Macrophages use mitochondrial lactate metabolism for ACLY-dependent histone acetylation, immune suppression, and tumor progression.
Abstract
Lactate accumulation is a hallmark of solid cancers and is linked to the immune suppressive phenotypes of tumor-infiltrating immune cells. We report herein that interleukin-4 (IL-4)–induced M0 → M2 macrophage polarization is accompanied by interchangeable glucose- or lactate-dependent tricarboxylic acid (TCA) cycle metabolism that directly drives histone acetylation, M2 gene transcription, and functional immune suppression. Lactate-dependent M0 → M2 polarization requires both mitochondrial pyruvate uptake and adenosine triphosphate–citrate lyase (ACLY) enzymatic activity. Notably, exogenous acetate rescues defective M2 polarization and histone acetylation following mitochondrial pyruvate carrier 1 (MPC1) inhibition or ACLY deficiency. Lastly, M2 macrophage–dependent tumor progression is impaired by conditional macrophage ACLY deficiency, further supporting a dominant role for glucose/lactate mitochondrial metabolism and histone acetylation in driving immune evasion. This work adds to our understanding of how mitochondrial metabolism affects macrophage functional phenotypes and identifies a unique tumor microenvironment (TME)–driven metabolic-epigenetic link in M2 macrophages.
INTRODUCTION
Highly glycolytic “Warburg-like” cancer cells take up glucose and produce lactate, which fundamentally changes the metabolic landscape of tumor microenvironments (TMEs) (1, 2). Although lactate has long been considered just a metabolic waste product of glycolytic tumors, it is becoming increasingly recognized as an important TME signal responsible for regulating the effector functions of a variety of tumor-infiltrating immune cells (3–5). A recent paradigm-shifting study revealed that circulating lactate—derived directly from glucose metabolism following glycolysis and lactate secretion—is actively metabolized through the mitochondrial tricarboxylic acid (TCA) cycle (6). Perhaps relatedly, tumor-derived lactate was shown to be sufficient to drive macrophage M2 polarization in a hypoxia-inducible factor–1α (HIF-1α)–dependent manner (7), although the metabolic mechanism(s) involved has not yet been elucidated. Given that M2-polarized macrophages occupy a low-glucose/high-lactate tumor stromal environment (8) and rely on mitochondrial metabolism (9, 10), an understanding of how lactate is functionally metabolized and whether/how it regulates gene expression patterns will help to further elucidate the mechanisms by which the TME influences tumor-infiltrating macrophage functions.
Macrophages carry out both physiologic and pathophysiologic functions leading to health and disease (11). For example, anti-inflammatory, proangiogenic M2-polarized macrophages (TAMs) promote wound healing and resolution of immune responses (12), but in late-stage cancers, this phenotype is co-opted by tumors to evade antitumor immunity (13). Conversely, while proinflammatory M1 macrophages help eradicate tumors by enhancing antitumor immune responses, they also drive tumor development by contributing to chronic inflammation–associated cell damage and DNA mutations (14). Accumulating evidence demonstrates that macrophage polarization phenotypes are sensitive to local metabolites within the TME, which influence their immunophenotypes and effector functions (15–17). Notably, the nomenclature of in vitro and in vivo macrophage polarization is nuanced (11). Previous studies demonstrate that in vitro interleukin-4 (IL-4)–polarized M2 bone marrow–derived macrophages (BMDMs) have similar phenotypic characteristics to in vivo immune-suppressive M2-like TAMs (18). In acknowledgement of previously established nomenclature guidelines (19), in vitro IL-4–polarized macrophages are M(IL-4) polarized and, for readability, will be referred to as M2 macrophages, and immunosuppressive TAMs will be referred to as M2-like TAMs.
Adenosine triphosphate (ATP)–citrate lyase (ACLY) enzymatic activity forms an important functional link between the TCA cycle and epigenetic histone acetylation (20). Specifically, glucose is first converted to pyruvate through glycolysis, which is then incorporated into the mitochondrial TCA cycle to produce citrate (21). A subfraction of this glucose-derived citrate is extruded out of the mitochondria and cleaved by ACLY, resulting in increased nucleo-cytoplasmic pools of acetyl–coenzyme A (CoA) that can then be used for nuclear histone acetylation (20).
ACLY has pleiotropic functions in macrophage polarization as acetyl-CoA can support histone acetylation as well as de novo lipogenesis (22). Initial studies in macrophages found that pharmacologic ACLY inhibitors suppress M1 macrophage–mediated cytokine and prostaglandin E2 production (23, 24). In contrast, IL-4–dependent M2 macrophage polarization triggers Akt-mTORC1–dependent ACLY phosphorylation and activation (25). More recently, a conditional macrophage ACLY-knockout transgenic model was used to identify an ACLY contribution to macrophage-dependent atherosclerotic plaque formation (26). However, the absolute requirements for ACLY, and the metabolic substrates involved, in direct M0 → M2–like TAM polarization and effector functions have not been fully elucidated. Using gene expression analyses, chromatin immunoprecipitation (ChIP), and metabolomic approaches, we now report that direct IL-4–induced M0 → M2 macrophage polarization interchangeably uses glucose or lactate as TCA cycle carbon sources to maximally drive ACLY-dependent histone acetylation at M2 gene-specific promoters, resulting in T cell suppressive functionality.
RESULTS
Lactate supports direct M0 → M2 macrophage polarization through mitochondrial pyruvate metabolism
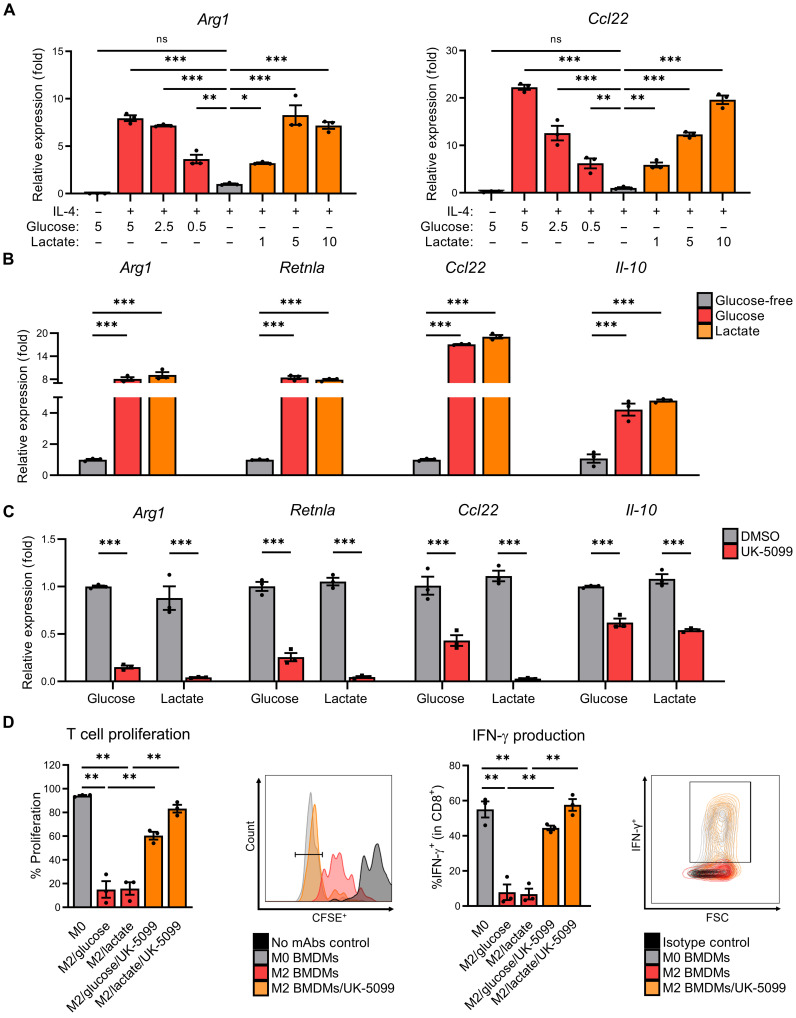
To investigate how changing TME metabolite concentrations differentially affect M2 macrophage polarization, we first treated primary murine BMDMs with the T helper 2 cytokine IL-4 in the presence/absence of various concentrations of glucose and lactate. Removal of extracellular glucose significantly reduced the expression of canonical M2-associated gene products arginase 1 (Arg1) and C-C motif chemokine 22 (Ccl22)—an effect that was rescued in a dose-dependent manner with molar equivalent additions of exogenous lactate as assessed by quantitative polymerase chain reaction (qPCR) (Fig. 1A) and immunoblotting (fig. S1A). The concentrations of exogenous lactate added are comparable to lactate levels found in the TME, which has been reported to be as high as 10 to 30 mM concentrations (27). In addition to Arg1 and Ccl22, exogenous lactate also rescued the reduced expression of M2 macrophage–associated gene Retnla and Il-10 in glucose-free culture conditions (Fig. 1B). Because low pH (i.e., high acidity) has been shown to promote M2 polarization (28), we confirmed that sodium lactate does not result in significant reductions of pH (fig. S1B).
