. S., Kim H. T., Lee J. W., Chung C. H., Ahn T., Lim J. M., Kim I. K., Chae H. J., Kim H. R., Enhanced lysosomal activity is involved in Bax inhibitor-1-induced regulation of the endoplasmic reticulum (ER) stress response and cell death against ER stress: Involvement of vacuolar H+-ATPase (V-ATPase). J. Biol. Chem. 286, 24743–24753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballabio A., Bonifacino J. S., Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Soyombo A. A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K., TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281, 7294–7301 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Colletti G. A., Miedel M. T., Quinn J., Andharia N., Weisz O. A., Kiselyov K., Loss of lysosomal ion channel transient receptor potential channel mucolipin-1 (TRPML1) leads to cathepsin B-dependent apoptosis. J. Biol. Chem. 287, 8082–8091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldstein A. E., Werneburg N. W., Li Z., Bronk S. F., Gores G. J., Bax inhibition protects against free fatty acid-induced lysosomal permeabilization. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G1339–G1346 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H., Ito A., Sakai N., Matsuzawa Y., Yamashita S., Nojima H., RECS1 is a negative regulator of matrix metalloproteinase-9 production and aged RECS1 knockout mice are prone to aortic dilation. Circ. J. 70, 615–624 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Gong F., Gu J., Wang H., Up regulated Tmbim1 activation promotes high fat diet (HFD)-induced cardiomyopathy by enhancement of inflammation and oxidative stress. Biochem. Biophys. Res. Commun. 504, 797–804 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Zhang J., Fu Y., Chen J., Li Q., Guo H., Yang B., Genetic variant of TMBIM1 is associated with the susceptibility of colorectal cancer in the Chinese population. Clin. Res. Hepatol. Gastroenterol. 43, 324–329 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Orlando G., Law P. J., Palin K., Tuupanen S., Gylfe A., Hänninen U. A., Cajuso T., Tanskanen T., Kondelin J., Kaasinen E., Sarin A.-P., Kaprio J., Eriksson J. G., Rissanen H., Knekt P., Pukkala E., Jousilahti P., Salomaa V., Ripatti S., Palotie A., Järvinen H., Renkonen-Sinisalo L., Lepistö A., Böhm J., Mecklin J.-P., Al-Tassan N. A., Palles C., Martin L., Barclay E., Tenesa A., Farrington S., Timofeeva M. N., Meyer B. F., Wakil S. M., Campbell H., Smith C. G., Idziaszczyk S., Maughan T. S., Kaplan R., Kerr R., Kerr D., Buchanan D. D., Win A. K., Hopper J., Jenkins M., Lindor N. M., Newcomb P. A., Gallinger S., Conti D., Schumacher F., Casey G., Taipale J., Cheadle J. P., Dunlop M. G., Tomlinson I. P., Aaltonen L. A., Houlston R. S., Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Hum. Mol. Genet. 25, 2349–2359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez D. A., Zamorano S., Lisbona F., Rojas-Rivera D., Urra H., Cubillos-Ruiz J. R., Armisen R., Henriquez D. R., Cheng E. H., Letek M., Vaisar T., Irrazabal T., Gonzalez-Billault C., Letai A., Pimentel-Muiños F. X., Kroemer G., Hetz C., BH3-only proteins are part of a regulatory network that control the sustained signalling of the unfolded protein response sensor IRE1α. EMBO J. 31, 2322–2335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hetz C., Bernasconi P., Fisher J., Lee A. H., Bassik M. C., Antonsson B., Brandt G. S., Iwakoshi N. N., Schinzel A., Glimcher L. H., Korsmeyer S. J., Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1α. Science 312, 572–576 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Ramirez O., Garcia A., Rojas R., Couve A., Hartel S., Confined displacement algorithm determines true and random colocalization in fluorescence microscopy. J. Microsc. 239, 173–183 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Deng J., Carlson N., Takeyama K., Dal Cin P., Shipp M., Letai A., BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell 12, 171–185 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Zhang H., Nimmer P., Rosenberg S. H., Ng S.-C., Joseph M., Development of a high-throughput fluorescence polarization assay for Bcl-x(L). Anal. Biochem. 307, 70–75 (2002). [DOI] [PubMed] [Google Scholar]
- 66.P. N.-H. P. Pihán, N. Demaurex, C. Hetz, Simultaneous determination of intraluminal lysosomal calcium and pH by dextran-conjugated fluorescent dyes, in Methods in Cell Biology, O. Kepp, Ed. (Elsevier, 2021), pp. 199–208. [DOI] [PubMed] [Google Scholar]
- 67.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stefani E., Bezanilla F., Cut-open oocyte voltage-clamp technique. Methods Enzymol. 293, 300–318 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Armstrong C. M., Bezanilla F., Currents related to movement of the gating particles of the sodium channels. Nature 242, 459–461 (1973). [DOI] [PubMed] [Google Scholar]
- 72.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 73.Tucker B., Lardelli M., A rapid apoptosis assay measuring relative acridine orange fluorescence in zebrafish embryos. Zebrafish 4, 113–116 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Franken N. A., Rodermond H. M., Stap J., Haveman J., van Bree C., Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Bezu L., Sauvat A., Humeau J., Gomes-da-Silva L. C., Iribarren K., Forveille S., Garcia P., Zhao L., Liu P., Zitvogel L., Senovilla L., Kepp O., Kroemer G., eIF2α phosphorylation is pathognomonic for immunogenic cell death. Cell Death Differ. 25, 1375–1393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nunes P., Guido D., Demaurex N., Measuring phagosome pH by ratiometric fluorescence microscopy. J. Vis. Exp. , e53402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bers D. M., Patton C. W., Nuccitelli R., A practical guide to the preparation of Ca2+ buffers. Methods Cell Biol. 99, 1–26 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S9
References
Movie S1