Fig. 1. Macrophages maintain M2 polarization in TME-like conditions through mitochondrial pyruvate/lactate metabolism.
(A) Arg1 and Ccl22 expression of BMDMs starved in glucose-free (GF) media + 10% dialyzed fetal bovine serum (dFBS) (4 hours) before supplementation with glucose or lactate (as indicated), and then polarization with IL-4 (20 ng/ml) for 48 hours. ns, not significant. (B) M2-associated gene expression of BMDMs starved in GF media + 10% dFBS (4 hours) before supplementation with glucose (5 mM) or lactate (10 mM) and polarization with IL-4 (20 ng/ml) for 48 hours. (C) M2-associated gene expression of BMDMs starved in GF media + 10% dFBS (4 hours) before pretreatment ± UK-5099 (25 μM), supplementation with glucose (5 mM) or lactate (10 mM), and polarization with IL-4 (20 ng/ml) for 48 hours. DMSO, dimethyl sulfoxide; mAbs, monoclonal antibodies. (D) Proliferation (left) and interferon-γ (IFN-γ) production (right), with representative images, of CD8+ T cells from splenocytes cocultured for 3 days with BMDMs that were pretreated ± UK-5099 (25 μM) and polarized with IL-4 (20 ng/ml) or vehicle control [phosphate-buffered saline (PBS)] for 24 hours. Data are presented as means ± SEM and represent at least three independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by one-way analysis of variance (ANOVA) (A, B, and D) or two-way ANOVA (C) with Tukey’s post-test. CFSE+, carboxyfluorescein diacetate succinimidyl ester–positive.
M2 macrophages rely on mitochondrial metabolism (9, 10), and lactate is readily converted to pyruvate, which can then enter the TCA cycle by mitochondrial import via the MPC1 transporter (21, 29). Hence, we next asked whether inhibition of MPC1 by UK-5099 (30) blocks lactate- and/or glucose-dependent M2 polarization. As shown in Fig. 1C, UK-5099 strongly inhibited IL-4–induced expression of M2-associated genes in primary BMDMs cultured in both glucose and lactate and, as shown in fig. S1 (C and D), pyruvate. To assess whether M1 polarization is similarly affected by inhibition of mitochondrial pyruvate uptake, we evaluated the expression of the canonical M1-associated gene product, Nos2, in lipopolysaccharide (LPS)/interferon-γ (IFN-γ)–polarized BMDMs pretreated with UK-5099 and found significant, but much more modest, reductions in Nos2 expression (fig. S1E) suggesting that M1 polarization is less dependent on mitochondrial metabolism consistent with previous reports (10). To assess whether loss of M2-associated gene expression is simply due to loss of cellular viability following inhibition of mitochondrial lactate metabolism, we conducted trypan blue exclusion test and found that UK-5099 has no significant effects on overall macrophage viability (fig. S1F). Last, to ensure that the inhibitory effects observed with UK-5099 on M2 polarization were due to specific MPC1 inhibition, we also tested mitoglitazone, a molecularly distinct MPC inhibitor (31), which inhibited Arg1 expression in a dose-dependent manner (fig. S1G) and phenocopied the inhibition of the same UK-5099–sensitive M2-associated genes (fig. S1H).
Previous work by Colegio et al. determined that lactate-transporting monocarboxylate transporters (MCTs) are required for the induction of M2-associated genes (7). To better understand this mechanism, we determined the expression of the predominant lactate-exporting MCT1 and lactate-importing MCT4 in BMDMs cultured in exogenous lactate or Lewis lung carcinoma (LLC)–conditioned supernatants. The expression of MCT1 was reduced following lactate or LLC-conditioned media even in unpolarized BMDMs (fig. S2A, left), whereas the expression of MCT4 was induced in BMDMs polarized with IL-4 and cultured in either exogenous lactate or LLC supernatants (fig. S2A, middle). We also observed that the ratio of MCT4/MCT1 was generally elevated (i.e., high MCT4/MCT1 suggests overall lactate import) in both lactate- and LLC supernatant–treated BMDMs even in naïve macrophages (fig. S2A, right) suggesting that macrophages use MCTs in sensing extracellular/tumor-derived lactate to then drive M2 polarization.
Arguably, the most important functional consequence of M2-like TAMs is the acquisition of immune-suppressive activity that negatively controls activation and antitumor cytolytic function of tumor-infiltrating CD8+ T cells (13). Hence, we next tested whether lactate functionally supports M2-polarized macrophage immunosuppression and whether it similarly requires mitochondrial pyruvate uptake. As shown in Fig. 1D, M2 macrophage polarization in the presence of glucose or lactate is equally sufficient to induce significant suppressive activity against anti-CD3/anti-CD28–activated CD8+ splenocyte proliferation (Fig. 1D, two left panels) and IFN-γ production (Fig. 1D, two right panels). In addition, and consistent with the observed requirement for mitochondrial pyruvate uptake in glucose and lactate-dependent M2 gene expression (Fig. 1C), UK-5099 abrogated M2 macrophage–mediated immune-suppressive activity allowing for improved CD8+ T cell proliferation and IFN-γ production in both glucose and lactate culture conditions (Fig. 1D). Together, these data suggest that both glucose and lactate-derived mitochondrial pyruvate metabolism are necessary for maximal IL-4–induced M0 → M2 macrophage polarization and immunosuppressive function.
Mitochondrial pyruvate metabolism supports metabolic reprogramming in M2 macrophages
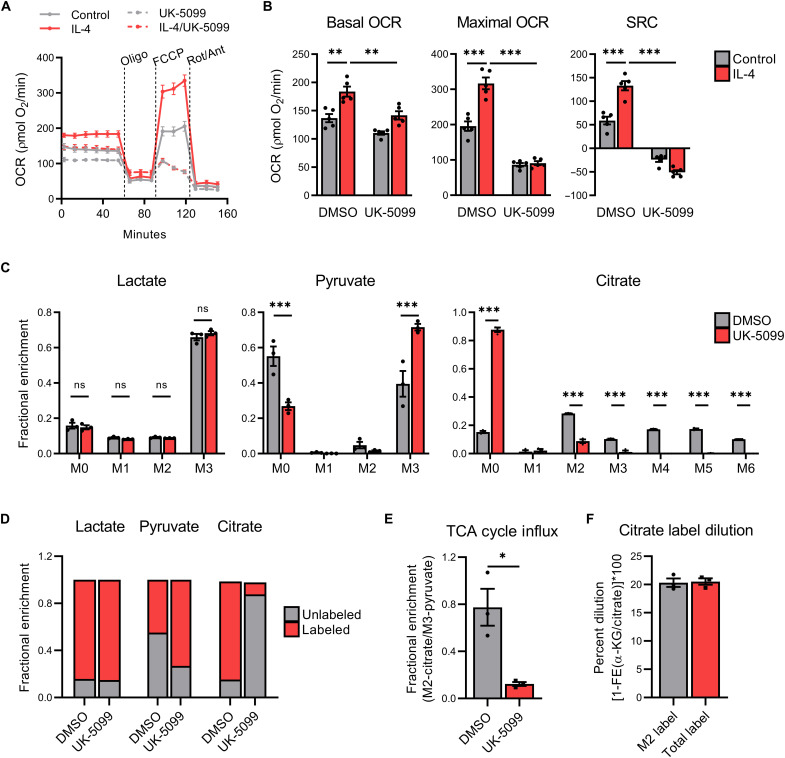
While M1 macrophage polarization relies primarily on glycolytic metabolism and terminal lactate secretion (10), M2 polarization is accompanied by a metabolic shift toward mitochondrial metabolism, TCA cycle activity, and oxidative phosphorylation (OXPHOS) (9, 10). However, the relative requirements, fate, and mechanism by which mitochondrial pyruvate metabolism controls gene expression programs and functional outcomes in M2 macrophage polarization are largely unresolved. Using extracellular flux analysis of BMDMs, we confirmed earlier studies (9, 10) showing that IL-4 increases basal and maximal oxygen consumption rates (OCRs) and spare respiratory capacity (SRC) (Fig. 2, A and B). In contrast, pretreatment with UK-5099 reduces both steady-state and IL-4–induced OCR and SRC (Fig. 2, A and B). As expected, UK-5099 modestly enhanced basal extracellular acidification rate (ECAR), but this difference was largely attenuated following inhibition of OXPHOS (fig. S2B) with oligomycin A injection. Last, we identified that lactate can support the IL-4–mediated enhancement of OCR to a similar extent as glucose (fig. S2C). Combined, these findings suggest that exogenous lactate supports metabolic reprogramming during M2 polarization and that lactate may be undergoing metabolism within the TCA cycle in M2 macrophages.
Fig. 2. M2 macrophages actively metabolize lactate within the mitochondrial TCA cycle.
(A) Trace, (B) (left) basal OCR, (middle) maximal OCR, and (right) SRC of BMDMs pretreated ± UK-5099 (25 μM) and then polarized with IL-4 (20 ng/ml) or vehicle control (PBS) for 24 hours before extracellular flux analysis with oligomycin (oligo), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone plus antimycin A (Rot/Ant). Fractional enrichment of (C) lactate (left), pyruvate (middle), and citrate (right) in BMDMs starved in GF media + 10% dFBS (4 hours) before pretreatment with UK-5099 (25 μM), supplementation with 13C-lactate (10 mM), and then polarization with IL-4 (20 ng/ml) for 6 hours. (D) Cumulative comparison of unlabeled (M0) versus labeled lactate (M1 to M2), pyruvate (M1 to M2), and citrate (M1 to M6). (E) Metabolic influx of lactate derived carbons into the TCA cycle and (F) percent dilution of M2- and total- (M2 to M6) citrate labeling into α-ketoglutarate (α-KG), as determined by 1–fractional enrichment (FE) of α-KG/citrate, in BMDMs treated as previously indicated. Data are presented as means ± SEM of four (A and B) or three (C to F) replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by two-way ANOVA (A to C) or Student’s t test (E).
Tumor-derived lactate is sufficient to induce M2-associated gene expression (7), and lactate can be catabolized in the TCA cycle within specific organs (6) but whether, and if so how, macrophage polarization-specific lactate metabolism occurs is largely unknown. To address this, we conducted isotope tracing using uniformly labeled lactate ([U-13C]-lactate) in IL-4–polarized M2 BMDMs treated with ±UK-5099 in glucose-free RPMI. As shown in Fig. 2C, 13C-lactate accumulates as fully labeled lactate (M + 3 isotopolog) in M2-polarized macrophages and is not affected by UK-5099 treatment (Fig. 2C, left). In untreated M2-polarized BMDMs, 13C-lactate is converted to fully labeled pyruvate (M+3) (Fig. 2C, middle) and is also incorporated into the first round of the TCA cycle as (M + 2) citrate and (M + 3 – M + 6) citrate following subsequent TCA cycling (Fig. 2C, right). As expected, UK-5099 treatment reduces the fractional enrichment of M+2 citrate isotopolog with a corresponding increase in M+3 pyruvate, indicating that 13C-lactate–derived carbons are not being efficiently incorporated into the TCA cycle. Comparison of unlabeled (M+0) versus labeled lactate (M + 1 – M + 3), pyruvate (M + 1 – M + 3), and citrate (M + 1 – M + 6) shows the shifts in isotopolog labeling following UK-5099 treatment (Fig. 2D), which results in significant decreases in TCA cycle incorporation of 13C-lactate–derived carbons (Fig. 2E). These findings suggest that lactate-derived mitochondrial pyruvate is sufficient for IL-4–induced metabolic reprogramming and that lactate is efficiently taken up in M2 macrophages and catabolized, via pyruvate, within the TCA cycle.
Resident TAMs actively take up exogenous glucose in the TME in vivo, which directly contributes to their protumorigenic activities (32). To determine whether IL-4–polarized M2 BMDMs preferentially use glucose or lactate, we conducted isotope tracing studies on M2 BMDMs using uniformly labeled lactate ([U-13C]-lactate) in the presence or absence of equimolar amounts of unlabeled glucose for 3 hours. Unexpectedly, we found that the accumulation of fully labeled lactate (M+3) and incorporation of 13C-lactate derived carbons into citrate (M+2) are not affected by the presence of unlabeled glucose (fig. S2D). Given that these findings may be solely attributed to a temporal effect as the incorporation of lactate-derived carbons into citrate requires fewer enzymatic steps than glycolysis, and glucose-derived incorporation into citrate achieves steady state at approximately 4 hours (33), we repeated this experiment for 6 hours. We observed that while exogenous unlabeled glucose slightly reduced the accumulation of 13C-lactate (M + 3) with a concomitant increase in glucose-derived lactate (M + 0), there was no significant decrease in the incorporation of 13C-lactate–derived carbons into citrate (M + 2) (fig. S2E). M + 2 citrate was the most enriched isotopolog at this time point (fig. S2E, right), suggesting that lactate was preferentially used for citrate production in this model.
Mitochondrial lactate metabolism does not affect signal transducer and activator of transcription 6 phosphorylation, HIF-1α stabilization, and protein lactylation during IL-4–induced M2 polarization
Initial studies designed to identify a mechanistic link between lactate-derived mitochondrial pyruvate metabolism and IL-4–induced expression of M2 macrophage–associated genes ruled out signal transducer and activator of transcription 6 (STAT6) phosphorylation (fig. S3A) and HIF-1α stabilization (17). Using HIF-oxygen dependent degradation domain (ODD)–luciferase BMDMs to quantify HIF-1α stability demonstrated that, while IL-4 induced a modest increase in luciferase activity (i.e., HIF-1α stability), no effect was observed with either UK-5099 or a succinate dehydrogenase inhibitor (34), Atpenin A5 (fig. S3B).
A recent study described an intriguing functional role for endogenous, macrophage-derived, lactate being metabolized into an acyl-CoA (i.e., lactyl-CoA) and resulting in direct histone lactylation thus promoting functional LPS/IFN-γ–induced M1 → M2 macrophage repolarization (5). However, in our studies investigating direct IL-4–induced M0 → M2 polarization, we found no fractional enrichment of the M3 isotopolog (fig. S3C) and a very low relative abundance of lactate in the protein fraction (fig. S3D). Last, we determined whether lactate-derived α-ketoglutarate may serve as a substrate for histone demethylation–dependent M0 → M2 macrophage polarization (35). Analysis of the 13C-lactate tracer studies revealed no discernable changes in the relative abundance (fig. S3D) or fractional enrichment of α-ketoglutarate (fig. S3E).
ACLY supports M0 → M2 macrophage polarization and histone acetylation
Citrate can undergo TCA cycle mitochondrial export and subsequent cleavage by ACLY to form acetyl-CoA substrates for nuclear histone acetylation (20, 22). Analysis of our isotope tracing studies indicated that 20.32 and 20.52% of 13C- lactate–derived labeling of citrate isotopologs from the first TCA cycle round (M+2 citrate) and subsequent TCA cycling (M+3 – M+6 citrate), respectively, were not incorporated into α-ketoglutarate (Fig. 2F). This dilution of 13C-citrate labeling into α-ketoglutarate suggests that TCA cycle efflux of citrate may be occurring, which would, in theory, then be available for ACLY-dependent cleavage.
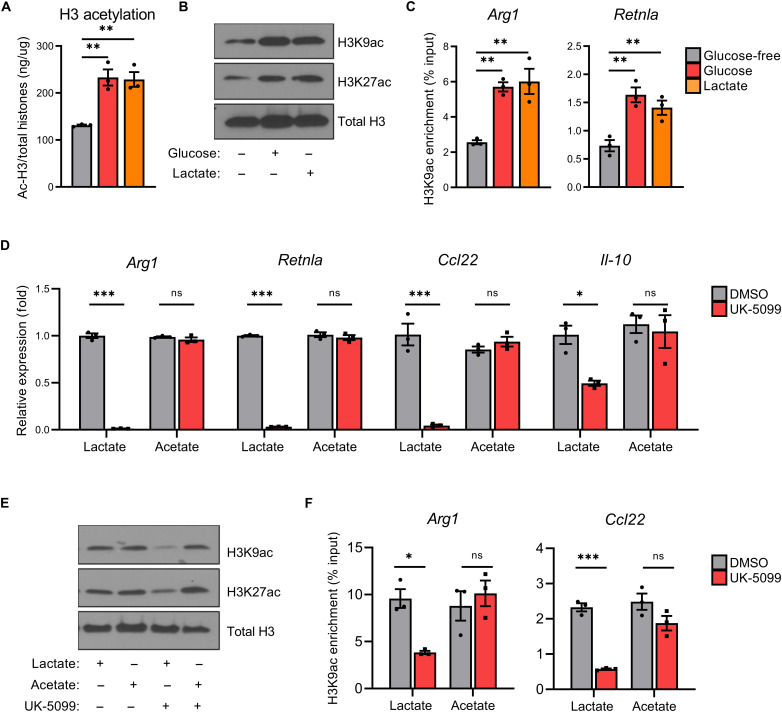
Consistent with this hypothesized role for mitochondrial pyruvate-dependent histone acetylation, glucose and lactate independently increase total H3 (Fig. 3A), lysine residue–specific H3 (Fig. 3B), and M2 gene promoter–specific (Fig. 3C) histone acetylation. Taking advantage of the fact that anabolism of acetate by acetyl-CoA synthetase short-chain family member 2 (ACSS2) can provide an alternative, mitochondrial-independent source of acetyl-CoA (36), we found that the addition of exogenous acetate very effectively rescues UK-5099–dependent inhibition of M2 macrophage–associated gene product expression (Fig. 3D), lysine residue–specific H3 (Fig. 3E), and M2 gene promoter–specific histone acetylation (Fig. 3F). Conversely, small-molecule inhibition of ACSS2 reduces the expression of M2-specific gene products in BMDMs polarized with exogenous acetate, but not lactate (fig. S3, F and G). To determine whether inhibition of mitochondrial lactate metabolism with UK-5099 and/or exogenous acetate supplementation nonspecifically induces alterations in global gene expression, we analyzed the expression of several “house-keeping” gene products. As seen in fig. S3H, while UK-5099 reduces the expression of IL-4–induced M2-associated gene products, the expression of the several house-keeping gene products was largely unaffected, consistent with prior observations that highly expressed/induced genes are more specifically regulated by histone acetylation compared to basally expressed genes (37). Overall, these findings indicate a requirement for lactate-derived acetyl-CoA in IL-4–induced M2 macrophage histone acetylation/gene expression and suggest a potential role for ACLY in this process.
Fig. 3. Mitochondrial lactate metabolism supports M2 polarization through ACLY-dependent histone acetylation.
(A) Total acetylated histone H3 enzyme-linked immunosorbent assay, (B) lysine residue–specific acetylation immunoblot, and (C) H3K9ac ChIP of BMDMs starved in GF media + 10% dFBS (4 hours) before supplementation with glucose (5 mM) or lactate (10 mM) and polarization with IL-4 (20 ng/ml) for 6 hours. (D) M2 gene expression, (E) lysine residue–specific acetylation, and (F) H3K9ac ChIP of BMDMs starved in GF media + 10% dFBS (4 hours) before pretreatment ± UK-5099 (25 μM), supplementation with lactate (10 mM) or acetate (10 mM), and polarization with IL-4 (20 ng/ml) for 48 (D) or 6 hours (E and F). Data are presented as means ± SEM and represent at least three experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by one-way ANOVA (A and C) or two-way ANOVA (D and F) with Tukey’s post-test.
In addition to lactate-derived acetyl-CoA production, lactate can be used to support OXPHOS. Following depletion of oxidizable metabolites (i.e., glucose and glutamine), cells can undergo a “lactate metabolic shift” where lactate is oxidized to maintain bioenergetics needed for cellular viability (38, 39). Notably, in all experiments in this investigation, BMDMs were supplemented with fresh glutamine for use as an alternative oxidizable metabolic substrate before IL-4–induced M2 polarization. In addition, because lactate oxidation supports ATP production, we conducted an ATP assay in BMDMs polarized in glucose-free media supplemented with glucose or lactate. As shown in fig. S3I, as compared to oligomycin A treatment, which blocks ATP production (40), ATP levels were not significantly affected by depletion of glucose or lactate. To determine whether inhibition of mitochondrial lactate metabolism with UK-5099 treatment and/or exogenous acetate supplementation affects cellular bioenergetics, we next conducted an ATP assay and found no discernable changes in ATP levels with UK-5099 in contrast to the depletion of ATP following oligomycin A pretreatment (fig. S4A). Furthermore, oligomycin A pretreatment in IL-4–polarized M2 BMDMs significantly decreased the concentrations of transcribed RNA, while total RNA concentrations were largely unaffected by UK-5099 treatment (fig. S4B). Notably, to limit generalized metabolic dysfunction, minimal concentrations of oligomycin A were used to inhibit ATP synthase (40). Given that inhibition of cellular bioenergetics with oligomycin A induced significant generalized cellular dysfunction in BMDMs, in contrast to UK-5099 treatment, this suggests that while lactate oxidation is likely occurring, the predominant contribution of mitochondrial lactate metabolism in IL-4–induced M2 gene expression is for lactate-derived acetyl-CoA production and subsequent ACLY-dependent histone acetylation.
ACLY is necessary for M2-associated gene expression, immune-suppressive activity, and M2 TAM–dependent tumor progression
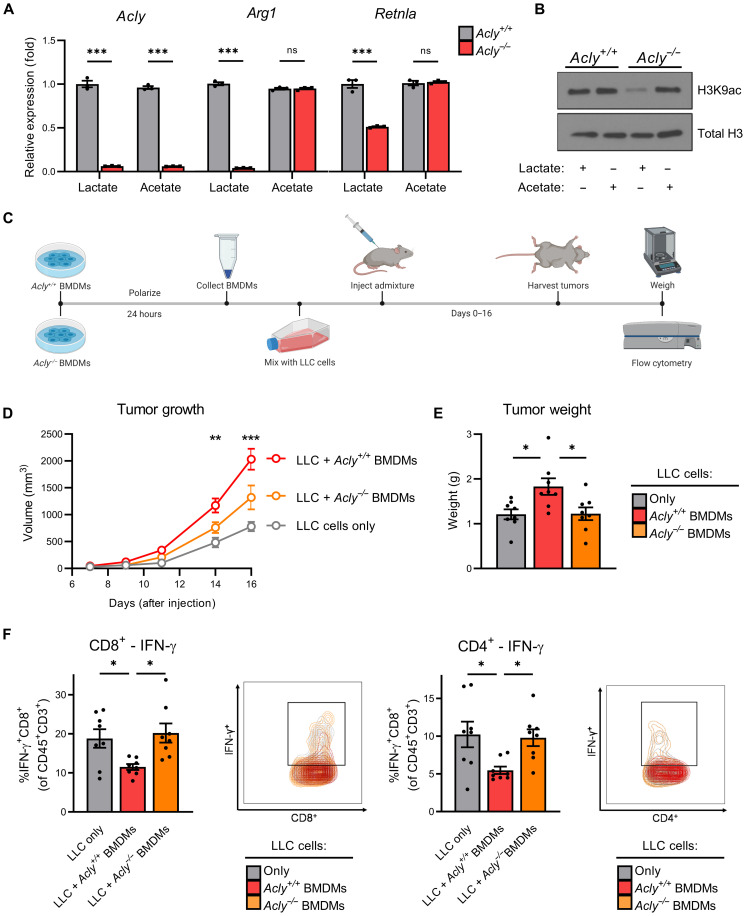
To validate a requirement for ACLY in M0 → M2 macrophage polarization, we used an inducible ACLY-knockout transgenic mouse model (UBC-Cre-ERT2 Aclyf/f mice). As shown in Fig. 4A, ACLY deficiency phenocopies UK-5099–mediated inhibition of mitochondrial pyruvate uptake on reducing M2-associated gene expression in macrophages polarized in lactate but did not affect acetate-dependent polarization. In addition, ACLY deficiency reduces lysine residue–specific histone acetylation—an effect that can be rescued with exogenous acetate (Fig. 4B). ACLY-deficient BMDMs maintained high viability (fig. S4C) and expression of the macrophage terminal differentiation marker F4/80 (fig. S4D), suggesting that loss of M2 polarization following ACLY depletion is not due to these factors.
Fig. 4. ACLY is required for maximal M2 macrophage polarization and tumor progression.
(A) M2 gene expression and (B) lysine residue–specific acetylation of Acly+/+ and Acly−/− BMDMs starved in GF media + 10% dFBS (4 hours) before supplementation with lactate (10 mM) or acetate (10 mM) and polarization with IL-4 (20 ng/ml) for 48 (A) or 6 hours (B). (C) Schematic works flow of in vivo tumor admixture model. (D) Growth and (E) weight of tumors consisting of LLC cells only, LLC cells + M2-polarized Acly+/+ BMDMs, or LLC cells + M2-polarized Acly−/− BMDMs. (F) IFN-γ production in CD8+ cytotoxic T cells (left) and CD4+ helper T cells (right) in the respective tumors. Data are presented as means ± SEM of three (A and B) or eight (D to F) replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by two-way ANOVA (B) or one-way ANOVA (D to F) with Tukey’s post-test.
Given that the defining effector function hallmark of M2 macrophages is their ability to suppress antitumor immunity leading to enhanced tumor growth, we next used an in vivo tumor admixture murine model (41) to investigate a potential requirement for ACLY in M2 macrophage–mediated tumor progression. LLC cells were co-injected with Acly+/+ or Acly−/− M2-polarized BMDMs (CD45.2+) into congenic B6.SJL mice (CD45.1+) (Fig. 4C). While tumors bearing Acly+/+ M2-polarized BMDMs had increased tumor growth (Fig. 4D), end point tumor weight (Fig. 4E), and gross tumor burden (fig. S4E) versus LLC cells alone, tumors bearing Acly−/− M2-polarized BMDMs exhibited marked reductions in tumor outgrowth and end point tumor burden compared to tumors bearing Acly+/+ M2 BMDMs.
Flow cytometric analyses of immune effector cells within tumors revealed a lack of residual Acly+/+ or Acly−/− CD45.2+ TAMs (fig. S4F), suggesting nearly complete loss of initiator TAMs at the time of our end point analysis. No discernable differences were found between percentages of infiltrating CD4+ T cells (fig. S4G, left), CD8+ T cells (fig. S4G, middle), or CD4+/CD8+ T cell ratio (fig. S4G, right), suggesting that TAM-derived ACLY did not directly affect total intratumoral T cell infiltration per se in this model. However, tumors from LLC/Acly−/− M2 BMDMs co-injection had significantly fewer IFN-γ–expressing CD8+ T cells (Fig. 4F, left) and CD4+ helper T cells (Fig. 4F, right) compared to LLC/Acly+/+ M2 BMDM co-injection tumors. These findings suggest that ACLY expression/activity in intratumoral M2 macrophages is necessary for maximal TAM immunosuppressive activity and that loss of ACLY allows for a more robust antitumor immune response resulting, ultimately, in reduced tumor growth and progression.
DISCUSSION
Solid TMEs represent a unique, physiologic paradigm of a high-lactate/low-glucose metabolic compartment in human pathology (1, 3). Several prior studies have shown that higher lactate concentrations in the TME are sufficient to drive both innate and adaptive immune responses (4, 7), but whether, and if so how, lactate specifically controls gene expression patterns and functional immune phenotypes is largely unknown.
Our studies now reveal that IL-4–induced M2 macrophages functionally use extracellular lactate to fuel mitochondrial TCA cycling and oxidative metabolism. Lactate fully compensates for low or absent glucose in driving M2 macrophage–associated gene expression patterns and functional immune suppressive activity. 13C-lactate tracing metabolomics demonstrates that extracellular lactate is efficiently converted to pyruvate, imported into mitochondria, and further metabolized by the TCA cycle to citrate. Our data further suggest that this lactate-derived citrate is effluxed out of the mitochondria to specifically drive histone acetylation in a manner that requires acetyl-CoA generation by ACLY, which is necessary for maximal M2 gene expression, immune-suppressive activity, and tumor progression.
Recently, a very novel metabolic-epigenetic link was identified where M1 macrophages were found to use glycolysis for lactate anabolism to generate lactyl-CoA and resulting in histone lactylation–dependent LPS/IFN-γ–induced M1 → M2 macrophage transition (5). Our findings, however, indicate that IL-4–induced M0 → M2 macrophage metabolic reprogramming preferentially allows for lactate catabolism—via mitochondrial pyruvate (42)—to generate sufficient acetyl-CoA for use in histone acetylation–dependent gene expression. It is not unreasonable to speculate that, in early developing tumors where glucose supplies are high and lactate is low, M1 macrophages with high glycolytic/low TCA activity (i.e., low citrate levels) can afford to use terminal glycolysis–derived lactate as a direct source of epigenetic lactylation and initiation of M1 → M2 transition. As these same tumors grow and expand (with concomitant reductions in glucose and increases in extracellular lactate), M2 macrophage metabolic reprogramming occurs resulting in high TCA activity and, because glucose is scarce, these early M2 macrophages can begin to use lactate both as a source of oxidative fuel and, via citrate efflux, as a source of histone acetylation.
Although our studies did not find a direct link between lactate- or glucose-dependent M2 polarization and HIF-1α stabilization, they do support the previously described requirement for HIF-1α in driving tumor-derived lactate-induced M2 polarization that was done in the absence of IL-4 (7). The differences between our observations and Colegio et al. might be explained by different ex vivo macrophage polarization models (i.e., tumor-derived lactic acid versus IL-4–induced polarization requiring sodium lactate). Nonetheless, we envision that in vivo these mechanisms coexist by which lactate-dependent histone acetylation allows for maximally efficient HIF-1α–dependent transcription at a subset of HIF-1α–sensitive M2-associated gene promoters (7).
ACLY serves as a metabolic nexus to supply nucleo-cytosolic pools of acetyl-CoA from mitochondrial citrate to be used for lipid synthesis, prostaglandin production, and histone acetylation (22, 43). We now show that ACLY is necessary to support histone acetylation and subsequent maximal expression of M2 macrophage–associated gene products. In a very recent study, Van den Bossche and colleagues (26) used an independent, macrophage-specific, conditional ACLY transgenic model to show that chronic loss of ACLY in vivo stabilizes atherosclerotic plaques and suggested that ACLY contributes to fatty acid metabolism, efferocytosis, and M2 polarization phenotypes. In our current study, we used BMDMs from an inducible ACLY knockout transgenic model that allowed us to assess acute ACLY deletion on direct IL-4–induced M0 → M2 polarization and to validate defective histone acetylation using exogenous acetate. Using in vitro M2-polarized Acly+/+ and Acly−/− primary macrophages, we identified that, in a more chronic model of macrophage ACLY deficiency, M2 macrophage–dependent tumor growth and progression were significantly impaired, coincident with increased numbers of infiltrating IFN-γ+CD8+ and IFN-γ+CD4+ T lymphocytes. We observed that TAM ACLY deficiency led to a more marked reduction in tumor weight compared to volume. While we do not have any experimental explanation for this currently, we suspect that this discrepancy likely lies in changes in tumor density due to specific differences in cellularity and/or composition of the tumor stromal matrix, as would be expected with greater T cell–dependent oncolytic activities in the TAM ACLY-deficient tumors.
Future studies are certainly warranted to better and more completely understand the following unanswered questions: (i) Do ACLY-expressing M2-like TAMs use lactate in vivo for the observed phenotypic effects on tumor growth and antitumor immunity? (ii) How much, if any, does TAM-derived lactate (secondary to glycolysis-mediated lactate excretion) contribute to subsequent TAM uptake of lactate? (iii) What is the relative contribution of lactate-dependent oxidation to bioenergetic metabolism (i.e., OXPHOS, and amino acid synthesis) versus lactate-derived acetyl-CoA production in supporting M2 polarization? With respect to the latter question, our current data identified that most of the basal OCR is resistant to UK-5099 treatment, which indicates that mitochondrial lactate uptake only partially supports oxidative metabolism and suggests that other metabolic substrates are predominantly used for bioenergetic support. In addition, approximately 20% of 13C-lactate–derived carbons were lost during the conversion from citrate to α-ketoglutarate, which suggests to us that this portion of lactate-derived carbons may be undergoing export out of the mitochondria while the rest of the lactate-derived carbons are retained in the TCA cycle to help maintain bioenergetic homeostasis.
Our metabolomic analysis of IL-4–polarized M2 BMDMs suggests that if both glucose and lactate are sufficiently abundant in the TME, then M2-like TAMs may preferentially use lactate-derived carbons for citrate production. Potential reasons for this include the fact that pyruvate production from lactate requires fewer enzymatic steps and the fact that lactate to pyruvate conversion generates NADH (reduced form of nicotinamide adenine dinucleotide), which has a feedback inhibitory effect on glycolytic enzymes like glyceraldehyde-3-phosphate dehydrogenase (44). Given the recent findings that TAMs in vivo take up a substantial amount of glucose in glucose-replete TMEs (32), future studies into the relative utilization of glucose versus lactate in vivo will help to better clarify whether/how nutrient partitioning in the TME changes in a spatiotemporal and/or polarization-specific manner.
Numerous ACLY inhibitory compounds have been identified and are being developed as anticancer agents (45). Although the initial rationale for ACLY antagonists as antitumor agents was based primarily on disrupting tumor cell fatty acid biosynthesis secondary to glucose addiction, it will be important in future translational studies to also assess collateral effects on tumor-infiltrating immune effector cells and the relative disruption of transcriptional reprogramming, polarization/ differentiation phenotypes, and immune-suppressive functions. Because infiltrating M2-like TAMs represent a dominant source of antitumor T cell–suppressive activity in late-stage malignant tumors, which can functionally skew immune checkpoint blockade (ICB) clinical efficacy (13), ACLY targeting may represent a unique immunotherapeutic approach to alleviate ICB resistance in the future.
MATERIALS AND METHODS
Mice
Wild-type C57BL/6 mice were obtained from Harlan Laboratories (Dublin, VA). B6/SJL mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Aclyf/f mice have been previously reported (36), and bones from UBC-Cre ERT2; Aclyf/f mice were provided by K. Wellen. Animals were maintained under specific pathogen–free conditions and handled in accordance with the Association for Assessment and Accreditation of Laboratory Animals Care international guidelines. The Institutional Animal Care and Use Committee at the University of Louisville approved the experiments. Six- to 16-week-old mice were used in all experiments.
Cell culture and BMDM differentiation
Mice were euthanized by CO2 asphyxiation, and death was confirmed by cervical dislocation. Bone marrow cells from the tibias and femurs were differentiated in RPMI 1640 supplemented with fetal bovine serum (FBS) (5%) and recombinant murine macrophage colony-stimulating factor (M-CSF) (25 ng/ml; PeproTech) for 7 days. For the inducible ACLY depletion experiments, Aclyfl/fl UBC Cre ERT2 BMDMs were treated with 4-hydroxytamoxifen (5 μM) or vehicle control on day 4 of differentiation. Following differentiation, the cells were counted and plated with RPMI 1640 supplemented with 10% FBS (without M-CSF) overnight. The following day, the BMDMs were washed with phosphate-buffered saline (PBS) and starved of glucose by addition of glucose-free RPMI 1640 (Gibco) supplemented with 10% dialyzed FBS (dFBS) and 2 mM l-glutamine for 4 to 6 hours before treatment with the indicated compounds and stimulated with recombinant murine IL-4 (20 ng/ml; PeproTech) for 4 to 48 hours.
RNA purification and real-time qPCR
Total RNA was extracted using RNeasy Mini Kit (QIAGEN) following the manufacturer’s instructions. The resulting RNA was quantified using a NanoDrop 8000 ultraviolet-visible spectrophotometer (Thermo Fisher Scientific), and the complementary DNA (cDNA) was synthesized with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative measurement of cDNA levels was performed using TaqMan Fast Advanced Master Mix (Applied Biosystems) with TaqMan Gene Expression Primers (Applied Biosystems) on the 7500 Fast Real-Time PCR System (Applied Biosystems). Relative expression profiles of mRNA levels were calculated using the comparative Ct method [2−(ΔΔCt)] using 18S ribosomal RNA levels as an endogenous reference control.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors and homogenized, and samples were denatured in LB sample buffer at 98°C. Five to 20 μg of protein were loaded into a 4 to 20% Mini-PROTEAN TGX Gel (Bio-Rad Laboratories) and separated by electrophoresis before being transferred onto an Immobilon-P polyvinylidene difluoride membrane (EMD Millipore). After blocking, membranes were probed overnight at 4°C with primary antibodies (Abs) and then for 1 hour at room temperature with secondary Abs. The blots were developed using Pierce ECL Plus Western Blotting Substrate (Thermo Fisher Scientific).
In vitro BMDM–T cell coculture assay
BMDMs were treated with UK-5099 (25 μM) or dimethyl sulfoxide (DMSO) in the presence of the indicated metabolites ± IL-4 (20 ng/ml) for 24 hours before being collected, washed, and counted via trypan blue exclusion, and live cells were plated in 96-well plates. BMDMs were then cocultured with carboxyfluorescein diacetate succinimidyl ester–labeled splenocytes from syngeneic mice in the presence of anti-CD3/anti-CD28 agonistic Abs for 3 days. The cells were stimulated with phorbol 12-myristate 13-acetate (PMA)/ionomycin plus GolgiPlug for 6 hours and then stained with anti-CD8 or anti-CD4 monoclonal Abs (mAbs) and fixed/permeabilized for intracellular cytokines staining. The data were acquired using a FACSCanto cytometer (BD Biosciences) and analyzed using FlowJo V10 software (Tree Star, Ashland, OR).
Extracellular flux analysis
For extracellular flux assays, BMDMs were plated in Seahorse XF96 cell culture microplates (Seahorse Biosciences, Agilent) overnight and then pretreated with UK-5099 (25 μM) or DMSO for 30 min followed by polarization with IL-4 (20 ng/ml) for 16 hours. The OCR and ECAR were measured using the XF96 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA, USA) according to the manufacturer’s instructions. During the assay, wells were injected with oligomycin (5 μM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (2 μM), and rotenone (1 μM)/antimycin A (5 μM). Each condition was performed in four to six replicates, and data were analyzed using Seahorse Wave 2.6 Desktop Software (Agilent).
CellTiter-Glo luminescent ATP assay
A total of 1 × 105 BMDMs were plated for 24 hours in 100 μl of complete RPMI 1640 into opaque-walled 96-well plates. The next day, the cells were switched to glucose-free RPMI 1640 + 10% dFBS for 4 hours before being pretreated with UK-5099 (25 μM) or oligomycin A (10 nM; positive control); supplemented with glucose (5 mM), lactate (10 mM), or acetate (10 mM); and polarized with IL-4 (20 ng/ml) for 24 hours. The plate was then allowed to equilibrate to room temperature for 30 min before 100 μl of the CellTiter-Glo Reagent was added to each well. The plates were then shaken on an orbital shaker for 5 min and then allowed to incubate for 10 min at room temperature before the luminescence was measured on a SpectraMax iD3 plate reader (Molecular Devices).
13C-lactate labeling
A total of 1 × 107 BMDMs were plated for 24 hours in complete RPMI 1640. The next day, the cells were switched to glucose-free RPMI 1640 + 10% dFBS for 4 hours. The BMDMs were then pretreated with UK-5099 (25 μM) or DMSO for 30 min before IL-4 (20 ng/ml) polarization in 13C-Lactate (10 mM). In the preferential utilization experiments, unlabeled 12C-Glucose (5 mM) was added simultaneously. After 3 or 6 hours of labeling, cells were washed twice with ice-cold 0.1× PBS and extracted with 1 ml of 50% methanol containing 20 μM l-norvaline (internal control). Polar (aqueous layer) and insoluble fractions (protein) were separated by centrifugation at 4°C and 15,000 rpm for 10 min. The polar fraction was dried by SpeedVac (Thermo Fisher Scientific) followed by derivatization. The protein pellet was subsequently washed four times with 50% methanol and once with 100% methanol to remove polar contaminants and dried by SpeedVac.
Pellet hydrolysis
Hydrolysis of the protein pellet was performed by first resuspending the dried pellet in deionized H2O followed by the addition of equal part 6 N HCl. The samples were vortexed thoroughly and incubated at 95°C for 2 hours. All reactions were quenched with 100% methanol with 200 μM l-norvaline and then incubated on ice for 30 min. The supernatant was collected after centrifugation at 15,000 rpm at 4°C for 10 min and was subsequently dried by SpeedVac followed by derivatization.
Sample derivatization
Dried polar and hydrolyzed pellet samples were derivatized by the addition of 50 μl of methoxyamine hydrochloride (20 mg/ml) in pyridine. Samples were incubated at 30°C for 90 min followed by centrifugation at 15,000 rpm for 10 min. The supernatant was transferred to a v-shaped amber glass chromatography vial, followed by the addition of 80 μl of N-methyl-trimethylsilyl-trifluoroacetamide and incubation for 30 min at 37°C. The derivatized samples were then analyzed by gas chromatography–mass spectrometry (GC-MS).
GC-MS quantitation
An Agilent 7800B GC coupled to a 7010A triple quadrupole MS detector equipped with a high-efficiency source was used for this study. GC-MS protocols were similar to those described previously (46), except that a modified temperature gradient was used for GC: Initial temperature was 130°C, held for 4 min, rising at 6°C/min to 243°C, rising at 60°C/min to 280°C, and held for 2 min. The electron ionization energy was set to 70 eV. 13C isotopologs were identified by selected ion monitoring of the following ions at known retention times: α-ketoglutarate, 304 to 209; citrate, 465 to 471; lactate, 219 to 222; pyruvate, 174 to 177 and lactate, 219 to 222. Scan [mass/charge ratio: 50 to 800] and full-scan mode were used for target metabolite analysis. For determination of protein lactylation, lactate, 219 to 222 was used to identify lactate in the hydrolyzed protein pellet sample. Metabolites were identified using the FiehnLib metabolomics library (available through Agilent) by retention time and fragmentation pattern, and quantitation was performed using Agilent MassHunter Workstation Software. Natural abundance correction was performed using IsoCorrectoR software, and relative abundance was corrected for recovery using the l-norvaline standard and adjusted to protein input (47).
Histone acetylation enzyme-linked immunosorbent assay
Histones were extracted using the EpiQuick Total Histone Extraction Kit (EpiGentek) and analyzed using the EpiQuick Total Histone H3 Acetylation Detection Fast Kit (EpiGentek) according to the manufacturer’s protocols. Briefly, 1 × 107 BMDMs were polarized for 4 to 6 hours in the indicated conditions before collection using Cell Striper (Corning), incubation with prelysis, and then lysis buffer. The histone containing supernatant fraction was collected and treated with the balance buffer before 150 ng of the histone extract was added to the enzyme-linked immunosorbent assay wells for 2 hours at room temperature. The wells were then washed and incubated with the secondary Ab-containing solution for 1 hour on an orbital shaker. The wells were then developed with the enzyme reaction solution for 5 min before being quenched and measured for absorbance at 450 nm.
Chromatin immunoprecipitation
ChIP was performed using the SimpleChip Enzymatic Chromatin IP Kit [Cell Signaling Technology (CST)] according to the manufacturer’s instructions. Briefly, 1.2 × 107 BMDMs were fixed with 1% formaldehyde and lysed, and then the nuclei were isolated. The chromatin was digested with Micrococcal Nuclease and then sonicated to lyse the nuclear membrane. The cross-linked chromatin was immunoprecipitated with anti-H3K9ac Ab (CST, C5B11) overnight at 4°C. The chromatin was recovered with Protein G Agarose Beads and eluted, and the cross-links were reversed with Elution Buffer and Proteinase K overnight at 65°C. Input and immunoprecipitated DNA were analyzed by real-time qPCR, and the data are presented as percent of the total input chromatin.
In vivo tumor admixture model
The tumor admixture model was performed as previously described (41). Briefly, Acly+/+ and Acly−/− BMDMs (CD45.2+) were polarized with IL-4 for 24 hours before being mixed with LLC cells (CD45.2+) at a 1:2.5 ratio, respectively, in Matrigel (Corning) and injected subcutaneously into the flanks of congenic B6.SJL mice (CD45.1+). Beginning on day 7 after injection, palpable tumors were measured three times a week, and the tumor volume was calculated by the following formula: (length × width2)/2. At the experimental end point on day 16 after injection, the mice were euthanized, and the tumors were resected, weighed, and digested in RPMI 1640 containing collagenase IV, hyaluronidase, and deoxyribonuclease I for 30 min at 37°C. Aliquots of the single-cell suspensions were either stimulated with PMA/ionomycin plus GolgiPlug for 6 hours before staining or directly stained with the indicated mAbs.
Statistical analysis
All result representative data are presented as the means ± SEM and analyzed for statistical significance using GraphPad Prism 8.3 (GraphPad Software, La Jolla, California, USA).
Acknowledgments
Funding: This work was supported by National Institutes of Health (NIH) Predoctoral Fellowship Award F30CA232550 (to J.T.N.), NIH Pre- to Postdoctoral Transition Award 4K00CA212455 (to H.C.A.), NIH Research Grant R35NS116824 (to M.S.G.), NIH Research Grant R01AG060056 (to L.A.J.), NIH Research Grant R01AG062550 (to L.A.J.), NIH Research Grant R01CA213990 (to J.Y.), NIH Research Grant R01DK116005 (to K.E.W.), NIH Research Grant R01CA174761 (to K.E.W.), NIH Research Grant R01AG066653 (to R.C.S.), V Scholar Grant (to R.C.S.), NIH Research Grant R01CA186661 (to R.A.M.), NIH Research Grant GB130096P3 (to R.A.M.), and NIH Project Grant P20GM135004 (to J.Y. and R.A.M.).
Author contributions: Conceptualization: J.T.N. and R.A.M. Methodology: J.T.N., B.F.C., J.Y., K.E.W., R.C.S., and R.A.M. Investigation: J.T.N., B.E.R., A.E.G., L.R.C., S.M.M., L.E.A.Y., R.C.B., E.J.K., A.W.-M., M.B.d.S.R., E.R.R., H.C.A., and B.V.B. Funding acquisition: J.T.N., K.M.M., and R.A.M., Resources: J.T.N., L.A.J., M.S.G., B.F.C., J.Y., K.E.W., R.C.S., and R.A.M. Supervision: J.T.N. and R.A.M. Writing—original draft: J.T.N. and R.A.M. Writing—review and editing: J.T.N., K.M.M., J.Y., K.E.W., R.C.S., and R.A.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Aclyf/f mice were generated using a targeting vector made by the NIH Knockout Mouse Project (KOMP: 80097). Reagents derived from the targeting vector may be provided via pending scientific review and a completed material transfer agreement. Requests should be submitted to K.E.W. (wellenk@upenn.edu). Aclyf/f mice were deposited in the Mutant Mouse Resource and Research Center (MMRRC stock no: 43555) and are available through the Jackson Laboratory.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Warburg O., On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- 2.Warburg O., On respiratory impairment in cancer cells. Science 124, 269–270 (1956). [PubMed] [Google Scholar]
- 3.Certo M., Tsai C.-H., Pucino V., Ho P.-C., Mauro C., Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 21, 151–161 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Angelin A., Gil-de-Gómez L., Dahiya S., Jiao J., Guo L., Levine M. H., Wang Z., Quinn W. J. III, Kopinski P. K., Wang L., Akimova T., Liu Y., Bhatti T. R., Han R., Laskin B. L., Baur J. A., Blair I. A., Wallace D. C., Hancock W. W., Beier U. H., Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M., Ding J., Czyz D., Hu R., Ye Z., He M., Zheng Y. G., Shuman H. A., Dai L., Ren B., Roeder R. G., Becker L., Zhao Y., Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui S., Ghergurovich J. M., Morscher R. J., Jang C., Teng X., Lu W., Esparza L. A., Reya T., Zhan L., Guo J. Y., White E., Rabinowitz J. D., Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colegio O. R., Chu N. Q., Szabo A. L., Chu T., Rhebergen A. M., Jairam V., Cyrus N., Brokowski C. E., Eisenbarth S. C., Phillips G. M., Cline G. W., Phillips A. J., Medzhitov R., Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmona-Fontaine C., Deforet M., Akkari L., Thompson C. B., Joyce J. A., Xavier J. B., Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl. Acad. Sci. U.S.A. 114, 2934–2939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van den Bossche J., Baardman J., Otto N. A., van der Velden S., Neele A. E., van den Berg S. M., Luque-Martin R., Chen H.-J., Boshuizen M. C. S., Ahmed M., Hoeksema M. A., de Vos A. F., de Winther M. P. J., Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 17, 684–696 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Van den Bossche J., Baardman J., de Winther M. P. J., metabolic characterization of polarized M1 and M2 bone marrow-derived macrophages using real-time extracellular flux analysis. J. Vis. Exp. 105, 53424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sica A., Mantovani A., Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 122, 787–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe S., Alexander M., Misharin A. V., Budinger G. R. S., The role of macrophages in the resolution of inflammation. J. Clin. Invest. 129, 2619–2628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P., Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biswas S. K., Mantovani A., Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 11, 889–896 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Jha A. K., Huang S. C.-C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K. M., Ashall J., Everts B., Pearce E. J., Driggers E. M., Artyomov M. N., Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Viola A., Munari F., Sanchez-Rodríguez R., Scolaro T., Castegna A., The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noe J. T., Mitchell R. A., Tricarboxylic acid cycle metabolites in the control of macrophage activation and effector phenotypes. J. Leukoc. Biol. 106, 359–367 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A., Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Murray P. J., Allen J. E., Biswas S. K., Fisher E. A., Gilroy D. W., Goerdt S., Gordon S., Hamilton J. A., Ivashkiv L. B., Lawrence T., Locati M., Mantovani A., Martinez F. O., Mege J. L., Mosser D. M., Natoli G., Saeij J. P., Schultze J. L., Shirey K. A., Sica A., Suttles J., Udalova I., van Ginderachter J. A., Vogel S. N., Wynn T. A., Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B., ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bricker D. K., Taylor E. B., Schell J. C., Orsak T., Boutron A., Chen Y.-C., Cox J. E., Cardon C. M., Van Vranken J. G., Dephoure N., Redin C., Boudina S., Gygi S. P., Brivet M., Thummel C. S., Rutter J., A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietrocola F., Galluzzi L., Bravo-San Pedro J. M., Madeo F., Kroemer G., Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 21, 805–821 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Infantino V., Iacobazzi V., Palmieri F., Menga A., ATP-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 440, 105–111 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Langston P. K., Nambu A., Jung J., Shibata M., Aksoylar H. I., Lei J., Xu P., Doan M. T., Jiang H., MacArthus M. R., Gao X., Kong Y., Chouchani E. T., Locasale J. W., Snyder N. W., Horng T., Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat. Immunol. 20, 1186–1195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covarrubias A. J., Aksoylar H. I., Yu J., Snyder N. W., Worth A. J., Iyer S. S., Wang J., Ben-Sahra I., Byles V., Polynne-Stapornkul T., Espinosa E. C., Lamming D., Manning B. D., Zhang Y., Blair I. A., Horng T., Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. eLife 5, e11612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baardman J., Verberk S. G. S., van der Velden S., Gijbels M. J. J., van Roomen C. P. P. A., Sluimer J. C., Broos J. Y., Griffith G. R., Prange K. H. M., van Weeghel M., Lakbir S., Molenaar D., Meinster E., Neele A. E., Kooij G., de Vries H. E., Lutgens E., Wellen K. E., de Winther M. P. J., Van den Bossche J., Macrophage ATP citrate lyase deficiency stabilizes atherosclerotic plaques. Nat. Commun. 11, 6296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Cruz-López K. G., Castro-Muñoz L. J., Reyes-Hernandez D. O., Garcia-Carrancá A., Manzo-Merino J., Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front. Oncol. 9, 1143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Kenawi A., Gatenbee C., Robertson-Tessi M., Bravo R., Dhillon J., Balagurunathan Y., Berglund A., Vishvakarma N., Ibrahim-Hashin A., Choi J., Luddy K., Gatenby R., Pilon-Thomas S., Anderson A., Ruffell B., Gillies R., Acidity promotes tumour progression by altering macrophage phenotype in prostate cancer. Br. J. Cancer 121, 556–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herzig S., Raemy E., Montessuit S., Veuthey J. L., Zamboni N., Westermann B., Kunji E. R. S., Martinou J. C., Identification and functional expression of the mitochondrial pyruvate carrier. Science 337, 93–96 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Halestrap A. P., The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. Biochem. J. 148, 85–96 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Divakaruni A. S., Wiley S. E., Rogers G. W., Andreyev A. Y., Petrosyan S., Loviscach M., Wall E. A., Yadava N., Heuck A. P., Ferrick D. A., Henry R. R., McDonald W. G., Colca J. R., Simon M. I., Ciaraldi T. P., Murphy A. N., Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc. Natl. Acad. Sci. U.S.A. 110, 5422–5427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinfeld B. I., Madden M. Z., Wolf M. M., Chytil A., Bader J. E., Patterson A. R., Sugiura A., Cohen A. S., Ali A., Do B. T., Muir A., Lewis C. A., Hongo R. A., Young K. L., Brown R. E., Todd V. M., Huffstater T., Abraham A., O’Neil R. T., Wilson M. H., Xin F., Tantawy M. N., Merryman W. D., Johnson R. W., Williams C. S., Mason E. F., Mason F. M., Beckermann K. E., Heiden M. G. V., Manning H. C., Rathmell J. C., Rathmell W. K., Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alves T. C., Pongratz R. L., Zhao X., Yarborough O., Sereda S., Shirihai O., Cline G. W., Mason G., Kibbey R. G., Integrated, step-wise, mass-isotopomeric flux analysis of the TCA cycle. Cell Metab. 22, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kluckova K., Sticha M., Cerny J., Mracek T., Dong L., Drahote Z., Gottlieb E., Neuzil J., Rohlena J., Ubiquinone-binding site mutagenesis reveals the role of mitochondrial complex II in cell death initiation. Cell Death Dis. 6, e1749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P.-S., Wang H., Li X., Chao T., Teav T., Christen S., Di Conza G., Cheng W.-C., Chou C.-H., Vavakova M., Muret C., Debackere K., Mazzone M., Huang H.-D., Fendt S.-M., Ivanisevic J., Ho P.-C., α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Zhao S., Torres A., Henry R. A., Trefely S., Wallace M., Lee J. V., Carrer A., Sengupta A., Campbell S. L., Kuo Y. M., Frey A. J., Meurs N., Viola J. M., Blair I. A., Weljie A. M., Metallo C. M., Snyder N. W., Andrews A. J., Wellen K. E., ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. 17, 1037–1052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karmodiya K., Krebs A. R., Oulad-Abdelghani M., Kimura H., Tora L., H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zagari F., Jordan M., Stettler M., Broly H., Wurm F. M., Lactate metabolism shift in CHO cell culture: The role of mitochondrial oxidative activity. N. Biotechnol. 30, 238–245 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Wurm F. M., Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 22, 1393–1398 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Ruas J. S., Siqueira-Santos E. S., Rodrigues-Silva E., Castilho R. F., High glycolytic activity of tumor cells leads to underestimation of electron transport system capacity when mitochondrial ATP synthase is inhibited. Sci. Rep. 8, 17383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M., Tong Z., Ding C., Luo F., Wu S., Wu C., Albeituni S., He L., Hu X., Tieri D., Rouchka E. C., Hamada M., Takahashi S., Gibb A. A., Kloecker G., Zhang H. G., Bousamra M. II, Hill B. G., Zhang X., Yan J., Transcription factor c-Maf is a checkpoint that programs macrophages in lung cancer. J. Clin. Invest. 130, 2081–2096 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F., Zhang S., Vuckovic I., Jeon R., Lerman A., Folmes C. D., Dzeja P. P., Herrman J., Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab. 28, 463–475.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Icard P., Wu Z., Fournel L., Coquerel A., Lincet H., Alifano M., ATP citrate lyase: A central metabolic enzyme in cancer. Cancer Lett. 471, 125–134 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Svedruzic Z. M., Odorcic I., Chang C. H., Svedruzic D., Substrate channeling via a transient protein-protein complex: The case of D-Glyceraldehyde-3-phosphate dehydrogenase and L-lactate dehydrogenase. Sci. Rep. 10, 10404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granchi C., ATP citrate lyase (ACLY) inhibitors: An anti-cancer strategy at the crossroads of glucose and lipid metabolism. Eur. J. Med. Chem. 157, 1276–1291 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Sun R. C., Dukhande V. V., Zhou Z., Young L. E. A., Emanuelle S., Gentry M. S., Nuclear glycogenolysis modulates histone acetylation in human non-small cell lung cancers. Cell Metab. 30, 903–916.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andres D. A., Young L. E. A., Veeranki S., Hawkinson T. R., Levitan B. M., He D., Wang C., Satin J., Sun R. C., Improved workflow for mass spectrometry-based metabolomics analysis of the heart. J. Biol. Chem. 295, 2676–2686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4